Abstract
Chemotherapy and radiation are unable to eliminate all cancer cells, particularly apoptosis-resistant cancer cells, despite their ability to kill cancer cluster cells. Thus, it is important to identify methods that eliminate all cancer cells in order to prevent relapse. Salinomycin has the ability to control and eradicate different types of cancer, including breast cancer; however, its molecular mechanism remains unclear. The main difficulty in testing salinomycin activity and understanding the governing mechanisms is its low solubility in water (17 mg/l), which can hinder convenient delivery of salinomycin to the protein receptor at the cell surface of stem cells. In the present study, salinomycin was conjugated to the trans-activator of transcription-protein in order to facilitate its delivery to the cancer cells. Conjugated salinomycin demonstrated improved solubility in both in vitro. Salinomycin was tested in breast cancer cells (MCF7 and JIMT-1) by the cleavage of the linker through photolysis at l≥365 nm during in vitro analysis, in the present study.
Keywords: salinomycin, trans-activator of transcription protein, cancer cells, cancer, photo-cleavable, drug delivery, photolysis
Introduction
The last two decades have witnessed a change in the views towards chemotherapy and carcinogenesis. The discovery that cancer stem cells can be used to treat solid tumors has changed the way of thinking with regards to cancer treatment. The self-renewal capacity of cancer cells remains their most important feature and function (1–3) as it allows for renewal of cancer cells and cancer clusters following radiation and chemotherapy treatment (4).
Salinomycin, a polyketide organic compound extracted from Streptomyces albus (5,6) and its derivatives are the first promising compounds that have demonstrated activity against cancer stem cells (7). They can selectively inhibit seeding, proliferation and metastasis of breast cancer cells both in vitro and in vivo (8). Salinomycin and its derivatives have been reported to possess anti-cancer properties and they can act against cancer stem cells, thereby preventing the seeding of previously destroyed cancer cells (8–10). With a solubility of 17 mg/l, salinomycin is considered insoluble in water (11). However, such a poor aqueous solubility can hinder its delivery to the active site. Solubility is the most important physicochemical property for the successful delivery of oral drugs. Poor solubility of drug candidates and lead compounds may cause inefficient absorption at the active site, resulting in the loss of activity or clinical failure due to poor pharmacokinetics (12–14). For example, salinomycin is known to act on the binding site of P-glycoprotein (P-gp), a well-established plasma membrane protein that is used to facilitate drug movement out of cells. Binding salinomycin to the P-gp active site may result in cell sensitivity to drugs (15).
There are a number of methods that improve the solubility of organic compounds and drugs in order to enhance their delivery to active sites (16). The most common methods include using surfactants and co-solvents, regulating the pH, employing precipitation inhibitors, formation of complex salts and dissolution in an organic solvent before making a suspension in water (17). The majority of these methods pose several drawbacks and may negatively affect the experiment reproducibility. However, the method of solubilizing drugs using carriers, such as acids, sugars, polymeric forms, surfactants and urea has demonstrated better results. The best carrier is the one in which safety and stability are confirmed, and it does not interfere with the biological functions of the drug. Ideally, the carrier must be able to improve drug solubility and facilitate its delivery to the active site, thereby improving the activity of the drug. Thus, carriers based on short peptides or sugars, or a combination of both are considered to be the best options (18).
To the best of our knowledge, most of the current biological studies on salinomycin are performed by dissolving it in dimethyl sulfoxide, and then using the solution to make a suspension in water (6,9–11,15). In the present study, a new method was proposed in order to improve the solubility and enhance the delivery of salinomycin. Several chemical tools were used in order to improve the delivery of salinomycin to its active site. However, suspended samples that may lead to a wide range of precipitation products were not used in the present study when assessing improved delivery of the compound. In the present study, the main components of the chemical tools included a cell-penetrating protein, with or without a sugar component, and a photo-cleavable linker.
In order to overcome the abundant limitations of the aforementioned methods and increase the number of tools for improving the solubility and delivery of a drug to the active site, a photo-cleavable element, based on a modification of the salinomycin carboxylic acid, was sought and developed in the present study (19). Nevertheless, further work is required to develop novel tools in order to improve the solubility and facilitate the cell penetration of salinomycin.
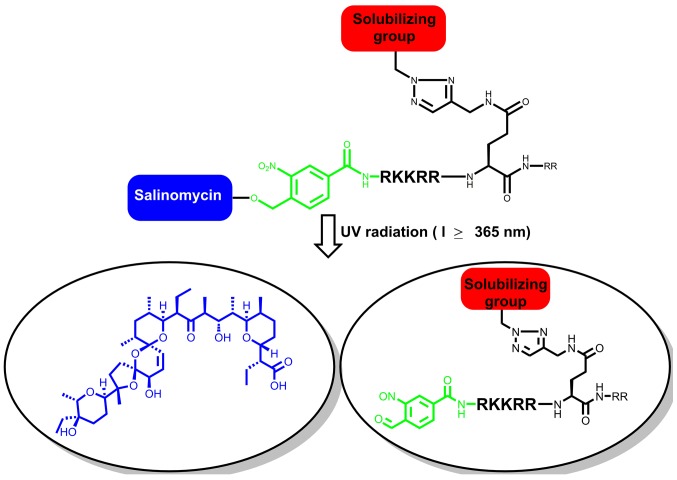
In the present study, salinomycin was conjugated to the trans-activator of transcription (TAT)-protein, as this sequence is rich in arginine that forms multiple cationic sites, under physiological pH (20). The initial contact between the TAT-protein and the cell surface occurs via electrostatic binding to the negatively charged glycosaminoglycans (GAGs) (20). The design constructed in the present study expected to facilitate cell penetration of salinomycin, and is as follows: A novel photo-cleavable linker-caged salinomycin conjugated to a TAT-peptide segment (RKKRRQRR). In the system implemented in the present study, the solubility of the TAT-peptide sequence was further improved by attaching a sugar moiety to the side chain of glutamine (Fig. 1). Besides improving drug solubility, the sugar moiety served as a tool for penetrating cells (21). The present study investigated methods to improve the solubility and cell penetration of salinomycin, in order to improve its activity against cancer cells. Biological interference was avoided by introducing a photolinker, 4-(hyroxymethyl)-3-nitro-benzoic acid (22), between salinomycin and the N-terminus of the peptide. The chemically stable photolinker, the sugar moiety and salinomycin were sequentially introduced to the TAT-protein segment using click chemistry (23,24). Salinomycin was recovered by mild irradiation at l≥365 nm during in vitro analysis.
Figure 1.
Schematic depiction of the caged-salinomycin design with a peptide and a solubilizing group on glutamine for enhancing cell penetration and the release of salinomycin through irradiation. UV, ultraviolet.
Materials and methods
Preparation of samples and instruments
Fluorenyl-methyloxycarbonyl (Fmoc)-AA-Wang resin, (2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexaf luorophosphate (HBTU) and (1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate were purchased from Merck KGaA (cat. no. 851006) or AnaSpec (cat. no. AS-21001). All other commercial reagents, including solvents (HPLC grade or higher), dimethyl sulfoxide-d6 (6d-DMSO) and chloroform-d (CDCl3), were purchased from Acros; Sigma Aldrich; Merck KGaA and VWR; Avantor, and used without further purification. All solvents used for extraction and chromatography were distilled prior to use. Anhydrous tetrahydrofuran (THF) (Acros; cat. no. ACR32697-0010), diethylether (Et2O) (Acros; cat. no. ACR32686-0010), dichloromethane (DCM) (Merck KGaA; cat. no. 1.06051.1001), methanol (Acros; cat. no. ACR41377-0025) and toluene (Sigma Aldrich; Merck KGaA; cat. no. 320552-1L) were purified using a PureSolv filtration system (PureSolv; Sigma Aldrich; Merck KGaA). Nitromethane (Sigma Aldrich; Merck KGaA; cat. no. 73478-100ML), potassium hydroxide (Sigma Aldrich; Merck KGaA; cat. no. P5958-250G), citric acid (Merck KGaA; cat. no. 8.18707.1000), 1,8-Diazabicyclo[5.4.0]undec-7-ene (Sigma Aldrich; Merck KGaA; cat. no. 139009-100G). Double distilled water was obtained using a purification system (MilliQ system; Merck Millipore). Glassware was flame- or oven-dried prior to use. The reactions were followed and monitored on thin-layer chromatography (TLC) silica gel plates (Merck KGaA; cat. no. 60F254) and visualized under a short-wave UV lamp (at 215, 254 and 280 nm), after heating the plates dipped in ammonium molybdate/cerium (IV) sulfate solution, and/or staining with a KMnO4 solution (at 100°C for 60 sec). Flash column chromatography was performed with silica gel (230–400 mesh; Merck KGaA), using a solvent with a polarity associated with the TLC mobility. Melting points were measured with an OptiMelt MPA100 melting point apparatus (Stanford Research System; cat. no. MPA100) and the values were used without correction. Routine electro-spray spectra were recorded on an 8040-LC-MS (Shimadzu Corporation). High-resolution mass spectra were recorded on a Micromass-Q-T of Ultima spectrometer in positive ion mode or on an Axima-CFRTM plus high-performance linear/reflectron Matrix-Assisted Laser Desorption/Ionization-Time Of Flight (MALDI-TOF) mass spectrometer (Shimadzu Corporation). High-performance liquid chromatography (HPLC) purification was performed with an LC-20AD XR system (Shimadzu Corporation) and analytical HPLC was performed using an 8040-LC-MS Triple Quadrupole Mass Spectrometer (Shimadzu Corporation). Peptide synthesis was performed using Biotage-Syro II (Biotage). Proton nuclear magnetic resonance (1H-NMR) spectra were recorded on Bruker DPX-400 FT or Bruker ARX-400 FT spectrometers (Bruker Corporation). All 1H signal assignments were confirmed by COSY spectra. Carbon-13 nuclear magnetic resonance (13C-NMR) spectra were recorded on Bruker DPX-400 FT (100.61 MHz) or Bruker ARX-400 FT (100.61 MHz) instruments (Bruker Corporation). All 13C signal assignments were confirmed by heteronuclear single quantum coherence (HSQC) spectroscopy using CDCl3 (99.9% D) or 6d-DMSO as the solvent (Cambridge Isotope Laboratories, Inc.). Chemical shifts are referenced to the deuterated solvent peak and coupling constants are reported in hertz. Photolysis was performed using an Aicure system (UJ 35; Panasonic Corporation).
Protected salinomycin
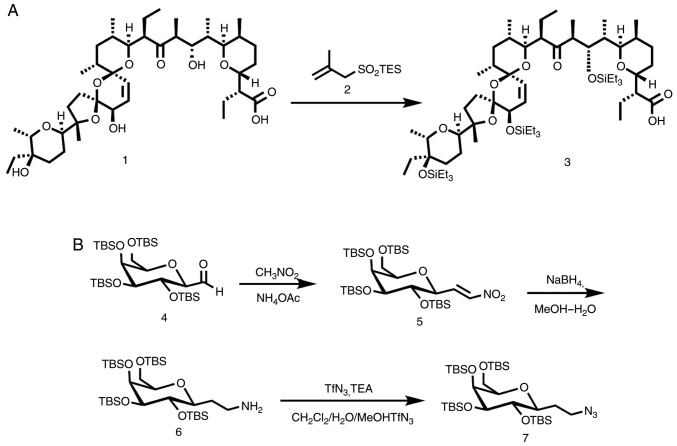
Salinomycin (Lucerna-Chem AG; cat. no. MCE-HY-17439-100MG) 1 (0.5 g; 0.666 mmol) (Rf, 0.16 at 3:2:1 Ether/pentane/CH2Cl2 system) was dissolved with triethylsilyl 2-methylprop-2-ene-1-sulfinate (1 g; 4.300 mmol) without solvent at −20°C (25). The mixture was stirred using a teflon-coated magnetic stir bar at 25°C for 30 min and subsequently cooled to −20°C, following which 5 ml of methanol (99.8%) was added. The sample was then allowed to reach 25°C following which the volatiles were evaporated in vacuo, to obtain pure, protected salinomycin 3 (0.72 g, 0.659 mmol, 99%) (Rf, 0.63 at 3:2:1 Ether/pentane/CH2Cl2 system), without requiring any further purification. TLC indicate complete conversion and pure product. The structure of protected salinomycin is presented in Fig. 2A.
Figure 2.
(A) Synthesis of fully protected salinomycin-free alcohols with triethylsilyl protection groups under mild conditions. (B) Synthesis of the azido sugar 7, the solubilizing group.
(((2S,3S,4R,5S,6R)-2-(2-azidoethyl)-6-(((tert-butyldimethy lsilyl)oxy)methyl)tetrahydro-2H-pyran-3,4,5-triyl)tris(oxy)) tris(tert-butyldimethylsilane). A total of 15 ml of THF containing tetra((tert-butyldimethylsilyl)oxy)-carbaldehyde 4 (2 g, 3.1 mmol; Rf, 0.20 at 9:1, light petroleum ether/ether) and nitromethane (0.625 ml, 11.500 mmol) were added to a stirred suspension of KOH (56 mg, 1.000 mmol), in 10 ml of THF cooled to 5°C. The mixture was subsequently heated to 20°C and stirred for 3 h. Ethyl acetate (EtOAc) (50 ml) and tap water (50 ml) were added to the mixture, the pH was adjusted to 3.0 using 10% citric acid and then extracted with EtOAc (3×30 ml). The combined EtOAc phases were washed with 4% NaCl (2×30 ml), dried over MgSO4 (10 g) and the volatiles were evaporated in vacuo. The crude product was subsequently dissolved in CH2Cl2 (2 ml). Subsequently, 1,8-Diazabicyclo[5.4.0]undec-7-ene (DBU) (0.8 ml; 5.000 mmol) was added to the solution of the crude product and stirred for 30 min, after which the mixture was diluted with Et2O (50 ml), quenched with 1M HCl (15 ml), washed with saturated NaHCO3 (10 ml) and resuspended over MgSO4 (5 g) to dry. The mixture was filtered and evaporated to give 2.2 g of crude 5 (Rf, 0.60 at 9:1, light petroleum ether/ether). The crude product was dissolved in 10 ml of methanol-water (5:1) and sodium borohydride (0.57 g; 15.00 mmol) was added. The mixture was stirred for 30 min at 0°C, and subsequently heated to 25°C. A saturated aqueous solution of sodium potassium tartarate (25 ml) was added and the aqueous phase was extracted with EtOAc (3×30 ml). The combined organic phase was dried over MgSO4 (5 g), filtered and concentrated in vacuo to give 1.8 g of crude 6 (Rf, 0.10 at 9:1, light petroleum ether/ether). The residue was dissolved in 1 ml of CH2Cl2 and 3 mg of CuSO4 in 6 ml of double distilled H2O, followed by the addition of triethylamine (390 µl; 2.930 mmol) and methanol (18 ml). A freshly prepared dichloromethane solution of trifluoromethanosulfonyl azide (3 ml; 1.780 mmol) was immediately added in a single portion. The reaction mixture was stirred at 0°C until TLC indicated that the reaction was complete (after ~90 min). The mixture was then extracted with CH2Cl2 (3×50 ml). The combined organic phase was dried over MgSO4 (5 g) and concentrated in vacuo. Flash chromatography (9:1, light petroleum ether/ether) yielded 1.4 g (68%, 4 steps) of the compound ‘7’ (Fig. 2B; Rf, 0.65 at 9:1, light petroleum ether/ether), a pure colorless oil (26).
1H NMR (400 MHz, CDCl3), δ 4.17 (d, J=10.0 Hz, 1H); 3.91 (td, J=3.0, 1.3 Hz, 1H); 3.86 (dd, J=10.0, 4.6 Hz, 1H); 3.74–3.71 (m, 1H); 3.69 (d, J=1.2 Hz, 1H); 3.66–3.57 (m, 2H); 3.40 (d, J=11.4 Hz, 2H); 1.94 (dt, J=14.1, 6.9 Hz, 1H); 1.62 (dtd, J=14.2, 7.0, 5.0 Hz, 1H); 0.93–0.82 (m, 36H); 0.17–0.05 (m, 24H).13C NMR (100.6 MHz, CDCl3), δ 77.60 (d, 1 J (C,H)=160); 77.26 (d, 1 J (C,H)=155); 76.67 (d, 1 J (C,H)=150); 73.15 (d, 1 J (C,H)=165); 72.82 (d, 1 J (C,H)=155); 67.85 (d, 1 J (C,H)=165); 61.44 (t, 1 J (C,H)=150); 48.35 (t, 1 J (C,H)=155); 31.37 (t, 1 J (C,H)=145); 26.10, 25.95, 25.91, 25.44, (4q, 1 J (C,H)=125; (CH3)3CSi); 19.60, 18.60, 18.40, 18.10 (4s, (CH3)3CSi); −4.05, −4.29, −4.67, −5.15 (8q, 1 J (C,H)=118, CH3Si); MALDI-HRMS calculated for (M + Na) C32H71N3O5Si4Na 712.4368 demonstrated 712.4372. Elementary analysis calculated for C32H71N3O5Si4 (689.4471): C 55.68, H 10.37; demonstrated: C 55.75, H 10.40. The structure of (((2S,3S, 4R,5S,6R)-2-(2-azidoethyl)-6-(((tert-butyldimethylsilyl) oxy)methyl)tetrahydro-2H-pyran- 3,4,5- triyl)tris(oxy))tris(tert-butyldimethylsilane) is presented in Fig. 2B.
Peptide synthesis
All peptides were synthesized at room temperature using a 0.20 mmol scale on the Fmoc-Gly-Wang resin (AnaSpec, Inc.; cat. no. AS-20057) (0.41 mmol/g loading), and an automated CS 336X peptide synthesizer from CSBio. The Fmoc-NH-Gly-Wang resin (0.41 mmol/g, 100–200 mesh) was used for all peptide syntheses. The procedure for a typical 0.2 mmol-scale synthesis is described below.
Resin swelling
Approximately 0.49 g of the Fmoc-NH-Gly-Wang resin was swollen for 15 min at 25°C in dimethylformamide (DMF)/CH2Cl2 (1:1).
Fmoc deprotection cycles
Fmoc deprotection was accomplished by adding 8.0 ml of 20% (v/v) piperidine in DMF to the resin and rocking the mixture for 20 min (two 10 min cycles).
Coupling cycles
A total of five equivalents of Fmoc-AA-OH and five equivalents of HBTU were dissolved in 5 ml of DMF, and then six equivalents of N,N-diisopropylethylamine (DIEA) were added in 3 ml of DMF. The mixture was added to the resin after shaking for 30 min at 25°C.
Washing cycles
Washing was performed after each reaction (deprotection or coupling) with 10 ml of DMF (five times) as previously described (27). Resin swelling, Fmoc deprotection, coupling cycle and washing cycle were performed using an automated Biotage syro II peptide synthesizer (Biotage) at 25°C.
Protocol for click chemistry
The resin (0.07 mmol) was suspended in DMF (1.5 ml) and ammonium azide (32 mg, 0.13 mmol) was added followed by the addition of a suspension of DIEA (113 µl, 0.66 mmol), CuI (24 mg, 10 mmol) and sodium ascorbate (200 mg, 1.00 mmol) in DMF/H2O (1.25:0.15 v/v). The mixture was shaken at 20°C for 8 h, and the reaction was monitored by liquid chromatography-mass spectrometry (LC-MS) (cat. no. LCMS-8040; Shimadzu Corporation). The resin was filtered and washed sequentially with: i) 5×10 ml of an imidazole solution in DMF; ii) 5×10 ml of DMF; iii) 5×10 ml of 10% double distilled water in DMF; iv) 5×10 ml of methanol; v) 20% piperidine in DMF and vi) 5×10 ml of CH2Cl2.
Washing
The resin was rinsed with DMF, DCM, methanol and diethyl ether, and subsequently dried under vacuum.
Peptide cleavage
The peptide resin was placed in a 30 ml glass solid-phase peptide synthesis (SPPS) reactor (Biotage) and was shaken with 10 ml of the freshly prepared cleavage cocktail for 2 h at 25°C. The resin cleavage cocktail contained trifluoroacetic acid (TFA):H2O: thioanisole:1,2-ethanedithiol:anisole (85:5:5:3:2 v/v). The cleavage solution was subsequently drained into a 50 ml glass vial and the resin was washed five times with TFA. The collected washings were then concentrated in vacuo in a water bath at 24°C for 5 min to reach a volume of 4–5 ml in order to remove TFA. The deprotected peptide was transferred to a 50 ml falcon tube and the peptide was subsequently precipitated using 30 ml of cold ether. The milky mixture was centrifuged (5,600 × g; −20°C; 10 min) to a pellet and the ether layer was decanted. The crude peptide thioester was dissolved in MeCN:H2O (1:1 v/v) with 0.05% TFA, lyophilized, re-dissolved with MeCN:H2O (1:1 v/v) with 0.05% TFA, filtered and lyophilized to provide the crude peptide for HPLC purification.
HPLC conditions
Preparative HPLC
Purification was performed at 25°C with a HPLC system (600 HPLC system; model no. 6 CE; Waters Corporation) and the peptides were detected at 215 and 280 nm. The peptides were eluted from the column using mobile phases A (0.1% TFA in double distilled H2O) and B (0.1% TFA in MeCN), at 25°C for 60 min using a Vydac-packed 25×250 mm reversed phase column (218TP C18 SPRING Col 250×25 mm 10 µm column; cat. no. DRM-S218TP1025 (BGB Analytik SA) and a flow rate of 10 ml/min were used.
Analytical HPLC
Analytical HPLC was performed at 25°C for 25 min with a 8040-LC-MS Triple Quadrupole Mass Spectrometer (Shimadzu Corporation), with detection from 210–450 nm, using a 4.6×250 mm Cosmosil C18 column and a flow rate of 0.5 ml/min. Fractions containing the pure target peptides were collected and lyophilized. Peptide purity and identity were confirmed by analytical HPLC and mass spectrometry, either by MALDI-TOF or electrospray ionization-MS.
Photolysis cleavage
In total, 100 µl of peptide was added to a 1 cm-thick quartz cell that was placed 5 cm away from the UV light source at ≥365 nm (Aicure UJ30/35 LED Spot Type UV Curing Systems; Panasonic Corporation). A total of 5 µl was taken from the sample each 20 sec for 3 min. Then, the samples were taken without any further treatment and the cleavage was monitored by RP-LC-MS. RP-HPLC analysis of the photocleavage of the peptide and the release of salinomycin by photolysis at ≥365 nm were plotted as a function of time. The compounds were detected using a tandem mass spectrometry (MS-MS) technique, and quantified by comparing the spectra with those of standard compounds at the same concentration.
MS-MS conditions
A stock solution of salinomycin (Lucerna-Chem AG; cat. no. MCE-HY-17439-100MG) was prepared at a concentration of 5 µmol/ml. Linearity was investigated over 10 points of calibrations with the concentration of the sample ranging between 0.1 and 20 µmol/ml. Samples were injected in increasing order and in decreasing order with 3 min washing between each injection, calibration curves solutions were prepared by serial dilution of acetonitrile. The expected concentration of salinomycin was of ~5 µmol/ml following photolysis cleavage of the peptide; therefore, the calibration curve samples concentrations were selected based on the expected concentration of salinomycin. Then, 5 µmol/ml of peptide was prepared, the photolysis was performed and samples were collected at various times (at 0, 20, 40, 60, 80 and 100 sec). The solution was then injected to a LC-MS system, and the MS-MS peaks were analyzed using a software (LabSolutions LLC; version 2.02; Shimadzu Corporation) to obtain the corresponding spectra. All experiments were performed as follows: System, LC-MS-8040; columns, Column raptor ARC-18 LC (Restek); column temperature, 40°C; mobile phase, A (0.1% TFA in double distilled water) and B (0.1% TFA in MeCN); gradients, 0–5 min 95% A, 18–22 min 5% A and 22–25 min 95% A; flow rate, 0.5 ml/min; injection volume, 30 µl; mass spectrometer, LCMS-8040 (Shimadzu Corporation; triple quadruple mass spectrometer); ionization, ESI positive mode; heating block temperature 400°C; drying gas, nitrogen (15 l/min).
Dose response assay
The dose-response of peptide 1 (with the solubilizing group), peptide 2 (without the solubilizing group) and salinomycin was evaluated using the MTT assay. The 50% inhibitory concentration (IC50) was used in order to assess the potency. Peptide 1, peptide 2 and salinomycin were initially dissolved in DMSO (Sigma Aldrich; Merck KGaA; cat. no. D8418-100ML), and then diluted with PBS to make the concentration of the three compounds in the range of 0.1–2 µM and the concentration of DMSO 0.2%. The control sample was prepared with 0.2% DMSO in PBS without adding the compounds (peptide 1, peptide 2 and salinomycin). For the assay, cells were seeded in 96-well plates (7,500 cells/well for MCF-7 and 5,500 cells/well for JIMT-1, 200 µl). MCF-7 cells were purchased from the American Type Culture Collection (cat. no. HTB-22). JIMT-1 cells were purchased from DSMZ (cat. no. ACC 589). The 96-well plates were incubated for 24 h at 37°C, after which peptide 1, peptide 2 and salinomycin were added. After 72 h of treatment, the plates were removed from the incubator (37°C) and the samples were exposed to radiation at l≥365 for 3 min. Subsequently, 20 µl of MTT (5 mg/ml) stock solution was added to each well. Following incubation at 37°C, with a 5% CO2 overlay for 4 h, the plates were removed from the incubator and the solutions were aspirated. The purple formazan product was dissolved by adding 100 µl of DMSO to each well. The plates were rotated for 5 min to distribute the DMSO evenly, and the absorbance was measured at 540 nm using a BioTek microplate reader. For each compound, eight independent dose-response experiments were performed with five replicates in each experiment. The software OriginPro (version 8.5; OriginLab Corporation) was used to plot the dose-response curves.
DEP assay
The DEP assays were conducted on dielectrophoretic reader (3DEP reader; DEPTech Ltd.), the analysis was performed by analyzing the radial motion of cells in the well for 10 sec, the DEP response was measured using 1–45 MHz. Electrical frequencies applied to the wells induce the cells to move inside the well due to the phenomenon of dielectrophoresis (28). In total, ~80 µl of prepared cell suspension was pipetted into a 3DEP disposable chip, inserted into the reader and exposed to 10–20 MHz to produce a full DEP spectrum. This process was repeated for each treatment group ≥3 times. Data were acquired over 15 sec (for JIMT-1) or 20 sec (for MCF-7) intervals (28).
Results
The synthesis of the caged glutamine model was designed in the present study using the TAT-protein sequence. In this sequence, tools for smoothly introducing the solubilizing groups were used. Another tool was used to attach the photo-cleavable linker to salinomycin, such that it was released at the suitable time during the in vitro analysis.
A number of methods have been used in order to protect salinomycin; however, all the common methods have failed to protect salinomycin from epimerization or elimination. In the present study, the triethylsilyl ether (TES) group was successfully incorporated to fully protect salinomycin, as depicted in (Fig. 2). The structure of salinomycin is presented in Fig. S1A. The MTT dose-response curves were analyzed in JIMT-1 and in MCF-7 cells (Fig. S1B and C). LC-MS-MS was performed to analyze the purity of salinomycin (Fig. S1D). Commercially available salinomycin was treated with triethylsilyl 2-methylprop-2-ene-1-sulfinate 2 at 0°C for 30 min in order to produce the fully silylated salinomycin with a quantifiable yield (25). Furthermore, the crude salinomycin-linker 3 was attached to the TAT-protein segment through a peptide bond (Fig. 2A).
The solubilizing group was selected from among the sugars of the carbohydrate family in order to avoid any toxicity interfering with the biological function. The synthesis of the solubilizing group is outlined in (Fig. 2B). A known aldehyde 4 (25,29–37) was treated under Henry reaction conditions (38) with nitromethane, in the presence of ammonium acetate to form the dehydrated product 5. The crude product 5 was subsequently treated with sodium borohydride in the presence of double distilled water and methanol for complete reduction in order to produce the amine form 6, which was treated with trifluoromethanesulfonyl azide in order to convert it to the azido sugar 7. The overall yield of the three steps was 55% (26).
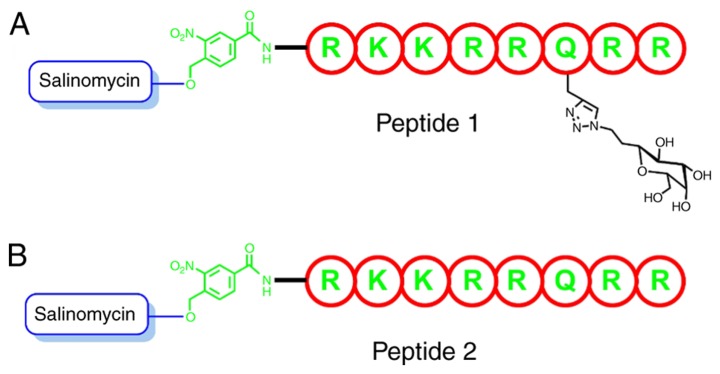
A total of two analogs were prepared in order to test the assumption and model design of the present study, one without the solubilizing sugar group and the other with the solubilizing group (Fig. 3). Peptides were synthesized using a standard Fmoc-SPPS protocol (39). Furthermore, the N-terminus of the peptide was coupled with (4-tert-butyldimethylsilyl (hydroxymethyl)-3-nitrobenzoic acid. The silyl group was subsequently removed using tetra-n-butylammonium fluoride (TBAF), followed by the esterification of salinomycin using DIC/DMA (40). The solubilizing group 7 was subsequently attached to glutamine in the TAT-protein segment using click chemistry on a solid support (24,41–43). Finally, the silylated groups were removed using TBAF, the peptides were cleaved from the solid support and the side chains were deprotected using dichloromethane/acetic acid/trifluoroethanol solution (3:1:1) treatment. The peptides were then purified using HPLC (Figs. S2 and S3).
Figure 3.
Fmoc-solid-phase peptide synthesis (SPPS). (A) Peptide with attached salinomycin and an extra sugar-solubilizing group. (B) Peptide with attached salinomycin without the solubilizing group.
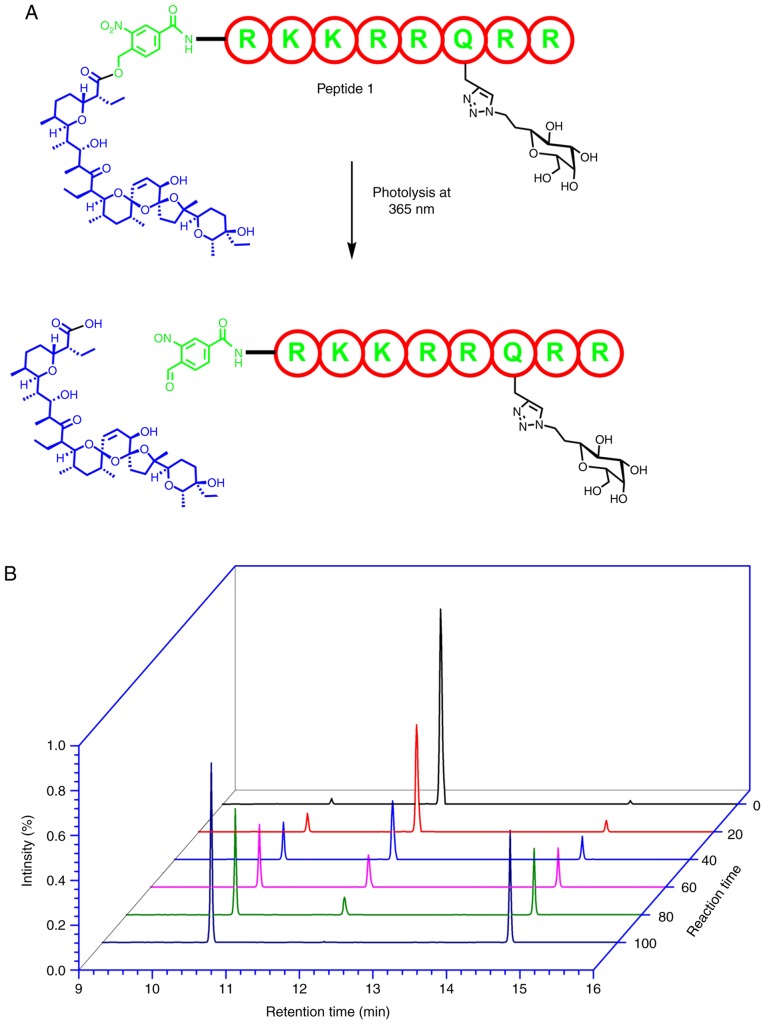
The nitrobenzyl ester groups of the two peptides are highly stable under physiological conditions (44). This comes as no surprise given that the cleavage of this group requires the peptides to be subjected to conditions of pH 12 and 75°C (45). In order to determine whether the cage can be cleaved to release the drug smoothly, a kinetic analysis was performed in the present study. The kinetic study was conducted under physiological conditions in order to control the cleavage conditions and optimize the procedure. As presented in Fig. 4A, complete and quantifiable conversion was achieved with UV irradiation at ≥365 nm within 80–100 sec.
Figure 4.
(A) Schematic depiction of the release of salinomycin by photolysis at 365 nm and a concentration of 2 µM. (B) RP-HPLC analysis of the photocleavage of peptide 1 and the release of salinomycin by photolysis at ≥365 nm, plotted as a function of time. The compounds were detected using the tandem mass spectrometry technique and quantified by comparing the spectra with those of standard compounds at the same concentration.
The kinetics of the cleavage was analyzed using an MS-MS technique, and a UV detector was implemented in order to follow the photolytic cleavage products. As a strong chromophore is not present, salinomycin does not yield a strong absorbance value after cleavage and is not well detected by the UV detector. RP-HPLC analysis of the photocleavage of peptide 1 and the release of salinomycin after photolysis at ≥365 nm, as a function of time is presented in (Fig. 4B). The kinetic study of the cleavage of the designed peptide 1 demonstrated a quick, clean and smooth cleavage process. The results of the present study indicate that an in vitro analysis can be performed conveniently as salinomycin can be released quickly during biological experiments.
In order to determine the biological effects of the solubilizing group, the peptide without the sugar stabilizing group (peptide 2) was synthesized following the same SPPS procedure used to synthesize peptide 1, and subsequently analyzed. The peptide without the solubilizing group was evaluated based on the dose-response curves obtained from the MTT assay.
The antiproliferative activity of the salinomycin analogs in MCF-7 and JIMT-1 breast cancer cells was evaluated using an MTT assay (Table I). Salinomycin, with the protein carrier and solubilizing group (peptide 1), exhibited an IC50 value three times lower than that of salinomycin in both cell lines. Similarly, the second analog, with the protein carrier but without the solubilizing group (peptide 2), exhibited an IC50 value lower than that of salinomycin in both cell lines. However, the IC50 value of peptide 2 was slightly higher than that of peptide 1. The results of the present study confirm that conjugating salinomycin to a protein carrier increases the activity of salinomycin, most likely due to an increase in cell penetration. Increasing the solubility, by attaching a sugar to the peptide, also yields increased activity (46). In order to quantify the cell viability, MTT was used, the cell viability change is presented in (Fig. S4A, C and E). In order to quantify apoptosis progression, the dielectrophoretic (DEP) method was used, according to the approach by Henslee et al (28), the progress in cancer cells is presented in (Fig. S4B, D and F).
Table I.
Antiproliferative activity of the peptide-conjugated analogs of salinomycin evaluated by an MTT-based dose-response assay.
| Group | Cell, IC50, mean ± SE, µM |
|---|---|
| Salinomycin SA, 1 | JIMI-1, 0.442±0.079 |
| MCF-7, 0.577±0.033 | |
| Peptide 1, with sugar solubilizing | JIMI-1, 0.195±0.032 |
| group | MCF-7, 0.134±0.016 |
| Peptide 2, without sugar solubi | JIMI-1, 0.382±0.052 |
| lizing group | MCF-7, 0.308±0.041 |
MTT reduction is assumed to be directly proportional to the cell number. All the biological results for each compound are ploted as figures and added to the supplementary information. IC50, 50% inhibitory concentration; SE, standard error.
Discussion
Many previous studies tried to improve salinomycin activity against cancer cells. Borgström et al (47) showed promising results, following the construction of a library of salinomycin analogs. Borgström et al (47) selectively modified alcohols on salinomycin. According to this previous study (47), the modifications of the allylic alcohol on C20 to smaller and less bulky groups of ester, carbonates and carbamate improved significantly the activity of native salinomycin. In addition, most of the groups used to modify salinomycin can be cleaved via esterases enzyme in the cells. This previous study demonstrated that these groups are temporary groups and can act as carriers by enhancing the lipophilic penetration, and the activity in the cell derives from the native salinomycin form.
The results of the present study confirm that the activity of salinomycin alone suffers due to its poor penetration of cancer cells. Salinomycin exhibits an improved activity level when conjugated to a protein carrier, such as TAT-protein, and a solubilizing group that allows greater penetration of the cells. In order to ensure that the maximum quantity of salinomycin reached the active site, the technique used in the present study employed photolysis in order to release salinomycin following cell penetration. As the ester bond is sensitive to X-ray radiation, this method can be used in cells, in vivo and in humans during radiotherapy of cancer cells. When machines producing mild X-rays, such as when a linear accelerator or cyberknife are used, exposure of the cancer cells to radiation is expected to release salinomycin inside the tumor cells. The same idea can be applied to other drugs that suffer from low solubility or the inability to penetrate cells.
Collectively, the present study improved the activity of salinomycin by increasing the penetration and the solubility using small TAT-peptides, which allowed the release of salinomycin inside the cells by photolysis. Using peptide carrier to improve salinomycin penetration resulted in IC50 values against cancer cells that were >4 times lower compared with the native salinomycin.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the Deanship of Scientific Research at Imam Abdulrahman Bin Faisal University (grant no. 2014255).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LA designed, synthesized, purified and analyzed the chemical compounds, and wrote and reviewed the published manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that he has no competing interests.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Becker MW, Wichal M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. 1997;124:1929–1939. doi: 10.1242/dev.124.10.1929. [DOI] [PubMed] [Google Scholar]
- 4.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 5.Ruben GC. The iso-competition point, a new concept for characterizing multivalent versus monovalent counterion competition, successfully describes cation binding to DNA. Biophys J. 1999;77:1–2. doi: 10.1016/S0006-3495(99)76867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitani M, Yamanishi T, Miyazaki Y, Ōtake N. Salinomycin effects on mitochondrial ion translocation and respiration. Antimicrob Agents Chemother. 1976;9:655–660. doi: 10.1128/AAC.9.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugschies A, Gässlein U, Rommel M. Comparative efficacy of anticoccidials under the conditions of commercial broiler production and in battery trials. Veterinary Parasitology. 1998;76:163–171. doi: 10.1016/S0304-4017(97)00203-3. [DOI] [PubMed] [Google Scholar]
- 8.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou S, Wang F, Wong ET, Fonkem E, Hsieh TC, Wu JM, Wu E. Salinomycin: A novel anti-cancer agent with known anti-coccidial activities. Curr Med Chem. 2013;20:4095–4101. doi: 10.2174/15672050113109990199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoszczak M, Huczyński A. Anticancer activity of polyether ionophore-salinomycin. Anticancer Agents Med Chem. 2015;15:575–591. doi: 10.2174/1871520615666150101130209. [DOI] [PubMed] [Google Scholar]
- 11.Huczynski A. Salinomycin: A new cancer drug candidate. Chem Biol Drug Des. 2012;79:235–238. doi: 10.1111/j.1747-0285.2011.01287.x. [DOI] [PubMed] [Google Scholar]
- 12.Caldwell GW, Ritchie DM, Masucci JA, Hageman W, Yan Z. The new pre-preclinical paradigm: Compound optimization in early and late phase drug discovery. Curr Top Med Chem. 2001;1:353–366. doi: 10.2174/1568026013394949. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann T, Schmitt J, Röhring C, Nimptsch D, Nöller J, Mohr C. ADME related profiling in 96 and 384 well plate format-a novel and robust HT-assay for the determination of lipophilicity and serum albumin binding. Curr Drug Deliv. 2006;3:181–192. doi: 10.2174/156720106776359177. [DOI] [PubMed] [Google Scholar]
- 14.Di L, Kerns EH, Li SQ, Petusky SL. High throughput microsomal stability assay for insoluble compounds. Int J Pharm. 2006;317:54–60. doi: 10.1016/j.ijpharm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Dewangan J, Srivastava S, Rath SK. Salinomycin: A new paradigm in cancer therapy. Tumor Biol. 2017;39:1010428317695035. doi: 10.1177/1010428317695035. [DOI] [PubMed] [Google Scholar]
- 16.Savjani KT, Gajjar AK, Savjani JK. Drug solubility: Importance and enhancement techniques. ISRN Pharm. 2012;2012:195727. doi: 10.5402/2012/195727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasinu P, Pillay V, Ndesendo VMK, du Toit LC, Choonara YE. Diverse approaches for the enhancement of oral drug bioavailability. Biopharm Drug Dispos. 2011;32:185–209. doi: 10.1002/bdd.750. [DOI] [PubMed] [Google Scholar]
- 18.Patel SG, Sayers EJ, He L, Narayan R, Williams TL, Mills EM, Allemann RK, Luk LYP, Jones AT, Tsai YH. Cell-penetrating peptide sequence and modification dependent uptake and subcellular distribution of green florescent protein in different cell lines. Sci Rep. 2019;9:6298. doi: 10.1038/s41598-019-42456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löwik DW, Meijer JT, Minten IJ, van Kalkeren H, Heckenmüller L, Schulten I, Sliepen K, Smittenaar P, van Hest JC. Controlled disassembly of peptide amphiphile fibres. J Pept Sci. 2008;14:127–133. doi: 10.1002/psc.969. [DOI] [PubMed] [Google Scholar]
- 20.Borrelli A, Tornesello A, Tornesello M, Buonaguro F. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules. 2018;23:E295. doi: 10.3390/molecules23020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallego I, Rioboo A, Reina JJ, Díaz B, Canales Á, Cañada FJ, Guerra-Varela J, Sánchez L, Montenegro J. Glycosylated cell-penetrating peptides (GCPPs) ChemBioChem. 2019;20:1400–1409. doi: 10.1002/cbic.201800720. [DOI] [PubMed] [Google Scholar]
- 22.Abramova TV, Silnikov VN. 4-Aminometyl-3-nitrobenzoic acid-a photocleavable linker for oligonucleotides containing combinatorial libraries. Nucleosides Nucleotides Nucleic Acids. 2005;24:1333–1343. doi: 10.1080/15257770500230509. [DOI] [PubMed] [Google Scholar]
- 23.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Sharpless KB, Kolb HC. Click chemistry A concept for merging process and discovery chemistry. Abstr Pap Am Chem Soc. 1999;217:U95–U. [Google Scholar]
- 25.Huang X, Craita C, Awad L, Vogel P. Silyl methallylsulfinates: Efficient and powerful agents for the chemoselective silylation of alcohols, polyols, phenols and carboxylic acids. Chem Commun (Camb) 2005:1297–1299. doi: 10.1039/b417894g. [DOI] [PubMed] [Google Scholar]
- 26.Levy DE, Tang C. Elsevier; 1995. The Chemistry of C-glycosides. [Google Scholar]
- 27.Amblard M, Fehrentz JA, Martinez J, Subra G. Methods and protocols of modern solid phase peptide synthesis. Mol Biotechnol. 2006;33:239–254. doi: 10.1385/MB:33:3:239. [DOI] [PubMed] [Google Scholar]
- 28.Henslee EA, Torcal Serrano RM, Labeed FH, Jabr RI, Fry CH, Hughes MP, Hoettges KF. Accurate quantification of apoptosis progression and toxicity using a dielectrophoretic approach. Analyst. 2016;141:6408–6415. doi: 10.1039/C6AN01596D. [DOI] [PubMed] [Google Scholar]
- 29.Vogel P, Awad LK. Disaccharide mimics as drugs against cancer and epitopes for anti-cancer vaccine candidates. Immun Res. 2016;12:57. [Google Scholar]
- 30.Awad L, Madani R, Gillig A, Kolympadi M, Philgren M, Muhs A, Gérard C, Vogel P. A C-linked disaccharide analogue of thomsen-friedenreich epitope induces a strong immune response in mice. Chemistry. 2012;18:8578–8582. doi: 10.1002/chem.201200364. [DOI] [PubMed] [Google Scholar]
- 31.Vogel P, Gerber-Lemaire S, Awad L, Bello C, Fiaux H, Juillerat-Jeanneret L, Gillig A, Kolympadi M, CARB 29-C-Disaccharides and Analogs, corp-author, editors. Amer Chemical Soc; Washington, DC: 2007. The Search for Anticancer Agents; pp. 493–493. [Google Scholar]
- 32.Awad L, Demange R, Zhu YH, Vogel P. The use of levoglucosenone and isolevoglucosenone as templates for the construction of C-linked disaccharides. Carbohydr Res. 2006;341:1235–1252. doi: 10.1016/j.carres.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Awad L, Riedner J, Vogel P. C-linked disaccharide analogue of the Thomsen-Friedenreich (T)-epitope alpha-O-conjugated to L-serine. Chemistry. 2005;11:3565–3573. doi: 10.1002/chem.200401301. [DOI] [PubMed] [Google Scholar]
- 34.Awad L. Synthesis of a C-linked disaccharide analogue of the Thomsen Friedenreich (TF)-epitope a-O-conjugated to L-serine and formation of a cluster as potential anticancer vaccine. 2005 doi: 10.1002/chem.200401301. [DOI] [PubMed] [Google Scholar]
- 35.Loay A. Synthesis of a C-linked disaccharide analogue of the Thomsen Friedenreich (TF)-epitope α-O-conjugated to l-serine and formation of a cluster as potential anticancer vaccine. École Polytechnique Fédérale De Lausanne. 2005 [Google Scholar]
- 36.Demange R, Awad L, Vogel P. Synthesis of C-linked analogues of β-d-galactopyranosyl-(1→3)-d-galactopyranosides and of β-d-galactopyranosyl-(1→3)-d-galactal. Tetrahedron: Asymmetry. 2004;15:3573–3585. doi: 10.1016/j.tetasy.2004.09.029. [DOI] [Google Scholar]
- 37.Awad L. Synthesis of C-linked and glycopeptide towards non-hydrozable T epitopes and artificial Anticancer Vaccines. DEA. 2001 [Google Scholar]
- 38.Hallinan EA, Kramer SW, Houdek SC, Moore WM, Jerome GM, Spangler DP, Stevens AM, Shieh HS, Manning PT, Pitzele BS. 4-Fluorinated L-lysine analogs as selective i-NOS inhibitors: Methodology for introducing fluorine into the lysine side chain. Org Biomol Chem. 2003;1:3527–3534. doi: 10.1039/B307563J. [DOI] [PubMed] [Google Scholar]
- 39.Hansen PR, editor. Methods Mol Biol. Humana Press; New York, NY: 2017. Antimicrobial peptides. [DOI] [Google Scholar]
- 40.Lécaillon J, Gilles P, Subra G, Martinez J, Amblard M. Synthesis of cyclic peptides via O-N-acyl migration. Tetrahedron Lett. 2008;49:4674–4676. doi: 10.1016/j.tetlet.2008.05.049. [DOI] [Google Scholar]
- 41.Awad L, Jejelava N, Burai R, Lashuel HA. A new caged-glutamine derivative as a tool to control the assembly of glutamine-containing amyloidogenic peptides. ChemBioChem. 2016;17:2353–2360. doi: 10.1002/cbic.201600474. [DOI] [PubMed] [Google Scholar]
- 42.Butterfield S, Hejjaoui M, Fauvet B, Awad L, Lashuel HA. Chemical strategies for controlling protein folding and elucidating the molecular mechanisms of amyloid formation and toxicity. J Mol Biol. 2012;421:204–236. doi: 10.1016/j.jmb.2012.01.051. [DOI] [PubMed] [Google Scholar]
- 43.Awad L, Jejelava N, Brik A, Lashuel H, editors. Novel chemical tools to facilitate the synthesis and control the folding and Self-assembly of amyloid-forming polypeptides. Biopolymers. 2011 [Google Scholar]
- 44.Klán P, Šolomek T, Bochet CG, Blanc A, Givens R, Rubina M, Popik V, Kostikov A, Wirz J. Photoremovable protecting groups in chemistry and biology: Reaction mechanisms and efficacy. Chem Rev. 2013;113:119–191. doi: 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene TW, Wuts PG. Wiley; 1999. Protective groups in organic synthesis. [DOI] [Google Scholar]
- 46.Borgström B, Huang X, Pošta M, Hegardt C, Oredsson S, Strand D. Synthetic modification of salinomycin: Selective O-acylation and biological evaluation. Chem Commun (Camb) 2013;49:9944–9946. doi: 10.1039/c3cc45983g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.