Abstract
Overexpression of α-methylacyl-coenzyme A racemase (AMACR/P504S) is a major abnormality that has been observed in prostate cancer, whereas microRNA (miRNA/miR) 200c, is downregulated. The aim of the present study was to explore whether miR200c was able to exert any regulatory effects on AMACR. To meet this aim, bioinformatics analysis was performed to identify potential binding sites for miR200c in the 3′-untranslated region (3′-UTR) of AMACR. Recombinant adenoviral and dual reporter gene assays were designed to examine the binding of miR200c to the potential seed sequences in the AMACR 3′-UTR. Conventional reverse transcription (RT)-PCR, RT-quantitative (q)PCR and western blotting were also used to examine the regulatory effects of miR200c on AMACR at the mRNA and protein levels. Furthermore, Cell Counting Kit-8, wound healing and Transwell assays were performed to investigate the biological effects of miR200c-AMACR deregulation on prostate cancer cell proliferation, migration and invasion. It was revealed that miR200c post-transcriptionally suppressed AMACR expression by interacting with the 90–97 nucleotide sequence of the AMACR mRNA 3′-UTR. Artificial overexpression of miR200c significantly downregulated the mRNA and protein levels of AMACR in DU145 and PC-3 prostate cancer cells. Knockdown of AMACR by RNA interference, or overexpression of miR200c by recombinant adenoviral Ad-miR200c, inhibited prostate cancer cell proliferation, migration and invasiveness. Taken together, the results of the present study revealed that miR200c may suppress the AMACR expression level post-transcriptionally. The results also indicate that perturbation of the miR200c-AMACR regulatory mechanism may be involved in prostate carcinogenesis and that this may be exploited in future therapeutic approaches to prostate cancer.
Keywords: AMACR, miR200c, prostate cancer, post-transcriptional regulation
Introduction
MicroRNAs (miRNAs/miRs) are small, non-coding RNA molecules that negatively regulate gene expression either by messenger RNA (mRNA) degradation, or translational repression via binding to the 3′-untranslated region (3′-UTR) of target genes. miRNA dysfunction can play an important role in tumorigenesis and tumor progression. miR200c is an important member of the miR200 family, which functions as a tumor suppressor and is aberrantly expressed in various types of malignant tumors, including breast (1,2), gastric (3,4), colorectal (5) and ovarian cancer (6), clear cell renal cell carcinoma (7) and prostate cancer (8). Furthermore, miR200c has been shown to be a key regulator of epithelial-mesenchymal transition (EMT) (9,10) in breast and prostate cancers. A number of genes, for example, those encoding BMI1 proto-oncogene, polycomb ring finger (BMI1) (11), E2F transcription factor 3 (11), fibronectin 1 (FN1) (12), insulin receptor substrate 1 (IRS1) (8), PTEN (13), fascin actin-bundling protein 1 (FSCN1) (14) and platelet-derived growth factor receptor (PDGFR) (15), have been identified as distinct miR200c target genes, which are involved in diverse biological processes, including tumor cell proliferation, metabolism, metastasis and angiogenesis. Downregulation of miR200c has been shown to promote cell proliferation and metastasis, as well as to induce stem-cell like phenotype, in various types of cancer (16,17). Tang et al (18) reported reduced miR200c level in prostate cancer cells by microarray analysis. A previous study published by the authors' research group also demonstrated that miR200c is downregulated in prostate cancer and that it was associated with the growth and progression of prostate cancer cells (8).
α-methylacyl-coenzyme A racemase (AMACR/P504S), an enzyme involved in fatty acid metabolism, was shown to be overexpressed in prostate cancer, which led to the promotion of cell proliferation (18). The same study also revealed that artificial suppression of AMACR with RNA interference significantly inhibited the proliferation of prostate cancer cells (19). It has also been reported that AMACR is overexpressed in other tumor types, including gastric (20) and colon (21) adenocarcinoma, breast cancer (1), gastrointestinal stromal tumors (22), and myxofibrosarcomas (23). Potential mechanisms of AMACR overexpression in prostate cancer and other tumors include transcriptional activation, for example, by CCAAT-enhancer-binding protein (C/EBP, and gene amplification (21–26). On the basis of the reduced expression of miR200c in prostate cancer cells and the authors' pilot bioinformatics analysis, the present study proposed the hypothesis that miR200c may be a negative regulator of AMACR.
The aim of the present study was to investigate the regulatory effect of miR200c on AMACR and to explore the biological functions of the miR200c-AMACR axis. In addition, experimental evidence is presented to show how miR200c directly targets AMACR by binding to the 90–97 nucleotide (nt) seed sequence of the AMACR mRNA 3′-UTR and that artificial upregulation of miR200c and/or downregulation of AMACR suppresses prostate cancer cell proliferation, migration and invasiveness.
Materials and methods
Cell lines and tissue samples
The human prostate cancer cell lines PC-3, DU145, CL1, LNCaP and C4-2B were obtained from the American Type Culture Collection. The cell lines were maintained in RPMI-1640 medium (Life Technologies; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal calf serum (FCS; Life Technologies; Thermo Fisher Scientific, Inc.) and 100 µg/ml penicillin and 100 µg/ml streptomycin. The 293 cell line was obtained from the American Type Culture Collection and maintained in DMEM (Life Technologies; Thermo Fisher Scientific, Inc.) with 10% FCS. Cells were cultured at 37°C with 5% CO2. Benign prostatic hyperplasia (BPH) tissue (n=3; patient age 65–77) and prostate cancer tissue samples (n=3; patient age 72–81; Gleason score 7–9) were collected from March to May 2018 by transurethral resection of the prostate. All tissue samples were collected in the West China Hospital according to the ethical guidelines and procedures approved by the institutional supervisory committee. Informed consent was obtained according to the institutional guidelines. Fresh tissue samples were stored immediately at −80°C.
RNA isolation, stem-loop reverse transcription (RT) and conventional RT-PCR
Total RNA was isolated using TRIzol® reagent (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. SMART® MMLV reverse transcriptase (Takara Biotechnology Co., Ltd.) was used for RT (temperature protocol: 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and 10°C for 10 min). Stem-loop RT-PCR was employed to examine the mature miR200c, using the primer for miR200c designed with the sequence: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCCATC-3′. The random RT primer, 5′-(dN)9−3′ (Takara Biotechnology Co., Ltd.), was used for other genes. The PCR primers sequences were as follows: miR200c (forward, 5′-GCATAGCCCGTAATACTGCCGGGTA-3′; reverse, 5′-GTGCAGGGTCCGAGGT-3′, 67 bp); U6 (forward, 5′-TGGAACGATACAGAGAAGATTAGCA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′, 66 bp); AMACR (forward, 5′-gcttatttatgccaggctgag-3′; reverse, 5′-cttcccacagactcaatttctg-3′, 314 bp); and β-actin (forward, 5′-CTGGCACCACACCTTCTACAATG-3′; reverse, 5′-CCTCGTAGATGGGCACAGTGTG-3′, 248 bp). Thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 30 sec, annealing at 56°C for 30 sec and extension at 72°C before a final extension at 72°C for 10 min.
The amplification products were resolved by 2% agarose gel electrophoresis and visualized by staining with the fluorescent dye, Goldview™ (Beijing Solarbio Science & Technology Co., Ltd.).
RT-quantitative (q)PCR
RT-qPCR was performed on a Light Cycler 2.0 RT-qPCR instrument (Roche Diagnostics GmbH) using SYBRGreen as fluorescence (Bio-Rad Laboratories, Inc.) and data were analyzed with Light Cycler software, version 4.05 (Roche Diagnostics GmbH) as previously described (27). The β-actin gene was used as control. The copy number of target genes (relative to β-actin) was determined using the 2−ΔΔCq method (28), where ΔΔCq=ΔCqexp-ΔCqcon=(Cqexp-target-Cqexp-actin)-(Cqcon-target-Cqcon-actin), in which ‘exp’ represents the experimental group, ‘con’ the control group and ‘target’ represents the gene of interest.
Western blot analysis
The primary antibodies used were as follows: AMACR (rabbit monoclonal, cat. no. ab175280; 1:1,000; Abcam) and GAPDH (mouse monoclonal, 1:5,000; cat. no. KC-5G4; Kangchen Bio-tech, Inc.). Horseradish peroxidase-labeled secondary antibodies (Both used at 1:10,000; goat anti-mouse IgG secondary antibody, cat. no. 31430 and goat anti-rabbit IgG secondary antibody, cat. no. 31460) were from Zymed; Thermo Fisher Scientific, Inc. Western blotting was performed as previously described (29). Briefly, after incubation, cells were trypsinized and lysed with RIPA cell lysis buffer containing PMSF (Beyotime Institute of Biotechnology) at 4°C for 10 min. The total protein was obtained by centrifugation at 12,000 g at 4°C for 10 min and quantified with a Bicinchoninic acid protein assay kit (Tiangen Biotech Co., Ltd.). Then total protein (40 µg/sample) was separated on a 10% SDS-PAGE gel and transferred to polyvinylidene difluoride membrane. After blocking with 5% nonfat milk at 37°C for 2 h, the membrane was incubated with antibodies against AMACR at 4°C overnight. After washing, the membrane was incubated with HRP-secondary antibody at 37°C for 1 h. Then the membrane was washed and analyzed with ECL detection system (EMD Millipore). The expression of GAPDH was used as internal reference. Images were captured using the ChemiDox XRS system (Bio-Rad Laboratories, Inc.). Quantification was performed using ImageJ software (version 1.47; National Institutes of Health).
Recombinant adenoviral vectors for overexpression of miR200c
The recombinant adenoviral vector for overexpression of miR200c, Ad-miR200c, was constructed according to the protocol previously reported (30). Briefly, the pri-miR200c sequence was amplified from 293 cell genomic DNA. PCR products were cloned into plasmid pMD19-T (Takara Biotechnology Co., Ltd.), verified by sequencing and subcloned into shuttle plasmid pAdTrack-CMV to construct pAdTrack-miR200c. pAdTrack-miR200c, linearized with PmeI, was used to transform BJ5183-AD-1 cells harboring the adenoviral pAdeasy-1 vector (Stratagene; Agilent Technologies, Inc.) for homologous recombination. Colonies were screened by plasmid miniprep and PacI restriction analysis to obtain clones with recombinant miR200c (designated ‘pAdeasy-miR200c’). PacI-linearized pAdeasy-miR200c was used to transfect 293 cells to obtain packaged recombinant miR200c adenovirus (designated Ad-miR200c). Ad-miR200c was amplified by repeated infection and verified by PCR. The pAdTrack-CMV empty vector was used as control (designated ‘Ad-control’). The titers and the multiplicity of infection were determined according to the manufacturer's protocols. After transfection for 48 h, green fluorescence was observed using an inverted fluorescence microscope (1X71; Olympus Corporation).
Luciferase reporter constructs and site-directed mutagenesis
The seed sequences of AMACR 3′-UTR (90–97 nt) with flanking sequences were amplified from the genomic DNA of 293 cells. The 3′-UTR of the AMACR mRNA was analyzed using TargetScan 6.2 (http://www.targetscan.org/). PCR products were cloned into plasmid pMD19-T (Takara Biotechnology Co., Ltd.) and subsequently subcloned into the vector pGL3-promoter (Promega Corporation) and inserted into the 3′-UTR downstream of the luciferase coding sequence. Overlapping PCR was used for site-directed mutagenesis of the seed sequences (CAGTATTA mutated to GGGGTTT) to construct pGL3-AMACR-UTRmutant (mut), the PCR primers for which were 5′-ATGGAGGAAGGGGTTTCAGT-3′ (forward) and 5′-ACTGAAACCCCTTCCTCCAT-3′ (reverse).
Dual reporter gene assays
Cells were cultured in 24-well plates (1×105 cells/well) and transfected with the reporter constructs (0.8 µg) using Novagen® NanoJuice™ transfection reagent (EMD Millipore). The pRL-CMV vector (0.02 µg; Promega Corporation) was co-transfected as the internal control. The luciferase expression of pGL3-promoter plasmid was used as baseline. Cells were infected with Ad-miR200c and subsequently trypsinized with 0.25% trypsin at 48 h post-transfection. The firefly and Renilla luciferase activities were determined using a Luminometer TD-20/20 (Turner BioSystems) with the Dual Luciferase® Reporter Assay system (Promega Corporation). The relative luciferase activity was determined by calculating the ratios of firefly/Renilla luciferase activities.
RNA interference
Double-stranded small-interfering RNAs (siRNAs) and controls (si-CON, 5′- GCGCGCTTTGTAGGATTCG-3′) were designed, synthesized, and purified by Guangzhou RiboBio Co., Ltd. The three siRNAs targeting AMACR were designed as follows: 5′-GCCACGATATCAACTATTT-3′ (si-AMACR1), 5′-GCACCTTTCTATACGACTT-3′ (si-AMACR2) and 5′-GGAGGTTGTTCATCATGAT-3′ (si-AMACR3). Cells were transfected with 0.1 nmol siRNA or si-CON using Novagen® NanoJuice™ transfection reagent (EMD Millipore) and collected 24 h later.
Artificial overexpression of AMACR in PC-3 and DU145 cells infected by Ad-miR200c
The full-length cDNA of the AMACR coding sequence was cloned into the thymine and adenine clone vector (pMD19-T) and subcloned into the pcDNA3.1 vector (Clontech Laboratories, Inc.). The primers used for cloning were as follows: 5′-AGATCTcatggcactgcagggca-3′ (forward) and 5′-GGTACCtagagactagcttttacctt-3′ (reverse). PC-3 and DU145 cells infected by Ad-miR200c were transfected with AMACR expression plasmid (pcDNA3.1-AMACR) or pcDNA3.1 blank vector (pcDNA3.1-blank) using Novagen® NanoJuice™ transfection reagent (EMD Millipore). After incubation for 4 h in serum-free medium, the medium was replaced with DMEM medium containing 10% FCS and cells were incubated for a further 44 h before analysis.
Cell Counting Kit-8 (CCK-8) assay
The cell suspension was seeded in 96-well plates (3,000 cells/well) and pre-incubated for 24 h with RPMI-1640 media containing 10% FCS, penicillin and streptomycin. Cells were subsequently treated with si-AMACR or si-CON for 0, 24, 48, or 72 h, respectively. Cells were also treated with Ad-CON, Ad-miR200c, or Ad-miR200c + pcDNA3.1-AMACR for 0, 24 or 48 h, respectively. Subsequently, 10 µl CCK-8 (Dojindo Molecular Technologies, Inc.) was added to each well and incubated for 2 h at 37°C. The absorbance was examined at 450 nm using a scanning multi-well spectrophotometer (Thermo Fisher Scientific, Inc.). The proliferation index (PI) was calculated as PI=[(Asample-Ablank)/(Acontrol-Ablank)] ×100%.
Wound-healing assay
DU145 and PC-3 cells were seeded in 12-well plates (3×105 cells/well) and incubated with PRMI-1640 media at 37°C with 5% CO2 until the cells reached ~95–100% confluence. A wound was created using a 200 µl pipette tip to scratch a vertical wound on the cell monolayer of each well, which was subsequently washed with PBS twice to remove the floating cells. The widths of the wounds were measured under an inverted light microscope (DM IL; Leica Microsystems GmbH) at 0 h and at 24 h after incubation in serum free medium.
Transwell assays
Transwell cell migration and invasion assays were performed using a Transwell insert (8-µm pore size) with or without BD Matrigel™ (Becton, Dickinson and Company). A total of 100 µl Matrigel was added to the upper chamber surface of the membrane for the invasion assay. A total of 1×105 cells in 200 µl serum free medium were plated in the upper chambers and 800 µl medium containing 10% serum was added to the lower chambers. After 48 h incubation, the membrane was washed with PBS, fixed in 4% paraformaldehyde at room temperature for 30 min and stained with 0.1% crystal violet at room temperature for 10 min. Cells on the upper side of the membrane were removed using cotton swabs and cells that had migrated to the lower side were counted using an inverted light microscope at 100× magnification.
Statistical analysis
All experiments were performed at least in triplicate, unless otherwise stated. Data are presented as the mean ± standard deviation. SPSS version 17 software (SPSS, Inc.) was used for general analysis of the statistical data. All quantitative data were analyzed using Student's t-test or one-way analysis of variance with Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Low expression of miR200c and overexpression of AMACR in prostate cancer cells and prostate cancer tissue
The expression of mature miR200c and AMACR mRNA was investigated by stem loop RT-PCR and conventional RT-PCR. The results revealed high expression levels of miR200c and low levels of AMACR, in BPH tissue samples (BPH-1, BPH-2 and BPH-3; Fig. 1A). In contrast, in prostate cancer tissue samples and prostate cancer cell lines (CL1, C4-2B, LNCaP, PC-3 and DU145 cell lines), decreased levels of miR200c and increased levels of AMACR were observed compared with (Fig. 1A). The RT-qPCR results showed that the expression levels of miR200c in the prostate cancer tissue samples and cell lines were ~25% compared with that in the BPH samples, whereas the AMACR level was ~3-4-fold higher in the prostate cancer tissue samples and cell lines compared with that of BPH (Fig. 1B).
Figure 1.
Expression levels of mature miR200c and AMACR in 3 BPH tissues, 3 PCa tissues and 5 prostate cancer cell lines. (A) RT-PCR of mature miR200c and AMACR in BPH tissues, PCa tissues and cell lines. (B) The results of RT-qPCR of mature miR200c and AMACR in BPH tissues, PCa tissues and cell lines. All experiments were performed in triplicate. Data are presented as the mean ± standard deviation *P<0.05 vs. all PCa tissues and prostate cancer cell lines. AMACR, α-methylacyl-coenzyme A racemase; BPH, benign prostate hyperplasia; PCa, prostate cancer; RT-q, reverse transcription-quantitative; miR, microRNA.
Artificial overexpression of miR200c by adenoviral vectors leads to downregulation of AMACR
The effect of miR200c on AMACR expression level is shown in Fig. 2. Concomitantly with the artificial overexpression of mature miR200c, the mRNA and protein levels of AMACR were both significantly downregulated in DU145 and PC-3 cells compared with the control (P<0.05; Fig. 2A and B).
Figure 2.
Effects of overexpression of miR200c on AMACR levels in DU145 and PC-3 cells. (A) The efficiency of Ad-miR200c and Ad-control infection is shown by the homogenously distributed green fluorescent protein expressed by the infected cells. The effects of overexpression of miR200c on miR200c and AMACR levels were demonstrated in RT-PCR and RT-qPCR experiments. Data are presented as the mean ± standard deviation. *P<0.05 vs. Ad-control. (B) Western blot analysis of AMACR protein level following overexpression of miR200c. All experiments were performed in triplicate. Data are presented as the mean ± standard deviation. *P<0.05 vs. Ad-control. Ad, adenovirus; AMACR, α-methylacyl coenzyme A racemase; RT-q, reverse transcription-quantitative; miR, microRNA.
Identification of potential miR200c seed sequences in the AMACR 3′-UTR
The 2,107-nt 3′-UTR of the AMACR mRNA was analyzed, which identified 90–97 nts of the 3′-UTR to be the potential seed sequence targeted by miR200c. Sequence analysis of the genomic DNA revealed no deletions or mutations in the AMACR 3′-UTR in DU145 and PC-3 cells (Fig. 3A).
Figure 3.
miR200c targets the 3′ UTR of AMACR. (A) Identification of the miR200c seed sequence in the AMACR 3′-UTR. The 90–97 nt of the AMACR 3′-UTR were predicted as potential seed sequence for miR200c. (B) Firefly/Renilla dual reporter gene assays. The activity of the basic pGL3 construct was used as a control (pGL3-Promoter). All experiments were performed in triplicate. Data are presented as the mean ± standard deviation. *P<0.05 vs. Ad-miR200c + pGL3-AMACR-UTR. Ad, adenovirus; AMACR, α-methylacyl coenzyme A racemase; 3′-UTR, 3′-untranslated region; miR, microRNA; mut, mutant.
miR200c targets the 3′-UTR of AMACR, as revealed by dual-reporter gene assay
Luciferase reporter gene constructs (pGL3-AMACR-UTR) were prepared, in which the potential seed sequence for miR200c in the AMACR 3′-UTR was inserted downstream of the luciferase coding sequence, as were constructs in which the seed sequence was mutated (pGL3-AMACR-UTRmut). Following artificial overexpression of miR200c by Ad-miR200c, dual-reporter assay revealed a significant decrease in luciferase reporter gene activity in cells treated with the pGL3-AMACR-UTR construct compared with controls, whereas mutations of the seed sequence significantly restored the luciferase gene activity in constructs bearing mutated 3′-UTR (P<0.05; Fig. 3B).
Knockdown of AMACR by RNAi or artificial overexpression of miR200c suppresses PC-3 cell proliferation, migration and invasiveness
siRNAs for AMACR were designed, which knocked down the AMACR levels in DU145 and PC-3 cells. Additionally, transfection of pcDNA3.1-AMACR also led to significant knockdown in both cell lines (P<0.05; Fig. 4A and B). The decrease in the mRNA expression level of AMACR in DU145 and PC-3 cells infected with Ad-miR200c could be reversed by artificial overexpression transfection of the pcDNA3.1-AMACR vector (P<0.05; Fig. 4C). The CCK-8 cell proliferation assay revealed that DU145 and PC-3 cell proliferation was inhibited by si-AMACR or by artificial overexpression of miR200c, whereas artificial AMACR overexpression by pcDNA3.1-AMACR transfection reversed the effects elicited by Ad-miR200c (P<0.01 or P<0.05; Fig. 4D).
Figure 4.
Effects of AMACR and miR200c on cell proliferation. (A) RT-PCR and RT-qPCR indicate the efficiency of AMACR downregulation by three different siRNAs. Si-AMACR-3 exhibited the highest efficacy and was chosen for subsequent experiments. (B) Successful transfection of the pcDNA3.1-AMACR plasmid was seen in DU145 and PC-3. (C) Overexpression of AMACR using pcDNA3.1-AMACR plasmid restored AMACR expression in DU145 and PC-3 cells. All cells were infected with Ad-miR200c. *P<0.05 vs. pcDNA3.1-blank (D) Cell Counting Kit-8 assay. The proliferation of PC-3 and DU145 cells was inhibited by downregulation of AMACR with siRNA of miRNA200c at 48 and 72 h. Overexpression of AMACR by pcDNA3.1-AMACR in PC-3 and DU145 cells partially reversed the effects of Ad-miR200c infection at 72 h. All experiments were performed in triplicate. Data are presented as the mean ± standard deviation. All quantitative data were analyzed using Student's t-test or one-way analysis of variance with Tukey's post-hoc test for group comparison (*P<0.05 and **P<0.01 vs. si-Control). Ad, adenovirus; AMACR, α-methylacyl coenzyme A racemase; siRNA, small interfering RNA; RT-q, reverse transcription-quantitative.
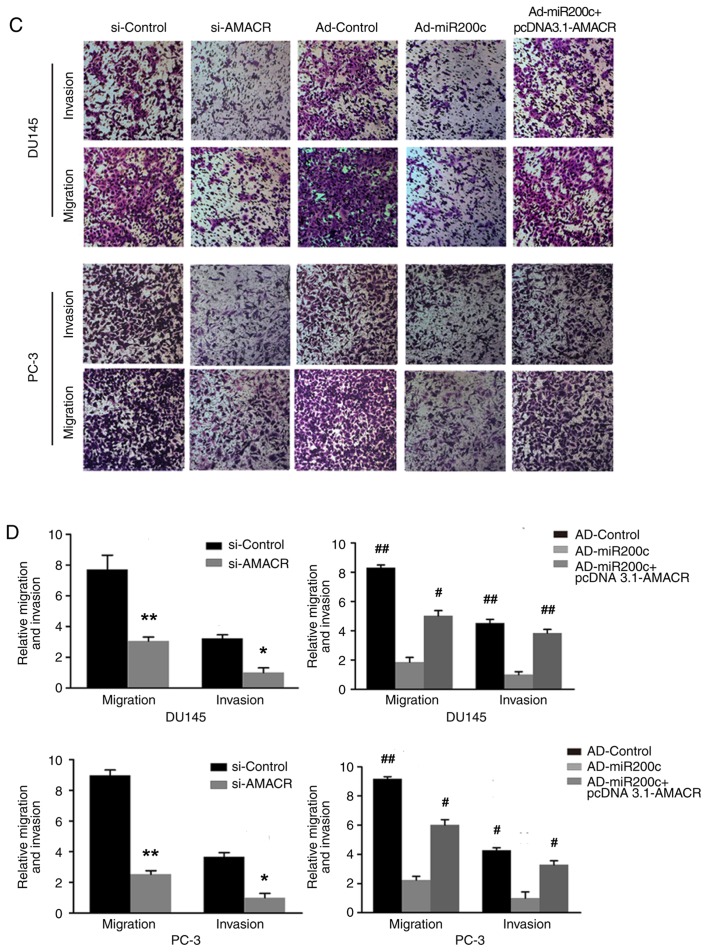
The wound-healing assays demonstrated decreased levels of cell migration following treatment of the prostate cancer cells with si-AMACR or artificial overexpression of miR200c compared with si-Control, a result that was reversed by artificial overexpression of AMACR using pcDNA3.1-AMACR (P<0.05; Fig. 5A and B). Similarly, Transwell assays revealed that si-AMACR or artificial overexpression of miR200c inhibited prostate cancer cell migration and invasiveness compared with controls, whereas artificial AMACR overexpression by pcDNA3.1-AMACR reversed the effects of Ad-miR200c (P<0.05; Fig. 5C and D).
Figure 5.
Effects of AMACR and miR200c on cell migration and invasion. (A) Wound-healing assay images and (B) quantitative analysis. si-knockdown of AMACR or overexpression of miR200c by Ad-miR200c inhibited wound-healing in DU145 and PC-3 cells. Restoration of AMACR using pcDNA3.1-AMACR reversed those inhibitory effects. (C) Transwell assay images and (D) quantitative analysis. Si-knockdown of AMACR or overexpression of miR200c by Ad-miR200c inhibited migration and invasiveness of the prostate cancer cells, whereas overexpression of AMACR reversed this inhibition. All experiments were performed in triplicate. Data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. si-Control, #P<0.05 and ##P<0.01 vs. Ad-miR200c. AD, adenovirus; AMACR, α-methylacyl coenzyme A racemase; siRNA, small interfering RNA; miR, microRNA.
Discussion
The present study aimed to investigate the regulatory effects of miR200c on AMACR and to explore the biological effects of miR200c-AMACR deregulation on prostate cancer cell proliferation, migration and invasion. The authors' pilot studies and bioinformatics analysis (data not shown) indicated that miR200c may regulate AMACR at post-transcriptional level. The present study showed that artificial overexpression of miR200c downregulated AMACR by interacting with the 3′-UTR of AMACR.
Previous studies have shown that miR-200c serves a critical role in inhibiting EMT, cell motility and tumor metastasis of various cancer types (9,10). The present results showed that knockdown of AMACR and artificial overexpression of miR200c led to marked inhibition of proliferation, migration and invasion of prostate cancer cells (9).
AMACR is an enzyme associated with lipid metabolism; specifically, the β-oxidation of dietary branched-chain fatty acids and bile acid synthesis (30). AMACR may be upregulated via different mechanisms in tumors, including an increase in the amount of branched-chain fatty acids in the diet (31), transcriptional activation by C/EBP (which may function as an enhancer) (26), Sp1 (21) or Brachyury (24), or by downregulation of inhibitory miRNAs (32). The branched-chain fatty acid, phytanic acid, predominantly exists in red meat and dairy products, an excessive intake of which is associated with both an increased risk of prostate cancer and an increase in the expression level of AMACR in prostate cancer cells (31). To the best of the authors' knowledge, the present study is the first to identify AMACR as a novel target of post-transcriptional regulation by miR200c.
The expression of AMACR is regulated by several transcription factors. The C/EBP family member, C/EBP, activates the expression of AMACR transcription (23), whereas the promoter activity of AMACR was shown to be downregulated by C/EBP, p53 and the p50 subunit of nuclear factor-κB, which may interact with the basal transcription machinery or other transcription factors (25). Recently, Brachyury, a T-box transcription factor, was shown to directly bind to and promote the promoter activity of AMACR in prostate cancer (24). Sp1 may lead to overexpression of AMACR by binding to the CpG island of the promoter region, whereas ZNF202 may inhibit AMACR expression (21). It has also been reported that, in gastrointestinal stromal tumors (22) and myxofibrosarcomas (23), two distinct soft tissue tumors, AMACR gene amplification resulted in its overexpression, which promoted tumor cell proliferation and cell cycle progression.
MiR200c, a major member of the miR200 family, has been shown to promote expression of the cell-adhesion molecule E-cadherin by suppressing the oncogenic transcription factor ZEB1, thus inhibiting EMT and metastatic potential of tumor cells (9,10). A variety of genes have been identified as distinct miR200c target genes, which are involved in diverse biological processes. For example, miR200c was shown to suppress renal cancer cell growth and metastasis by targeting BMI1 and E2F3, which are well recognized as oncogenes involved in cell cycle regulation and cell proliferation (11). miR200c was also shown to bind to FN1, the gene encoding fibronectin-1, a cell-adhesion molecule involved in multiple cellular processes, including embryogenesis, blood coagulation, wound healing and tumor metastasis, which also suppresses the proliferation, migration and invasion of gastric cancer cells (12). The present research group previously identified FSCN1 as a target inhibited by miR200c, the loss of which promoted expression of the actin-binding protein, fascin-1 and urothelial carcinoma invasiveness (14). PDGFR-β has also been shown to be an miR200c target, which mediates endothelial differentiation in triple-negative breast cancer (15). The present authors have previously reported that the gene for IRS1 is a target of miR200c, the downregulation of which, in prostate cancer, results in overexpression of IRS1, which promotes the proliferation of prostate cancer cells (8). The multi-targeting function of miR200c as a tumor-suppressor miRNA indicates that it has a pivotal role in inhibiting cell growth and migration.
The biological effects of AMACR overexpression may be manifold. As discussed above, AMACR overexpression has been associated with tumor cell proliferation in various tumor types, including prostate cancer (21–24). Interference of AMACR was shown to reduce cell proliferation and in vivo tumor growth in prostate cancer xenografts (19,33). In the authors' future research, they will study other regulatory mechanisms related with miR200c or AMACR more deeply and thoroughly. Taken together, the experiments in the present study have indicated that knockdown of AMACR expression, or artificial overexpression of miR200c may suppress prostate cancer cell proliferation, migration and invasiveness. The miR200c-AMACR regulatory mechanism may be involved in prostate cancer carcinogenesis and progression and could be exploited as a putative therapeutic target for prostate cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was support by grants from the Natural Science Foundation of China (grant nos. NSFC81572540, 81572541, 81272820, 81272848 and 81302225).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HX performed experiments and analyzed data, and drafted the manuscript. LN contributed to design of study, miR200C experiments, and data analysis. MZ contributed to data analysis and interpretation. JG contributed to CCK-8 assays and analyzed data. XC interpreted data and revised the manuscript. MX contributed to design of study and analysis of data. ZS contributed to the wound healing and transwell assays and analyzed data. NC and QZ designed the study, analyzed data, revised and finalized the manuscript. All authors have given approval of the final version of the mauscript.
Ethics approval and consent to participate
The present study followed institutional ethical guidelines and was supported by grants approved by ethics committee of the authors' institution. All tissue samples were collected in the West China Hospital according to the ethical guidelines and procedures approved by the institutional supervisory committee. Informed written consent was obtained according to the institutional guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Knezevic J, Pfefferle AD, Petrovic I, Greene SB, Perou CM, Rosen JM. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene. 2015;34:5997–6006. doi: 10.1038/onc.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, Zhang JD, Wiemann S, Sahin Ö. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633–651. doi: 10.1128/MCB.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X, Wang Y, Shan B, Han J, Zhu H, Lv Y, Fan X, Sang M, Liu XD, Liu W. The downregulation of miR-200c/141 promotes ZEB1/2 expression and gastric cancer progression. Med Oncol. 2015;32:428. doi: 10.1007/s12032-014-0428-3. [DOI] [PubMed] [Google Scholar]
- 4.Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, Santamarina I, Blanco M, Fernández-Tajes J, Quindós M, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186. doi: 10.1186/1479-5876-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao YC, Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015;36:4843–4850. doi: 10.1007/s13277-015-3138-3. [DOI] [PubMed] [Google Scholar]
- 7.Butz H, Szabó PM, Khella HW, Nofech-Mozes R, Patocs A, Yousef GM. miRNA-target network reveals miR-124as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget. 2015;6:12543–12557. doi: 10.18632/oncotarget.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su W, Xu M, Chen X, Nie L, Chen N, Gong J, Zhang M, Su Z, Huang L, Zhou Q. MiR200c targets IRS1 and suppresses prostate cancer cell growth. Prostate. 2015;75:855–862. doi: 10.1002/pros.22968. [DOI] [PubMed] [Google Scholar]
- 9.Puhr M, Hoefer J, Schäfer G, Erb HH, Oh SJ, Klocker H, Heidegger I, Neuwirt H, Culig Z. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am J Pathol. 2012;181:2188–2201. doi: 10.1016/j.ajpath.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Radisky DC. miR-200c at the nexus of epithelial-mesenchymal transition, resistance to apoptosis, and the breast cancer stem cell phenotype. Breast Cancer Res. 2011;13:110. doi: 10.1186/bcr2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu M, Liang Z, Chen L, Tan G, Liu L, Wang K, Chen H, Liu J. MicroRNA-200c suppresses cell growth and metastasis by targeting Bmi-1 and E2F3 in renal cancer cells. Exp Ther Med. 2017;13:1329–1336. doi: 10.3892/etm.2017.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed Pharmacother. 2017;88:285–292. doi: 10.1016/j.biopha.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Guo X, Zhang L, Zhang W, Zhou Q, Tian Z, Zheng Y, Liao Q, Wang H, Li G, et al. MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget. 2017;8:5206–5218. doi: 10.18632/oncotarget.14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Nie L, Su Z, Xu M, Chen N, Gong J, Xie H, Zhong J, Tan J, Xu Y, et al. MicroRNA200c suppresses urothelial carcinoma invasiveness by targeting FSCN1. Int J Clin Exp Pathol. 2017;10:5665–5674. [Google Scholar]
- 15.D'Ippolito E, Plantamura I, Bongiovanni L, Casalini P, Baroni S, Piovan C, Orlandi R, Gualeni AV, Gloghini A, Rossini A, et al. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res. 2016;76:5562–5572. doi: 10.1158/0008-5472.CAN-16-0140. [DOI] [PubMed] [Google Scholar]
- 16.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 18.Tang X, Tang X, Gal J, Kyprianou N, Zhu H, Tang G. Detection of microRNAs in prostate cancer cells by microRNA array. Methods Mol Biol. 2011;732:69–88. doi: 10.1007/978-1-61779-083-6_6. [DOI] [PubMed] [Google Scholar]
- 19.Zha S, Ferdinandusse S, Denis S, Wanders RJ, Ewing CM, Luo J, De Marzo AM, Isaacs WB. Alpha-methylacyl-CoA racemase as an androgen-independent growth modifier in prostate cancer. Cancer Res. 2003;63:7365–7376. [PubMed] [Google Scholar]
- 20.Jindal Y, Singh A, Kumar R, Varma K, Misra V, Misra SP, Dwivedi M. Expression of alpha methylacyl CoA racemase (AMACR) in gastric adenocarcinoma and its correlation with helicobacter pylori infection. J Clin Diagn Res. 2016;10:EC10–EC12. doi: 10.7860/JCDR/2016/19539.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Leav I, Revelo MP, Deka R, Medvedovic M, Jiang Z, Ho SM. Deletion hotspots in AMACR promoter CpG island are cis-regulatory elements controlling the gene expression in the colon. PLoS Genet. 2009;5:e1000334. doi: 10.1371/journal.pgen.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li CF, Chen LT, Lan J, Chou FF, Lin CY, Chen YY, Chen TJ, Li SH, Yu SC, Fang FM, et al. AMACR amplification and overexpression in primary imatinib-naïve gastrointestinal stromal tumors: A driver of cell proliferation indicating adverse prognosis. Oncotarget. 2014;5:11588–11603. doi: 10.18632/oncotarget.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li CF, Fang FM, Lan J, Wang JW, Kung HJ, Chen LT, Chen TJ, Li SH, Wang YH, Tai HC, et al. AMACR amplification in myxofibrosarcomas: A mechanism of overexpression that promotes cell proliferation with therapeutic relevance. Clin Cancer Res. 2014;20:6141–6152. doi: 10.1158/1078-0432.CCR-14-1182. [DOI] [PubMed] [Google Scholar]
- 24.Pinto F, Pértega-Gomes N, Vizcaíno JR, Andrade RP, Cárcano FM, Reis RM. Brachyury as a potential modulator of androgen receptor activity and a key player in therapy resistance in prostate cancer. Oncotarget. 2016;7:28891–28902. doi: 10.18632/oncotarget.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, Wu W, Zhao J, Yu C, Liu W, Jiang A, Zhang J. Molecular cloning and preliminary analysis of the human alpha-methylacyl-CoA racemase promoter. Mol Biol Rep. 2009;36:423–430. doi: 10.1007/s11033-007-9196-x. [DOI] [PubMed] [Google Scholar]
- 26.Zha S, Isaacs WB. A nonclassic CCAAT enhancer element binding protein binding site contributes to alpha-methylacyl-CoA racemase expression in prostate cancer. Mol Cancer Res. 2005;3:110–118. doi: 10.1158/1541-7786.MCR-04-0178. [DOI] [PubMed] [Google Scholar]
- 27.Chen N, Chen X, Huang R, Zeng H, Gong J, Meng W, Lu Y, Zhao F, Wang L, Zhou Q. BCL-xL is a target gene regulated by hypoxia-inducible factor-1{alpha} J Biol Chem. 2009;284:10004–10012. doi: 10.1074/jbc.M805997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Gong J, Zeng H, Chen N, Huang R, Huang Y, Nie L, Xu M, Xia J, Zhao F, et al. MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res. 2010;70:2728–2738. doi: 10.1158/0008-5472.CAN-09-3718. [DOI] [PubMed] [Google Scholar]
- 30.Ferdinandusse S, Denis S, IJlst L, Dacremont G, Waterham HR, Wanders RJ. Subcellular localization and physiological role of alpha-methylacyl-CoA racemase. J Lipid Res. 2000;41:1890–1896. [PubMed] [Google Scholar]
- 31.Mobley JA, Leav I, Zielie P, Wotkowitz C, Evans J, Lam YW, L'Esperance BS, Jiang Z, Ho SM. Branched fatty acids in dairy and beef products markedly enhance alpha-methylacyl-CoA racemase expression in prostate cancer cells in vitro. Cancer Epidemiol Biomarkers Prev. 2003;12:775–783. [PubMed] [Google Scholar]
- 32.Erdmann K, Kaulke K, Thomae C, Huebner D, Sergon M, Froehner M, Wirth MP, Fuessel S. Elevated expression of prostate cancer-associated genes is linked to down-regulation of microRNAs. BMC Cancer. 2014;14:82. doi: 10.1186/1471-2407-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Festuccia C, Gravina GL, Mancini A, Muzi P, Cesare ED, Kirk R, Smith M, Hughes S, Gibson R, Lian LY, et al. Trifluoroibuprofen inhibits α-methylacyl coenzyme A racemase (AMACR/P504S), reduces cancer cell proliferation and inhibits in vivo tumor growth in aggressive prostate cancer models. Anticancer Agents Med Chem. 2014;14:1031–1041. doi: 10.2174/1871520614666140327152607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.