Summary
The γ-tubulin ring complex (γ-TuRC) is an essential regulator of centrosomal and acentrosomal microtubule formation, yet its structure is not known. Here, we present a cryo-EM reconstruction of the native human γ-TuRC at ~3.8 Å resolution, revealing an asymmetric, cone-shaped structure. Pseudo-atomic models indicate that GCP4, GCP5 and GCP6 form distinct Y-shaped assemblies that structurally mimic GCP2/GCP3 subcomplexes distal to the γ-TuRC “seam”. We also identify an unanticipated structural bridge that includes an actin-like protein and spans the γ-TuRC lumen. Despite its asymmetric architecture, the γ-TuRC arranges γ-tubulins into a helical geometry poised to nucleate microtubules. Diversity in the γ-TuRC subunits introduces large (>100,000 Å2) surfaces in the complex that allow for interactions with different regulatory factors. The observed compositional complexity of the γ-TuRC could self-regulate its assembly into a cone-shaped structure to control microtubule formation across diverse contexts, e.g. within biological condensates or alongside existing filaments.
Keywords: γ-tubulin ring complex, microtubule, microtubule nucleation, single particle cryo-EM structure, GCP2, GCP3, GCP4, GCP5, GCP6, actin
Graphical Abstract
eTOC blurb
A high resolution cryo-EM reconstruction of the cell’s microtubule nucleating machinery reveals an unusual cone-shaped structure that mimics the microtubule lattice.
Introduction
Microtubules are dynamic, μm-scale polymers of α/β-tubulin heterodimers. In addition to forming a structural scaffold that defines cellular morphology (Kirschner and Mitchison, 1986), microtubules perform a multitude of important intracellular functions, such as providing tracks for motor proteins to transport cargos and generating the pushing and pulling forces required to faithfully segregate chromosomes in dividing cells (Forth and Kapoor, 2017; Vale, 2003). Although the control of microtubule growth and shrinkage (also termed dynamic instability (Mitchison and Kirschner, 1984)) has been thoroughly characterized (Brouhard and Rice, 2018), the factors that generate de novo microtubules in cells are less understood.
The γ-tubulin ring complex (γ-TuRC) is a macromolecular assembly that nucleates microtubules in vitro and is essential for proper microtubule network formation in eukaryotes (Knop et al., 1997a; Raff et al., 1993; Stearns and Kirschner, 1994; Zheng et al., 1995). The tubulin-like GTPase γ-tubulin, initially discovered in Aspergillus nidulans (Oakley and Oakley, 1989), localizes to the centrosome and associates with at least five other proteins to form ~32S (~2 MDa) “ring”-shaped complexes (Stearns et al., 1991; Zheng et al., 1995), but the precise identity and stoichiometry of components within the native γ-TuRC are still debated (Kollman et al., 2011). Thus, despite ~30 years of research since the discovery of γ-tubulin (Oakley and Oakley, 1989), the composition of γ-TuRCs, as well as how this complex assembles from a heterogeneous set of components to facilitate microtubule nucleation, remains unclear.
The question of composition is further complicated by the finding that cytoplasmic γ-TuRCs are not only recruited to canonical sites of microtubule assembly, such as the centrosome (Stearns et al., 1991), but also to diverse acentrosomal locations, such as meiotic spindles (Schuh and Ellenberg, 2007), Golgi outposts (Rivero et al., 2009), and other regions of the cytoplasm (Moutinho-Pereira et al., 2009). This recruitment depends on a variety of γ-TuRC-associated “attachment factors”, which can copurify with native γ-TuRCs (Hutchins et al., 2010; Lüders et al., 2006; Teixidó-Travesa et al., 2010). Several attachment factors have been identified, including the pericentriolar protein CDK5Rap2 (Fong et al., 2008), which enhances the microtubule nucleating activity of purified γ-TuRCs in vitro (Choi et al., 2010). Based on these observations, it has been proposed that recruitment of γ-TuRCs to sites of microtubule assembly leads to γ-TuRC “activation” (Kollman et al., 2011), but such models have been difficult to test given the current lack of structural data for the native complex.
Our understanding of γ-TuRC structure comes mainly from studies of heterotetramers of γ-tubulin complex protein 2, or GCP2, GCP3, and γ-tubulin, also termed the γ-tubulin small complex (γ-TuSC) (Oegema et al., 1999). In S. cerevisiae, the γ-TuSC is a Y-shaped complex whose base is comprised of laterally associated N-terminal domains of GCP2 and GCP3 (Kollman et al., 2008). Notably, and in the presence of a fragment of the spindle-pole body factor Spc110, recombinant S. cerevisiae γ-TuSCs have a propensity to self-assemble into helical oligomers of varying length in vitro (Kollman et al., 2010). In contrast, negative stain electron tomography and metal shadowing studies have indicated that D. melanogaster γ-TuRCs, which are biochemically similar to the human complex, assemble into finite, lock washer-shaped structures (Moritz et al., 2000). In addition to GCP2 and GCP3, human γ-TuRCs also contain the proteins GCP4, GCP5, and GCP6 (Hutchins et al., 2010; Murphy et al., 2001; Teixidó-Travesa et al., 2010). However, structural models for human γ-TuRC subcomponents in the context of the native complex are not available. Whether the human γ-TuRC assembles from a helical oligomer of γ-TuSCs, how the finite size of the complex might be, are all open questions.
Here, we present a cryo-EM reconstruction of the native human γ-TuRC at ~3.8 Å resolution. Our reconstruction reveals an asymmetric cone-shaped structure assembled from at least 26 polypeptides. We build pseudo-atomic models for the core γ-TuRC components, including GCP4, GCP5 and GCP6, which we find form distinct Y-shaped assemblies of GCP4/GCP5 and GCP4/GCP6 that structurally mimic human GCP2/GCP3 subcomplexes. We also identify an unanticipated structural bridge that includes an actin-like protein and spans >300 Å across the γ-TuRC lumen. Our structure reveals how the human γ-TuRC arranges γ-tubulin into a helical geometry poised to nucleate microtubules.
Results
The native human γ-TuRC is an asymmetric, cone-shaped structure
To examine the architecture of human γ-TuRCs, we first sought to purify the native complex. Several previously reported purification strategies were explored (Murphy et al., 2001; Zheng et al., 1995). We found that isolation of the native γ-TuRC from HeLa S3 cytoplasmic extracts via its affinity for the CM1 domain of CDK5Rap2, also termed the γ-TuRC-mediated Nucleation Activator (γ-TuNA) (Choi et al., 2010), reliably yielded complex containing core components whose proportions matched previous qualitative estimates (Murphy et al., 2001), as judged by SDS-PAGE and iBAQ analysis of mass spectrometry data (Figures S1A and S1B; Tables S1 and S2; see STAR Methods for purification details) (Schwanhäusser et al., 2011). γ-TuRCs purified in this manner were also capable of nucleating microtubule assembly in vitro in a standard turbidity-based assay (Figure S1C), consistent with previous results (Murphy et al., 2001).
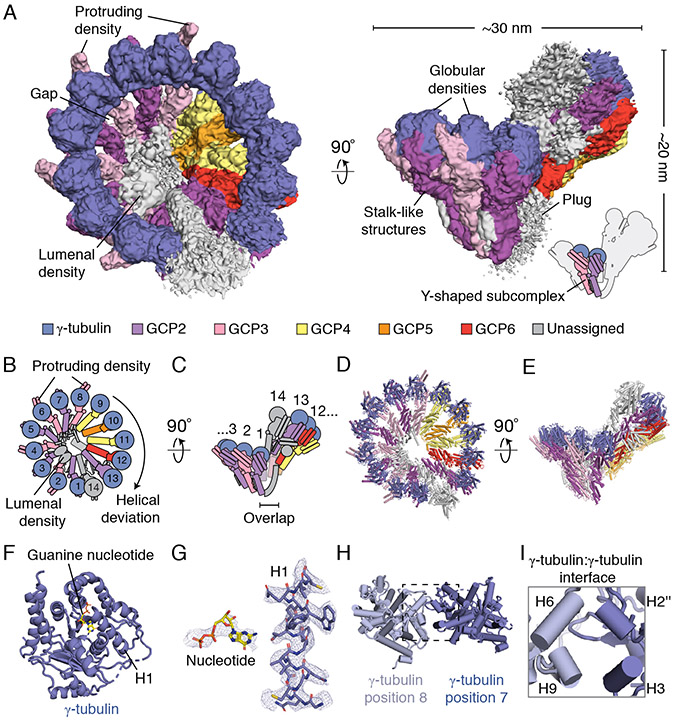
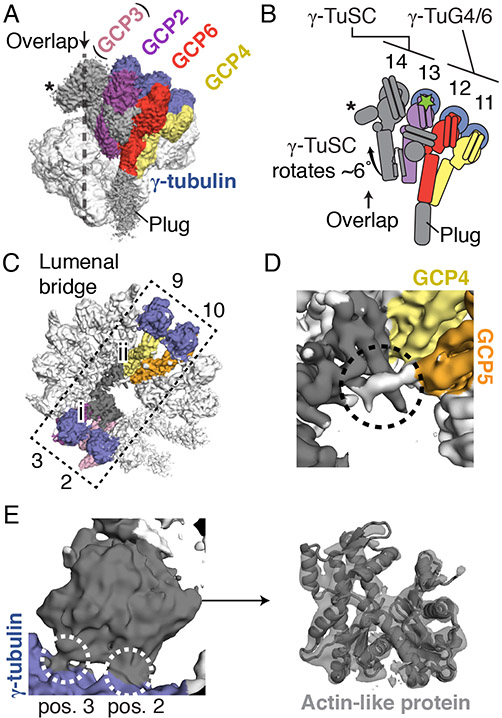
We determined the structure of the native human γ-TuRC using cryo-electron microscopy (cryo-EM) to an overall resolution of ~3.8 Å (Figure 1A and Figure S1D-S1G; see STAR Methods for details). The γ-TuRC structure revealed a conical shape ~30 nm in diameter and ~20 nm in height (Figure 1A). Fourteen similarly-sized (~4 nm-wide) globular densities line the open face of the cone, each supported by a stalk-like structure. Both the globular domains and the lower parts of the stalk-like structures make substantial lateral contacts with one another, while leaving several narrow gaps in the conical wall of the complex (Figure 1A). Additionally, a substantial density appears to extend down and obstruct the opening found at the base of the γ-TuRC (the “plug” in Figure 1A). Each pair of neighboring stalk-like densities along with their two associated globular domains closely resembles previous description of the Y-shaped γ-TuSC (Kollman et al., 2008) (Figure 1A). The human γ-TuRC cone shape is therefore made of 14 individual subunits that could form 7 γ-TuSC-like subcomplexes (Figure 1A-1C).
Figure 1. Cryo-EM reconstruction of the native human γ-TuRC.
A) Two views of the overall γ-TuRC density map (surface representation). Structural features and dimensions of the γ-TuRC are indicated. Schematic of the γ-TuRC highlighting a Y-shaped subcomplex (indicated) is shown in the bottom right. B) and C) Schematics of the γ-TuRC highlighting proposed subunit numbering, features of asymmetry (both compositional and structural), and the “overlap” region, viewed from the top (B) and side (C). D) and E) de novo molecular model for the γ-TuRC (cartoon representation), viewed from the top (D) and side (E). F) Refined γ-tubulin model (cartoon representation) with guanine nucleotide (stick representation) and helix H1 indicated. G) Examples of γ-tubulin density quality. Refined models for GDP (left, stick representation) and helix H1 of γ-tubulin (right, stick representation) shown in the corresponding γ-tubulin density (blue mesh). H) View of two γ-tubulin models at positions 8 (light blue cartoon representation) and 7 (blue cartoon representation), highlighting the γ-tubulin:γ-tubulin interface (dashed rectangle). I) Inset from H) showing interacting helices (labeled cylinders). Except for H) and I), γ-TuRC subunits are colored according to the legend in A). See also Figures S1-S3 and Tables S1-S5.
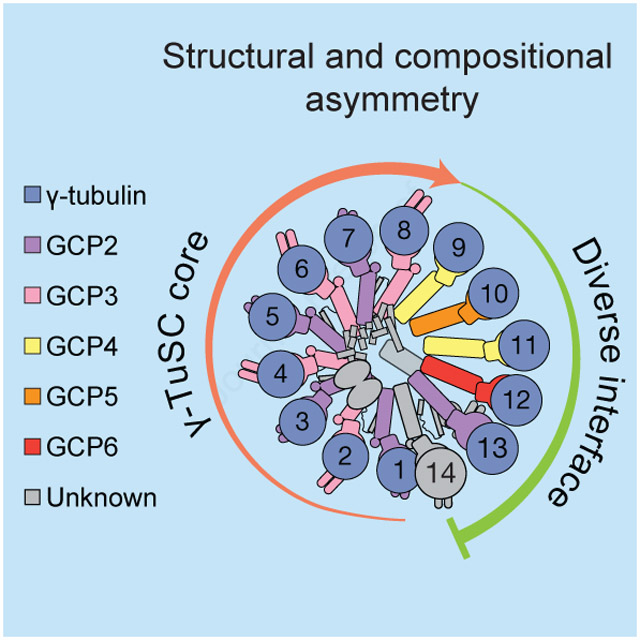
We numbered the individual stalk-like subunits, starting at the base of the γ-TuRC cone and finishing at subunit 14, which partially overlaps with the first subunit (the γ-TuRC “overlap”) (Figure 1B-C). This organization suggests apparent helical symmetry in the complex. However, the γ-TuRC exhibits at least three features that reveal structural asymmetry. First, an externally-protruding density appears only at positions 2, 4, 6, and 8 of the complex (“protruding density”; Figure 1A), suggesting that a set of four similar γ-TuSC-like subcomplexes, each with one protruding density, occupies these positions. Second, the remaining three γ-TuSC-like subunits at positions 9 to 14, which do not exhibit protruding densities, deviate from a helically symmetric geometry, conferring ellipticity to the γ-TuRC cone shape (“helical deviation”; Figures 1A-B). Third, a substantial (~10 × 2 × 4 nm) density is situated in the lumen of the cone structure and contacts the stalk-like densities from positions 2-10 (“lumenal density”; Figures 1A-B). These results suggest that the human γ-TuRC adopts an asymmetric conical structure comprised of heterogeneous γ-TuSC-like subunits.
The γ-TuRC density map, which was generated from ~460,000 particles (Figure S2 and Table S3), displayed a broad range of local resolution (3.0 Å - 12 Å; Figure S1F), revealing a degree of structural “breathing” in this large complex (further discussed below). To improve the local resolution across the γ-TuRC and gain detailed insight into its molecular features, we processed different parts of the complex separately in a “divide and conquer” strategy that employed the use of particle subtraction (Scheres, 2016) (Figure S2; see STAR Methods for processing details). In brief, signal in raw particles not corresponding to desired segmentation regions in the γ-TuRC was subtracted. Signal-subtracted particles were then subjected to focused 3D classification and refinement. The resulting locally refined, segmented maps had more homogeneous resolution (FSC0.146 ranged from 3.4 to 4.5 Å for 6 of the 7 maps, and 6.6 Å for the one map corresponding to the “overlap” region; Figure S2). These maps revealed clearly-defined side chain densities across a majority of the complex and were used to generate a molecular model of the core γ-TuRC structure, which included 26 assigned and at least 5 tentatively assigned subunits (Figures 1D-1E, Table S4 and S5, and Video S1; see STAR Methods for details regarding model building and subunit assignment criteria).
The native γ-TuRC presents at least 13 γ-tubulin molecules on the face of the cone
The structure of γ-tubulin in the context of the γ-TuRC is not currently known. Previous high-resolution structural models for γ-tubulin come from crystallographic studies of the monomeric human protein (Aldaz et al., 2005; Rice et al., 2008). We rigid-body fitted a crystallographic model for human γ-tubulin into globular densities at positions 1-13 at the open face of the cone (Figure 1A; PDB: 3CB2 (Rice et al., 2008)). After refinement, these densities clearly accommodated γ-tubulin side chains, as well as a guanine nucleotide (Figures 1F and 1G). A 14th γ-tubulin subunit could be placed into the globular density at position 14 (Figures 1D and 1E), although it did not meet our criteria for assignment (see STAR Methods). The conformations of hallmark structural features (α-helices H6-H7 and the intermediate β-sheet) of the refined γ-tubulin models at positions 1-13 closely resembled the original crystallographic model (root-mean-square-deviation RMSD = 1.0 Å; Figure S3A-C) (Aldaz et al., 2005; Rice et al., 2008). These data suggest that, under our purification conditions (see STAR Methods), assembly into the native human γ-TuRC does not induce significant structural changes in γ-tubulin, in contrast with a recent study of a recombinant yeast γ-TuSC (Brilot and Agard, 2018). Such structural differences could reflect the possibility of multiple conformational states of γ-tubulin within different γ-TuRC assemblies.
Our results also provide details about the γ-tubulin:γ-tubulin lateral interface, which includes interactions between helix H2’’ and H3 of one γ-tubulin with helix H6 and H9 of the subsequent γ-tubulin (Figures 1H and 1I). We observed two repeating arrangements of these interfaces; in one, the neighboring γ-tubulins are ~9 Å apart (between positions 1 and 2, 3 and 4, 5 and 6, 7 and 8, 9 and 10, and 11 and 12), and in the other, neighboring γ-tubulins are farther (~15 Å) apart (between positions 2 and 3, 4 and 5, 6 and 7, 8 and 9, 10 and 11, and 12 and 13) (Figure S3D and S3E). We note that this alternating variation in the inter-γ-tubulin interfaces is consistent with different spacings within and between Y-shaped subcomplexes in the γ-TuRC.
Two copies of GCP4 are distal to the “overlap” region
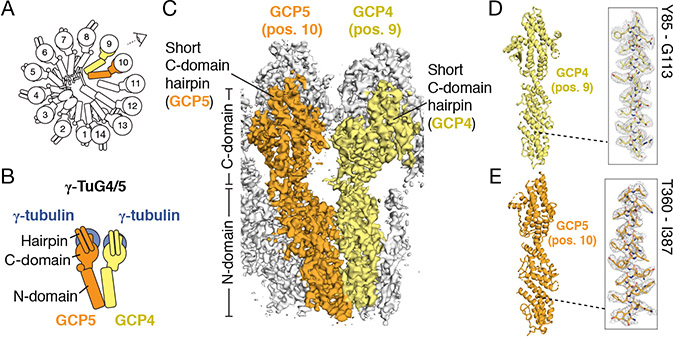
Next, we turned our attention to the stalk-like densities that are located below each γ-tubulin and together wrap around the cone-shaped structure. These densities have a characteristic overall organization comprising two bundles each built from ~7 consecutive α-helices. The lower (N-domain) bundle adopts an elongated (~8 × ~2 nm) fold while the upper (C-domain) bundle, which contacts γ-tubulin, is more compact (~5 nm × ~4 nm) and displays a prominent (>2 nm) two-helix hairpin on the outer face of the γ-TuRC cone (Figures 2A and 2C). The general topology of the stalk-like structures resembles a previously described crystallographic model for GCP4, the only human GCP subunit with available structural data. GCP4 is organized into two α-helical bundles. One corresponds to the conserved γ-TuRC interacting protein motif 1 (GRIP1, corresponding to the N-domain) domain; the other corresponds to the γ-TuRC interacting protein motif 2 (GRIP2; C-domain) domain (Guillet et al., 2011). Notably, GCP4 has also been proposed to form part of a γ-TuSC-like subcomplex at the γ-TuRC overlap (i.e., at or near positions 1 or 14) (Farache et al., 2016; Kollman et al., 2011; Tovey and Conduit, 2018). Surprisingly, however, the crystallographic model for GCP4 instead fit into the densities at positions 9 and 11 (Figures 2C, 2D, 3C, and 3D; Table S4; ~82% GCP4 sequence coverage). The resolution of our density map enabled side chain identification that confirmed the identity of GCP4 at these positions of the γ-TuRC (Figures 2D and 3D). Further, we aligned our refined GCP4 model to the monomeric, crystallographic GCP4 model via their N-domains (Guillet et al., 2011), which revealed that within the γ-TuRC the GCP4 C-domain is rotated by ~10° towards the preceding subunit (Figure S3F), and helix 21 is rotated by ~90° towards the GCP4-associated γ-tubulin (Figure S3G). Together, these results indicate that complexed GCP4 is structurally different from the monomeric protein and that the γ-TuRC contains two copies of GCP4, both of which are located distal to the “overlap” region.
Figure 2. Positioning of GCP4 and GCP5 in the γ-TuRC.
A) Schematic of the γ-TuRC establishing the viewing angle and highlighting the locations of GCP4 (yellow) and GCP5 (orange). B) Schematic of the γ-tubulin/GCP4/GCP5 heterotetramer that forms a Y-shaped γ-TuSC-like assembly, γ-TuG4/5. C) The density map of γ-TuRC positions 9-11 used to model N- and C-domains and short C-domain hairpins of GCP4 (yellow surface) and GCP5 (orange surface), viewed from the angle indicated in A). D) and E) Molecular models (left) of GCP4 and GCP5. Insets (right) show the fit of a region of the models in the density maps (mesh). See also Figures S2-S4 and Tables S1, S4 & S5.
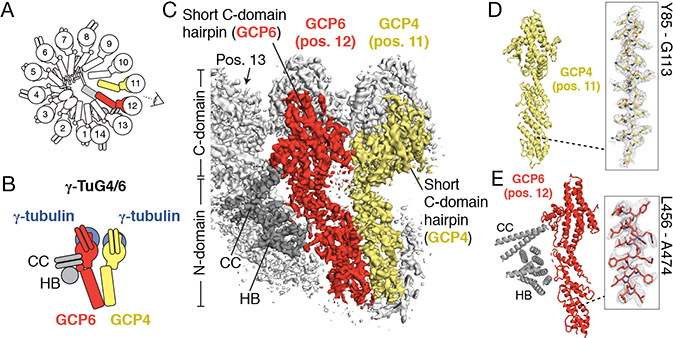
Figure 3. Positioning of GCP4 and GCP6 in the γ-TuRC.
A) Schematic of the γ-TuRC highlighting the locations of GCP4 (yellow) & GCP6 (red). B) Schematic of a γ-tubulin/GCP4/GCP6 heterotetramer that forms the γ-TuSC-like assembly, γ-TuG4/6 and interacting coiled-coil (CC) and helical bundle (HB) of the unassigned densities (dark gray). C) Density at positions 11-13 used to model N- and C-domains and short C-domain hairpins (indicated) of the second GCP4 (yellow surface) and GCP6 (red surface), viewed from the angle indicated in A). CC and HB densities interacting with position 13 are indicated. D) Molecular model of GCP4 at position 11 (left). Inset (right) shows the fit of GCP4 residues Y85-G113 (indicated; stick representation) in the density map (mesh). E) Molecular model of GCP6 at position 12 (left). Poly-alanine models of the CC and HB densities are shown for consistency with the observed density (indicated). Inset (right) shows the fit of GCP6 residues L456-A474 (indicated; stick representation) in the density map (mesh). See also Figures S2 & S4 and Tables S1, S2, S4 & S5.
GCP4 and GCP5 form a unique Y-shaped γ-TuSC-like subcomplex
GCP5 and GCP6 have been proposed to be form Y-shaped assemblies like the γ-TuSC (Kollman et al., 2011), but their structure, position and stoichiometry within the native human complex are not known. In addition to the conserved GRIP1 (N-domain) and GRIP2 (C-domain) motifs that define the GCP family (Gunawardane et al., 2000), GCP5 and GCP6 also feature large sequence inserts (Figure S4A and Figure S4G) (Murphy et al., 2001), making it difficult to make predictions about their precise folds without structural data. We first focused on building a model for GCP5 because it has previously been shown to biochemically interact with GCP4 (Farache et al., 2016). Consistent with this report, we found that a de novo molecular model for human GCP5 N- and C-domains could be built into the stalk-like density at position 10 of the γ-TuRC, neighboring GCP4 at positions 9 and 11 (Figures 2E and S4A and Table S4). As above, analysis of amino acid side chain registries in α-helical densities enabled assignment of GCP5 to this position of the complex (Figure 2E). Our model revealed that, consistent with conservation of the GRIP1/GRIP2 motifs, the core GCP5 model is similar in overall topology to GCP4, except for slight conformational variability in the C-domain (Figure S3H). Following the naming convention of the γ-TuSC (Oegema et al., 1999), we propose that GCP4 at position 9 and GCP5 at position 10, along with their associated γ-tubulins, constitute a Y-shaped γ-TuSC-like subcomplex (γ-tubulin subcomplex containing GCP4 and GCP5, or “γ-TuG4/5”) (Figure 2B).
The GCP5 (position 10) density had two additional features that differed from other γ-TuRC stalk-like densities. First, we observed unassigned density below the GCP5 N-domain that interacts with the lumenal density (see below) as well as the neighboring GCP4s at positions 9 and 11 (Figure S4B). This “N-terminal density” may correspond in part to the ~250 N-terminal amino acids currently not modeled in the GCP5 N-domain (Figure S4A). Second, 3D classification of the overall γ-TuRC density map revealed two subclasses that differ mainly in the conformation of the GCP5 C-domain. The first subclass corresponds to the characteristic “elongated” stalk-like conformation that was used to build the GCP5 model described above (Figure S4C). In the second subclass, the GCP5 density adopts a “compacted” conformation within the complex, in which the GCP5-associated γ-tubulin was displaced ~5-10 Å towards the base of the γ-TuRC (Figure S4D). Further, in the “compacted” GCP5 subclass, the proximal Y-shaped subcomplexes occupying positions 11-14 are also displaced slightly away from the center of the γ-TuRC cone (Figures S4E and S4F).
GCP4 and GCP6 form a unique Y-shaped γ-TuSC-like subcomplex
We next focused on building a molecular model for GCP6, the largest γ-TuRC subunit (Murphy et al., 2001). We found that a de novo model for the GCP6 N-domain could be built and refined only into the stalk-like density at position 12 of the γ-TuRC (Figure 3C, E and Figure S4G). The registry of model side chains matched high-resolution densities in the N- and C-domains (Figure 3E and Figure S4H; see Methods for details), which, combined with the generally low sequence identity across the GCP family (Murphy et al., 2001), further validated the identity of GCP6 at this position. Our model for GCP6 also defined the boundaries of the large (>800 amino acid) insertion found between the N- and C-domains unique to this γ-TuRC subunit. Given the organizational similarity to the γ-TuG4/5 described above, we propose that the second GCP4 at position 11 and GCP6 at position 12, along with their associated γ-tubulins, constitute a distinct Y-shaped γ-TuSC-like subcomplex, the “γ-TuG4/6” (Figure 3B).
We also observed additional GCP6-associated densities at position 12. First, we noticed that the “plug” density in the overall map contacts the lower part of the GCP6 N-domain (Figures 1A and S4I). The current resolution of the “plug” limits our ability to build a model in this region, but we note that the N-terminal ~350 amino acids not accounted for in our GCP6 N-domain model could in part account for this density. The “plug” may alternatively correspond to one of the unaccounted-for γ-TuRC-associated proteins identified in our mass spectrometry analysis, such as NEDD1 or NME7 (Table S2) (Haren et al., 2006; Liu et al., 2014; Lüders et al., 2006). Second, we observed a coiled coil density (CC) that stems from between the N- and C-domains of GCP6 at position 12 and contacts a small (~2 nm wide) α-helical bundle (HB). Both the CC and HB in turn interact with the neighboring stalk density at position 13 (Figures 3C, 3E, and S6B). Though these densities currently lack an assignment, the large (~800 residue) GCP6 insertion sequence located between its N- and C-domains is predicted to contain a coiled coil and may in part account for the CC and HB features (Figures 3E and S4G; Table S4). The γ-TuNA peptide used to purify γ-TuRCs here is predicted to form a short (~30 amino acid) coiled-coil and is therefore also a plausible candidate for the CC (Figure S5G; see below). Together, our data provide structural models for GCP4, GCP5 and GCP6, and suggest that these proteins form two distinct subcomplexes, γ-TuG4/5 and γ-TuG4/6, within the γ-TuRC.
GCP2 and GCP3 are arranged into a γ-TuSC core comprising roughly half of the γ-TuRC
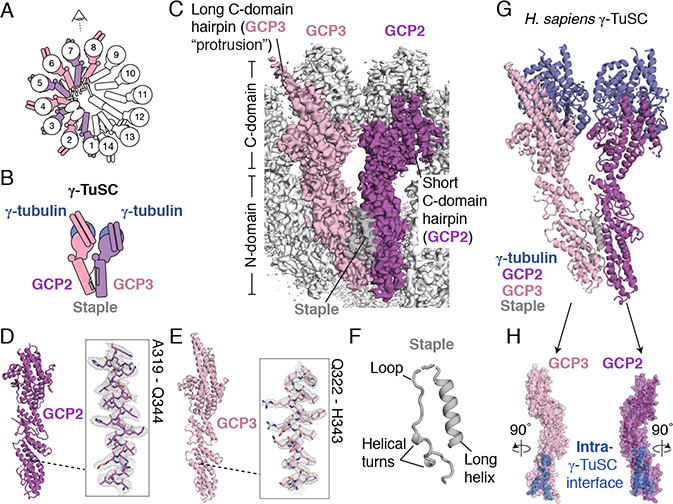
We next sought to build models for the evolutionarily-conserved proteins GCP2 and GCP3, which form γ-TuSCs in other species (Knop et al., 1997b; Oegema et al., 1999), but whose structures in the context of the native γ-TuRC are not known. We found that near complete (~72% full sequence coverage) de novo models for human GCP2 could be built into the stalk-like densities at positions 1, 3, 5 and 7 (Figures 4A, 4C, 4D, and , and S5A; Table S4). Similarly, GCP3 could be built into positions 2, 4, 6, and 8 (Figures 4A, 4C, 4E, and S5B; Table S4). Side chain registries in these high-resolution densities confirmed the alternating positioning of these two proteins in the γ-TuRC (Figures 4D, 4E, and S5C). Between each pair of GCP2 and GCP3 N-domains is a small α-helical density resembling a “staple” that forms several contacts with both proteins (Figures 4C and 4F). Given our modeling criteria (see STAR Methods), we could not confidently assign this density, though we note that the staple could correspond to either the γ-TuNA peptide used to purify γ-TuRCs in this study (Figure S5G), or an extension of the GCP2 N-terminus via a poorly resolved connecting density (Figure S5D). Following historical convention (Oegema et al., 1999), we refer to the Y-shaped arrangement of GCP2, GCP3, and their corresponding γ-tubulin molecules as the human “γ-TuSC” (Figures 4B and 4G).
Figure 4. Structures and arrangement of human GCP2 and GCP3.
A) Schematic of the γ-TuRC highlighting locations of GCP2 (purple) and GCP3 (pink). B) Schematic of a γ-tubulin/GCP2/GCP3 heterotetramer that forms the human γ-TuSC. C) Overall density map used to model GCP2 (position 7, purple surface), GCP3 (position 8, pink surface), and the staple (grey surface) viewed from the angle indicated in A). N- and C-domains, long (GCP3) and short (GCP2) C-domain hairpins, and the staple density, are indicated. D) Molecular model of GCP2 (left). Inset shows the fit of GCP2 residues A319-Q344 (stick representation) in the density map (mesh). E) Molecular model of GCP3 (left). Inset shows the fit of GCP3 residues Q322-H343 (stick representation) in the density map (mesh). F) Poly-alanine model of the staple (cartoon representation). Secondary structure features are indicated. G) Molecular model of the human γ-TuSC comprising GCP2, GCP3, and two copies of γ-tubulin. The staple is also shown to be consistent with the density map in C). H) Rotated GCP2 (purple surface) and GCP3 (pink surface) models highlighting the intra-γ-TuSC interface (blue surface). See also Figures S2 & S5 and Tables S1, S4 & S5.
The GCP2 and GCP3 models are largely similar to one another the exception is the C-domain hairpin, which is ~10 Å longer in GCP3 vs. GCP2 (Figure S7C) and forms the basis of the repeating “protruding density” described in Figure 1. Further, our high-resolution models reveal details of a ~3,000 Å2 interface between GCP2 and GCP3 (intra-γ-TuSC; Figure 4H), and between GCP3 and GCP2 (inter-γ-TuSC; Figure S7F). In addition, on the lumenal side of the γ-TuRC, a long (>50 Å) unassigned α-helix forms additional contacts with the N-domains of GCP2 and GCP3 at positions 3-6 (Figure S5H), which may stabilize interactions between multiple γ-TuSCs. Together, our results provide near-atomic structural models for the human γ-TuSC, and indicate that four Y-shaped γ-TuSCs comprise the stalk-like densities at positions 1-8 of the human γ-TuRC.
The γ-TuRC “overlap” region contains a conformationally altered terminal γ-TuSC
We next sought to characterize the remaining subunits found at the γ-TuRC “overlap”. We found that position 13 of the γ-TuRC corresponds to an additional GCP2 based on the fit of side chains from our GCP2 model into the C-domain density (Figures 5A, S6B, and S6C), as well as the presence of a small α-helical density between the N-domains of positions 13 and 14 (Figures S6B and S6F) resembling the γ-TuSC “staple” described above. We reasoned that the less-resolved stalk-like density at the subsequent position 14 likely corresponds to GCP3 based on a rigid body fit of our GCP3 model and the observation that GCP2 and GCP3 exist in pairs as part of the γ-TuSC (Oegema et al., 1999; Vinh et al., 2002) (Figures 5A and S6E). Notably, the γ-TuSC and γ-TuSC-like subcomplexes at positions 1-12 are organized into a continuous lattice-like arrangement mediated by the lateral associations between GCP N-domains (Figures 1A, 1D, and 1E). In contrast, the tentative γ-TuSC at positions 13-14 is in a displaced configuration relative to the neighboring stalk-like subunits in the complex, in which the GCP2-associated γ-tubulin acts as a hingepoint for a ~6° rotation of this subcomplex towards the conical axis (Figures 5A and 5B). The rotation allows this γ-TuSC to avoid steric clashes that would otherwise occur in part between the N-domain of our tentative GCP3 model at position 14 and the bi-lobed structure in the lumenal density (described below). Additionally, we observed a compact (~2 × 2 × 3 nm), unassigned density that stems from the C-domain of GCP3 and is situated at the “overlap” region, just above the γ-tubulin at position 1 (Figures 5A. 5B, and S6F). Together, our results indicate that the γ-TuRC “overlap” region is likely composed of two γ-TuSCs (one at positions 1-2, and another at positions 13-14).
Figure 5. Composition of the γ-TuRC “overlap” and lumenal bridge regions.
A) Side view of the γ-TuRC cryo-EM density map highlighting subunits leading up to the γ-TuRC “overlap” (indicated). A density at position 14 (asterisk) extending into the “overlap” region (dashed line) from an arrangement of GCP2 & the tentatively-assigned GCP3 (the “terminal γ-TuSC”) is indicated. B) Schematic of the γ-TuSC at the overlap adjacent to γ-TuG4/6 indicating its ~6° rotation relative to a hinge point in the γ-tubulin at position 13 (green star). The GCP6 “plug” is omitted for clarity. C) Top view of the overall γ-TuRC density map highlighting the lumenal bridge (dashed rectangle, spanning from positions 2 and 3 to positions 9 and 10) and its two sub-domains (i: bi-lobed density domain; ii: α-helical bundle domain). D) Zoomed-in view of the α-helical bundle domain ((ii) in C); grey surface) that interacts (dashed circle) with density (white surface) stemming from GCP5 (orange surface). E) Left: Zoomed-in view of the interactions (white dashed circles) between the bi-lobed density ((i) in C); grey surface) and γ-tubulins at positions 2 and 3 (blue surface). Right: A rigid-body fit of β-actin (grey cartoon representation; PDB ID 2HF3; (Rould et al., 2006)) in the bi-lobed density (transparent grey surface). See also Figures S5-S6 and Tables S1, S2, S4 & S5.
A bridge structure containing an actin-like protein spans across the γ-TuRC lumen
Lastly, we examined the substantial but unexpected density spanning the γ-TuRC lumen, which we name the “lumenal bridge”. This density can be divided into two sections: 1) a bi-lobed density located near positions 2 (GCP3) and 3 (GCP2) (Figure 5C); and 2) ~16 α-helices arranged in extended bundles that reach across the lumen and contact positions 9 and 10 (Figure 5C). The bi-lobed density measures ~5 × 5 × 2 nm and is comprised of a mixture of α-helical and β-sheet secondary structures. One end of this density contacts the γ-tubulins at positions 2 and 3 in the γ-TuRC (Figures 5C and 5E). Remarkably, a model for β-actin (Rould et al., 2006) could be rigid-body fitted into this density (Figure 5E), suggesting that this protein adopts an actin-like fold. The presence of an actin-like protein in purified γ-TuRCs has previously been reported (Choi et al., 2010; Oegema et al., 1999). Our data reveal that an actin-like protein is a structural component of the human γ-TuRC, which further exemplifies the diverse structural role for actin-like proteins in large macromolecular assemblies, as in, e.g., the dynactin complex (Schafer et al., 1994).
The second section of the lumenal bridge measured ~9 × 3 × 2 nm and was built from several bundles of consecutive, short (10-20 residues) α-helices loosely organized in a chain (Figure S6G). Interestingly, an α-helix at one end of this domain contacts the actin-like protein at the barbed end groove (Figures S6G and S6H), a common interaction site for diverse actin-binding proteins (Pollard, 2016). The other end of the α-helical bundle domain was in close contact with GCP4 at position 9 and GCP5 at position 10 (FigureS 5C and 5D). An unassigned density for a loop stemming from the N-domain of GCP5 (described above) contacts several α-helices in these bundles (Figure 5D). Our mass spectrometry data indicated the presence of the small proteins MZT1 and MZT2A/B (Table S2) (Hutchins et al., 2010). Their propensity to form multiple short α-helices (Figures S6I and S6J) leads us to hypothesize that these proteins may constitute at least part of the α-helical bundles in this domain. Together, our results identify a structural motif, the lumenal bridge, that spans from the γ-tubulins at positions 2-3 via an actin-like protein in the γ-TuRC lumen, through a total of >300 Å to the γ-tubulins at positions 9-10 on the opposite end of the cone-shaped structure (Figure 5C).
Discussion
In this study, we have determined a cryo-EM reconstruction of the native human γ-TuRC, which we find is built from at least 26 distinct polypeptides organized into an asymmetric cone-shaped structure. Our data lead to structural models for human GCP2, GCP3, GCP5, and GCP6 and provide, at near-atomic resolution, many of the critical contacts needed to form this unusual macromolecular complex from a defined set of components. These results provide insight into biological mechanisms of γ-TuRC recruitment, regulation, and the control of microtubule numbers in cells.
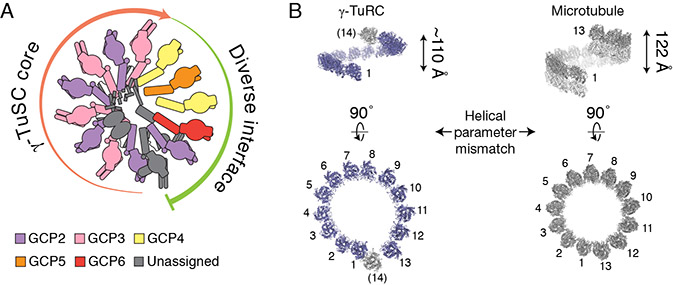
Our structure reveals that the ~2.2 MDa γ-TuRC is built from two types of Y-shaped building blocks: 1) the γ-TuSC; and 2) subunits that structurally mimic the γ-TuSC (i.e., γ-TuG4/5 and γ-TuG4/6; Figures 1A, 2B, 3B and 4B). Together, four GCP2/GCP3 γ-TuSCs make up one half of the cone, and the γ-TuSC-like subunits γ-TuG4/5 and γ-TuG4/6, likely ending with a γ-TuSC in an altered conformation at the “overlap” (also referred to as the “seam”), constitute the other half of the γ-TuRC structure. Our data also provide structural evidence supporting the hypothesis that the conserved GRIP1 (N-domain) and GRIP2 (C-domain) motifs of GCP2, GCP3, GCP5 and GCP6 should adopt similar protein folds (Guillet et al., 2011; Kollman et al., 2011). In addition to the evolutionarily conserved GCP2 and GCP3 proteins found in the γ-TuSCs, the GCP4, GCP5 and GCP6 subunits diversify the γ-TuRC (Figure 6A), introducing additional, large (>100,000 Å2; significantly greater than a typical protein-protein interface: ~1,000 Å2 (DeLano et al., 2000)), non γ-TuSC protein interaction interfaces.
Figure 6. Asymmetric molecular architecture of the γ-TuRC.
A) Schematic of γ-TuRC organization. The γ-TuRC consists of two highly asymmetric, but similarly-sized halves: 1) a γ-TuSC oligomer-like “core” (salmon arrow); and 2) an arrangement of divergent γ-TuSC-like subunits (γ-TuG4/5 & γ-TuG4/6) capped by a terminal γ-TuSC that, together, form a large and diverse binding surface for regulatory factors (green arc). The γ-tubulins are not shown for clarity. B) Left: Two views of the quasi-helical arrangement of γ-tubulin models in the human γ-TuRC. Right: Two views of the helical arrangement of α- (grey) and β- (white) tubulin dimers within the microtubule lattice (PDB: 6E7B; Ti et al., 2018). The helical rise (indicated) and diameter of the γ-tubulin ring do not perfectly match the microtubule lattice.
The γ-TuRC may use its extensive and diverse surface area to regulate the formation of properly oriented microtubules in cells by recruiting accessory factors. The γ-TuNA peptide comprising the conserved CM1 motif of CDK5Rap2 (Choi et al., 2010) could constitute 1) the “staple” motif found on the outer face of each Y-shaped γ-TuSC subcomplex (Figures 4C and 4F), or 2) the CC domain at position 13 (Figures 3C, 3E, and S6B). Notably, both locations are found on the exposed outer surface of the conical γ-TuRC. Our data suggest that CDK5Rap2 and other CM1 motif-containing proteins may form a “tether” via this motif that could link the outer face of the γ-TuRC to, e.g. the centrosome, consistent with current models (Kollman et al., 2011; Tovey and Conduit, 2018). Other attachment factors, such as NEDD1 (Haren et al., 2006; Lüders et al., 2006), may bind to the γ-TuG4/5 or γ-TuG4/6 to similarly orient γ-TuRCs in, e.g., the augmin/HAUS-mediated branched microtubule networks found in plant cortical arrays and the spindle apparatus (Murata et al., 2005; Petry et al., 2013; Uehara et al., 2009). Of note is the “plug” region at the base of our γ-TuRC model, which, though currently unassigned, could itself correspond to an attachment factor such as NEDD1, a protein species we detect at stoichiometric levels (Table S2). In either case, our structure suggests that sites of γ-TuRC attachment may form anchor points that orient the γ-tubulin ring away from diverse microtubule organizing centers in order to establish and maintain the polarity of cellular microtubule networks (Muroyama et al., 2016; Sánchez-Huertas et al., 2016).
Our work also identifies a previously unanticipated structural feature within the γ-TuRC - the lumenal bridge (Figure 5C). This structure spans >300 Å from the γ-tubulins at positions 2 (GCP3) and 3 (GCP2) to γ-tubulins at positions 9 (GCP4) and 10 (GCP5), and is built from several unexpected protein densities in the γ-TuRC lumen, including an actin-like protein. The lumenal bridge provides an extended set of protein-protein contacts that could stabilize the association between the γ-TuSC core at positions 1-8 and the γ-TuG4/5 at positions 9-10 (Figures 5C-5E, S6G, and S6H). The actin-like protein may also be a “stopper” that sterically blocks further oligomerization of γ-TuSC-like subunits past position 14 of the complex. Evidence for this second hypothesis comes from manual docking of an extra γ-TuSC model into the theoretical positions 15 and 16 (data not shown), as well as the observation that the γ-TuSC at positions 13-14 is rotated in a manner that avoids steric clashes between the N-domain of GCP3 and the actin-like protein (Figures 1A, Figure 5A, and 5B). Interestingly, recent studies have identified a role for the centrosome in regulating actin filament formation in cells and in vitro (Farina et al., 2016). We note that the barbed end of the γ-TuRC actin-like protein points into the lumen of the complex and associates with a helix in domain ii of the lumenal bridge (Figures 5C, S6G, and S6H), an orientation that likely requires a large conformational change to be compatible with templating actin filament formation (Pollard, 2016).
We found that the native human γ-TuRC likely arranges 14 γ-tubulins into a single-turn helix closely resembling, but not perfectly matching, the 13-protofilament, 3-start geometry of α/β-tubulin dimers in the microtubule lattice (Figures 1A and 6B) (Ti et al., 2018). Specifically, the 13 well-resolved γ-tubulins at positions 1-13 adopt a helical rise that is at least 10 Å smaller than the ~120 Å observed in the microtubule (meaning the γ-tubulin-ring is too “flat”; Figure 6B, top), and there is a gap of almost a full γ-tubulin subunit between positions 1 and 13 (meaning the γ-tubulin-ring is also too “wide”; Figure 6B, bottom). Our data suggest that in order to match the microtubule lattice, the γ-TuRC needs to undergo conformational changes to expand the helical rise and shrink the gap at the overlap such that the γ-tubulin at position 14 is situated directly above the γ-tubulin at position 1 (Kollman et al., 2010).
Despite this structural mismatch, our γ-TuRCs retain microtubule nucleating activity (Figure S1C). Currently, the precise mechanism of microtubule assembly from soluble tubulin dimers is poorly understood (Roostalu and Surrey, 2017), but it has been speculated that γ-tubulin “ring closure” is required for efficient microtubule nucleation from γ-TuRCs (Kollman et al., 2015). Our findings imply that the native human γ-TuRC could achieve γ-tubulin “ring closure” via several avenues, including: 1) long-range conformational changes (Figures S1F and S4C-S4F); 2) potential mechanical work generated by GTP-hydrolysis within γ-tubulin; or 3) coupling to the binding and GTP-hydrolysis cycle of its substrate, α/β-tubulin, which we note can stably associate with the complex under certain purification conditions (Zheng et al., 1995). Our structural data suggests that such long-range conformational changes could be focused at the γ-TuSC-like side of the complex (positions ~9-14), which we find deviates substantially from the ideal helical parameters compared to the γ-TuSC core (positions 1-8) (the “helical deviation” in Figure 1A; Figure 6B). Consistent with this observation, we found that GCP5 can adopt an altered conformation that propagates to the subsequent γ-TuG4/6 and possibly to the tentative γ-TuSC at positions 13-14 (Figure S4C-F). Notably, however, variability in the lateral spacing between γ-tubulins persists even within the γ-TuSC core (Figures S3D and S3E). Such conformational variability in lateral contacts is not observed in the microtubule lattice, indicating that the entire γ-TuRC, including the γ-TuSCs, must also undergo conformational changes for γ-tubulin “ring closure” to occur.
Alternatively, our structure raises the possibility that a ring of 13 tubulin-binding sites with helical symmetry that perfectly matches the microtubule lattice may not necessarily be a prerequisite for the microtubule-nucleating function of the γ-TuRC. The native complex used here contains an “activating” peptide, the γ-TuNA (Choi et al., 2010), and exhibits demonstrable nucleating activity in vitro (Figure S1C). Previous studies using recombinant S. cerevisiae γ-TuSC oligomers also showed that they are also capable of nucleating microtubules in vitro despite a substantial structural mismatch between the γ-tubulin ring and the microtubule lattice (Kollman et al., 2010). Furthermore, as few as 2-3 laterally-associated tubulin binding sites are predicted to overcome a substantial portion of the kinetic barrier facing a microtubule during nucleation (Rice et al., 2019). More work will be needed to determine whether human γ-TuRCs must undergo conformational changes to facilitate microtubule nucleation, and how this would affect other proposed functions of the complex, such as microtubule minus end-capping, anchoring, and protofilament number specification (Dammermann et al., 2003; Martin and Akhmanova, 2018).
The asymmetric, cone-shaped structure of the γ-TuRC has a distinct shape compared to similarly-sized, soluble macromolecular complexes, such as the 60S human ribosome (~25 nm-wide) (Khatter et al., 2015), or viral capsids like CCMV (~20 nm-diameter) (Fox et al., 1998). As demonstrated with their budding yeast orthologs (Kollman et al., 2010), the human γ-TuSC and γ-TuSC-like subunits may have a propensity to self-assemble in a manner analogous to viral capsids. How then might the finite structure of the γ-TuRC be established? We note that γ-TuRC subcomponents represent some of the least abundant proteins in cultured human cells (<<500 copies per cell (Beck et al., 2011)). We propose that a low concentration of γ-TuSC-like subunits, possibly in combination with the lumenal bridge, could “cap” the self-assembly of four γ-TuSCs. This would provide a self-regulating mechanism for limiting the size and total number of active γ-TuRC assemblies, and, therefore, microtubule initiation sites in cells (Brinkley et al., 1981). Such microtubule “book-keeping” could be important for controlling microtubule architecture and numbers in, e.g., branched microtubule networks (Goshima et al., 2008), or in biomolecular condensates in the centrosome (Woodruff et al., 2017) and liquid-like spindle domains (So et al., 2019), which can concentrate tubulin and associated factors to incredibly high levels that would be difficult to regulate (Baumgart et al., 2019). Our study provides a foundation for dissecting how the γ-TuRC assembles and functions in diverse cellular contexts.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tarun M. Kapoor (kapoor@rockefeller.edu). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HeLa S3 cell cytoplasmic extracts were prepared according to (Abmayr and Carrozza, 2001) and were a generous gift from Dr. Robert Roeder. GFP-γ-TuNA and GFP nanobody constructs were expressed in Escherichia coli BL21(DE3) pRIL cells cultured in LB medium supplemented with appropriate antibiotics at 37 °C and at 230 rpm until induction, at which point the temperature was shifted to 16-18 °C. Tubulin was purified from bovine brains through three cycles of polymerization/depolymerization followed by phosphocellulose chromatography, similar to (Gell et al., 2011).
METHOD DETAILS
Purification of native human γ-TuRCs
Native γ-TuRCs were isolated from HeLa S3 cells based on the reported affinity of human γ-TuRCs for CDK5RAP2 (Choi et al., 2010). First, a bacterial expression construct for CDK5Rap2’s CM1 domain, also known as the “γ-TuNA” (γ-TuRC-mediated Nucleation Activator) region, comprising amino acids 51-100 fused to GFP as an affinity tag (GFP-γ-TuNA), was generated by PCR amplifying CDK5Rap251-100 from pRcCMV Cep215 (NCBI accession BAB13459.3), a gift from Erich Nigg (Addgene plasmid # 41152; http://n2t.net/addgene:41152; RRID:Addgene_41152). The PCR product was inserted into the multiple cloning site of a modified pET-SUMO vector (pET His6 Sumo TEV LIC cloning vector (1S), a gift from Scott Gradia (Addgene plasmid # 29659; http://n2t.net/addgene:29659; RRID:Addgene_29659) containing an N-terminal EGFP-PreScission fusion tag using standard restriction cloning techniques. The plasmid was transformed into BL21(DE3) pRIL cells (Stratagene) and His6-SUMO-TEV-GFP-PreScission-γ-TuNA expression was induced with 0.5 mM IPTG for 16 hours at 18 °C. Cell pellets from 6 L culture were resuspended in 120 mL Ni-NTA lysis buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 15 mM imidazole, 0.1% (v/v) Tween-20 and 1 mM 2-mercaptoethanol) and lysed by two passes through an Emulsiflex C-5 (Avestin). Lysate was clarified at 35,000 rpm in a Type 45 Ti rotor (Beckman) for 45 min at 4 °C and the supernatant was mixed with 3 mL His60 (Takara Biosciences) resin pre-equilibrated in lysis buffer. The resin was then washed extensively with lysis buffer. Protein was eluted with Ni-NTA elution buffer (lysis buffer containing an additional 485 mM imidazole). Peak fractions were identified by Bradford assay, pooled, concentrated to ~1 mL with a 10 kDa cutoff spin filter (Millipore), and incubated with 1 mg TEV protease for 2 hr at 4 °C. GFP-γ-TuNA was then further purified over a Superdex 75 10/300 column pre-equilibrated in gel filtration buffer (40 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 2 mM 2-mercaptoethanol). Peak fractions were identified by SDS-PAGE, pooled, supplemented with sucrose to 10% (w/v), aliquoted, snap frozen in liquid nitrogen, and stored at −80 °C.
The construct for a recombinant GFP nanobody (clone LG-16 (Fridy et al., 2014)) was a kind gift from Dr. Michael Rout. The plasmid was transformed into BL21(DE3) pRIL cells (Stratagene) and periplasmic expression was induced with 0.1 mM IPTG for 16 hr at 16 °C. Cells from 6 L of culture were harvested at 5,0 00 X g for 10 min and resuspended in TES buffer (200 mM Tris-HCl, pH 8.0, 0.5 mM EDTA and 500 mM sucrose). The cells were osmotically shocked on ice for 30 min by diluting them 5-fold in 1:4 (v/v) water:TES buffer. Periplasmic extract was separated from cell debris by centrifugation for 10 min at 6,000 × g, and this supernatant was further clarified at 20,000 × g for 20 min at 4 °C. This final supernatant was supplemente d with NaCl to 150 mM and incubated with 3 mL His60 resin (Takara Biosciences) for 30 min. The resin was washed in batch with wash buffer 1 (20 mM Tris-HCl, pH 8.0 and 900 mM NaCl) followed by wash buffer 2 (20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 10 mM imidazole). The resin was then washed further with wash buffer 2, and the protein was eluted with Ni-NTA elution buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 250 mM imidazole). Peak fractions were identified by Bradford assay, pooled, and concentrated with a 10 kDa cutoff spin filter. The nanobody was further purified over a Superdex 75 10/300 column equilibrated in coupling buffer (150 mM sodium bicarbonate, pH 8.0 and 150 mM NaCl). Approximately 10 mg of GFP nanobody was coupled to a 1 mL NHS HiTrap column (GE Healthcare) according to the manufacturer’s instructions.
Cytoplasmic extract from ~10 L HeLa cell S3 suspension cultures, a kind gift from Dr. Robert Roeder (Abmayr and Carrozza, 2001), was thawed on ice and diluted 1:1 with ice cold γ-TuRC buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.1 % (v/v) IGEPAL and 0.1 mM GTP) containing one complete EDTA-free Protease Inhibitor Cocktail tablet (Roche). 5 mg of purified γ-TuNA GFP was added directly to the extract, which was then incubated on ice for 1 hr. The extract was then passed over the GFP nanobody column. The column was washed with 20 mL of γ-TuRC buffer lacking protease inhibitors. 100 μg of PreScission protease was diluted into 1 mL γ-TuRC buffer and this mixture was injected onto the column. Proteolytic digestion of the GFP tag was allowed to proceed for 6 hr, after which γ-TuRCs were eluted from the column with γ-TuRC buffer. The peak fraction was layered onto a 2 mL sucrose step gradient made up of gradient buffer (40 mM Na-HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 2 mM 2-mercaptoethanol, 0.01% (v/v) IGEPAL and 0.1 mM GTP) containing 5%, 16.7%, 28.3%, and 40% (w/v) sucrose layered in TLS-55 centrifuge tubes (Beckman). The gradient was spun in a TLS-55 rotor (Beckman) at 50,000 rpm for 3 hr at 4 °C and fractionated into 150 μL fractions with a cut-off pipette tip. Fractions were analyzed by immunoblotting against γ-tubulin and peak γ-tubulin-containing fractions, typically found around ~30% sucrose, were pooled, aliquoted, snap frozen in liquid N2, and stored at −80 °C SDS-PAGE analysis of our purified γ-TuRCs is shown in Figure S1B. For this analysis, protein in each fraction was precipitated with 10% (w/v) TCA and re-dissolved in 1X SDS sample buffer after centrifugation before running on a 4-20% Tris-glycine-SDS pre-cast gel (Novex).
Mass spectrometry
A 40 μL aliquot of purified native γ-TuRCs was thawed and mixed with 10 μL of 5X sample buffer (Tris-HCl, pH 6.8, 10% (w/v) SDS, 50% (w/v) glycerol, 700 mM 2-mercaptoethanol, and 0.25% (w/v) bromophenol blue). After boiling, the sample was loaded into a single lane of a 4-20% Tris-glycine pre-cast gel with “wide wells” (Novex) and allowed to migrate ~1 cm into the stacking gel. A corresponding ~1 cm × 1 cm gel plug was cut out of the gel, further cut into 1 mm cubes, and destained with 33% (v/v) acetonitrile in 50 mM ammonium bicarbonate. To reduce disulfide bonds, gel pieces were incubated with 200 μL 20 mM TCEP in 50 mM ammonium bicarbonate at 56 °C for 45 minutes. Following removal of the TCEP solution, cysteines were alkylated with 200 μL 55 mM 2-chloroacetamide in 50 mM ammonium bicarbonate at room temperature in the dark for 30 minutes, then washed briefly with 50 mM ammonium bicarbonate. Proteins were digested with 80 μL 3.1 ng/uL trypsin in 50 mM ammonium bicarbonate for 4 h at 37°C, followed by addition of a further 125 ng trypsin and incubation overnight. After removing the solution to a clean tube, peptides were extracted from the gel with 100 μL 70% acetonitrile, 1.7% formic acid, 0.1% TFA at 37 “C for 1 h. The supernatant was collected and combined with the initial pool of tryptic peptides. Extraction was repeated sequentially with 100 mM triethyl ammonium bicarbonate and acetonitrile, each with 100 μL at 37 °C for 1 h. Peptides were dried by vacuum centrifugation, resuspended in 5% formic acid, 0.1% TFA, and desalted on C18 membranes before being loaded onto an EASY-Spray column (Thermo Fisher Scientific, 15cm × 75μm ID, PepMap C18, 3μm) via an EASY-nLC 1200 (Thermo Fisher Scientific). Column temperature was set to 35°C. Using a flow rate of 3 00 nL/min, peptides were eluted with a gradient of 2-32% solvent B in 33 minutes, followed by 32-80% B in 5 minutes, where solvent B was 0.1% formic acid in 95% acetonitrile and solvent A was 0.1% formic acid in water. A spray voltage of 1.8 kV ionized peptides as they eluted into an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) acquiring online data-dependent CID fragmentation spectra. Raw data were analyzed with MaxQuant 1.6.2.10.
Turbidity-based microtubule nucleation assay
Tubulin was purified from bovine brains through three cycles of polymerization/depolymerization followed by phosphocellulose chromatography and stored at −80 °C, as described previously (Gell et al., 2011). Before the assay, tubulin was quickly thawed at 37 °C and centrifuged for 10 min at 4 °C and at 350,000 × g. The concentration of the supernatant was measured spectrophotometrically using an extinction coefficient of 115,000 M−1cm−1 and a molecular weight of 110 kDa for the tubulin dimer.
Microtubule nucleation was assayed by preparing 75 μL reactions containing 7.5% (v/v) native γ-TuRCs (final concentration estimated to be in the pM range), 10 μM tubulin, 1 mM GTP, and 3.4 M glycerol in BRB80 (80 mM K-PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA). An equivalent volume of control buffer (40 mM Na-HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 2 mM 2-mercaptoethanol, 0.01% (v/v) IGEPAL, 0.1 mM GTP, and 30% (w/v) sucrose) was included for tubulin-only controls. Components were mixed and incubated on ice for 5 min. Reactions were split and loaded into a pre-warmed 384-well, clear-bottomed microplate in duplicate (~32 uL reaction per well), and 15 uL of fluorescence-free immersion oil (Cat# 16212, Cargille Laboratories) was layered over each reaction. This loading time was limited to 5 minutes. The microplate was loaded onto a BioTek Synergy Neo microplate reader pre-warmed to 37 °C and absorbance measurements at 350 nm were initiated at a frequency of 1-2 per min for up to 4 hours. A γ-TuRC-only control (data not shown) was performed to confirm that purified γ-TuRCs alone do not contribute to an increase in turbidity.
Cryoelectron microscopy of native γ-TuRCs
Due to very low (sub-nM) sample concentrations, a modification of a previously described multiple application technique was used to obtain grids with densely packed γ-TuRC particles (Snijder et al., 2017). The following procedure was conducted on ice. 2 μL of frozen sample was applied to plasma treated Quantifoil R2/2 300-square-mesh copper grids coated with a continuous carbon film. After 5-10 min the sample was blotted away and replaced with another 2 μL. This was repeated for a total of 5-8 applications. As γ-TuRC storage buffer contained ~30 % (w/v) sucrose, 20 μL of washing buffer (40 mM HEPES-KOH pH 7.5, 150 mM KCl, 1 mM MgCl2, 0.01% Tween-20 (v/v), 0.1 mM GTP, and 1 mM 2-mercaptoethanol) was used to rinse the grid twice with 1-2 min incubation in between. Finally, 3 μL of washing buffer was applied to the grid, transferred to a Vitroblot IV (FEI), blotted for 4-5 s at 100% humidity and 4 °C, and then plunged into liquid ethane.
Micrographs were recorded on an FEI Titan Krios equipped with a Gatan K2 Summit detector using Serial EM automated data collection (Mastronarde, 2005). Data was collected in 3 separate sessions (Table S3). The first dataset was exploratory and employed the following settings: 300kV, 70 μm C2 aperture, 33 frames, 45 e−/Å2, 6.6 s exposure, and 1.335 Å per pixel. In order to acquire as much data as possible, data acquisition was optimized for the subsequent two sessions and the following settings were used: 300kV, 100 μm C2 aperture, 21 frames, 45 e−/Å2, 4.1s exposure, 1.036 Å per pixel. In these sessions, 8 non-overlapping micrographs were recorded per hole, and a 3 × 3 grid of 9 holes was collected for every stage shift by applying an appropriate beam tilt (i.e., 72 images were recorded per stage shift). This enabled the acquisition of ~4,500 micrographs per 24 hours in the second and third datasets.
Images for each dataset were corrected for beam-induced drift using MotionCor2 (Zheng et al., 2017). CTF parameters for drift corrected micrographs were estimated using CTFFIND4 (Rohou and Grigorieff, 2015). Subsequent processing used Relion v.3.0 (Zivanov et al., 2018). Initial particle cleanup was conducted using dose-weighted and drift corrected micrographs binned to a pixel size of 5.18 Å (Figure S2). For the first dataset, a small set of particles were manually picked and subjected to reference-free 2D classification. The obtained 2D-class averages that represented particles of different size and shape were used as references for autopicking all binned micrographs from dataset 1. The autopicked particles were cleaned by several cycles of 2D classification. These cleaned particles were used to generate an initial model. The initial model was low-pass filtered to 200 Å and used as a reference for 3D refinement followed by 3D classification. 99,524 particles corresponding to the major 3D class were selected. These particles were subjected to another round of 2D classification in order to select balanced number of particles of different orientations: 35k particles corresponding to the dominant 2D class averages and 35k that corresponded to the rarer views. The obtained 2D class averages were used as references for autopicking from datasets 2 and 3.
As a large number of micrographs were recorded for datasets 2 and 3 (Table S3), the autopicked particles were randomly split into subsets of ~230k particles. In order to avoid losing particles of less abundant orientations, each 230k particle subset was merged with a set of 70k cleaned and balanced particles from dataset 1 (see above). This yielded well resolved 2D class averages during each cycle that pooled particles from the new dataset that might otherwise be lost during classification. After 2 cycles of 2D classification and cleanup, and after removing the 70k particles from dataset 1, the remaining particles (typically ~100k - 150k) were subjected to 3D refinement. The output translational information from the 3D refinement was used to re-extract particles with the “re-center refined coordinates” option selected in Relion. Re-centered particles were subjected to another round of 3D refinement followed by 3D classification. Particles from 3D classes corresponding to γ-TuRC were selected and yielded a set of “good” particles (Table S3).
“Good” particles from each dataset were re-extracted as before from the micrographs binned to a pixel size of 1.335 Å. The particles from dataset 1 and 2 were pooled and subjected to global 3D refinement. Due to “breathing” of the complex (Figure S1F), the particles were subjected to another round of local 3D refinement but with a mask including only the stalk densities (excluding globular densities on the open face of the cone). This resulted in better alignment accuracies and, subsequently, higher resolution which in turn yielded better CTF refinement and Bayesian polishing. The CTF-refined and Bayesian-polished particles of datasets 1 and 2 were pooled with “good” particles from the latest dataset (dataset 3) and subjected to another round of 3D classification. A final 467,803 particles were selected from the 3D classes of highest resolution. These “final” particles were used for the overall refinement that yielded 3.81 Å resolution.
In order to improve local resolution, the “final” particles were subjected to 3D focused classification and refinement as described before (Nguyen et al., 2015). In short, signal of the density outside region of interest (e.g. positions 1 to 3) were subtracted from raw-particles. This significantly increased alignment accuracy during subsequent 3D refinement and sensitivity during alignment-free 3D classification. Focused 3D classification yielded the most homogenous particles in selected regions that were then used to generate the final refined maps for those regions.
Model building
Generation of the molecular model for the γ-TuRC is summarized in Table S4. Because of varying resolution across the structure, even after local focused refinements, we designated three levels of assignment confidence to each domain with specific criteria: assigned = >50 contiguous amino acids with well-defined Cα secondary structure and matching side chain densities. Only these portions of the structure are colored in Figures 1-6; tentatively assigned = rigid-body docking of an existing model, or de novo poly-Ala building based on secondary structure predictions and/or prior interaction information from literature; and unassigned = no identification, de novo poly-Ala builds only if secondary structures are resolvable. Tentative and unassigned models/densities are colored in grey throughout the paper.
γ-tubulin (PDB ID: 3CB2 (Rice et al., 2008)) was rigid-body fitted into the overall density map (Figure 1A) using UCSF Chimera’s “Fit in map” function (Pettersen et al., 2004). γ-tubulin models in positions 1-13 were refined into the corresponding density maps using the “Real space refine zone” feature in COOT with torsion, planar peptide, trans peptide and Ramachandran restraints enforced and using a refinement weighting of ≤10 (Emsley et al., 2010). Models were further refined using phenix.real_space_refine and problem areas were fixed manually in COOT. This procedure was iterated until acceptable model parameters were achieved. When necessary, higher resolution segmented maps (Figure S2) were used to further refine the models and to build missing coordinates in the density and/or remove overextended model coordinates lacking clear density for side chains, especially in loop regions. GDP was similarly modeled into the γ-tubulin nucleotide binding site density.
Models for GCP4 were built by dividing the crystal structure for GCP4 (PDB ID: 3RIP (Guillet et al., 2011)) into N- (a.a. 1 - 346) and C- (a.a. 349 - 657) domains, which were independently rigid-body fitted into stalk-like density maps at positions 9 and 11. Models were refined into the density maps as described above for γ-tubulin. N- and C-domain models were then joined together and the connecting residues were manually refined in COOT and with phenix.real_space_refine to ensure model fidelity.
Sequences corresponding to the N- and C-domains of GCP2, GCP3, GCP5 and GCP6 were determined from multiple sequence alignments with GCP4 combined with secondary structure predictions using JPred (Drozdetskiy et al., 2015) and visual inspection of our density maps. Initial models for GCP2, GCP3, and GCP5 N- and C-domains were generated with the Phyre2 server (Kelley et al., 2015) using the GCP4 crystal structure (Guillet et al., 2011) as a template. Models were fitted, corrected where necessary using secondary structure predictions and α-helical registries of the densities as a guide, and refined into the γ-TuRC density as described above for γ-tubulin and GCP4. All assigned γ-TuRC components were subjected to a final round of refinement as above to ensure fidelity of protein-protein contacts in our model of the complex. Refinement statistics for one of each of the assigned γ-TuRC subunits, as well as all assigned subunits from positions 1-13, are summarized in Table S5.
Poly-alanine models for the “staple” densities, the α-helical bundle in the lumenal bridge, the CC, the HB, and the γ-TuSC-associated helix were all built de novo and refined with the “real-space refine” function in COOT. The final γ-TuRC model includes all assigned and unassigned but built subunits (PDB 6V6S).
Visualization and analysis
Figures were generated in PyMol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311 (Pettersen et al. 2004). γ-tubulin movement associated with compaction of the GCP5 C-domain (Figure S4E-F) was visualized and quantified within PyMol using the ColorByRMSD (http://pymolwiki.org/index.php/ColorByRMSD) script modified by Thomas Holder and Nicolas Coudray.
QUANTIFICATION AND STATISTICAL ANALYSIS
Resolution estimations of cryo-EM density maps are based on the 0.143 Fourier Shell Correlation (FSC) criterion (Rosenthal and Henderson, 2003). All statistical validation performed on the deposited models (PDB ID 6V5V, 6V69, 6V6B, 6V6C, and 6V6S) was done using the Phenix package (Table S5) (Williams et al., 2018).
DATA AND CODE AVAILABILITY
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/): EMD-21054, EMD-21060, EMD-21063, EMD-21067, EMD-21068, EMD-21070, EMD-21073, and EMD-21074. Structural models of the γ-TuRC and one of each assigned subunit has been deposited in the Protein Data Bank (PDB; https://www.rcsb.org/): PDB ID 6V5V, 6V69, 6V6B, 6V6C, and 6V6S.
Supplementary Material
Figure S1. Purification, characterization, and cryo-EM reconstruction of the native human γ-TuRC, related to Figure 1. A) Native human γ-TuRC purification scheme. B) SDS-PAGE analysis of purified native human γ-TuRC. Bands corresponding to core γ-TuRC components are indicated. Bands for GCP2 and GCP3 are very close to each other due to similar molecular weights. Contaminating bands were observed at ~25 kDa and ~10 kDa, likely corresponding to uncleaved GFP-γ-TuNA (*) and cleaved γ-TuNA (**) based on size. Double asterisk indicates a contaminant at <10 kDa that likely corresponds to cleaved γ-TuNA. C) Turbidity-based nucleation assay of reactions containing tubulin (10 μM) and GTP (1 mM) in the presence (blue circles) or absence (black squares) of purified native γ-TuRC (7.5% (v/v)). D) Cryo-EM micrograph of native human γ-TuRCs. Example particles selected for further processing are indicated (boxes). E) 2D-class averages of the γ-TuRC. F) Two views of the overall γ-TuRC density map analyzed by ResMap (Kucukelbir et al., 2014), showing a resolution distribution ranging from 3 to 12 Å. G) Gold-standard Fourier shell correlation (FSC) curve of the overall γ-TuRC density map. The FSC at 0.143 is indicated.
Figure S2. Cryo-EM data processing of the γ-TuRC, related to Figure 1. Focused 3D classification and refinement procedures used to improve the resolution in the overall γ-TuRC density map.
Figure S3. Comparison of γ-tubulin and GCP4 to known structures and alignment of GCP5 to GCP4, related to Figure 1 and 2. A) - C) Alignment of the γ-tubulin model from this study (blue cartoon representation) to a previously-reported crystal structure of γ-tubulin:GDP ((Rice et al., 2008); white cartoon representation). Alignment was performed as in (Rice et al., 2008). RMSD of the overall alignment is indicated in A). B) and C) RMSDs of hallmark γ-tubulin secondary structure features are indicated. D) View of γ-tubulin models at position 3-5 highlighting a displacement (red dashed lines, distance indicated) in the γ-tubulin:γ-tubulin interface at position 4-5 (light blue - dark blue) compared to at position 3-4 (dark blue - light blue). E) Schematic of the γ-TuRC highlighting locations of the two types of γ-tubulin:γ-tubulin interfaces shown in using the same γ-tubulin coloring scheme in D). F) Alignment of GCP4 model from this study (yellow cartoon representation) with a previously reported crystal structure (PDB ID: 3RIP; (Guillet et al., 2011); grey cartoon representation) via their N-domains. A ~10° rotation in the C-domain of our GCP4 model relative to the aligned N-domains is indicated (arrow). G) Rotated view of F) focusing on the C-domain. A ~90° rotation in helix 21 relative to the C-domain is indicated. H) Alignment of our GCP5 model (orange cartoon representation) with our GCP4 model (yellow cartoon representation) via their N-domains. A ~20° rotation in the GCP5 C-domain rel ative to the aligned N-domains indicated (arrow).
Figure S4. GCP5 and GCP6 extra densities and model details, related to Figure 2 and 3. A) Schematic of the human GCP5 sequence highlighting regions modeled in this study (orange rectangles). B) View of the GCP5-associated N-terminal density (grey surface representation) that interacts with N-domains of GCP4 (position 9), GCP5 (position 10) and possibly GCP4 at position 11 (cartoon representations). C) and D) Lumenal view of positions 9-11 of the γ-TuRC with the GCP5 C-domain in the extended (C) and compacted (D) conformation. Downward displacement in γ-tubulin at position 10 is indicated (arrow). E) and F) Compaction of the GCP5 C-domain at position 10 results in a displacement of the γ-TuG4/6 subcomplex away from the conical axis relative to the “elongated” GCP5 subclass, represented by the broken arc with arrows showing displacement directions (E). The direction and extent of the resulting individual γ-tubulin displacement is illustrated by vector plot and average translation of all the atoms (F). Maps were low-pass filtered to 8 Å (C-D) and 13 Å (E-F). G) Schematic of the human GCP6 sequence highlighting regions modeled in this study (red rectangles). The ~800 residue insertion sequence is indicated, with a highly α-helical region containing coiled coils (see Methods) shown as a white rectangle. H) Fit of GCP6 residues K1615-K1641 (indicated; red stick representation) in a density map for the GCP6 C-domain (mesh). I) Left: Schematic of the γ-TuRC highlighting the location of GCP6 and establishing the viewing angle. Right: γ-TuRC model (surface representation) with GCP6 highlighted (red) overlayed with a segmented, 13 Å low-pass filtered density map of the “plug” density (light red surface representation, indicated). In A) and G), some short (<~25-50 residue) connecting loops not currently modeled are omitted for clarity.
Figure S5. GCP2, GCP3, “staple”, and γ-TuSC-associated helix features, related to Figure 4. A) Schematic of the human GCP2 sequence highlighting regions modeled in this study (purple rectangles). B) Schematic of the human GCP3 sequence highlighting regions modeled in this study (pink rectangles). C) Panels showing the fit of residues A319-Q344 (purple stick representation) from GCP2 models at positions 1, 3, and 5, as well as residues Q322-H343 (pink stick representation) from GCP3 models at positions 2, 4, and 6, within the density map (mesh). D) Cartoon (top) and surface (bottom) representations of the staple domain bound to N-domains of GCP2 and GCP3. The white portion of the surface represents a currently unassigned region possibly connecting the staple to the N-terminus (a.a. 232) of GCP2. E) Alignment of GCP2 and GCP3 models to one another focusing on the C-domain hairpin. The ~10 Å extension of the GCP3 vs. the GCP2 hairpin is indicated. F) GCP2 (purple surface) and GCP3 (pink surface) models rotated along their long axes (indicated) highlighting the inter-γ-TuSC interface (blue surface). An asterisk indicates a small interface formed between C-domains that is missing in the intra-γ-TuSC interface (Figure 4H). G) Secondary structure prediction of the γ-TuNA peptide generated using Jpred (Drozdetskiy et al., 2015). α-helices (grey cylinders), β-sheets (grey arrow), and residues predicted to form a coiled coil (red letters) are indicated. H) A long unassigned α-helix density (grey surface representation) spans along the N-domains of γ-TuSCs at positions 3 to 6 (surface representation colored according to Figure 1).
Figure S6. Identification and organization of a terminal γ-TuSC at the “overlap” region and details of the lumenal bridge, related to Figure 5. A) Schematic of the γ-TuRC highlighting the locations of GCP6 (red) adjacent to a GCP2 subunit at position 13 (purple) that is associated with a γ-TuNA (light blue). B) Density map for positions 11-13 used to model GCP2 (position 13, purple surface) with a staple (grey surface) viewed from the angle indicated in A). C) Refined model of GCP2 C-domain residues M725 - D764 (stick representation) in the corresponding α-helical density (mesh) at position 13. D) Schematic of the γ-TuRC highlighting the location of GCP2 (purple) and a staple domain (grey) adjacent to a tentative GCP3 subunit (grey) at position 14. E) Rigid-body fit of the GCP3 model (grey cartoon representation) into the corresponding density (grey transparent surface) at position 14. F) View of GCP2 (purple surface), GCP3 (grey surface), and a staple (grey surface) density constituting the tentative terminal γ-TuSC at position 13-14. Indicated is an extra density associated with GCP3 (dashed black circle around grey surface representation; see also Figure 4A) that is situated above the γ-tubulin in position 1 (the “overlap”). Views in E - F) are from the angle specified in D). G) A poly-alanine model (cartoon representation) built into the α-helical bundle densities comprising domain (ii) of the γ-TuRC lumenal bridge. An interacting helix that contacts the actin-like protein is indicated. H) α-helical bundles of domain (ii) (grey cartoon representation) interacting with a model for the actin-like protein in domain (i) (grey surface representation) in the lumenal bridge. An interacting α-helix contacts the actin-like protein at the barbed end groove (indicated). I) and J) Secondary structure predictions of MZT1 and MZT2A/B (α-helical regions shown as red bars).
Move S1, related to Figure 1. Surface representation of the native human γ-TuRC model. Subunits are colored according to the legend in Figure 1A.
Table S1, related to Figure 1. Mass spectrometry analysis of native human γ-TuRC “core” components.
Table S2, related to Figure 1. Mass spectrometry analysis of native human γ-TuRC associated factors.
Table S3, related to Figure 1. Cryo-EM data collection statistics.
Table S4, related to Figure 1. Model building and assignments
Table S5, related to Figure 1. γ-TuRC model refinement statistics.
Highlights.
High resolution cryo-EM structure of the native human γ-tubulin ring complex (γ-TuRC)
Identification and structural models for γ-tubulin, GCP2, GCP3, GCP4, GCP5 and GCP6
Identification of an actin-like protein in the γ-TuRC lumen
Model for γ-TuRC assembly and regulation of its microtubule nucleating activity
Acknowledgments:
The authors are grateful to Dr. G. Alushin for critical reading of the manuscript, Dr. R. Roeder for the gift of HeLa cell extracts, and Dr. M. Rout for the gift of a GFP nanobody expression construct, as well as M. Ebrahim, J. Sotiris, and H. Ng and the Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center for fantastic cryo-EM support. This work was funded by an NIH grant to T.M.K. (R35 GM130234). M.W. was supported by an HFSP Fellowship (LT000025/18-L1) and a Merck Postdoctoral Fellowship. LU is supported by the Rockefeller University’s Pels Center for Biochemistry and Structural Biology.
Footnotes
Declaration of interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abmayr SM, and Carrozza MJ (2001). Preparation of Nuclear and Cytoplasmic Extracts from Mammalian Cells In Current Protocols in Pharmacology, Enna SJ, Williams M, Ferkany JW, Kenakin T, Porsolt RD, and Sullivan JP, eds. (Hoboken, NJ, USA: John Wiley & Sons, Inc.),. [Google Scholar]
- Aldaz H, Rice LM, Stearns T, and Agard DA (2005). Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature 435, 523–527. [DOI] [PubMed] [Google Scholar]
- Baumgart J, Kirchner M, Redemann S, Woodruff J, Verbavatz J-M, Julicher F, Hyman A, Mueller-Reichert T, and Brugues J (2019). Soluble tubulin is locally enriched at mitotic centrosomes in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, and Aebersold R (2011). The quantitative proteome of a human cell line. Mol. Syst. Biol 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, and Agard DA (2018). The Atomic Structure of the Microtubule-Nucleating γ-Tubulin Small Complex and its Implications for Regulation. [Google Scholar]
- Brinkley BR, Cox SM, Pepper DA, Wible L, Brenner SL, and Pardue RL (1981). Tubulin assembly sites and the organization of cytoplasmic microtubules in cultured mammalian cells. J. Cell Biol 90, 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, and Rice LM (2018). Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol 19, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y-K, Liu P, Sze SK, Dai C, and Qi RZ (2010). CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol 191, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol, 26, 1367–72. [DOI] [PubMed] [Google Scholar]
- Cretu C, Schmitzová J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Lührmann R, and Pena V (2016). Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol. Cell 64, 307–319. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Desai A, and Oegema K (2003). The minus end in sight. Curr. Biol 13, R614–R624. [DOI] [PubMed] [Google Scholar]
- DeLano WL, Ultsch MH, de Vos AM, and Wells JA (2000). Convergent solutions to binding at a protein-protein interface. Science 287, 1279–1283. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, and Barton GJ (2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farache D, Jauneau A, Chemin C, Chartrain M, Rémy M-H, Merdes A, and Haren L (2016). Functional Analysis of γ-Tubulin Complex Proteins Indicates Specific Lateral Association via Their N-terminal Domains. J. Biol. Chem 291, 23112–23125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina F, Gaillard J, Guérin C, Couté Y, Sillibourne J, Blanchoin L, and Théry M (2016). The centrosome is an actin-organizing centre. Nat. Cell Biol 18, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K-W, Choi Y-K, Rattner JB, and Qi RZ (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth S, and Kapoor TM (2017). The mechanics of microtubule networks in cell division. J. Cell Biol 216, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Wang G, Speir JA, Olson NH, Johnson JE, Baker TS, and Young MJ (1998). Comparison of the native CCMV virion with in vitro assembled CCMV virions by cryoelectron microscopy and image reconstruction. Virology 244, 212–218. [DOI] [PubMed] [Google Scholar]
- Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyö D, Chait BT, et al. (2014). A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell Christopher, Friel Claire T., Borgonovo Barbara, Drechsel David N., Hyman Anthony A., and Howard Jonathon. 2011. “Purification of Tubulin from Porcine Brain.” Methods in Molecular Biology, 777: 15–28. [DOI] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, and Vale RD (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol 181, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, and Nigg EA (2007). Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell. Sci 20, 4321–31. [DOI] [PubMed] [Google Scholar]
- Guillet V, Knibiehler M, Gregory-Pauron L, Remy M-H, Chemin C, Raynaud-Messina B, Bon C, Kollman JM, Agard DA, Merdes A, et al. (2011). Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol 18, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, and Zheng Y (2000). Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J. Cell Biol 151, 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Remy M-H, Bazin I, Callebaut I, Wright M, and Merdes A (2006). NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol 172, 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JRA, Toyoda Y, Hegemann B, Poser I, Hériché J-K, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. (2010). Systematic Localization and Purification of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science 1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, and Klaholz BP (2015). Structure of the human 80S ribosome. Nature 520, 640–645. [DOI] [PubMed] [Google Scholar]
- Kirschner M, and Mitchison T (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329–342. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, and Schiebel E (1997a). The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16, 1550–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, and Schiebel E (1997b). The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16, 1550–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Zelter A, Muller EGD, Fox B, Rice LM, Davis TN, Agard DA, and Stearns T (2008). The Structure of the γ-Tubulin Small Complex: Implications of Its Architecture and Flexibility for Microtubule Nucleation. MBoC 19, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, and Agard DA (2010). Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, and Agard DA (2011). Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol 12, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez J-J, Sali A, Kilmartin J, Davis TN, et al. (2015). Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat. Struct. Mol. Biol 22, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Choi Y-K, and Qi RZ (2014). NME7 is a functional component of the γ-tubulin ring complex. Mol. Biol. Cell 25, 2017–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J, Patel UK, and Stearns T (2006). GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol 8, 137–147. [DOI] [PubMed] [Google Scholar]
- Martin M, and Akhmanova A (2018). Coming into Focus: Mechanisms of Microtubule Minus-End Organization. Trends Cell Biol. 28, 574–588. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mitchison T, and Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guénebaut V, Heuser J, and Agard DA (2000). Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol 2, 365–370. [DOI] [PubMed] [Google Scholar]
- Moutinho-Pereira S, Debec A, and Maiato H (2009). Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol. Biol. Cell 20, 2796–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, and Hasebe M (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol 7, 961–968. [DOI] [PubMed] [Google Scholar]
- Muroyama A, Seldin L, and Lechler T (2016). Divergent regulation of functionally distinct γ-tubulin complexes during differentiation. J. Cell Biol 213, 679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Preble AM, Patel UK, O’Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T, and McIntosh JR (2001). GcP5 and GcP6: Two New Members of the Human γ-Tubulin Complex. MBoC 12, 3340–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Galej WP, Bai X-C, Savva CG, Newman AJ, Scheres SHW, and Nagai K (2015). The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CE, and Oakley BR (1989). Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338, 662–664. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, and Zheng Y (1999). Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol 144, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, and Vale RD (2013). Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pollard TD (2016). Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW, Kellogg DR, and Alberts BM (1993). Drosophila gamma-tubulin is part of a complex containing two previously identified centrosomal MAPs. J. Cell Biol 121, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L, Moritz M, and Agard DA (2019). Microtubules form by progressively faster tubulin accretion, not by nucleation-elongation. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Montabana EA, and Agard DA (2008). The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. U. S. A 105, 5378–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, and Rios RM (2009). Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, and Surrey T (2017). Microtubule nucleation: beyond the template. Nat. Rev. Mol. Cell Biol 18, 702–710. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Rould MA, Wan Q, Joel PB, Lowey S, and Trybus KM (2006). Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J. Biol. Chem 281, 31909–31919. [DOI] [PubMed] [Google Scholar]
- Sánchez-Huertas C, Freixo F, Viais R, Lacasa C, Soriano E, and Lüders J (2016). Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat. Commun 7, 12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, and Schroer TA (1994). Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol 126, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW (2016). Chapter Six - Processing of Structurally Heterogeneous Cryo-EM Data in RELION In Methods in Enzymology, Crowther RA, ed. (Academic Press; ), pp. 125–157. [DOI] [PubMed] [Google Scholar]
- Schuh M, and Ellenberg J (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Snijder J, Borst AJ, Dosey A, Walls AC, Burrell A, Reddy VS, Kollman JM, and Veesler D (2017). Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J. Struct. Biol 198, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C, Seres KB, Steyer AM, Mönnich E, Clift D, Pejkovska A, Möbius W, and Schuh M (2019). A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, and Kirschner M (1994). In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell 76, 623–637. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, and Kirschner M (1991). Gamma-tubulin is a highly conserved component of the centrosome. Cell 65, 825–836. [DOI] [PubMed] [Google Scholar]
- Tang Guang, Peng Liwei, Baldwin Philip R., Mann Deepinder S., Jiang Wen, Rees Ian, and Ludtke Steven J. 2007. “EMAN2: An Extensible Image Processing Suite for Electron Microscopy.” Journal of Structural Biology 157 (1): 38–46. [DOI] [PubMed] [Google Scholar]
- Teixidó-Travesa N, Villén J, Lacasa C, Bertran MT, Archinti M, Gygi SP, Caelles C, Roig J, and Lüders J (2010). The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21, 3963–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S-C, Alushin GM, and Kapoor TM (2018). Human β-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability. Dev. Cell 47, 175–190.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovey CA, and Conduit PT (2018). Microtubule nucleation by γ-tubulin complexes and beyond. Essays Biochem. 62, 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Nozawa R-S, Tomioka A, Petry S, Vale RD, Obuse C, and Goshima G (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. U. S. A 106, 6998–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD (2003). The molecular motor toolbox for intracellular transport. Cell 112, 467–480. [DOI] [PubMed] [Google Scholar]
- Vinh DBN, Kern JW, Hancock WO, Howard J, and Davis TN (2002). Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol. Biol. Cell 13, 1144–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryoelectron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, and Mitchison T (1995). Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–583. [DOI] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Purification, characterization, and cryo-EM reconstruction of the native human γ-TuRC, related to Figure 1. A) Native human γ-TuRC purification scheme. B) SDS-PAGE analysis of purified native human γ-TuRC. Bands corresponding to core γ-TuRC components are indicated. Bands for GCP2 and GCP3 are very close to each other due to similar molecular weights. Contaminating bands were observed at ~25 kDa and ~10 kDa, likely corresponding to uncleaved GFP-γ-TuNA (*) and cleaved γ-TuNA (**) based on size. Double asterisk indicates a contaminant at <10 kDa that likely corresponds to cleaved γ-TuNA. C) Turbidity-based nucleation assay of reactions containing tubulin (10 μM) and GTP (1 mM) in the presence (blue circles) or absence (black squares) of purified native γ-TuRC (7.5% (v/v)). D) Cryo-EM micrograph of native human γ-TuRCs. Example particles selected for further processing are indicated (boxes). E) 2D-class averages of the γ-TuRC. F) Two views of the overall γ-TuRC density map analyzed by ResMap (Kucukelbir et al., 2014), showing a resolution distribution ranging from 3 to 12 Å. G) Gold-standard Fourier shell correlation (FSC) curve of the overall γ-TuRC density map. The FSC at 0.143 is indicated.
Figure S2. Cryo-EM data processing of the γ-TuRC, related to Figure 1. Focused 3D classification and refinement procedures used to improve the resolution in the overall γ-TuRC density map.
Figure S3. Comparison of γ-tubulin and GCP4 to known structures and alignment of GCP5 to GCP4, related to Figure 1 and 2. A) - C) Alignment of the γ-tubulin model from this study (blue cartoon representation) to a previously-reported crystal structure of γ-tubulin:GDP ((Rice et al., 2008); white cartoon representation). Alignment was performed as in (Rice et al., 2008). RMSD of the overall alignment is indicated in A). B) and C) RMSDs of hallmark γ-tubulin secondary structure features are indicated. D) View of γ-tubulin models at position 3-5 highlighting a displacement (red dashed lines, distance indicated) in the γ-tubulin:γ-tubulin interface at position 4-5 (light blue - dark blue) compared to at position 3-4 (dark blue - light blue). E) Schematic of the γ-TuRC highlighting locations of the two types of γ-tubulin:γ-tubulin interfaces shown in using the same γ-tubulin coloring scheme in D). F) Alignment of GCP4 model from this study (yellow cartoon representation) with a previously reported crystal structure (PDB ID: 3RIP; (Guillet et al., 2011); grey cartoon representation) via their N-domains. A ~10° rotation in the C-domain of our GCP4 model relative to the aligned N-domains is indicated (arrow). G) Rotated view of F) focusing on the C-domain. A ~90° rotation in helix 21 relative to the C-domain is indicated. H) Alignment of our GCP5 model (orange cartoon representation) with our GCP4 model (yellow cartoon representation) via their N-domains. A ~20° rotation in the GCP5 C-domain rel ative to the aligned N-domains indicated (arrow).
Figure S4. GCP5 and GCP6 extra densities and model details, related to Figure 2 and 3. A) Schematic of the human GCP5 sequence highlighting regions modeled in this study (orange rectangles). B) View of the GCP5-associated N-terminal density (grey surface representation) that interacts with N-domains of GCP4 (position 9), GCP5 (position 10) and possibly GCP4 at position 11 (cartoon representations). C) and D) Lumenal view of positions 9-11 of the γ-TuRC with the GCP5 C-domain in the extended (C) and compacted (D) conformation. Downward displacement in γ-tubulin at position 10 is indicated (arrow). E) and F) Compaction of the GCP5 C-domain at position 10 results in a displacement of the γ-TuG4/6 subcomplex away from the conical axis relative to the “elongated” GCP5 subclass, represented by the broken arc with arrows showing displacement directions (E). The direction and extent of the resulting individual γ-tubulin displacement is illustrated by vector plot and average translation of all the atoms (F). Maps were low-pass filtered to 8 Å (C-D) and 13 Å (E-F). G) Schematic of the human GCP6 sequence highlighting regions modeled in this study (red rectangles). The ~800 residue insertion sequence is indicated, with a highly α-helical region containing coiled coils (see Methods) shown as a white rectangle. H) Fit of GCP6 residues K1615-K1641 (indicated; red stick representation) in a density map for the GCP6 C-domain (mesh). I) Left: Schematic of the γ-TuRC highlighting the location of GCP6 and establishing the viewing angle. Right: γ-TuRC model (surface representation) with GCP6 highlighted (red) overlayed with a segmented, 13 Å low-pass filtered density map of the “plug” density (light red surface representation, indicated). In A) and G), some short (<~25-50 residue) connecting loops not currently modeled are omitted for clarity.
Figure S5. GCP2, GCP3, “staple”, and γ-TuSC-associated helix features, related to Figure 4. A) Schematic of the human GCP2 sequence highlighting regions modeled in this study (purple rectangles). B) Schematic of the human GCP3 sequence highlighting regions modeled in this study (pink rectangles). C) Panels showing the fit of residues A319-Q344 (purple stick representation) from GCP2 models at positions 1, 3, and 5, as well as residues Q322-H343 (pink stick representation) from GCP3 models at positions 2, 4, and 6, within the density map (mesh). D) Cartoon (top) and surface (bottom) representations of the staple domain bound to N-domains of GCP2 and GCP3. The white portion of the surface represents a currently unassigned region possibly connecting the staple to the N-terminus (a.a. 232) of GCP2. E) Alignment of GCP2 and GCP3 models to one another focusing on the C-domain hairpin. The ~10 Å extension of the GCP3 vs. the GCP2 hairpin is indicated. F) GCP2 (purple surface) and GCP3 (pink surface) models rotated along their long axes (indicated) highlighting the inter-γ-TuSC interface (blue surface). An asterisk indicates a small interface formed between C-domains that is missing in the intra-γ-TuSC interface (Figure 4H). G) Secondary structure prediction of the γ-TuNA peptide generated using Jpred (Drozdetskiy et al., 2015). α-helices (grey cylinders), β-sheets (grey arrow), and residues predicted to form a coiled coil (red letters) are indicated. H) A long unassigned α-helix density (grey surface representation) spans along the N-domains of γ-TuSCs at positions 3 to 6 (surface representation colored according to Figure 1).
Figure S6. Identification and organization of a terminal γ-TuSC at the “overlap” region and details of the lumenal bridge, related to Figure 5. A) Schematic of the γ-TuRC highlighting the locations of GCP6 (red) adjacent to a GCP2 subunit at position 13 (purple) that is associated with a γ-TuNA (light blue). B) Density map for positions 11-13 used to model GCP2 (position 13, purple surface) with a staple (grey surface) viewed from the angle indicated in A). C) Refined model of GCP2 C-domain residues M725 - D764 (stick representation) in the corresponding α-helical density (mesh) at position 13. D) Schematic of the γ-TuRC highlighting the location of GCP2 (purple) and a staple domain (grey) adjacent to a tentative GCP3 subunit (grey) at position 14. E) Rigid-body fit of the GCP3 model (grey cartoon representation) into the corresponding density (grey transparent surface) at position 14. F) View of GCP2 (purple surface), GCP3 (grey surface), and a staple (grey surface) density constituting the tentative terminal γ-TuSC at position 13-14. Indicated is an extra density associated with GCP3 (dashed black circle around grey surface representation; see also Figure 4A) that is situated above the γ-tubulin in position 1 (the “overlap”). Views in E - F) are from the angle specified in D). G) A poly-alanine model (cartoon representation) built into the α-helical bundle densities comprising domain (ii) of the γ-TuRC lumenal bridge. An interacting helix that contacts the actin-like protein is indicated. H) α-helical bundles of domain (ii) (grey cartoon representation) interacting with a model for the actin-like protein in domain (i) (grey surface representation) in the lumenal bridge. An interacting α-helix contacts the actin-like protein at the barbed end groove (indicated). I) and J) Secondary structure predictions of MZT1 and MZT2A/B (α-helical regions shown as red bars).
Move S1, related to Figure 1. Surface representation of the native human γ-TuRC model. Subunits are colored according to the legend in Figure 1A.
Table S1, related to Figure 1. Mass spectrometry analysis of native human γ-TuRC “core” components.
Table S2, related to Figure 1. Mass spectrometry analysis of native human γ-TuRC associated factors.
Table S3, related to Figure 1. Cryo-EM data collection statistics.
Table S4, related to Figure 1. Model building and assignments
Table S5, related to Figure 1. γ-TuRC model refinement statistics.
Data Availability Statement
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/): EMD-21054, EMD-21060, EMD-21063, EMD-21067, EMD-21068, EMD-21070, EMD-21073, and EMD-21074. Structural models of the γ-TuRC and one of each assigned subunit has been deposited in the Protein Data Bank (PDB; https://www.rcsb.org/): PDB ID 6V5V, 6V69, 6V6B, 6V6C, and 6V6S.