Abstract
Background
Klebsiella pneumoniae is considered the most clinically relevant species of Enterobacteriaceae, known to cause severe infections including liver abscesses. To the best of our knowledge, a large proportion of iron in the human body is accumulated and stored in the liver. We hypothesize that increased iron availability is an important factor driving liver abscess formation and we therefore aim to understand the effects of iron on K. pneumoniae causing liver abscesses.
Results
All tested K. pneumoniae clinical isolates, including those isolated from liver abscesses and other abdominal invasive infection sites, grew optimally when cultured in LB broth supplemented with 50 μM iron and exhibited the strongest biofilm formation ability under those conditions. Decreased growth and biofilm formation ability were observed in all tested strains when cultured with an iron chelator (P < 0.05). The infection model of G. mellonella larvae indicated the virulence of liver abscess-causing K. pneumoniae (2/3) cultured in LB broth with additional iron was significantly higher than those under iron-restricted conditions (P < 0.05). The relative expression levels of the four siderophore genes (iucB, iroB, irp1, entB) in K. pneumoniae strains isolated from liver abscesses cultured with additional iron were lower than those under iron-restricted conditions (P < 0.05).
Conclusions
It is suggested by our research that iron in the environment can promote growth, biofilm formation and enhance virulence of K. pneumoniae causing liver abscesses. A lower expression of siderophore genes correlates with increased virulence of liver abscess-causing K. pneumoniae. Further deeper evaluation of these phenomena is warranted.
Keywords: Iron, Klebsiella pneumoniae, Growth, Biofilm, Virulence
Background
Klebsiella pneumoniae is considered the most clinically relevant species of Enterobacteriaceae, known to cause both community-acquired and nosocomial infections, including liver abscesses, pneumonia, urinary tract infections and bacteremia worldwide [1]. In the past two decades, a distinct hypervirulent variant of K. pneumoniae, characterized by its hypermucoviscous phenotype, was firstly isolated from liver abscesses in Asia and has emerged as a clinically significant pathogen responsible for highly invasive infections [2]. Unlike classical K. pneumoniae, hypervirulent K. pneumoniae (hvKp) can spread from the original site of infection to other organs. Once invasive dissemination occurs, patients often suffer severe and irreversible refractory sequelae, such as blindness and central nervous system damage [2, 3]. The conditions of patients infected with hypervirulent K. pneumoniae causing liver abscess are serious, posing a great threat to public health and has attracted the attention of clinicians.
The pathogenicity of K. pneumoniae mainly arise from various virulence factors which allow it to overcome innate host immunity and to maintain infection in a mammalian host. The main virulence factors that play an important role in pathogenicity are capsular polysaccharide, lipopolysaccharide, pili and siderophores [4]. Therein, K1 and K2 capsular types are considered to be highly pathogenic to human [5]. rmpA is an important activator of capsular production, resulting in the formation of hypermucoviscous phenotype and enhancement of virulence, while aerobactin (iucB), yersiniabactin (irp1), salmochelin (iroB) and enterobacterin (entB) are the four siderophores of K. pneumoniae [6, 7]. Therefore, understanding the virulence characteristics of K. pneumoniae and taking appropriate measures are critical for clinical assessment, controlling the prognosis and reducing mortality risk of patients with K. pneumoniae infection.
It is recognized that most organisms require iron for a variety of metabolic and informational cellular pathways [8]. However, ferric ion (Fe3+) and its derivatives are poorly soluble, and cannot be utilized directly by most organisms. It is essential to synthesize and secrete siderophores to meet the demand for iron required for bacterial growth and metabolism. To repress microbial growth, iron availability within the human body is strictly limited [9]. One notable exception is the liver, in which iron concentration is used for assessing body iron stores [10]. These increased iron levels probably promote growth of K. pneumoniae in the liver and provide an advantage for liver abscess formation. Therefore, our study was designed to evaluate the effects of iron on the growth, biofilm formation and virulence of liver abscess-causing K. pneumoniae.
Results
Antimicrobial susceptibility testing
Five K. pneumoniae isolates (FK3065, FK3170, FK3226, FK3992, FK4003) were sensitive to conventional antimicrobials (levofloxacin, ciprofloxacin, cefepime, ceftazidime, imipenem, gentamicin, amikacin, tobramycin, ampicillin/sulbactam, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, aztreonam, cefotetan, ertapenem, ceftriaxone). However, FK3087 was resistant to ampicillin/sulbactam, levofloxacin, ciprofloxacin, gentamicin and trimethoprim/sulfamethoxazole.
Effect of iron on growth and biofilm formation of the K. pneumoniae isolates
All K. pneumoniae isolates grew optimally in LB broth containing 50 μM iron, superior to the growth in LB broth containing 30 μM, 10 μM, and 0 μM iron, separately (P < 0.05). The growth of all tested strains in the iron-restricted environment caused by the iron chelating agent was worse (P < 0.05) (Fig. 1).
Fig. 1.
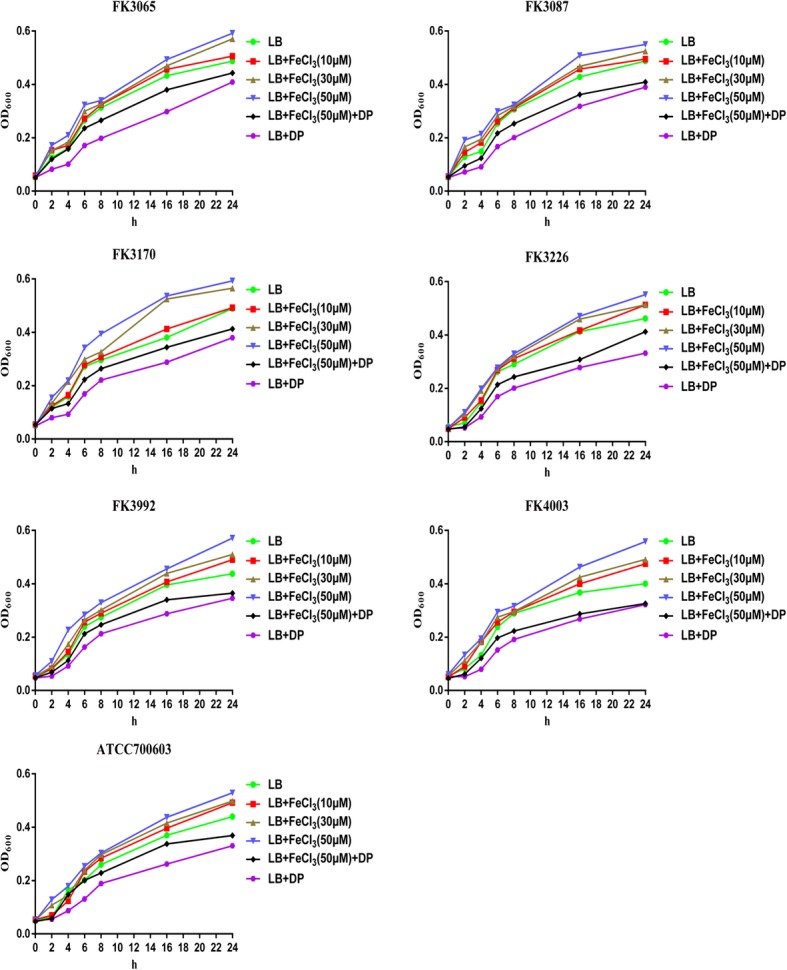
The growth curves of K. pneumoniae. All K. pneumoniae isolates grew optimally in LB broth containing 50 μM iron, superior to the growth in LB broth containing 30 μM, 10 μM, and 0 μM iron or 200 μM iron chelator +(−) 50 μM iron(P < 0.05). DP: 2,2′-Dipyridyl, added as an iron chelator. LB + 200 μM iron chelating agent group was used as control
In addition, we further found that iron promotes biofilm formation of K. pneumoniae in a concentration-dependent manner. All strains exhibited the strongest biofilm formation ability in LB broth supplemented with 50 μM iron, which was superior to the biofilm formation ability of the respective strains in LB broth containing 30 μM, 10 μM and 0 μM iron (P < 0.05). The strains showed a weaker biofilm formation ability in an iron-restricted environment (P < 0.05) (Fig. 2).
Fig. 2.
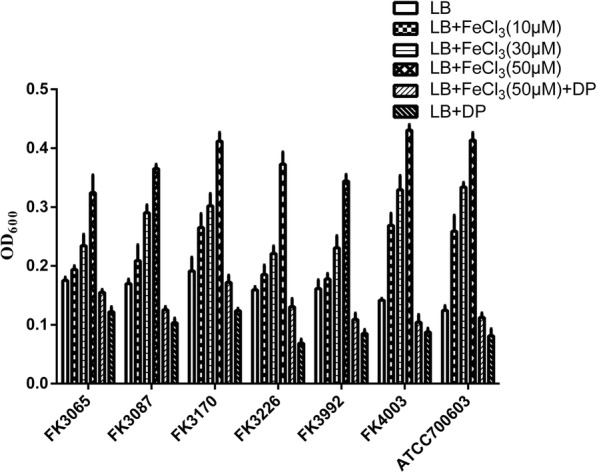
Biofilm-forming capabilities of K. pneumoniae. All strains exhibited the strongest biofilm formation ability in LB broth supplemented with 50 μM iron, which was superior to the biofilm formation ability of the respective strains in LB broth containing 30 μM, 10 μM and 0 μM iron or 200 μM iron chelator +(−) 50 μM iron (P < 0.05). DP: 2,2′-Dipyridyl, added as an iron chelator. LB + 200 μM iron chelating agent group was used as control
The virulence factors of K. pneumoniae isolates
The three liver abscess-causing K. pneumoniae isolates (FK3226, FK3992 and FK4003) carried at least nine virulence genes, including four kinds of siderophore genes (entB, iucB, iroB, irp1). The serotypes of these three isolates were K1, K2 and K1, respectively. The three non-liver abscess-causing K. pneumoniae isolates (FK3065, FK3087 and FK3170) carried four virulence genes, including the siderophore gene entB, and their serotypes did not belong to one of the common capsule types tested. The virulence genes of each strain were detailed in Table 1.
Table 1.
The virulence genes of K. pneumoniae
| Isolates | Virulence genes |
|---|---|
| FK3065 | entB, uge, mrkD, fimH |
| FK3087 | entB, uge, mrkD, fimH |
| FK3170 | entB, uge, mrkD, fimH |
| FK3226 | iucB, iroB, entB, irp1, rmpA, wcaG, ybtA, magA |
| ureA, uge, wabG, mrkD | |
| FK3992 | iucB, iroB, entB, irp1, rmpA, ybtA, ureA, uge, wabG, mrkD |
| FK4003 | iucB, iroB, entB, irp1, rmpA, ureA, uge, wabG, mrkD |
| ATCC700603 | iucB, iroB, entB, irp1, rmpA, ybtA, ureA, wabG, mrkD |
Infection model of G. mellonella larvae
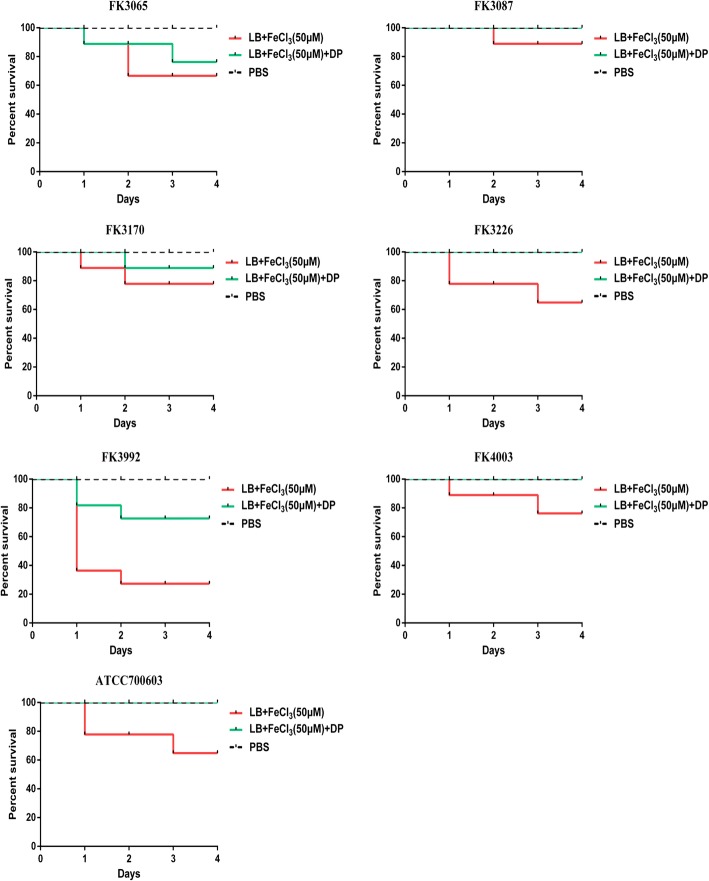
The mortality rates of G. mellonella after infection with the liver abscess-causing K. pneumoniae isolates (FK3226, FK3992) and K. pneumoniae ATCC700603 cultured with additional iron were significantly higher than those cultured with iron chelator (P < 0.05). While the mortality rates of G. mellonella after infection with the non-liver abscess-causing K. pneumoniae isolates (FK3065, FK3087 and FK3170) and liver abscess-causing K. pneumoniae (FK4003) were not significantly different under these two conditions (Fig. 3).
Fig. 3.
Models of Galleria mellonella infected with K. pneumoniae. The virulence of FK3226, FK3992 and ATCC 700603 with additional iron was significantly higher than that of the strains with iron chelator (P < 0.05). DP: 2,2′-Dipyridyl, added as an iron chelator. The PBS was control group
Quantification of siderophore gene expression with qRT-PCR
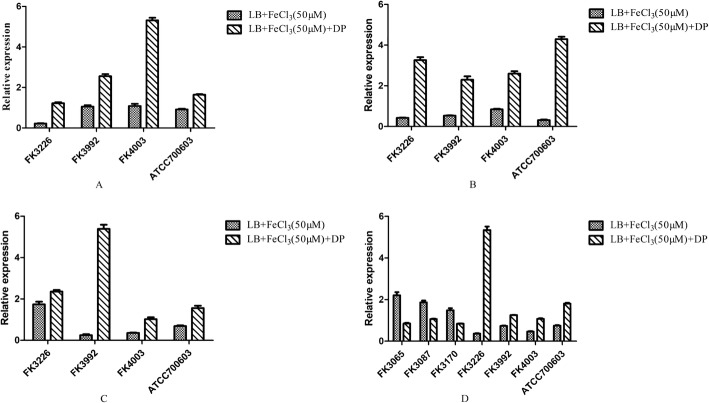
The relative expression levels of the four siderophore genes (iucB, iroB, irp1, entB) in the liver abscess-causing K. pneumoniae strains cultured with additional iron were lower than those cultured under iron-restricted conditions (P < 0.05). On the contrary, the relative expression levels of the siderophore gene entB in the non-liver abscess-causing K. pneumoniae isolates cultured with extra iron were higher than those cultured under iron-restricted conditions (P < 0.05) (Fig. 4).
Fig. 4.
Expression ratio of K. pneumoniae virulence genes. aiucB gene. biroB gene. cirp1 gene. dentB gene. DP: 2,2′-Dipyridyl, added as an iron chelator. LB control was used to calculate relative expression
Discussion
The Gram-negative bacillus K. pneumoniae is a leading cause of healthcare-associated infections, including urinary tract infections, surgical sites infections, soft tissues infections, bacteremia and pneumonia [11]. In the 1980s, case reports from Taiwan described community acquired liver abscesses caused by hvKp in patients with serious concomitant endorgan manifestations, such as meningitis and endophthalmitis [12]. Since the first report in Taiwan, hvKPs have been observed with increasing frequency in many countries in Asia, Europe and America [12]. It is well known that hvKp can invade many organs in patient, including liver. Considering the different iron content between the liver and other tissues, we speculated that this higher iron level may be an important factor to affect the virulence of K. pneumoniae in the liver and provide an advantage for liver abscess formation. In this study, three liver abscess-causing K. pneumoniae isolates and three non-liver abscess-causing K. pneumoniae isolates were selected to perform growth, biofilm and virulence investigations under different iron conditions.
In this research, we demonstrated that moderate amounts of iron promoted the growth of K. pneumoniae. A slower growth of K. pneumoniae isolates was discovered in iron-restricted environments. Furthermore, it was illustrated that iron promoted biofilm formation of K. pneumoniae, and the ability of bacterial biofilm formation was significantly attenuated by addition of an iron chelator. Previously, it has been demonstrated that the biofilm formation ability of Pseudomonas aeruginosa was significantly reduced in an iron-restricted environment, and biofilm formation increased significantly after iron complementation, indicating that iron plays an important role in the formation of P. aeruginosa biofilms [13]. The presence of an appropriate amount of iron is also conducive to the biofilm formation of Escherichia coli and Staphylococcus aureus [14, 15]. These findings, together with our investigation, indicate that iron plays a crucial role in the process of bacterial biofilm formation and growth.
In this study, the virulence genes and capsular serotypes of seven experimental strains were detected by PCR. These three liver abscess-causing K. pneumoniae isolates carried more virulence genes, including all four siderophore genes iucB, iroB, irp1, entB, while the strains isolated from the other sites only carried enterobactin (entB) among their four virulence factors. Bacteria secrete siderophores to bind and import iron, which is required for growth. In this investigation, the effect of iron concentration on the relative expression of siderophore genes was detected by qRT-PCR. As expected, the tested K. pneumoniae strains causing liver abscesses showed lower relative expression levels of the four siderophore genes (iucB, iroB, irp1, entB) in media with high iron levels, compared to iron-restricted media. In the presence of the iron chelator, high-affinity iron binding siderophores are needed to obtain sufficient iron for growth, while at high iron concentrations less metabolically costly uptake mechanisms can be employed. In contrast, the single siderophore gene entB of the non-liver abscess-causing K. pneumoniae isolates was expressed relatively higher after addition of iron than during iron deficiency caused by iron chelating agent. This is a surprising finding, which might indicate that these strains rely on other iron uptake mechanisms apart from siderophores, which warrants further study.
Previous studies have shown that different concentrations of iron could affect the surface characteristics of the pili and porins and further influence the virulence of Acinetobacter baumannii [16]. Therefore, We attempted to demonstrate that this higher iron level may be an important factor to affect the virulence of K. pneumoniae in the liver. To investigate the effect of iron on the virulence of K. pneumoniae, we determined the virulence of K. pneumoniae through the wax larvae infection model. Wax larvae are an invertebrate host model frequently used to detect the virulence of K. pneumoniae [17]. According to the results of the G. mellonella infection model, three strains caused higher mortality rate with extra iron than iron-restricted growth environment, indicating the potential virulence in vivo. The results of qRT-PCR indicate that a lower expression of siderophore genes correlates with increased virulence. Due to the various needs of different strains for iron, the expression of siderophore genes was not exactly the same. Other mechanisms by which iron affects the virulence of K. pneumoniae need to be further explored.
Conclusions
In summary, iron can promote the growth, biofilm formation and enhance virulence of K. pneumoniae causing liver abscess. Moreover, the diverse expression of siderophores genes may be one of the factors that regulate the variation in virulence. To further understand these phenomena, a deeper evaluation of the phenomena is warranted.
Methods
Bacterial strains and antimicrobial susceptibility profiling
Six K. pneumoniae clinical isolates (FK3065, FK3087, FK3170, FK3226, FK3992, FK4003) were collected from patients in the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, China during 2016 and 2017. Herein, FK3226, FK3992, FK4003 were isolated from patients with liver abscess, and FK3065, FK3087, FK3170 were isolated from other abdominal invasive infection sites of non-liver abscess patients (ascites, biliary drainage fluid). Identification was conducted on all isolates using VITEK MS system (bioMérieux, Marcy L’Etoile, France). Antimicrobial susceptibility testing was performed by VITEK2 system (bioMérieux, Marcy L’Etoile, France) with AST-GN13 card. K. pneumoniae ATCC 700603 served as the control strain.
Growth curves and biofilm formation
The effect of iron on the growth of K. pneumoniae was measured following previous methods with some modifications [18, 19]. In brief, overnight cultures of all K. pneumoniae clinical isolates (FK3065, FK3087, FK3170, FK3226, FK3992, FK4003) and K. pneumoniae ATCC 700603 were diluted 1:100 in Luria-Bertani (LB) broth supplemented with different iron concentrations (50 μM, 30 μM, 10 μM, 0 μM) and with 200 μM iron chelating agent (2,2′-Dipyridyl) + (−) 50 μM iron, respectively. Wherein, strains under LB + 200 μM iron chelating agent condition were used as control and LB + 200 μM iron chelating agent + 50 μM iron was set to produce an iron-restricted condition. The cultures were incubated at 37 °C with constant shaking at 180 rpm. Samples were collected at 0 h, 2 h, 4 h, 6 h, 8 h, 16 h, 24 h and the absorbance at 600 nm was determined. Each sample was measured in triplicates and averages of absorbance values were used for analysis. The growth of K. pneumoniae was evaluated by plotting the values of OD600 against time.
The biofilm assay was performed as published with some modifications [20]. Briefly, six K. pneumoniae clinical isolates and K. pneumoniae ATCC 700603 were grown overnight in LB broth. The overnight cultures were then diluted 1:100 in fresh LB broth supplemented with different iron concentrations (50 μM, 30 μM, 10 μM, 0 μM) and with 200 μM iron chelating agent +(−) 50 μM iron. A total of 100 μL of each dilution were added to a 96-well polystyrene microtiter plate and incubated at 37 °C for 24 h. Wells containing media alone were used as blank. Planktonic cells were removed and the wells were washed twice with sterile water, then the wells were stained with 150 μL 0.1% crystal violet for 10 min and rinsed twice with sterile water. Stained biofilms were solubilized with 95% ethanol and quantified by measuring the OD600 using a microplate reader. Each sample was measured in triplicates and averages of absorbance values were used for analysis.
Detection of virulence genes of isolates
Virulence genes (magA, iucB, iroB, entB, irp1, iroN, kfuBC, rmpA, wcaG, alls, ybtA, ureA, uge, wabG, fimH and mrkD) and capsular serotypes (K1, K2, K5, K20, K54 and K57) of the six K. pneumoniae clinical isolates and K. pneumoniae ATCC 700603 were amplified by Polymerase Chain Reaction (PCR). Primers for the aforementioned genes are listed in Supplementary Table S1 [21, 22]. Positive PCR products were sequenced by Beijing Genomics Institute Technology Co. Ltd. (Shanghai, China). Nucleotide sequences were compared using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Infection model of Galleria mellonella larvae
G. mellonella killing assays were carried out on the six clinical isolates and K. pneumoniae ATCC 700603 as described previously, with minor modifications [17]. Eight larvae weighing between 200 mg–250 mg were randomly selected for each strain. A 10 μL of bacterial suspension (108 CFU/mL) in phosphate-buffered saline (PBS) was injected into the last left proleg using a 25 μL Hamilton precision syringe. The bacterial suspension was prepared by culturing the strains in LB broth containing 50 μM iron, and 50 μM iron with 200 μM iron chelating agent for 24 h. Larvae injected with 10 μl PBS were used as control. The insects were incubated at 37 °C in the dark and observed after 24 h, 48 h and 72 h. Larvae were considered dead when they repeatedly failed to respond to physical stimuli. The primary outcome for the insect model was rapidity and extent of mortality of G. mellonella assessed with Kaplan-Meier analysis and log-rank test.
Quantitative reverse transcription PCR (qRT-PCR)
The effects of iron on the expression levels of K. pneumoniae siderophore genes (iucB, iroB, entB and irp1) were evaluated using quantitative reverse transcription PCR (qRT-PCR). For RNA extraction, K. pneumoniae isolates were grown in fresh LB medium with 50 μM iron, and 50 μM iron with 200 μM iron chelating agent at 37 °C for 24 h. K. pneumoniae isolates grown in fresh LB medium were used as control. Total RNA was extracted using a RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The extracted RNA samples were stored at − 80 °C. Purified RNA was reverse transcribed into cDNA for qRT-PCR analysis using a cDNA synthesis kit (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. Gene expression levels were measured with qRT-PCR using a 7500 RT-PGE system (TOYOBO, Osaka, Japan) and SYBR Green qRT-PCR Kit (TOYOBO) with the specific primers listed in Supplementary Table S2 [23]. The rpoB gene was used as an internal control to normalize the data. Each sample was measured in triplicates and averages of Ct values were used for analysis. Gene expression levels were calculated using 2− △△ Ct method.
Statistical analysis
All experiments were conducted independently with at least two replicates on different days, and results were expressed as mean ± standard deviation or average. The total area under the curve was calculated for analysis on growth. Unpaired or two-tailed paired t-tests were used to evaluate the significance of differences between two groups. One-way analysis of variance (ANOVA) was performed to analyze the significance among more groups. Statistical significance was determined at P < 0.05. Statistical analyses were performed using SPSS version 17.0 statistical software.
Supplementary information
Additional file 1: Table S1. Primers used for PCR amplification of virulence genes
Additional file 2: Table S2. Primers used for qRT-PCR of siderophore genes
Acknowledgments
The authors acknowledge the financial support of the National Natural Science Foundation of China (No.81741059) and the Health Department of Zhejiang Province of the People’s Republic of China (No.2019KY098). The authors also want to give thanks to Dr. Andrea Rocker from Monash University for her help in paper writing.
Abbreviations
- PCR
Polymerase Chain Reaction
- qRT-PCR
Quantitative Reverse Transcription Polymerase Chain Reaction
Authors’ contributions
TC and GFD contributed equally to this study. TC and GFD wrote the manuscript under supervision of JMC, TLZ and QW. TC, GFD, SQZ, XCZ and YJZ performed the research. TC and GFD performed the data analysis. The research plan for this project was conceived based on several rounds of discussions among all co-authors. All authors read and approved the final version of the manuscript.
Funding
This work was supported by research grants from the National Natural Science Foundation of China (No. 81971986) and the Health Department of Zhejiang Province of the People’s Republic of China (No.2019KY098). These two funding bodies provided funds for the purchase of consumption materials for the study but had no role in the design of the study and collection, analysis, and interpretation of data and writing of the manuscript.
Availability of data and materials
The datasets used and analysed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Ethics approval and consent is deemed unnecessary in this research according to the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tao Chen and Guofeng Dong contributed equally to this work.
Contributor Information
Tieli Zhou, Email: wyztli@163.com.
Qing Wu, Email: wuqing830@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01727-5.
References
- 1.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2016;16(9):e190–e195. doi: 10.1016/S1473-3099(16)30021-4. [DOI] [PubMed] [Google Scholar]
- 3.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8(7):1111–1123. doi: 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang CT, Chuang YP, Shun C, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced ironacquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2008;197(12):1717–1727. doi: 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 8.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13(5):509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kami M, Hamaki T, Kishi Y. Hepatic iron concentration and total body iron stores in thalassemia major. N Engl J Med. 2000;343(22):1657. doi: 10.1056/NEJM200011303432217. [DOI] [PubMed] [Google Scholar]
- 11.Corbella M, Caltagirone M, Gaiarsa S, et al. Characterization of an outbreak of extended-Spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit in Italy. Microb Drug Resist. 2018;24(8):1128–1136. doi: 10.1089/mdr.2017.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Lopez J, Garcia-Caballero A, Navarro-San Francisco C, et al. Hypermucoviscous Klebsiella pneumoniae: a challenge in community acquired infection. IDCases. 2019;17:e00547. doi: 10.1016/j.idcr.2019.e00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiens JR, Vasil AI, Schurr MJ, Vasil ML. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. MBio. 2014;5(1):e01010–e01013. doi: 10.1128/mBio.01010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alves JR, Pereira AC, Souza MC, et al. Iron-limited condition modulates biofilm formation and interaction with human epithelial cells of enteroaggregative Escherichia coli (EAEC) J Appl Microbiol. 2010;108(1):246–255. doi: 10.1111/j.1365-2672.2009.04417.x. [DOI] [PubMed] [Google Scholar]
- 15.Lin MH, Shu JC, Huang HY, Cheng YC. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS One. 2012;7(3):e34388. doi: 10.1371/journal.pone.0034388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics. 2011;12(1):126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Insua JL, Llobet E, Moranta D, et al. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth galleria mellonella. Infect Immun. 2013;81(10):3552–3565. doi: 10.1128/IAI.00391-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palacios M, Broberg CA, Walker KA, Miller VL. A Serendipitous Mutation Reveals the Severe Virulence Defect of a Klebsiella pneumoniae fepB Mutant. MSphere. 2017;2(4):e00341–e00317. doi: 10.1128/mSphere.00341-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasqua M, Visaggio D, Lo Sciuto A, et al. Ferric Uptake Regulator Fur Is Conditionally Essential in Pseudomonas aeruginosa. J Bacteriol. 2017;199(22):e00472–e00417. doi: 10.1128/JB.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilksch JJ, Yang J, Clements A, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7(8):e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi: 10.18388/abp.2015_1148. [DOI] [PubMed] [Google Scholar]
- 22.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929. doi: 10.1038/srep38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadasy KA, Domiati-Saad R, Tribble MA. Invasive Klebsiella pneumoniae syndrome in North America. Clin Infect Dis. 2007;45(3):e25–e28. doi: 10.1086/519424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used for PCR amplification of virulence genes
Additional file 2: Table S2. Primers used for qRT-PCR of siderophore genes
Data Availability Statement
The datasets used and analysed during the current study available from the corresponding author on reasonable request.