Abstract
Aims
KDM1A/LSD1 and ZNF217 are involved in a protein complex that participates in transcriptional regulation. ZNF217 has been analysed in numerous cancers and its amplification has been associated with advanced stages of disease; however, a similar role for KDM1A/LSD1 has not been uncovered. In this study, we estimated the number of KDM1A/LSD1 and ZNF217 gene copies in tissue samples from patients diagnosed with colorectal cancer (CRC), as well as its association with clinicopathological features in patients with CRC.
Methods
Paraffin-embedded tumour samples from 50 patients with CRC with a histopathological diagnosis of CRC were included. The number of copies of KDM1A/LSD1 and ZNF217 genes was determined by fluorescence in situ hybridisation (FISH). We also analysed the association between copy numbers of selected genes and clinicopathological data based on multivariate analysis.
Results
Deletion of the KDM1A/LSD1 gene occurred in 19 samples (38%), whereas ZNF217 gene amplification was identified in 11 samples (22%). We found a significant association between lymph node metastasis or advanced tumour stage and KDM1A/LSD1 gene deletion (p value=0.0003 and p value=0.011, respectively).
Conclusions
KDM1A/LSD1 gene deletion could be considered a novel prognostic biomarker of late-stage CRC.
Keywords: KDM1A/LSD1, ZNF217, colorectal cancer
Introduction
Worldwide, colorectal cancer (CRC) is the third most common neoplasm and the second leading cause of death related to cancer.1 Environmental, epigenetic and genetic factors have been associated with this disease.2 Molecular changes found in CRC can be classified into three main groups including microsatellite instability, CpG island methylator phenotype and chromosomal instability. The latter explains most CRC cases and is characterised by gains and losses of whole or partial chromosomes that could result in gene copy number variations.3
The chromosomal instability found in patients with CRC often includes gains in chromosomes 5, 7, 8q, 13, 20 and X, as well as losses in 18, 17 p, 14, 5q, 4 and 1 p.4 It has been documented that a 20q gain is associated with adenoma-carcinoma progression due to the selection of genes that confer an advantage for survival and growth,5 whereas 1 p deletion occurs in 50% of CRC cases and is related to tumour progression and metastasis.6 7 The KDM1A/LSD1 gene is located in 1p36.12 and encodes the lysine-specific demethylase 1A8; in turn, this protein is involved in different complexes and functions such as cellular proliferation and chromosome segregation.9 More specifically, KDM1A and the ZNF217/CoREST/CtBP1 complex co-operate to repress transcriptional activity.10 The ZNF217 gene, mapped to 20q13.2 and described as an oncogene, is frequently duplicated in CRC.11 Further, augmented gene copy numbers or amplification usually correlates with increased expression.12 13 Since ZNF217 gene amplification has been identified in approximately 60%–65% of patients with CRC, it has been proposed to be a biomarker for this disease and perhaps other human cancers.11
The transcriptional repressor complex KDM1A–ZNF217 regulates tumour suppressor genes and thus plays a critical role in cancer development. KDM1A functions in the repression of chromatin structure by demethylating lysine 4 of histone H3 (H3K4), but also promotes the activation of target genes through methyl modifications to lysine 9 of histone H3 (H3K9); whereas KDM1A recognises specific substrates that are added to the complex,14 15 ZNF217 protein acts as a transcriptional regulator of the target genes.16
Even though the amplification of ZNF217 has been well documented in CRC, the amplification status of KDM1A/LSD1 in any cancer type is not known. The aim of this work was to evaluate gene copy number variation in KDM1A/LSD1 and ZNF217 genes and its association with clinicopathological features in patients with CRC.
Materials and methods
Patients and tumour samples
Tumour samples and medical records from patients evaluated at the Histopathology Service of Civil Hospitals “Dr. Juan I. Menchaca” and “Fray Antonio Alcalde” at Guadalajara, Jalisco, Mexico, between 2003 and 2012 were included.
Fifty paraffin-embedded tumour samples of CRC were selected with a histopathological diagnosis of colorectal adenocarcinoma obtained before the patients received chemotherapy and/or radiation. Clinicopathological data were reviewed, such as age, sex, tumour location, tumour, node, metastases (TNM) stage, lymph node metastasis and distant metastasis. This work was a cross-sectional retrospective study.
Deparaffined and pretreatment samples
From sections comprising tumour cells that were defined histologically, 5 µm thick slices were cut and fixed on slides for fluorescence in situ hybridisation (FISH) analysis. Briefly, slides were heated at 58°C overnight on a HYBrite TM (VYSIS, Downers Grove, Illinois, USA), run through a series of three steps in Citrisolv clearing agent (FISHER brand) for 10 min each, incubated twice in 100% ethanol for 5 min each at room temperature and left to dry. Slides were then deparaffined with the pretreatment kit I (2J02-32 Abbott).
Interphase FISH
FISH analysis was performed according to manufacturer’s protocol by co-hybridising the sample and probes at 75°C in the HYBrite device. We used a probe that included the BAC clone RP11-152I19 (1p36.12, NCBI36/hg18, CHR1:23184031–23355672; Empire Genomics, USA) labelled with 5-fluorescein fluorophore and the reference CEP 1 ALPHA spectrum orange probe (D1Z5, Abbott Molecular 06J39-036, USA); these probes identify 100% of KDM1A/LSD1 gene sequences and the chromosome 1 centromere (p11.1q11.1), respectively. We also used the ZNF217/1061L1 (Cytocell LPS 005, UK) specific probe for ZNF217 in 20q13.2 (spectrum green) coupled with the reference probe 1061 L1 located in 20pter (spectrum red). FISH signals were analysed using an AxioImager.A1 epifluorescence microscope (Carl Zeiss, Jena, Germany) equipped with filters for DAPI (4’,6-diamidino-2-phenylindole), FITC (fluorescein isothiocyanate) and Texas Red.
FISH interpretation
Two investigators (RR-R and MLA-M) evaluated 100 non-overlapping interphase cells for each case. The ratio of gene deletion or amplification was assessed by dividing the total number of target gene probe signals by the total number of control probe signals. A ratio value ≤0.85 was considered a gene deletion,17 whereas a ratio value ≥2 was established as gene amplification.18 Additionally, apparent aneuploidies of chromosome 1 and 20 were identified when the signal proportion of the gene target versus the control probes was different from 2:2. According to Kuhn et al, 19 we interpreted these findings as polysomy (trisomy, tetrasomy or more) whenever ≥3 copies of the control probe were found in at least 40% of the cells analysed.
Statistical analysis
Statistical packages for data analysis were IBM SPSS Statistics Base V.22.0 and Microsoft Excel 2010, and we used Pearson correlation analysis to test for any association between FISH results for KDM1A/LSD1 and ZNF217 genes and CRC characteristics. To evaluate predictive factors, a multivariate analysis of age, sex, tumour location, TNM stage, lymph node metastasis and distant metastasis was performed. A p<0.05 was considered statistically significant.
Results
The 50 patients with CRC had a mean age of 61 years (range 28–89). Their characteristics are shown in table 1.
Table 1.
Clinical and histopathological characteristics of patients with colorectal cancer
| N (%) | |
| Age | |
| Over 50 | 38 (76) |
| Under 50 | 12 (24) |
| Sex | |
| Female | 22 (44) |
| Male | 28 (56) |
| Tumour location | |
| Colon | 38 (76) |
| Rectum | 12 (24) |
| TNM stage | |
| I–II | 26 (52) |
| III–IV | 24 (48) |
| Lymph node metastasis | 15 (30) |
| Distant metastasis | 9 (18) |
TNM, tumour/node/metastasis staging system.
KDM1A analysis
A loss of the KDM1A/LSD1 gene was detected in 38% (n=19) of the analysed samples (figure 1). With respect to clinical and histopathological characteristics, this deletion was only associated with metastasis to the lymph nodes (p=0.0003) and TNM stage III–IV (p<0.011). In addition, chromosome 1 gains were observed in 64% (n=32) of samples.
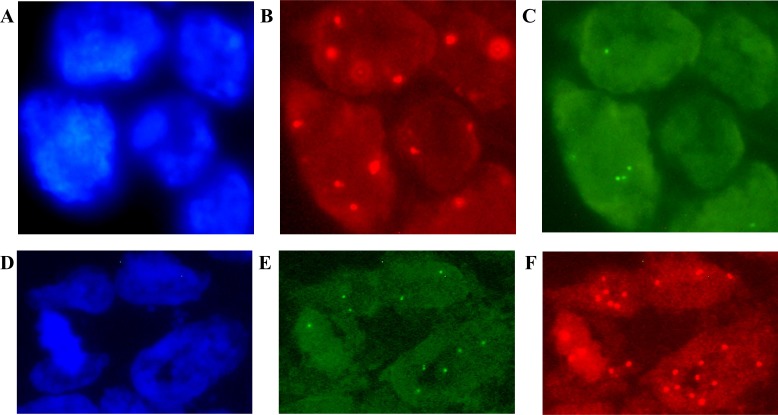
Figure 1.
Fluorescence in situ hybridisation (FISH) results for the KDM1A/LSD1 (A–C) and ZNF217 (D–F) genes in patients with colorectal cancer (CRC). Image (A) shows the cells stained with DAPI; (B–C) gene deletion is interpreted for (B) chromosome 1 centromere signals in red versus (C) green signals for the KDM1A/LSD1 gene. In addition, image (D) shows DAPI counterstaining; (E–F) gene amplification is considered according to (E) 20pter control signals in green versus (F) red signals of the ZNF217 gene. Original magnification ×100 using an AxioImager.A1 epifluorescence microscope (Carl Zeiss, Jena, Germany). Uneven illumination was corrected using a control image as described by Marty.41
ZNF217 analysis
The expected amplification of ZNF217 was observed in only 22% (n=11) of cases (figure 1), but it was not associated with any clinical or histopathological characteristics. Chromosome 20 polysomy was observed in 66% (n=33) of CRC cases.
The concurrence of KDM1A/LSD1 gene deletion and ZNF217 gene gain (either by gene amplification and/or polysomy) was found in 34% (n=17) of cases (table 2) and was associated with metastasis to the lymph node (p<0.0007) and TNM stage III–IV (p<0.025).
Table 2.
Findings for KDM1A/LSD1 and ZN217 gene copy number variation in patients with colorectal cancer
| Patient | Sex | Age |
KDM1A/LSD1
deletion |
ZNF217
amplification |
Chr 1 polysomy | Chr 20 polysomy |
| 2 | F | 62 | + | + | + | |
| 3 | M | 62 | ||||
| 4 | M | 76 | + | |||
| 6 | M | 48 | + | |||
| 7 | F | 47 | + | + | ||
| 8 | F | 67 | + | + | + | |
| 9 | F | 69 | + | + | + | |
| 10 | F | 55 | + | |||
| 11 | M | 78 | + | + | ||
| 12 | F | 89 | + | + | + | |
| 13 | M | 78 | + | + | ||
| 14 | F | 62 | + | + | + | |
| 15 | M | 84 | + | |||
| 16 | F | 47 | + | |||
| 18 | F | 69 | + | + | ||
| 19 | F | 75 | + | + | + | |
| 20 | M | 70 | + | + | + | + |
| 21 | M | 41 | + | + | ||
| 22 | F | 82 | + | + | ||
| 23 | F | 63 | + | + | + | |
| 24 | M | 77 | + | + | ||
| 25 | M | 53 | + | + | + | |
| 26 | M | 41 | + | + | + | |
| 27 | M | 35 | + | + | + | |
| 28 | M | 52 | + | + | + | |
| 29 | F | 76 | + | + | + | + |
| 31 | M | 58 | + | + | + | |
| 32 | M | 56 | + | + | + | |
| 33 | F | 57 | + | |||
| 34 | F | 48 | + | + | + | |
| 35 | M | 50 | + | |||
| 36 | M | 71 | + | + | ||
| 37 | M | 38 | + | + | ||
| 39 | M | 64 | + | + | ||
| 40 | F | 69 | + | + | ||
| 42 | M | 65 | + | + | ||
| 44 | F | 34 | + | + | ||
| 45 | M | 55 | + | + | ||
| 46 | F | 41 | + | |||
| 47 | M | 63 | + | + | + | |
| 48 | F | 62 | + | + | ||
| 49 | F | 85 | + | |||
| 50 | M | 52 | + | + | + |
+, positive alteration; F, female; M, male.
Copy number variation of KDM1A/LSD1 and ZNF217 was assessed in 43 of 50 CRC patient samples. The corresponding data including polysomies of chromosomes 1 and 20 are shown in table 2. It was found that 32% (n=16) of patients harboured both KDM1A/LSD1 gene deletion and chromosome 1 polysomy. Regarding the ZNF217 gene, only 10% (n=5) of the patient samples exhibited chromosome 20 polysomy and gene amplification.
Discussion
KDM1A and ZNF217 proteins are components of transcriptional complexes that positively modulate or co-repress the expression of target genes including some related to carcinogenesis.10 14 Actually, Sehrawat et al 20 demonstrated that KDM1A in association with ZNF217 stimulates prostate cancer cell survival, through activation of the cell cycle and embryonic stem cell genes that are enriched in lethal prostate tumours. This supports the fundamental role of both proteins in transcriptional regulation during cancer.
In the present assessment of copy number variations in KDM1A and ZNF217, we identified ZNF217 amplification in only 22% of CRC samples. Gene amplification has been associated with poor prognosis in different types of cancer.16 In CRC, several studies have reported that ZNF217 gene amplification is associated with increased metastatic potential,21 poorer survival11 and more aggressive clinical behaviour,22 whereas a poor response or resistance to treatment with 5-fluorouracil was found to be related to CRC microsatellite instability.23 However, we did not find any association between ZNF217 amplification and clinical or histopathological data. This lack of association could be ascribed to the lower ratio of gene amplification as compared with frequencies of 41%–100% found in studies supporting such associations11 21 22; however, it must be noted that Hidaka et al 21 classified samples according to liver metastasis and that Postma et al 22 analysed flat colorectal carcinomas, in addition to the fact that in both studies, the sample size was smaller than ours. Similarly, Huang et al 24 did not identify prognostic significance for ZNF217 amplification based on that observed in 31% of 68 patients with ovarian clear cell carcinoma.
Regarding KDM1A/LSD1 gene copy variation, we found gene deletion in 38% of patients. Even though no other marker on chromosome 1 p was analysed and the KDM1A/LSD1 gene is located on 1p36.12, our findings could still be related to the loss of chromosome 1 p, since this chromosomal region is deleted in up to 50% of colorectal carcinomas.7 Chromosome 1p36 contains several candidate tumour suppressor genes such as CHD5, CAMTA1, KIF1B and CASZ1, which are related to the modulation of chromatin structure, transcription (via interactions with histones), poor prognosis in cancer, apoptosis, cell migration, cell proliferation and tumorigenicity.25 Nevertheless, the relevance of KDM1A/LSD1 might be greater due to its participation in several cellular processes such as histone and DNA methylation,26 cell differentiation,27 cell proliferation,28 chromatid segregation during mitosis29 and epithelial–mesenchymal transition (EMT), a phenomenon well known to facilitate metastasis.30 Indeed, KDM1A interacts with SNAIL1 to recruit the KDM1A corepressor complex, which in turn leads to the H3K4me2 demethylation of E-cadherin-associated active promoters and the downregulation of SNAIL1 and/or E-cadherin, which promotes cell motility and ultimately EMT.31 32
By including results of KDM1A/LSD1 gene deletion and clinicopathological data in a multivariate analysis, we identified a relationship between lymph node metastasis or advanced tumour stages and KDM1A/LSD1 gene deletion (p values=0.0003 and 0.011, respectively). Hence, this deletion could be a useful biomarker for the late stages of cancer. In accordance, Wang et al 33 showed that KDM1A protein is part of the Mi-2/nucleosome remodelling and deacetylase corepressor complex and performed in vitro assays to assess the effect of this protein on breast cancer cells. These authors found that the overexpression of wild-type KDM1A led to a threefold decrease in cell invasion, whereas knockdown increased cell invasiveness by approximately fivefold. Moreover, Wang et al obtained the same results after analysing breast cancer metastasis in mice, wherein lung metastasis was suppressed in tumours with KDM1A overexpression and metastatic spread was found in the absence of KDM1A. Altogether, these data suggest that KDM1A/LSD1 gene suppression is key during metastasis. The apparent discrepancy between this conclusion and the previously annotated role for KDM1A in cancer, especially the association between KDM1A overexpression and poor prognosis and the proliferation and invasion of several neoplasms, such as lung, liver, oesophagus, and colon cancers (15), can be resolved if we consider the genetic background of the cells and the effect of environmental signals on KDM1A.34
Based on the analysis of gene copy numbers, we also inferred aneuploidies of chromosome 1 and 20 from the FISH patterns for gene and control signals. We found 32 (64%) and 33 cases (66%) with gains of chromosome 1 and 20, respectively. Comparable polysomies, mainly trisomies and tetrasomies, are common in cancer cells.35 Although aneuploidies, and specifically trisomies, can suppress tumour growth in the short term, they improve the long-term advantages of premalignant cells and generate a destabilising effect that contributes to the aggressive growth of tumours.36 It seems that aneuploidy, found in ~85% of different cancers,37 results in heterogeneity and facilitates the adaptation of cancer cells via the acquisition of advantageous features.38 In CRC, aneuploidy is observed in advanced stages probably as a consequence of genetic alterations in diploid or polyploid cells.39 40
Despite the fact that simultaneous KDM1A/LSD1 deletions and ZNF217 gains occurred in 34% of patients and was associated with metastasis to the lymph node (p<0.0007) and TNM stage III–IV (p<0.025), this was not found to comprise a synergistic effect of gene copy number variation, since ZNF217 gains and clinicopathological characteristics were not associated in patients with CRC. Since small sample size is a limitation of this study, the lack of association between ZNF217 amplification and CRC could also be related to the low number of advanced-stage tumour samples analysed. Notwithstanding, given the colocalisation of KDM1A/LSD1 and ZNF217 proteins in the transcriptional repression protein complex, the findings of deletion and amplification of KDM1A/LSD1 and ZNF217 genes could still be related to CRC progression, and further studies based on CRC patients with advanced tumour stages or metastasis are required.
In conclusion, our results show that KDM1A/LSD1 gene deletion is associated with lymph node metastasis and advanced stages of CRC and could be considered a biomarker of prognostic value in this common malignancy. Synergism between KDM1A/LSD1 gene losses and ZNF217 gene gains has also emerged as possible event relevant to the progression of CRC. Finally, to our knowledge, this is the first report to analyse KDM1A/LSD1 and ZNF217 copy numbers and their association with clinicopathological data in patients with CRC.
Take home messages.
Deletion of the KDM1A/LSD1 gene is associated with lymph node metastasis and advance tumour stages in patients with CRC.
KDM1A/LSD1 gene deletion could be a useful biomarker of late-stage colorectal carcinomas.
Deletion of KDM1A/LSD1 and amplification of ZNF217 genes could exert a synergistic effect on the progression of CRC.
Acknowledgments
We thank Dr. H. Rivera for the critical review of the manuscript.
Footnotes
Handling editor: Runjan Chetty.
Contributors: MLA-M and MG-A conceived and designed the study, RAF-T and FJC-C collected and prepared the samples; RR-R and JP-S performed the experiments. RR-R wrote the paper. MG-A, MLA-M and JMM-O reviewed and edited the manuscript. The final version of the manuscript has been read and approved by all authors.
Funding: The present study was supported by internal funds of Universidad de Guadalajara.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This is a cross-sectional and retrospective study approved by the committees of research, ethics and biosecurity, incorporated into the Research State Register 65/UG-JAL/2011. Informed consent was not obtained since the samples corresponded to residual material after pathological diagnosis. The ethics committee approved the material use for scientific research in accordance with the provisions of the ‘International Ethical Guidelines for Health Related Research Involving Humans’, prepared by the Council for International Organizations for Medical Sciences in collaboration with the WHO. The observance of the Mexican General Health Law was also considered. We obtained coded samples before use to maintain absolute confidentiality.
Provenance and peer review: Not commissioned; internally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1. Bray F, Ferlay J, Soerjomataram I, et al. . Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Obuch JC, Ahnen DJ. Colorectal cancer. genetics is changing everything. Gastroenterol Clin North 2016;45:459–76. [DOI] [PubMed] [Google Scholar]
- 3. Tariq K, Ghias K, Tariq K, et al. . Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med 2016;13:120–35. 10.20892/j.issn.2095-3941.2015.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Migliore L, Migheli F, Spisni R, et al. . Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol 2011;2011:1–19. 10.1155/2011/792362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett 2018;16:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thorstensen L, Qvist H, Heim S, et al. . Evaluation of 1p losses in primary carcinomas, local recurrences and peripheral metastases from colorectal cancer patients. Neoplasia 2000;2:514–22. 10.1038/sj.neo.7900111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knösel T, Schlüns K, Stein U, et al. . Chromosomal alterations during lymphatic and liver metastasis formation of colorectal cancer. Neoplasia 2004;6:23–8. 10.1016/S1476-5586(04)80050-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. NCBI Gene [Internet]. Bethesda (MD): National Center for Biotechnology Information (US), National Library of Medicine, 2007. Available: https://www.ncbi.nlm.nih.gov/gene/23028 [Accessed 22 Feb 2019].
- 9. Kozub MM, Carr RM, Lomberk GL, et al. . Lsd1, a double-edged sword, confers dynamic chromatin regulation but commonly promotes aberrant cell growth. F1000Res 2016;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banck MS, Li S, Nishio H, et al. . The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics 2009;4:100–6. 10.4161/epi.4.2.7953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rooney PH, Boonsong A, McFadyen MCE, et al. . The candidate oncogeneZNF217 is frequently amplified in colon cancer. J Pathol 2004;204:282–8. 10.1002/path.1632 [DOI] [PubMed] [Google Scholar]
- 12. Bagci O, Kurtgöz S. Amplification of cellular oncogenes in solid tumors. N Am J Med Sci 2015;7:341–6. 10.4103/1947-2714.163641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quinlan KG, Verger A, Yaswen P, et al. . Crossley M: amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim Biophys Acta 1775;2007:333–40. [DOI] [PubMed] [Google Scholar]
- 14. Thillainadesan G, Isovic M, Loney E, et al. . Genome analysis identifies the p15INK4B tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol 2008;28:6066–77. 10.1128/MCB.00246-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagasawa S, Sedukhina AS, Nakagawa Y, et al. . Lsd1 overexpression is associated with poor prognosis in basal-like breast cancer, and sensitivity to PARP inhibition. PLoS One 2015;10:e0118002 10.1371/journal.pone.0118002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohen PA, Donini CF, Nguyen NT, et al. . The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget 2015;6:41566–81. 10.18632/oncotarget.5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pinkham MB, Telford N, Whitfield GA, et al. . Fishing tips: what every clinician should know about 1p19q analysis in gliomas using fluorescence in situ hybridisation. Clin Oncol 2015;27:445–53. 10.1016/j.clon.2015.04.008 [DOI] [PubMed] [Google Scholar]
- 18. Geppert C-I, Rümmele P, Sarbia M, et al. . Multi-colour fish in oesophageal adenocarcinoma—predictors of prognosis independent of stage and grade. Br J Cancer 2014;110:2985–95. 10.1038/bjc.2014.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhn E, Bahadirli-Talbott A, Shih I-M. Frequent CCNE1 amplification in endometrial intraepithelial carcinoma and uterine serous carcinoma. Mod Pathol 2014;27:1014–9. 10.1038/modpathol.2013.209 [DOI] [PubMed] [Google Scholar]
- 20. Sehrawat A, Gao L, Wang Y, et al. . Lsd1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci U S A;20181:E4179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hidaka S, Yasutake T, Takeshita H, et al. . Differences in 20q13.2 copy number between colorectal cancers with and without liver metastasis. Clin Cancer Res 2000;6:2712–7. [PubMed] [Google Scholar]
- 22. Postma C, Hermsen M, Coffa J, et al. . Chromosomal instability in flat adenomas and carcinomas of the colon. J Pathol 2005;205:514–21. 10.1002/path.1733 [DOI] [PubMed] [Google Scholar]
- 23. Lassmann S, Weis R, Makowiec F, et al. . Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med 2007;85:293–304. 10.1007/s00109-006-0126-5 [DOI] [PubMed] [Google Scholar]
- 24. Huang H-N, Huang W-C, Lin C-H, et al. . Chromosome 20q13.2 ZNF217 locus amplification correlates with decreased E-cadherin expression in ovarian clear cell carcinoma with PI3K-Akt pathway alterations. Hum Pathol 2014;45:2318–25. 10.1016/j.humpath.2014.07.020 [DOI] [PubMed] [Google Scholar]
- 25. Henrich K-O, Schwab M, Westermann F. 1P36 tumor Suppression—A matter of dosage? Cancer Res 2012;72:6079–88. 10.1158/0008-5472.CAN-12-2230 [DOI] [PubMed] [Google Scholar]
- 26. Wang J, Hevi S, Kurash JK, et al. . The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 2009;41:125–9. 10.1038/ng.268 [DOI] [PubMed] [Google Scholar]
- 27. Amente S, Lania L, Majello B. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta 1829;2013:981–6. [DOI] [PubMed] [Google Scholar]
- 28. Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J Biol Chem 2007;282:15471–5. 10.1074/jbc.M701023200 [DOI] [PubMed] [Google Scholar]
- 29. Lv S, Bu W, Jiao H, et al. . Lsd1 is required for chromosome segregation during mitosis. Eur J Cell Biol 2010;89:557–63. 10.1016/j.ejcb.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 30. Ambrosio S, Sacc CD, Majello B. Epigenetic regulation of epithelial to mesenchymal transition by the lysine-specific demethylase LSD1/KDM1A. Biochim Biophys Acta Gene Regul Mech 1860;2017:905–10. [DOI] [PubMed] [Google Scholar]
- 31. Lin T, Ponn A, Hu X, et al. . Requirement of the histone demethylase LSD1 in SNAI1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010;29:4896–904. 10.1038/onc.2010.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ismail T, Lee H-K, Kim C, et al. . KDM1A microenvironment, its oncogenic potential, and therapeutic significance. Epigenetics Chromatin 2018;11:33 10.1186/s13072-018-0203-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Zhang H, Chen Y, et al. . Lsd1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009;138:660–72. 10.1016/j.cell.2009.05.050 [DOI] [PubMed] [Google Scholar]
- 34. Hino S, Kohrogi K, Nakao M. Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci 2016;107:1187–92. 10.1111/cas.13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitelman F, Johansson B, Mertens F. Mitelman database of chromosome aberrations and gene fusions in cancer, 2019. Available: http://cgap.nci.nih.gov/Chromosomes/Mitelman [Accessed 1 Apr 2019].
- 36. Sheltzer JM, Ko JH, Replogle JM, et al. . Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 2017;31:240–55. 10.1016/j.ccell.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol 2006;18:658–67. 10.1016/j.ceb.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 38. Cosenza MR, Krämer A. Centrosome amplification, chromosomal instability and cancer: mechanistic, clinical and therapeutic issues. Chromosome Res 2016;24:105–26. 10.1007/s10577-015-9505-5 [DOI] [PubMed] [Google Scholar]
- 39. Sugai T, Uesugi N, Nakamura S-ichi, et al. . Evolution of DNA ploidy state and DNA index in colorectal adenomas and carcinomas using the crypt isolation technique: new hypothesis in colorectal tumorigenesis. Pathol Int 2003;53:154–62. 10.1046/j.1440-1827.2003.01448.x [DOI] [PubMed] [Google Scholar]
- 40. De Angelis PM, Stokke T, Beigi M, et al. . Chromosomal 20q gain in the DNA diploid component of aneuploid colorectal carcinomas. Int J Cancer 2007;120:2734–8. 10.1002/ijc.22537 [DOI] [PubMed] [Google Scholar]
- 41. Marty GD. Blank-field correction for achieving a uniform white background in brightfield digital photomicrographs. Biotechniques 2007;42:716–20. 10.2144/000112488 [DOI] [PubMed] [Google Scholar]