Abstract
Development of cisplatin resistance in colorectal cancer is largely caused by dysregulation of signaling pathways, including the Wnt/β-catenin signaling pathway, in cancer cells. Further investigation into the molecular mechanism of chemoresistance could improve outcomes for patients with colorectal cancer. The present study determined that fibroblast growth factor 9 (FGF9) was overexpressed in tumor tissues compared with normal tissues from patients with colorectal cancer. Using the colorectal cancer cell line LoVo, transfection of recombinant FGF9 decreased cisplatin-induced cell apoptosis whilst FGF9 silencing increased cisplatin-induced apoptosis. Western blot analysis and reverse transcription-quantitative polymerase chain reaction demonstrated that FGF9 decreased adenomatous polyposis coli (APC) mRNA and protein expression and contributed to activation of the Wnt/β-catenin signaling pathway. Notably, an increase in FGF9 and β-catenin protein expression and a decrease in APC protein expression was observed in the established LoVo cisplatin resistant cell line (LoVo/cisplatin). Silencing of FGF9 reversed cisplatin resistance of LoVo/cisplatin cells. In conclusion, the present findings suggested that FGF9 activated the Wnt signaling pathway and was a mediator of cisplatin resistance in colorectal cancer.
Keywords: fibroblast growth factor 9, cisplatin resistance, colorectal cancer, Wnt/β-catenin signaling pathway
Introduction
Colorectal cancer is ranked as the second most commonly diagnosed cancer type in males and third most commonly diagnosed cancer type in females (1). Chemotherapy using agents such as cisplatin and oxaliplatin is the conventional treatment approach for colorectal cancer patients (2,3); however development of chemoresistance frequently occurs leading to a mortality rate of >33% in developed countries (4). Due to an increased understanding of colorectal cancer molecular pathogenesis, multiple target therapy agents have been used for colorectal cancer patients (5,6); however, the clinical beneficial rate is often unsatisfactory (7). Therefore, there is an urgent need for research into chemoresistance in order to provide novel targets for colorectal cancer therapy.
The fibroblast growth factor (FGF) signaling pathway is involved in regulation of homeostasis, angiogenesis and organogenesis (8,9). Aberrant activation of FGF signaling is observed in cancer cells and considered a critical step during carcinogenesis (10). During activation of FGF signaling, FGFs bind to high affinity tyrosine kinase FGF receptors (FGFRs) on the surface of cells (11). FGF9 is highly conserved and ubiquitously expressed in embryos (12,13). Overexpression of FGF9 is observed in several types of cancer and its expression is associated with prognosis (14,15). Recent research has determined that FGF9 exerts oncogenic activity in cancer cells via regulating expression of several key genes such as T-box 3 and vascular endothelial growth factor A (16,17). In colorectal cancer cells, FGF9 protein expression is maintained at a high level via translational activation (18). To date, whether FGF9 mediates cisplatin resistance in colorectal cancer and the underlying mechanisms remain not fully understood.
Overactivation of the Wnt signaling pathway is an important step during cancer initiation and development (19). Following Wnt binding to receptors, the signal is transduced to the nucleus leading to stabilization of transcription co-activator β-catenin and activation of Wnt target gene expression (20). Activity of β-catenin is tightly controlled in cells. Negative regulators of the Wnt/β-catenin pathway, such as tumor suppressor adenomatous polyposis coli (APC), determine the stability and cellular location of β-catenin (21). In particular, the Wnt/β-catenin pathway is a well-known mediator of cancer stemness and promotes chemotherapy resistance (22). In aldehyde dehydrogenase-positive colorectal cancer, activation of the Wnt/β-catenin pathway facilitates development of cisplatin resistance (23).
The present study determined that FGF9 was elevated in colorectal tumors compared with matched normal tissue. In colorectal cancer cells, FGF9 overexpression decreased cisplatin-induced cell apoptosis whilst FGF9 silencing increased cisplatin-induced cytotoxicity. Mechanistically, FGF9 repressed APC expression and activated the Wnt/β-catenin signaling pathway. Notably, FGF9 and β-catenin protein expression increased whilst APC protein expression decreased in the LoVo cisplatin resistant cell line (LoVo/cisplatin). FGF9 knockdown reversed cisplatin resistance of LoVo/cisplatin cells. In conclusion, the results demonstrated that FGF9 activated the Wnt signaling pathway and was a mediator of cisplatin resistance in colorectal cancer.
Materials and methods
Tissue samples from patients
Tumor tissue and matched normal tissue (5 cm away from the tumor) was collected from 20 patients with colorectal cancer (age, 47–64 years old; mean age, 54.3±7.2 years old; 14 male and 6 female) at the Shanghai Eighth People's Hospital between March 2015 and October 2016. Patients who received any prior radiotherapy or chemotherapy treatment were excluded from enrollment. Written consent was provided by all enrolled patients. All experiments were approved by the Ethics Committee of the Shanghai Eighth People's Hospital. Samples were immediately frozen at −80°C following collection. The stage of colon cancer was defined according to the TNM system classification of the American Joint Committee on Cancer (AJCC, 7th edition) (24).
Cell culture
The colorectal cancer cell line LoVo (parental) was purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Ham's F-12K medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) under standard conditions (37°C; 5% CO2).
Cisplatin was purchased from Selleck Chemicals (Houston, TX, USA). To determine cisplatin sensitivity, cells were treated for 48 h with various concentrations of cisplatin (1, 2, 4, 8 and 16 µM).
To establish the cisplatin resistant LoVo subline (LoVo/cisplatin), LoVo cells were divided into two groups and treated with gradually increasing concentrations of cisplatin (100, 200, 400 nm, 1, 2, 5 µM, for 1 month) or dimethyl sulfoxide (DMSO) for 6 months. The cell viability (data not shown) of LoVo cells which was determined by Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was initially significantly suppressed by cisplatin; however, following 6 months of cisplatin treatment, cells no longer responded to 5 µM cisplatin and were considered to be a cisplatin resistant subline (LoVo/cisplatin). The LoVo cells treated with DMSO were considered as the parental cell line (LoVo/parental).
Construction of plasmid and overexpression of FGF9
Full length FGF9 cDNA was amplified from LoVo cells by polymerase chain reaction (PCR) using a Taq DNA polymerase SuperMix kit (Invitrogen; Thermo Fisher Scientific, Inc.) under the following thermocycling conditions: 94°C for 2 min followed by 35 cycles of 94°C for 2 sec, 60°C for 60 sec and 72°C for 1 min and ligated into pcDNA3.1 vector (Addgene, Inc., Cambridge, MA, USA) using the restriction sites for HindIII and XhoI (New England Biolabs, Inc.). The primer sequences for FGF9 were as listed: Forward, 5′-AAGCTTATGGCTCCCTTAGGTGAAGT-3′, and reverse, 5′-CTGCAGTCAACTTTGGCTTAGAATAT-3′. The amplified sequence was verified by sequencing. For overexpression of FGF9, 2 µg pcDNA3.1-FGF9 vector was mixed with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) in serum-free Ham's F-12K medium for 15 min, then added to cells cultured in 6-well plates (1×106 cells/well). In the control group, 2 µg pcDNA3.1 was mixed with Lipofectamine 3000 in serum-free Ham's F-12K medium for 15 min, then added to cells cultured in 6-well plates (1×106 cells/well). The cells were maintained for 48 h, and transfection efficiency was confirmed by western blotting before experimentation.
Silencing of FGF9
Control small interfering RNA (siRNA, 5′-CAGUACUUUUGUGUAGUACAA-3′) and FGF9 siRNA (5′-GCGAUACUAUGUUGCAUUATT-3′, 3′-UAAUGCAACAUAGUAUCGCCT-5′) were purchased from GenePharma Co. Ltd. (Suzhou, China). For knockdown of FGF9, FGF9 siRNA was mixed with Lipofectamine RNAiMax (Invitrogen) in serum-free Ham's F-12K medium for 5 mins then the mixture was added into culture medium of 6-well plates (1×106 cells/well). Following incubation for 48 h, cells were collected and transfection efficiency was confirmed by western blot before the subsequent experiments.
Cell viability assay
LoVo cells were seeded in 96-well plates at a density of 1×103 cells/well. The cell viability of each well was determined by Cell Counting Kit-8 (Dojindo Molecular Technologies) according to manufacturer's protocol. Cell viability was evaluated by detection of the absorbance of each well at 450 nm using a plate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells were extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA concentration was detected using Nanodrop 1000 (Thermo Fisher). RNA was reverse transcribed into cDNA with PrimeScript RT reagent Kit (Takara Bio, Inc., Otsu, Japan). RT-qPCR was performed with a SYBR Premix Ex Taq Kit (Takara) on a CFX-96 Real-Time PCR Detection System (Bio-Rad). The following thermocycling conditions were used: Denaturing, 95°C for 30 sec; annealing, 95°C for 5 sec; elongation, 60°C for 30 sec, for 40 cycles. The primer sequences were as follows: FGF9, forward 5′-ATGGCTCCCTTAGGTGAAGTT-3′ and reverse 5′-CCCAGGTGGTCACTTAACAAAAC-3′; APC, forward 5′-GGAGACAGAATGGAGGTGCT-3′ and reverse 5′-TCTTCAGTGCCTCAACTTGC-3′; and GAPDH, forward 5′-GGACTCATGACCACAGTCCATGCC-3′ and reverse 5′-TCAGGGATGACCTTGCCCACAG-3′. Quantification was carried out by the 2−ΔΔCq method (25).
Western blot analysis
Lysates were prepared using radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) following the manufacturer's instruction. Protein concentration was determined by the BCA method. For western blot, proteins in lysates (20 µg/lane) were separated by 8% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, then blocked with 5% non-fat milk. Membranes were incubated with primary antibody against APC (cat. no. 2504; 1:1,000; Cell Signaling Technology, Danvers, MA, USA), β-catenin (cat. no. 848; 1:1,000; Cell Signaling Technology), GAPDH (cat. no. sc-47724; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and FGF9 (cat no. sc-8413; 1:1,000; Santa Cruz Biotechnology, Inc.) at 4°C overnight. Membranes were then incubated with horseradish peroxidase-labeled secondary antibodies against mouse (cat no. ab6289; 1:10,000; Abcam, Cambridge, UK) and rabbit (cat no. ab6721; 1:10,000; Abcam) at room temperature for 2 h. Protein bands were visualized using enhanced chemiluminescence western blot substrate (Pierce; Thermo Fisher Scientific, Inc.). The intensity of bands was calculated using Image J software version 1.8.0 (National Institutes of Health, Bethesda, MD) and normalized to GAPDH.
Immunohistochemical staining
The normal tissues and tumor tissues were fixed with 4% paraformaldehyde (PFA) solution at 4°C overnight, embedded in paraffin and sliced into 4-µm sections. Immunohistochemistry (IHC) was performed as follows: The sections were first deparaffinized and rehydrated in 0.03% H2O2 in 95% methanol for 20 min at room temperature to block the endogenous peroxidase activity; antigen retrieval was carried out by water bath (Immunosaver; Nisshin EM, Tokyo, Japan) for 45 min at 98°C; sections were blocked with normal horse serum (Thermo Fisher Scientific, Inc.) for 20 min at room temperature to eliminate non-specific staining prior to incubation with an anti-FGF9 antibody (cat no. ab71395; 1:1,000; Abcam) overnight at 4°C; the next day, sections were incubated with horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG (cat no. ab205718; 1:2,000; Abcam) for 1 h at room temperature; color was developed by 3, 3′-diaminobenzidine (ZsBio, Beijing, China), sections were counterstained with hematoxylin for 30 sec at room temperature. Images from 6 individual fields of view were captured using a light microscope and the attached camera (×40, Nikon, Tokyo, Japan).
Cell apoptosis assay
The Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection kit (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) was used to investigate apoptosis, according to manufacturer's protocol. Following treatment with siRNA and/or cisplatin, cells were harvested and suspended in 1X binding buffer. Annexin V-FITC and PI solution were then added into cell suspension and incubated for 15 min. Finally, the cells were analyzed using a FACSCalibur system (Becton, Dickinson and Company). The % apoptotic cells was calculated with FlowJo software version 10.2 (FlowJo LLC, Ashland, OR).
Statistical analysis
All data were analyzed using Graphpad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) and expressed as mean ± standard deviation. The two-tailed Student's t-test was used to compare two groups and determine the P-value. Three or more groups were compared with one-way analysis of variance followed by Newman Keul's test. P<0.05 was considered to indicate statistical significance.
Results
Overexpression of FGF9 in colorectal tumor tissues and normal tissues
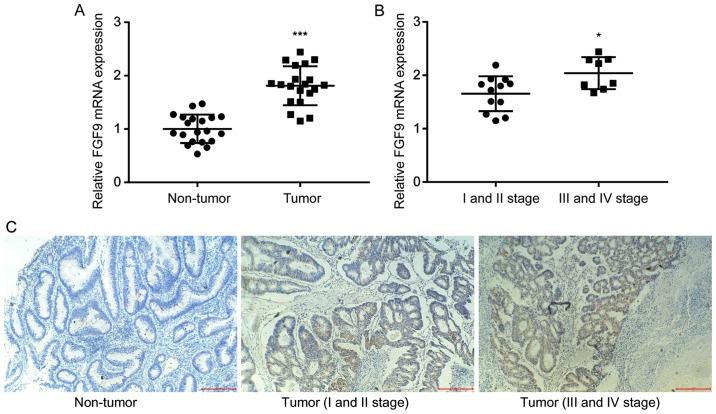
To investigate the role of FGF9 in colorectal cancer, RT-qPCR was used to detect FGF9 mRNA expression in tumor tissues and matched normal tissues from 20 colorectal cancer patients. There was a significant increase of FGF9 mRNA expression levels in colorectal tumor tissue compared with normal tissue (Fig. 1A), which was consistent with previous literature (18). In addition, clinicopathological analysis revealed that higher FGF9 expression was associated with late stage colorectal cancer (III and IV stage) rather than early stage colorectal cancer (I and II stage) (Fig. 1B). IHC staining demonstrated that FGF9 expression was markedly higher in tumor tissues (I and II stage; III and IV stage) compared with the non-tumor tissues. In addition, FGF9 expression was obviously increased in tumor tissues at III and IV stage compared with tissues at I and II stage (Fig. 1C). These results suggested that FGF9 overexpression might promote colorectal cancer progression.
Figure 1.
FGF9 expression is elevated in colorectal cancer. (A) In comparison with matched normal tissues, expression of FGF9 mRNA was increased in colorectal tumor tissues from 20 patients with colorectal cancer. (B) Overexpression of FGF9 mRNA was detected in late-stage colorectal tumor tissues (n=8) compared with early-stage colorectal tumor tissues (n=12). (C) FGF9 protein expression levels were examined in the normal and tumor tissues by immunohistochemistry. Representative images are shown (magnification, ×200). *P<0.05 and ***P<0.001. FGF9, fibroblast growth factor 9.
FGF9 regulates cisplatin sensitivity in colorectal cancer cells
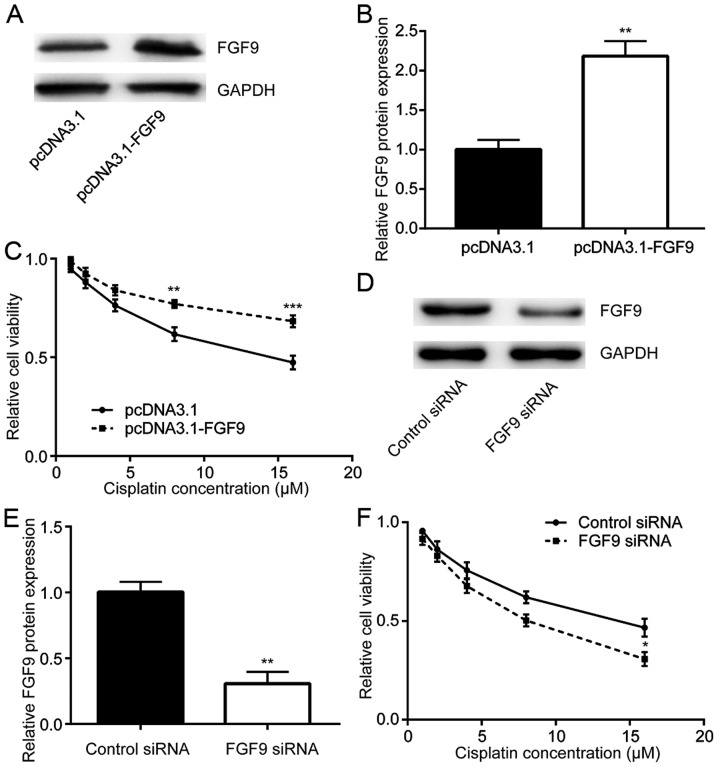
To study whether FGF9 expression was related to cisplatin resistance, FGF9 was overexpressed in LoVo cells then sensitivity to cisplatin was detected. Transfection of recombinant FGF9 increased FGF9 expression in LoVo cells (Fig. 2A and B). Cisplatin treatment decreased cell viability in a dose dependent manner whilst overexpression of FGF9 decreased this cytotoxic effect of cisplatin (Fig. 2C). By contrast, silencing of FGF9 expression by transfection of FGF9 siRNA enhanced the cytotoxic effect of cisplatin on LoVo cells (Fig. 2D-F). Therefore, FGF9 expression was associated with cisplatin sensitivity of colorectal cancer cells.
Figure 2.
FGF9 regulates cisplatin sensitivity in colorectal cancer cells. (A) Transfection of recombinant FGF9 elevated FGF9 protein expression in LoVo cells. (B) Quantitative analysis of relative FGF9 expression. (C) Treatment with cisplatin (1, 2, 4, 8, 16 µM) reduced cell viability of LoVo cells in a dose-dependent manner. FGF9 overexpression attenuated the cytotoxicity induced by cisplatin. (D) Transfection of FGF9 siRNA decreased FGF9 protein expression in LoVo cells. (E) Quantitative analysis of relative FGF9 expression. (F) Compared with LoVo cells transfected with control siRNA, silencing of FGF9 increased cytotoxicity induced by cisplatin. **P<0.01 and ***P<0.001 vs. control. FGF9, fibroblast growth factor 9; siRNA, small interfering RNA.
FGF9 inhibits cisplatin-induced cell apoptosis in colorectal cancer cells
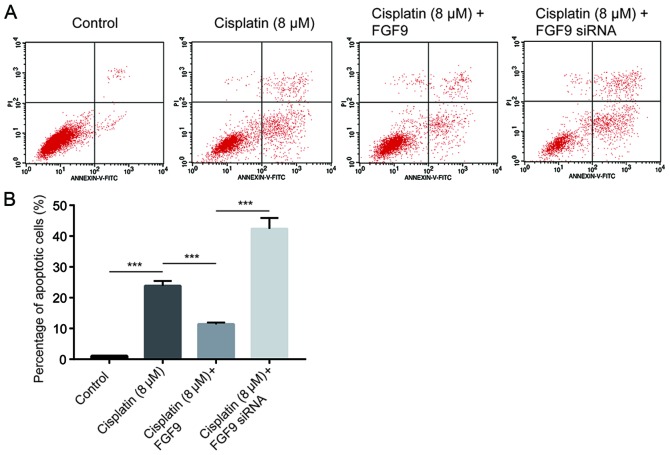
Cisplatin inhibits tumor cell growth via induction of cell apoptosis (26). Using flow cytometry analysis, it was determined that cisplatin (8 µM) significantly increased the % of apoptotic LoVo cells compared with the control (Fig. 3). Notably, overexpression of FGF9 decreased cisplatin-induced cell apoptosis. By contrast, silencing of FGF9 increased cell apoptosis (Fig. 3), which suggested that FGF9 sensitized colorectal cancer cells towards cisplatin and induced cell apoptosis.
Figure 3.
FGF9 promotes cell apoptosis induced by cisplatin. (A) Apoptosis was examined by flow cytometry in LoVo cells treated with cisplatin and FGF9 overexpression or silencing. Representative plots are shown. (B) Quantitative analysis of % of apoptotic cells. ***P<0.001 with comparisons indicated by lines. FGF9, fibroblast growth factor 9; siRNA, small interfering RNA.
FGF9 activates the Wnt/β-catenin pathway via repression of APC
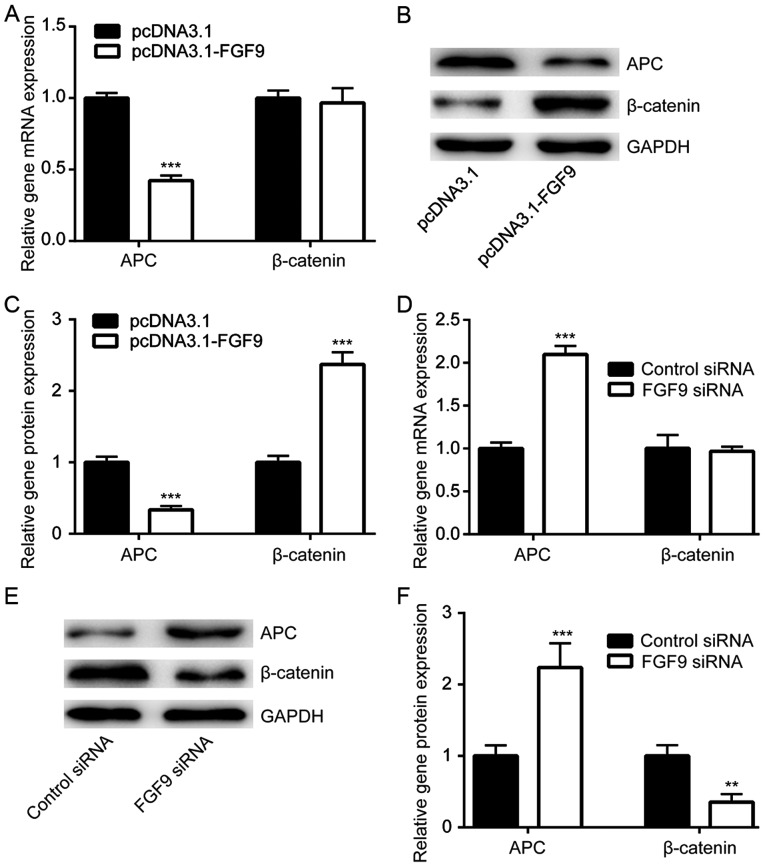
The Wnt/β-catenin signaling pathway is responsible for chemoresistance in cancer cells (27). APC decreases β-catenin expression and is a negative regulator of Wnt/β-catenin signaling (28). Using RT-qPCR, it was demonstrated that FGF9 overexpression significantly decreased APC mRNA expression, but not β-catenin mRNA expression in LoVo cells (Fig. 4A). Western blot analysis demonstrated that APC protein expression was downregulated and β-catenin protein expression was elevated upon transfection of pcDNA3.1-FGF9 (Fig. 4B and C). By contrast, silencing of FGF9 significantly increased APC mRNA expression and protein levels, whilst β-catenin protein levels decreased (Fig. 4D and F). These results suggested that FGF9 might regulate cisplatin sensitivity of colorectal cancer cells via activation of the Wnt/β-catenin pathway.
Figure 4.
FGF9 activates the Wnt/β-catenin signaling pathway in colorectal cancer cells. (A) Overexpression of FGF9 reduced APC mRNA expression but did not alter β-catenin mRNA expression in LoVo cells. (B) FGF9 overexpression reduced APC protein levels and elevated β-catenin protein levels. (C) Quantitative analysis of relative FGF9 and APC protein expression from panel B. (D) Knockdown of FGF9 increased APC mRNA expression but did not alter β-catenin mRNA levels in LoVo cells. (E) Knockdown of FGF9 increased APC protein levels and decreased β-catenin protein levels. (F) Quantitative analysis of relative FGF9 and APC protein expression from panel E. **P<0.01 and ***P<0.001 vs. control. FGF9, fibroblast growth factor 9; APC, adenomatous polyposis coli; siRNA, small interfering RNA.
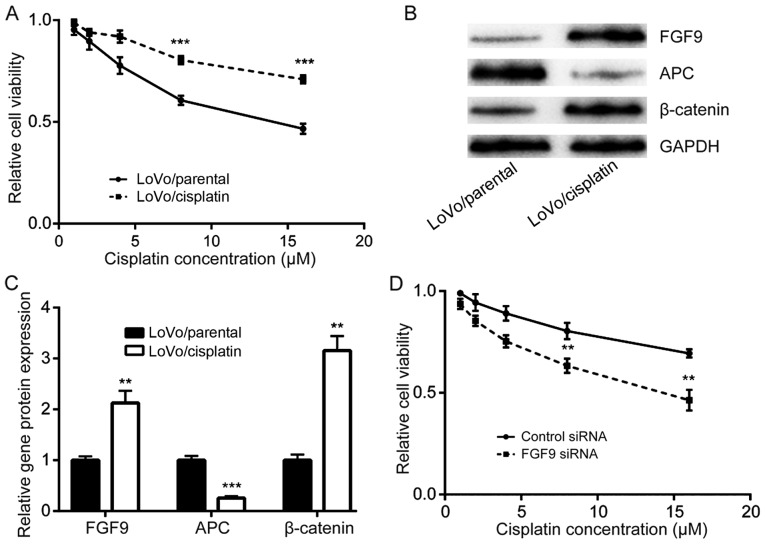
FGF9 overexpression is responsible for development of cisplatin resistance
The role of FGF9 in the development of cisplatin resistance in colorectal cancer cells was investigated. Via continuous exposure of LoVo cells with either DMSO or cisplatin, cisplatin resistant LoVo cells (LoVo/cisplatin) and parental LoVo cells (LoVo/parental) were established. Cell viability assays demonstrated that LoVo/cisplatin cells were less sensitive to cisplatin treatment compared with LoVo/parental cells (Fig. 5A). Western blot analysis determined that FGF9 and β-catenin protein expression was increased and APC protein expression was decreased in LoVo/cisplatin cells, compared with LoVo/parental cells (Fig. 5B and C). Notably, silencing of FGF9, enhanced cisplatin sensitivity of LoVo/cisplatin cells (Fig. 5D), which suggested that the FGF9/Wnt signaling pathway has a key role in cisplatin resistance development in colorectal cancer cells.
Figure 5.
Overexpression of FGF9 leads to development of cisplatin resistance in LoVo cells. (A) Compared with parental LoVo cells (LoVo/parental), cisplatin resistant LoVo cells (LoVo/cisplatin) were less sensitive to cisplatin treatment. ***P<0.001 vs. LoVo/parental. (B) Protein expression for FGF9 and β-catenin in LoVo/cisplatin cells increased and APC protein expression decreased compared with LoVo/parental cells. (C) Quantitative analysis of FGF9, APC and β-catenin protein expression from panel B. (D) Silencing of FGF9 enhanced cisplatin-induced cytotoxicity in LoVo/cisplatin cells. **P<0.01 vs. control siRNA. FGF9, fibroblast growth factor 9; APC, adenomatous polyposis coli; siRNA, small interfering RNA.
Discussion
Overexpression of FGF9 has been reported in colorectal cancer and is associated with poor clinical outcome in patients (29). A recent study demonstrated that FGF9-overexpressing colorectal cancer cells exhibited anti-epidermal growth factor receptor therapeutic properties via overactivation of FGFR signaling (30). To date, the role of FGF9 in mediating chemotherapy resistance in colorectal cancer remains not fully understood. The present study determined that FGF9 was a regulator of cisplatin sensitivity in colorectal cancer cells and a potential molecular mechanism was identified.
Several mechanisms have been identified to promote FGF9 expression in colorectal cancer including gene amplification, hypoxia stimulation and non-coding RNA elevation (18,30,31). The present study observed significant elevation of FGF9 in colorectal tumor tissues compared with normal tissues, which is consistent with a previous study (18). In addition, higher expression of FGF9 was associated with late-stage colorectal cancer compared with early-stage, suggesting that FGF9 may have a pivotal role in progression of colorectal cancer. FGF9 promotes colorectal cancer cell proliferation, migration and invasion (15). The present study determined that overexpression of FGF9 attenuated the cytotoxic effect of cisplatin in LoVo cells, whilst silencing of FGF9 enhanced the cytotoxic effect of cisplatin. Furthermore, FGF9 overexpression reduced cisplatin-induced cell apoptosis whilst FGF9 knockdown enhanced the apoptotic effect of cisplatin treatment. Notably, FGF9 expression was significantly elevated in the established cisplatin-resistant LoVo cells compared with parental LoVo cells. Silencing of FGF9 sensitized LoVo/cisplatin cells to cisplatin treatment. Taken together, these findings suggested that FGF9 was a mediator of cisplatin resistance in colorectal cancer cells, and might be a biomarker and treatment target for colorectal cancer.
It is well-known that the Wnt/β-catenin signaling pathway has a critical role in progression of colorectal cancer (28). In particular, activation of the Wnt/β-catenin signaling pathway is essential for maintenance of colorectal cancer cell stemness which contributes to development of chemotherapy resistance (27,32). The transcriptional control exerted by β-catenin is under stringent negative regulation mediated by various interacting factors, such as APC (21). It has been reported that FGF9 activates the Wnt/β-catenin signaling pathway in epithelial cells (33), and FGF9 has been identified as a target gene of Wnt/β-catenin signaling (34). The present study observed that FGF9 overexpression decreased APC expression and elevated β-catenin expression in LoVo cells. By contrast, FGF9 silencing increased APC expression and decreased β-catenin levels. In LoVo/cisplatin cells, an increase in APC expression and reduction of β-catenin expression was demonstrated. Taken together, these findings suggested that FGF9 might promote cisplatin resistance via activation of the Wnt/β-catenin signaling pathway in colorectal cancer cells.
In conclusion, the present study determined that FGF9 was an important mediator of cisplatin resistance in colorectal cancer cells via regulation of Wnt/β-catenin signaling, which suggested that FGF9 may be a potential biomarker and treatment target for colorectal cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of materials and data
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Author's contributions
ZZ, YZ, XQ and YW carried out the experiments and analyzed the data. JF conceived the experiments, supervised the present study and prepared the manuscript.
Ethics approval and consent to participate
Experiments involving patient tissues were approved by the Ethics Committee of the Shanghai Eighth People's Hospital. Written consent was provided by all enrolled patients.
Patient consent for publication
Not applicable.
Competing of interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Florea AM, Büsselberg D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers (Basel) 2011;3:1351–1371. doi: 10.3390/cancers3011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y, Coudert B, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- 4.Englinger B, Mair M, Miklos W, Pirker C, Mohr T, van Schoonhoven S, Lötsch D, Körner W, Ferk F, Knasmüller S, et al. Loss of CUL4A expression is underlying cisplatin hypersensitivity in colorectal carcinoma cells with acquired trabectedin resistance. Br J Cancer. 2017;116:489–500. doi: 10.1038/bjc.2016.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen MH, Gootenberg J, Keegan P, Pazdur R. FDA drug approval summary: Bevacizumab plus FOLFOX4 as second-line treatment of colorectal cancer. Oncologist. 2007;12:356–361. doi: 10.1634/theoncologist.12-6-713. [DOI] [PubMed] [Google Scholar]
- 6.Arnold D, Prager GW, Quintela A, Stein A, Moreno Vera S, Mounedji N, Taieb J. Beyond second-line therapy in patients with metastatic colorectal cancer: A systematic review. Ann Oncol. 2018;29:835–856. doi: 10.1093/annonc/mdy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stintzing S. Management of colorectal cancer. F1000Prime Rep. 2014;6:108. doi: 10.12703/P6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellman MB, Yan D, Ahmadinia K, Chen D, An HS, Im HJ. Fibroblast growth factor control of cartilage homeostasis. J Cell Biochem. 2013;114:735–742. doi: 10.1002/jcb.24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: From a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–279. doi: 10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]
- 11.Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, Nagano Y, Fujii S, Kunisaki C, Shiozawa M, Akaike M, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009;21:211–216. [PubMed] [Google Scholar]
- 12.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 13.Song J, Slack JM. XFGF-9: A new fibroblast growth factor from Xenopus embryos. Dev Dyn. 1996;206:427–436. doi: 10.1002/(SICI)1097-0177(199608)206:4<427::AID-AJA8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 14.Ohgino K, Soejima K, Yasuda H, Hayashi Y, Hamamoto J, Naoki K, Arai D, Ishioka K, Sato T, Terai H, et al. Expression of fibroblast growth factor 9 is associated with poor prognosis in patients with resected non-small cell lung cancer. Lung Cancer. 2014;83:90–96. doi: 10.1016/j.lungcan.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu YX, Liu JF, He ZM. miR-26a suppresses tumor growth and metastasis by targeting FGF9 in gastric cancer. PLoS One. 2013;8:e72662. doi: 10.1371/journal.pone.0072662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107:21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Jin C, Hamana T, Liu J, Wang C, An L, McKeehan WL, Wang F. Overexpression of FGF9 in prostate epithelial cells augments reactive stroma formation and promotes prostate cancer progression. Int J Biol Sci. 2015;11:948–960. doi: 10.7150/ijbs.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen TM, Shih YH, Tseng JT, Lai MC, Wu CH, Li YH, Tsai SJ, Sun HS. Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res. 2014;42:2932–2944. doi: 10.1093/nar/gkt1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 20.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–180. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 21.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang C, Yu M, Xie X, Huang G, Peng Y, Ren D, Lin M, Liu B, Liu M, Wang W, Kuang M. miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol Rep. 2017;38:2351–2359. doi: 10.3892/or.2017.5924. [DOI] [PubMed] [Google Scholar]
- 23.Chen B, Zhang D, Kuai J, Cheng M, Fang X, Li G. Upregulation of miR-199a/b contributes to cisplatin resistance via Wnt/β-catenin-ABCG2 signaling pathway in ALDHA1+colorectal cancer stem cells. Tumour Biol. 2017;39:1010428317715155. doi: 10.1177/1010428317715155. [DOI] [PubMed] [Google Scholar]
- 24.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 27.Chikazawa N, Tanaka H, Tasaka T, Nakamura M, Tanaka M, Onishi H, Katano M. Inhibition of Wnt signaling pathway decreases chemotherapy-resistant side-population colon cancer cells. Anticancer Res. 2010;30:2041–2048. [PubMed] [Google Scholar]
- 28.Schneikert J, Behrens J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. 2007;56:417–425. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leushacke M, Sporle R, Bernemann C, Brouwer-Lehmitz A, Fritzmann J, Theis M, Buchholz F, Herrmann BG, Morkel M. An RNA interference phenotypic screen identifies a role for FGF signals in colon cancer progression. PLoS One. 2011;6:e23381. doi: 10.1371/journal.pone.0023381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizukami T, Togashi Y, Naruki S, Banno E, Terashima M, de Velasco MA, Sakai K, Yoneshige A, Hayashi H, Fujita Y, et al. Significance of FGF9 gene in resistance to anti-EGFR therapies targeting colorectal cancer: A subset of colorectal cancer patients with FGF9 upregulation may be resistant to anti-EGFR therapies. Mol Carcinog. 2017;56:106–117. doi: 10.1002/mc.22476. [DOI] [PubMed] [Google Scholar]
- 31.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Z, Kang HY, Lee S, Kang SW, Goo B, Cho SB. Up-regulation of fibroblast growth factor (FGF) 9 expression and FGF-WNT/β-catenin signaling in laser-induced wound healing. Wound Repair Regen. 2014;22:660–665. doi: 10.1111/wrr.12212. [DOI] [PubMed] [Google Scholar]
- 34.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]