Abstract
Glioma is the most common malignant brain tumour in adults, and the aetiology and mechanism of this tumour remain largely unknown. Previous studies have demonstrated that the long non-coding RNA X-inactive specific transcript (XIST) is upregulated in many cancers, and a high expression level of XIST is associated with poor clinical outcome. In the present study, the expression and function of XIST were investigated in the glioma cell line U251. XIST and microRNA (miR)-133a levels in glioma cell lines were detected by reverse transcription-quantitative polymerase chain reaction. Small hairpin RNA XIST (sh-XIST) and mimics/inhibitor of miR-133a were transfected in glioma cell lines and cell proliferation, invasion, migration and epithelial-mesenchymal transition (EMT) were examined. Luciferase assays were used to verify the associations among XIST, miR-133a and SRY-box (SOX)4. When XIST was knocked down, the proliferation, metastasis and EMT of glioma cells decreased. Notably, downstream genes of SOX4 were also upregulated or downregulated upon sh-XIST treatment. Overexpression of miR-133a inhibited glioma proliferation, metastasis and EMT via reducing the expression of SOX4; in contrast, knockdown of miR-133a exhibited the opposite effect, which revealed that miR-133a negatively regulates glioma progression. Furthermore, using luciferase assays, it was demonstrated that XIST and SOX4 could bind miR-133a in the predicted binding site; XIST competed with SOX4 for miR-133a binding. In conclusion, a XIST/miR-133a/SOX4 axis and a mechanism of XIST glioma in promoting cell proliferation and metastasis were revealed. These findings revealed that XIST has an oncogenic role in the tumourigenesis of glioma and may serve as a potential therapeutic target for glioma.
Keywords: X-inactive specific transcript, glioma cancer, microRNA-133a, SRY-box 4
Introduction
Gliomas are the most pervasive and aggressive major type of primary tumour in the nervous system (1). The 5-year survival rate of glioblastomas patients is 5% (1). The development of the malignant phenotype is the result of complicated processes that influence the expression of genes associated with angiogenesis, invasion and proliferation of tumour cells (2). Further studies on gene regulation networks will lead to a better understanding of the molecular mechanisms in glioma and eventually find novel biomarkers and therapeutic targets for glioma (3).
Long non-coding RNAs (lncRNAs), which are a class of transcripts >200 nucleotides in length, have been demonstrated to serve important roles in gene transcription and post-transcription regulation (4). lncRNAs are frequently dysregulated in cancer progression; for instance, targeting the microRNA (miRNA or miR)-146b-5p/HuR/lincRNA-p21/β-catenin signalling pathway may be a valuable therapeutic strategy against glioma (5); the expression of the lncRNA-reprogramming was negatively correlated with Kruppel like factor 4 and positively correlated with SRY-box (SOX)11 in glioma (6). The present study focused on the lncRNA X-inactive specific transcript (XIST), which is required for transcriptional silencing of one X-chromosome during development in female mammals (7). It has been reported that the lncRNA XIST promotes glioma tumourigenicity and angiogenesis (8). A previous study demonstrated that XIST inhibited miR-133a in pancreatic cancer (PC) cell lines and further revealed a XIST/miR-133a/EGFR signalling axis and provided a potential mechanism for XIST in PC cell proliferation (9). miR-133a is a multicopy gene with two copies, one on chromosome 18 and one on chromosome 20 (10). In tumourigenesis, miR-133a is expressed at a low level in several cancer types, such as glioma and other solid tumours, the epithelial-mesenchymal transition (EMT)-related transcription factor SOX4 is a direct target gene of miR-133a, and miR-133a directly binds to the 3′untranslated region (UTR) of SOX4 (10).
SOX4 is a major member of the SOX family that is located on 6p22.3 and encodes a protein of 474 amino acids with three distinguishable domains: A high mobility group box, a glycine-rich region, and a serine-rich region (11). SOX4 is associated with many developmental processes, such as T cell differentiation and the development of thymocytes, the nervous system, and the embryonic cardiovascular system (12–14). In recent years, a number of studies have suggested that SOX4 may be associated with tumour development and progression (15–17).
In the present study, it was demonstrated that XIST influenced glioma cell proliferation and migration by regulating the miR-133a/SOX4 axis. These findings suggested that XIST is potentially a therapeutic target for glioma.
Materials and methods
Cell culture
U251 glioma cell lines, control cell line HEB and the 293 cells were obtained from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China) and cultured at 37°C in humidified 5% CO2. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All media contained 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). The 293 cells were used to investigate the regulatory mechanism between miR-133a and XIST or between miR-133a and SOX4.
XIST small hairpin RNA (shRNA) and miRNA transfection
On the day prior to transfection, U251 cells were digested by trypsin, counted and seeded in a 6-well plate at 1×105 cells/well. When the cell confluence reached 90%, the medium was changed with serum-free DMEM and incubated at 37°C overnight. An sh-XIST vector was used to achieve knockdown of XIST (GeneCopoeia, Inc.). miR-133a mimics (5′-UUUGGUCCCCUUCAACCAGCUG-3′ and 5′-GCUGGUUGAAGGGGACCAAAU-3′) or an miR-133a inhibitor (5′-CAGCUGGUUGAAGGGGACCAAA-3′; GeneCopoeia, Inc.) were used to achieve miR-133a overexpression and inhibition, respectively. Transient transfections were performed using Lipofectamine 2000 (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Sh-negative control (NC; sense, 5′-GATCCGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAACTTTTTG-3′; and antisense, 5′-AATTCAAAAAGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAACG-3′), mimics NC (5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′) and inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′; GeneCopoeia, Inc.) were used as respective controls. XIST shRNA (sense, 5′-CACCGCTCTTGAACAGTTAATTTGCTTCAAGAGAGCAAATTAACTGTTCAAGAGCTTTTTTG-3′ and antisense, 5′-GATCCAAAAAAGCTCTTGAACAGTTAATTTGCTCTCTTGAAGCAAATTAACTGTTCAAGAGC-3′) or miRNA (hsa-miR-133a mimics, 5′-UUUGGUCCCCUUCAACCAGCUG-3′ and 5′-GCUGGUUGAAGGGGACCAAAUU-3′; and hsa-miR-133a inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′) was transfected into the indicated cell lines for 24 h at 37°C. Transfection efficiency was determined via reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Cells were transfected with mimics or inhibitors at a final concentration of 100 nM.
RT-qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. cDNA was synthesized by reverse transcription of total RNA using the All-in-One™ First-Strand cDNA Synthesis kit (GeneCopoeia, Inc.) according to the manufacturers' instruction at room temperature. Then, using cDNA as the template, the gene expression levels were analysed by qPCR conducted using iTaq™ Universal One-Step SYBR RT-qPCR kits on a real-time PCR system (both Bio-Rad Laboratories, Inc.). The qPCR conditions were 95°C for 5 sec and 60°C for 15 sec, followed by 70°C for 15 sec, for 45 cycles. The relative gene expression level was calculated using the 2−ΔΔCq method (18). The primers used were as follows: XIST forward, 5′-ACGCTGCATGTGTCCTTAG-3′ and reverse, 5′-GAGCCTCTTATAGCTGTTTG-3′; miR-133a forward, 5′-TTTGGTCCCCTTCAACCAGCTG-3′ and reverse, 5′-TAAACCAAGGTAAAATGGTCGA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′ and GAPDH forward, 5′-CATCACCATCTTCCAGGAGCG-3′ and reverse, 5′-TGACCTTGCCCACAGCCTTG-3′. All results were obtained from at least three independent experiments.
Western blotting
Cells were washed with cold PBS twice and lysed with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) containing the protease inhibitor for 30 min on ice. Following centrifugation at 12,000 × g and 4°C for 15 min, the protein concentration was determined using the enhanced bicinchoninic acid BCA protein assay kit (Beyotime Institute of Biotechnology). The supernatant was loaded on a 10% SDS-PAGE gel (10 µg total protein/lane) and proteins were separated. Proteins were then transferred to a polyvinylidene difluoride membrane, blocked with 5% bovine serum albumin (Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at room temperature and incubated with primary antibody at 4°C overnight. The primary antibodies used in the present study were as follows: Anti-Sox4 antibody (1:1,000; sc-518016; Santa Cruz Biotechnology, Inc.), anti-E-cadherin antibody (1:1,000; sc-59778; Santa Cruz Biotechnology, Inc.), anti-vimentin antibody (1:1,000; sc-373717; Santa Cruz Biotechnology, Inc.), anti-N-cadherin antibody (1:1,000; sc-53488; Santa Cruz Biotechnology, Inc.), anti-α-catenin antibody (1:1,000; sc-9988; Santa Cruz Biotechnology, Inc.) and anti-β-actin antibody (1:5,000; #4970; Cell Signaling Technology, Inc.) Membranes were washed with TBS containing 0.1% Tween-20 five times and then incubated with donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody at room temperature (1:10,000; sc-2313; Santa Cruz Biotechnology, Inc.) for 1 h. Membranes were finally washed in TBS and developed with diaminobenzidine horseradish peroxidase color development kit (Beyotime Institute of Biotechnology) for chemiluminescence substrate. Densitometric analyses were performed using a chemiluminescence imaging system (model 5200; Tanon Science and Technology Co., Ltd.) and built-in Tanon MP software (version 2014).
MTT assay
U251 cells were suspended in DMEM culture medium containing 10% FBS and placed in a 96-well plate with approximately 2,000 cells/200 µl volume in each well. These cells were incubated under the condition of 5% CO2 and 37°C for 24 h. Then, 20 µl MTT (5 mg/ml) was added in every well, and these cells continued to be cultured for an additional 4 h. Culture medium was removed, and 150 µl DMSO was added for the dissolution of the crystals on the shaker for 10 min. Optical density values were measured at 490 nm.
Cell migration analysis
Migration ability was determined using a wound-healing assay. U251 cells were plated into 12-well plates without antibiotics, and cells were transfected with sh-XIST or the control or with miR-133a mimics, miR-133a inhibitor or the control. Then, 24 h later, transfected cells were wounded with a sterile plastic 100 µl micropipette tip, the floating debris were washed with PBS, and the remaining cells were cultured in serum-free medium at 37°C. The width of the wound was measured at 0 and 24 h.
Cell invasion analysis
First, 80 µl diluted Matrigel was put into the upper chamber of a 24-well Transwell chamber (Corning Inc.) and incubated at 37°C for 30 min for gelling. U251 cells were harvested, and the cells were suspended in serum-free DMEM/F12. Then, all cells in various groups were placed in the upper chamber with 200 µl serum-free DMEM, and 600 µl DMEM culture medium containing 10% FBS was placed in the bottom chamber. Cells were incubated at 37°C for 20 h, and cells that migrated or invaded the lower surface of the membrane were removed from the 24-well plates. Cells adhering to the membrane were stained with crystal violet for 10 min at room temperature, and each insert was counted at ×100 magnification using a bright field microscope (Model DC 300F; Leica Microsystems GmbH).
Luciferase reporter assay
Luciferase reporter gene assay was implemented using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's instructions. Cells were seeded into 24-well plates and transfected with a wild-type (wt)-XIST luciferase reporter gene vector, a mutant (mut)-XIST vector containing a 6-bp mutation on the predicted miR-133a binding site within XIST, a wt-SOX4 3′UTR vector, or a mut-SOX4 3′UTR vector (all from Shanghai GenePharma Co., Ltd.) containing a mutation in the predicted miR-133a binding site in the 3′UTR of SOX4, along with the miR-133a mimic or mimic NC using Lipofectamine 3000 (Life Technologies; Thermo Fisher Scientific, Inc.). Following 48 h, cells were lysed using passive lysis buffer (Promega Corporation) and the luciferase activity was detected. Luciferase activity was normalized against Renilla. All experiments were performed at least three times.
Statistical analysis
Data are presented as the mean ± standard deviation. Statistical analysis was performed using SPSS software 16.0 (SPSS, Inc.). Statistical significance was measured using Student's t-test or one-way analysis of variance in conjunction with Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of XIST and downregulation of miR-133a in glioma cell lines
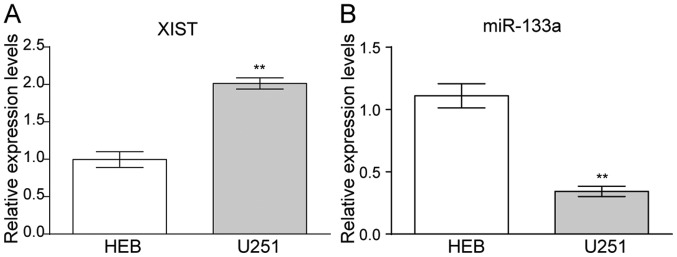
The relative expression levels of XIST and miR-133a were detected via RT-qPCR in the glioma cell line U251. The data revealed that XIST was upregulated in U251 compared with that in the controlled HEB cells (Fig. 1A). In contrast, miR-133a presented a lower expression in U251 compared with HEB cells (Fig. 1B).
Figure 1.
The expression of XIST and miR-133a in glioma cell lines. (A) XIST expression was higher in U251 compared with the control HEB cells. (B) miR-133a expression was lower in U251 compared with the control HEB cells. n=3. **P<0.01 vs. HEB. XIST, X-inactive specific transcript; miR, microRNA.
XIST suppresses miR-133a expression in glioma cell lines and enhances cell proliferation and invasion
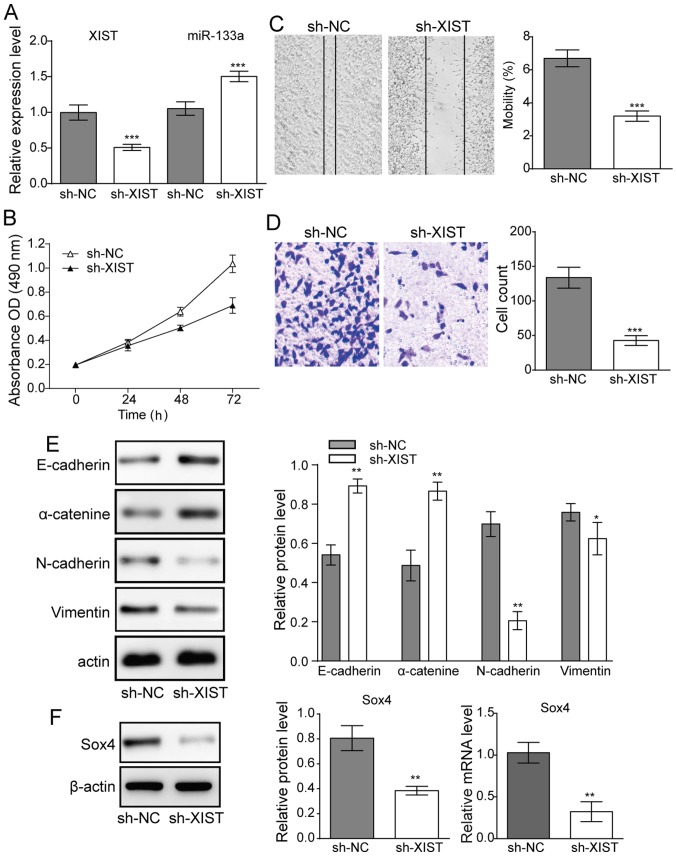
To evaluate the association of XIST expression with glioma cells' proliferation and metastasis. XIST knockdown was achieved by sh-XIST transfection, and the inhibitory efficiency was verified by RT-qPCR (Fig. 2A). The level of miR-133a was then detected by RT-qPCR. The results revealed that miR-133a expression was significantly enhanced when sh-XIST was transfected into U251 cells (Fig. 2A). These results indicated that XIST negatively regulated miR-133a in glioma cell lines.
Figure 2.
Effect of downregulation of XIST on glioma cell proliferation and metastasis. (A) Following transfection with sh-XIST, XIST was significantly downregulated. The transfection of sh-XIST further potentiated miR-133a in U251 cells. (B) Cell viability of U251 lines following sh-XIST transfection. (C) Cell migration (magnification, ×16) and (D) cell invasion assay of glioma cells (magnification, ×100). (E) Western blot results demonstrated the expression levels of E-cadherin N-cadherin, α-catenin and vimentin in glioma cells following sh-XIST transfection. (F) Expression levels of SOX4 transcription factor. n=3, *P<0.05, **P<0.01 and ***P<0.001 vs. sh-NC. XIST, X-inactive specific transcript; sh, small hairpin RNA; miR, microRNA; SOX, SRY-box; NC, negative control; OD, optical density.
MTT assays revealed that knocking down XIST reduced cell proliferation of U251 cell lines (Fig. 2B). Meanwhile, in the migration and invasion experiments, XIST knockdown also suppressed cell migration (Fig. 2C) and invasion activity (Fig. 2D) in glioma cells. Furthermore, the expression of EMT-related proteins E-cadherin, vimentin, N-cadherin and α-catenin, was investigated. Western blot results demonstrated that E-cadherin and α-catenin were significantly upregulated, whereas N-cadherin and vimentin were downregulated when XIST was knocked down in U251 cells (Fig. 2E), suggesting a suppression of EMT. Both mRNA and protein expression of the EMT-related transcription factor Sox4 were also significantly decreased upon knockdown of XIST (Fig. 2F), further supporting a significant reduction in EMT upon XIST knockdown.
Role of miR-133a in glioma cell proliferation, migration and EMT
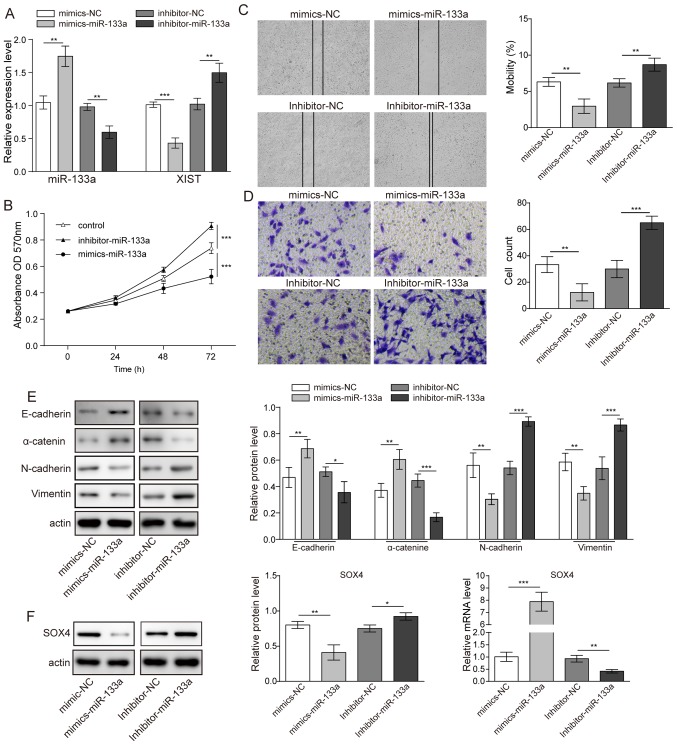
Mimics or an inhibitor of miR-133a were used to assess the role of miR-133a in glioma cell behaviour. It was first demonstrated that elevated miR-133a expression could lower XIST expression in U251 glioma cells. In contrast, inhibitor of miR-133a significantly suppressed the level of miR-133a and elevated the level of XIST (Fig. 3A). Using the MTT assay, it was demonstrated that when a mimic of miR-133a was used for transfection, the cell proliferation of U251 cells was inhibited compared with that of the control group; in contrast, U251 cells transfected with an inhibitor of miR-133a demonstrated increased proliferation (Fig. 3B). Consequently, in the migration and invasion test, the mimic of miR-133a notably decreased both migration and invasion abilities; whereas the inhibitor of miR-133a markedly increased migration and invasion (Fig. 3C and D). Furthermore, the EMT-related proteins E-cadherin, N-cadherin, α-catenin and vimentin were also detected when a mimic of miR-133a or inhibitor of miR-133a was transfected into U251 cell. Western blotting demonstrated that the mimic of miR-133a could increase the expression of E-cadherin and α-catenin but decrease the expression of N-cadherin and vimentin. The inhibitor of miR-133a exerted the opposite effect (Fig. 3E). These results collectively indicated that miR-133a acts as a tumour suppressor in glioma. The expression of the EMT-related transcription factor SOX4 was also detected by western blotting. The results revealed that both mRNA and protein expression of SOX4 was significantly reduced when the mimic of miR-133a was transfected and was significantly increased when the inhibitor of miR-133a was transfected (Fig. 3F).
Figure 3.
Effects of miR-133a on glioma cell proliferation and metastasis. (A) Relative expression levels of miR-133a and XIST in glioma cells following transfection with miR-133a mimics or inhibitor. (B) Cell viability following transfection with miR-133a mimics or inhibitor assessed by MTT assay. (C) Migration (magnification, ×16) and (D) invasion (magnification, ×100) of cell lines detected by scratch-wound healing and Transwell migration assays, respectively. (E) Western blotting showing the expression of E-cadherin, N-cadherin, α-catenin and vimentin in glioma cells following transfection with miR-133a inhibitor or mimics. (F) Expression level of SOX4 transcription factor. n=3. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; XIST, X-inactive specific transcript; SOX, SRY-box; NC, negative control; OD, optical density.
XIST competes with SOX4 for miR-133a binding
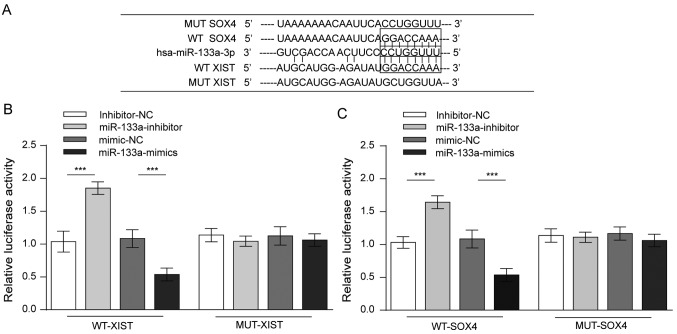
The above results collectively revealed that XIST and miR-133a served opposite roles in glioma cell proliferation, migration and invasion, as well as in the expression of SOX4 and EMT behaviour. A possible model was proposed in which XIST competed with SOX4 for miR-133a binding. To validate this hypothesis, four luciferase reporter gene vectors were constructed (wt-XIST luciferase reporter, mut-XIST vector containing a 6-bp mutation within the predicted miR-133a binding site within XIST, wt-SOX4 3′UTR, and mut-SOX4 3′UTR vector containing a 6-bp mutation within the predicted miR-133a binding site in the 3′UTR of SOX4; Fig. 4A). These indicated vectors were co-transfected into 293 cells along with the miR-133a mimics or the miR-133a inhibitor, and the luciferase activity was then determined by dual luciferase assays. The results demonstrated that the luciferase activity of either wt-XIST- or wt-SOX4-transfected cells was significantly suppressed by miR-133a mimics and was remarkably amplified by a miR-133a inhibitor. However, when the miR-133a binding site was mutated, such modulating effects on luciferase activity were abolished (Fig. 4B and C). These data strongly suggest that XIST competed with SOX4 for miR-133a binding.
Figure 4.
Binding affinity between SOX4, XIST and miR-133a. (A) Predicted complementary binding sites between SOX4 or XIST and miR-133a. (B) Binding affinity was detected via dual luciferase reporter assays using wild-type or mutant forms of XIST and miR-133a. (C) Dual luciferase activity demonstrating binding between miR-133a and SOX4 with wt or mut forms. n=3. ***P<0.001. SOX, SRY-box; XIST, X-inactive specific transcript; miR, microRNA; wt, wild-type; mut, mutant; NC, negative control.
Discussion
A previous study revealed that ~18% of lncRNAs are associated with human tumours, and that only 9% of human protein-coding genes perform this function (19). Thus, it is important to clarify the role of lncRNAs in tumours.
The lncRNA XIST is upregulated in many cancers (20), and a high expression level of XIST is associated with poor clinical outcome (9). For example, the expression level of lncRNA-XIST was significantly increased in both colorectal cancer tissues samples and colorectal cancer cells and promoted colorectal cancer cell proliferation by affecting the cell cycle (21); lncRNA-XIST was specifically upregulated in lung cancer cell lines and promoted lung cancer cell growth (22); lncRNA-XIST was specifically upregulated in pancreatic cancer tissues and cell lines, and high XIST expression in pancreatic cancer was associated with poorer prognosis (9). In the present study, it was demonstrated that XIST demonstrated high expression in the glioma cell line U251. Furthermore, when XIST was knocked down, the proliferation, migration and invasion of these cells were reduced. These results revealed that XIST may serve as a potential therapeutic target in glioma. For the mechanism through which XIST affects glioma, these assays demonstrated that lnc-XIST knockdown significantly upregulated E-cadherin and α-catenin, whilst N-cadherin and vimentin were downregulated. More in vitro and in vivo evidence is required to substantiate the regulation of XIST on glioma metastasis.
The SOX4 transcription factor belongs to a large family of proteins that serves a fundamental role during embryogenesis and controls cell fate and differentiation (23). SOX4 expression is increased in a wide variety of cancer types, including colorectal, breast and glioblastoma, and correlates with poor prognosis and disease progression (15,24). Several cancer-associated signalling pathways have been implicated in the activation of SOX4, including transforming growth factor-β, Wnt, and tumour necrosis factor-α (25–27). SOX4 activation controls various aspects of tumour development and progression, such as inhibition of apoptosis, induction of cell migration and metastasis, and the generation and maintenance of cancer stem cells (11,24). In the present study, it was demonstrated that knockdown of XIST could reduce the expression of SOX4, which revealed that XIST regulates the expression of SOX4.
Recently, an increasing number of studies (8–10) have focused on miRNA, as these small molecules affect many genetic pathways, including cell cycle checkpoint, cell proliferation, apoptosis, and altering the expression of miRNAs correlated with cancers by acting as tumour suppressors and oncogenes (28–30). Recently, miR-133a has been reported as a tumour suppressor in lung and osteosarcoma cancers, as well as head and neck squamous cell carcinomas, in which miR-133a reduces cell proliferation, migration, and invasion (31–33). However, previous study has explored the role of miR-133a in glioma (34). In the present study, it was demonstrated that overexpression of miR-133a promotes glioma proliferation, migration and invasion but reduces the expression of SOX4; in contrast, knockdown of miR-133a induces the opposite effect, revealing that miR-133a negatively regulates SOX4.
lncRNAs are widely associated with regulating gene expression networks at epigenetic, transcriptional and post-transcriptional levels, which may affect the growth, proliferation, differentiation, metabolism, apoptosis and other important physiological processes of cells (35). Sometimes, lncRNAs have miRNA response elements and act as natural miRNA sponges to reduce the binding of endogenous miRNAs to target genes (36). Following the indication that XIST and miR-133a had different effects on glioma cell proliferation, migration and invasion, it was further investigated whether XIST could mutually regulate miR-133a expression. In the present study, dual negative regulation between XIST and miR-133a in glioma cell lines was observed: Following XIST knock-down, miR-133a expression increased in glioma cell lines; XIST expression could be downregulated by miR-133a overexpression while being upregulated by miR-133a inhibition. Similar mechanisms were implicated when XIST was demonstrated to be regulated by miR-101, although further studies such as a binding assay are required for substantiation of this finding (37). In post-transcriptional regulation, lncRNAs act as competing endogenous RNAs to sponge miRNAs, consequently modulating the repression of miRNA targets (38,39). Using luciferase assays, it was demonstrated that XIST and SOX4 could bind miR-133a in the predicted binding site; thus XIST competed with SOX4 for miR-133a binding.
The present study still has some limitations at its current stage. For example, in vivo evidence was not sought to demonstrate the modulation of tumour metastasis by XIST. Furthermore, miR-133a mimic and inhibitor regulated XIST expression, however, the detailed mechanism requires further assays for elucidation. In conclusion, an XIST/miR-133a/SOX4 axis and a potential mechanism of XIST in glioma cell proliferation and metastasis were revealed. These findings suggest that XIST is potentially a therapeutic target for glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
CL substantially contributed to the conception of this study and drafting the manuscript. CT, ZQ and BaZ were involved in the study conception and design. MZ performed the literature research and experimental studies. BoZ and ZQ performed data acquisition and edited the manuscript. SW and XL performed data and statistical analysis. CT was involved in revising it critically for important intellectual content. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Prados MD, Levin V. Biology and treatment of malignant glioma. Semin Oncol. 2000;27(Suppl 6):S1–S10. [PubMed] [Google Scholar]
- 3.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: Genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 4.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Yu H, Shen Y, Liu Y, Yang Z, Sun T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/β-catenin pathway. Oncotarget. 2016;7:41505–41526. doi: 10.18632/oncotarget.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng S, Yao J, Chen Y, Geng P, Zhang H, Ma X, Zhao J, Yu X. Expression and functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in glioma. J Mol Neurosci. 2015;56:623–630. doi: 10.1007/s12031-014-0488-z. [DOI] [PubMed] [Google Scholar]
- 7.Wutz A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Yuan J, Li L, Yang Y, Xu X, Wang Y. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am J Transl Res. 2017;9:1845–1855. [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118:3349–3358. doi: 10.1002/jcb.25988. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Qin X, Li Y, Zhang X, Niu R, Zhang H, Cui A, An W, Wang X. MiR-133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. Am J Transl Res. 2015;7:1390–1403. [PMC free article] [PubMed] [Google Scholar]
- 11.Jafarnejad SM, Ardekani GS, Ghaffari M, Li G. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci. 2013;70:2677–2696. doi: 10.1007/s00018-012-1187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 13.Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83:986–994. doi: 10.1161/01.RES.83.10.986. [DOI] [PubMed] [Google Scholar]
- 14.Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res. 2000;79:180–191. doi: 10.1016/S0169-328X(00)00109-1. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Ju HL, Yuan XY, Wang TJ, Lai BQ. SOX4 is a potential prognostic factor in human cancers: A systematic review and meta-analysis. Clin Transl Oncol. 2016;18:65–72. doi: 10.1007/s12094-015-1337-4. [DOI] [PubMed] [Google Scholar]
- 16.Shi S, Cao X, Gu M, You B, Shan Y, You Y. Upregulated expression of SOX4 is associated with tumor growth and metastasis in nasopharyngeal carcinoma. Dis Markers. 2015;2015:658141. doi: 10.1155/2015/658141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foronda M, Martínez P, Schoeftner S, Gómez-López G, Schneider R, Flores JM, Pisano DG, Blasco MA. Sox4 links tumor suppression to accelerated aging in mice by modulating stem cell activation. Cell Rep. 2014;8:487–500. doi: 10.1016/j.celrep.2014.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS One. 2010;5:e10316. doi: 10.1371/journal.pone.0010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Cao X, Zhang L, Zhang X, Sheng H, Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemother Pharmacol. 2016;77:629–634. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 21.Song H, He P, Shao T, Li Y, Li J, Zhang Y. Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J BUON. 2017;22:696–703. [PubMed] [Google Scholar]
- 22.Tang Y, He R, An J, Deng P, Huang L, Yang W. lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol Rep. 2017;38:941–948. doi: 10.3892/or.2017.5751. [DOI] [PubMed] [Google Scholar]
- 23.Dai W, Xu X, Li S, Ma J, Shi Q, Guo S, Liu L, Guo W, Xu P, He Y, et al. SOX4 promotes proliferative signals by regulating glycolysis through AKT activation in melanoma cells. J Invest Dermatol. 2017;137:2407–2416. doi: 10.1016/j.jid.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 24.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: Friend or foe? Oncogene. 2013;32:3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 25.Vervoort SJ, Lourenco AR, van Boxtel R, Coffer PJ. SOX4 mediates TGF-β-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS One. 2013;8:e53238. doi: 10.1371/journal.pone.0053238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 27.Banno T, Gazel A, Blumenberg M. Effects of tumor necrosis factor-alpha (TNF alpha) in epidermal keratinocytes revealed using global transcriptional profiling. J Biol Chem. 2004;279:32633–32642. doi: 10.1074/jbc.M400642200. [DOI] [PubMed] [Google Scholar]
- 28.Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Boil. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei Q, Li X, Guo M, Fu X, Han W. The miRNA network: Micro-regulator of cell signalling in cancer. Expert Rev Anticancer Ther. 2014;14:1515–1527. doi: 10.1586/14737140.2014.953935. [DOI] [PubMed] [Google Scholar]
- 30.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Fujimura L, Yoshino H, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M, et al. Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int J Oncol. 2011;38:209–217. [PubMed] [Google Scholar]
- 32.Xu M, Wang YZ. miR133a suppresses cell proliferation, migration and invasion in human lung cancer by targeting MMP14. Oncol Rep. 2013;30:1398–1404. doi: 10.3892/or.2013.2548. [DOI] [PubMed] [Google Scholar]
- 33.Chen G, Fang T, Huang Z, Qi Y, Du S, Di T, Lei Z, Zhang X, Yan W. MicroRNA-133a inhibits osteosarcoma cells proliferation and invasion via targeting IGF-1R. Cell Physiol Biochem. 2016;38:598–608. doi: 10.1159/000438653. [DOI] [PubMed] [Google Scholar]
- 34.Sakr M, Takino T, Sabit H, Nakada M, Li Z, Sato H. miR-150-5p and miR-133a suppress glioma cell proliferation and migration through targeting membrane-type-1 matrix metalloproteinase. Gene. 2016;587:155–162. doi: 10.1016/j.gene.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 35.Ye ZQ, Wang T, Song W. Long noncoding RNAs in prostate cancer. Zhonghua Nan Ke Xue. 2014;20:963–968. (In Chinese) [PubMed] [Google Scholar]
- 36.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarver AL, Subramanian S. Competing endogenous RNA database. Bioinformation. 2012;8:731–733. doi: 10.6026/97320630008731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.