Abstract
LKB1 is a commonly mutated tumor suppressor in non-small cell lung cancer (NSCLC) that exerts complex effects on signal transduction and transcriptional regulation. To better understand the downstream impact of loss of functional LKB1, we developed a transcriptional fingerprint assay representing this phenotype. This assay was predictive of LKB1 functional loss in cell lines and clinical specimens, even those without detected sequence alterations in the gene. In silico screening of drug sensitivity data identified putative LKB1-selective drug candidates, revealing novel associations not apparent from analysis of LKB1 mutations alone. Among the candidates, MEK inhibitors showed robust association with signature expression in both training and testing datasets independent of RAS/RAF mutations. This susceptibility phenotype is directly altered by RNA interference-mediated LKB1 knockdown or by LKB1 re-expression into mutant cell lines and is readily observed in vivo using a xenograft model. MEK sensitivity is dependent on LKB1-induced changes in AKT and FOXO3 activation, consistent with genomic and proteomic analyses of LKB1-deficient lung adenocarcinomas. Our findings implicate the MEK pathway as a potential therapeutic target for LKB1-deficient cancers and define a practical NanoString biomarker to identify functional LKB1 loss.
Introduction:
Understanding molecular pathways responsible for key phenotypes such as tumor proliferation has allowed the development of targeted therapeutic strategies effective in the treatment of defined subsets of cancers. However, the development of therapies that target mutated tumor suppressors represent challenges, since these mutations lead to loss of function that cannot be easily directly targeted. Elucidating the consequences of tumor suppressor loss on signaling pathway activation or consistent changes in other tumor phenotypes such as immune evasion may inform the design of therapeutic strategies to target tumors with these alterations.
LKB1 is a serine-threonine kinase tumor suppressor that is among the most commonly mutated genes in non-small cell lung cancer (NSCLC), with loss occurring in approximately 30–35% of lung adenocarcinomas (1,2). It exhibits diverse regulatory roles, including control of energy homeostasis, metabolism, proliferation, the mTOR pathway (3–7), and maintenance of cellular polarity (4). LKB1 influences these phenotypes via phosphorylation of downstream effector kinases in the family of adenosine monophosphate activated protein kinase (AMPK). Given the complexity of LKB1-associated phenotypes, many approaches have been used to define pathway dependencies that may be exploited in treating these tumors. Molecular characterizations of human tumors, coupled with statistical approaches have identified dysregulated pathways and phenotypes (2,8–11). Genetically engineered mouse models link LKB1 loss to changes in gene and protein expression (1,12) and drug sensitivity (13,14). In vitro models allow study of cell lines in their basal state or after experimental manipulation of LKB1 or other factors (7,10,14–19). These approaches have identified additional strategies that may be useful for targeting LKB1 loss, including induction of metabolic stress, e.g. by phenformin, and inhibition of HSP90 stress response pathway (7,10,14,19).
We have recently analyzed integrated molecular data from the Cancer Genome Atlas (TCGA) and other sources to identify characteristic phenotypes associated with LKB1 loss in human lung adenocarcinomas (2). Other studies have taken similar approaches (9–11) and together our results demonstrate that LKB1 loss is associated with characteristic changes in gene and protein expression that reflect consistent alterations in intracellular signaling pathways. A transcriptional phenotype associated with LKB1 loss was used to derive a robust 16-gene signature of LKB1 loss that is highly predictive of LKB1 loss in validation sets, correctly identifying 97% of LKB1 mutations in the TCGA cohort. Moreover, expression of this signature identifies a subset of tumors that are wild-type by gene sequencing but demonstrate functional LKB1 loss comparable to the known mutant tumors.
Despite the wealth of knowledge derived from analysis of sophisticated molecular data, it is not straightforward to predict from such analyses resulting pathway dependences and clinical susceptibility to treatment. Therefore, in the current work we utilize studies of drug sensitivity data – the Cancer Cell Line Encyclopedia (CCLE) (15), Genomics of Drug Sensitivity in Cancer (GDSC) (16,17) and Cancer Therapeutics Resource Portal (CTRP) (18) – to empirically identify drug classes that may be effective in treating tumors with LKB1 loss. We then employ a panel of isogenic cell line derivatives in which experimental control of LKB1 activity allows us to study the direct effects of the tumor suppressor on drug sensitivity and pathway activity.
Methods and Materials:
Analysis of molecular data
Gene expression data from Affymetrix U133A microarrays was obtained for a total of 1231 cell lines characterized by the CCLE (15) and the Catalog of Somatic Mutations in Cancer (COSMIC) (20). The 16 genes corresponding to the LKB1-loss signature were used to calculate LKB1 loss score for each cell line, as described previously (2). For cell lines included in both CCLE and Sanger datasets, average of the two scores was used for subsequent analysis (Supplementary Table 1). LKB1, HRAS, NRAS, KRAS, and BRAF mutation status of cell lines were taken from characterization reported in CCLE (15), GDSC (16,17), and COSMIC (20) databases, and additional literature sources for LKB1 (Supplementary Tables 2,3). A cell line was considered to have LKB1-loss if reported as such in any study.
Statistical analysis
In vitro drug sensitivity data were obtained from the CCLE (15), GDSC (16,17) and CTRP (18). General linear models were used to determine association between drug sensitivity and LKB1-loss score for each of these datasets (Supplementary Tables 4.1–4.3). For analyses using binary classification, an LKB1-score cutoff of >0.2 was used to define positive signature expression, as described previously (2). Multivariate linear regression included LKB1 classifier score, HRAS, NRAS, KRAS, and BRAF mutations and MEK sensitivity. Statistical analysis was performed using the R statistical software package.
RNAseq of resected tumors
Sequencing followed standard protocol on RNA from frozen specimens of 48 lung adenocarcinomas resected at Vanderbilt University Medical Center. Denaturation of mRNA, elution, priming, and fragmentation followed TruSeq RNA Prep protocol. Double strand cDNA libraries were generated using 17ul fragmentation product input into first and second strand synthesis. After PolyA selection, samples were run using HiSeq PE 100 to generate 30 million pair end reads on an Illumina HiSeq 2500 instrument (Illumina, San Diego, California). Quality control followed recommendations in (21). Raw data and alignment QC utilized QC3 (22). Expression analysis employed MultiRankSeq (23). Raw data were aligned using TopHat 2 (24) against human HG19 reference genome. Gene expression was normalized to fragments per kilobase of of transcript per million reads (FPKM) by Cufflinks (25). Fifteen genes of the sixteen genes were used to calculate LKB1 loss signature (MUC5AC not reported) as previously described (2). Four missense mutations in LKB1 were detected by RNAseq analysis. Mutations and RNAseq gene expression data for the LKB1 signature genes are given in Supplementary Table 5.1. Tumor samples were collected with informed consent in accord with Institutional Review Board protocol (National Clinical Trial ID: ; Secondary Protocol No: VICCTHO0136).
NanoString nCounter assay
RNA expression of relevant genes was measured with nanoString nCounter system (nanoString Technologies, Seattle, WA). For each sample, 200 nanograms of total RNA was hybridized to a custom probeset according to the manufacturer’s instructions. Two sets of probes were generated, comprising genes in the LKB-loss signature, genes down-regulated in LKB1-deficient lung adenocarcinomas and housekeeping genes. The second set contained additional housekeeping genes and additional genes not directly used in this study. Raw count data were normalized by: (1) Background correction (2) positive control correction and (3) housekeeping gene correction. Genes were log2 transformed and the ‘nanoString’ signature score was generated by taking the average expression of the LKB1-loss signature genes and subtracting the average expression of the under-expressed genes. Complete probe lists and expression data are provided in Supplementary Tables 5.2–5.4.
Cell culture and gene transduction
HCC15, H2122, HCC515, H1395, H2126, H1355, H23, H157, H1993, H1435, HCC193, H520, HCC78, H292, HCC2935, Calu-1, H1299, H2085, Calu-3, and Calu-6 cell lines were generously shared by John Minna and Luc Girard (University of Texas, Southwestern). A549, H460, and H522 were purchased from ATCC (Rockville, MD). Cell lines were authenticated by DNA fingerprinting and tested for mycoplasma. Cells were cultured in RPMI1640 containing 5% FBS, without antibiotics. Empty pBABE viral plasmids, pBABE-LKB1 and pBABE-LKB1-K78I were obtained from Addgene and viruses generated according to standard protocol. After viral transduction, cells were selected under puromycin 1ug/ml for one to two weeks before performing subsequent experiments.
Proliferation and drug sensitivity assays
In vitro proliferation assays were performed in 96-well plates after seeding 1000 cells in each well. Trametinib, MK2206, paclitaxel, temsirolimus, dasatinib and BEZ-235 (Chemitek, Indianapolis, IN) were added after 24 hours. Quantitation of cell growth used Alamar Blue (Invitrogen) colorimetric assay.
Immunoblots
Cell lysates were harvested using RIPA lysis buffer containing phosphatase and protease inhibitors. Lysates were homogenized and run on pre-cast SDS-PAGE gels (BioRad). Nuclear and cytoplasmic extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents from Pierce. pAKT(S473), pAMPK(T172), pFOXO3 (T32), pERK1/2 (T202/Y204), ERK, FOXO3, LKB1, Cleaved PARP, and AKT, HDAC2, and GAPDH antibodies were obtained from Cell Signaling Technology.
RNA interference (RNAi) transfection
RNAi constructs LKB1 (s13579, s13580), FOXO3a (s5260) Silencer® Select small interfering RNA (siRNAs) and Stealth RNAi Negative Control Low GC Duplex no. 3 purchased from Invitrogen, (Carlsbad, CA, USA). Cells were reverse transfected using siRNA (250 pmol) or scramble RNA using Lipofectamine RNAiMAX (5 μl) with manufacturer’s instructions (Invitrogen). 24 hours after transfection cells were seeded onto 96 well plates for drug sensitivity assay or grown for harvesting protein lysates.
Lentivirus and infections
Short hairpin RNA (shRNA) experiments used MISSION pLKO.1 constructs (Sigma-Aldrich, St. Louis, MO) specific for LKB1 (clone NM_000455.x-548s1c1, NM_000455.x-1351s1c1), or scrambled control (SHC002). Polybrene was used for virus transduction into 1 × 106 Calu-1 cells, which were subsequently selected using puromycin (Sigma- Aldrich, St. Louis, MO) for one to two weeks.
Xenograft experiments
Suspensions of Calu-1/control and Calu-1/LKB1 shRNA (5×106 cells) were injected into the flanks of 5-week-old female non-obese diabetic severe combined immunodeficient (NOD-SCID) mice (the Jackson laboratories, Bar Harbor, ME). When tumor volumes reached 50 to 100 mm3, the mice were randomized to receive daily trametinib, 1 mg/kg, or saline injections. Tumor volume was calculated as 1/2 × length (mm) × width (mm)2. Tumor size and mouse body weight were measured twice per week. Mice were sacrificed on day 30 of treatment. The protocol was approved by Institutional Animal Care and Use Committee at The Ohio State University, Ohio (approval no. 2014A00000116), and carried out in accordance with institutional recommendations.
Results:
LKB1 loss is associated with a similar transcriptional phenotype across diverse tumor types.
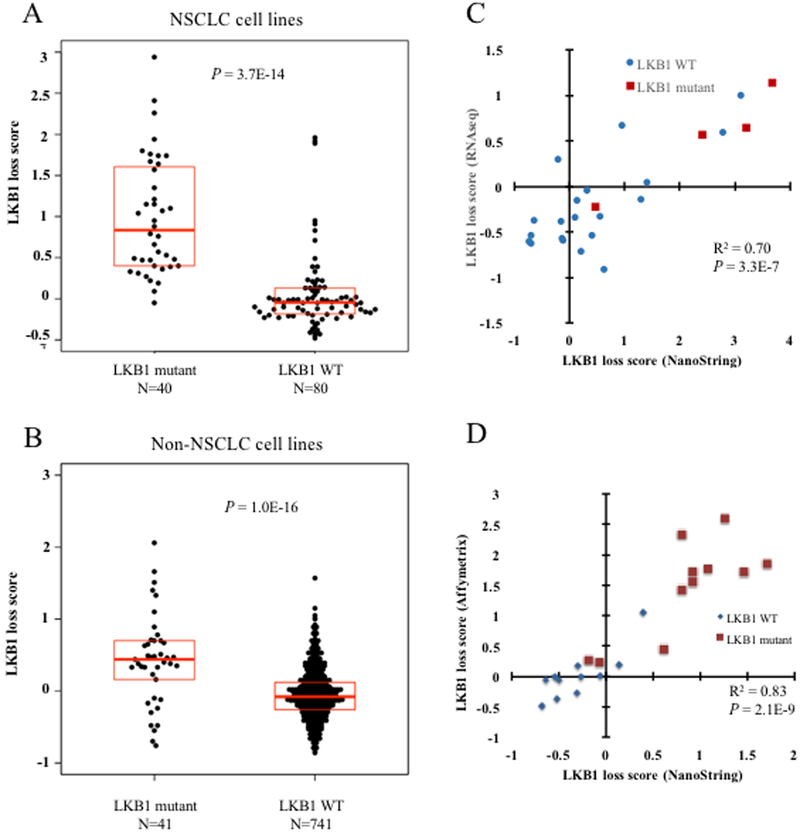
The transcriptional signature of LKB1 loss was derived and validated using resected human lung adenocarcinomas and NSCLC cell lines (2), but transcriptional effects of LKB1 loss have not been studied in other tumor types. Through previously published sequencing studies we have identified 81 cell lines that have both LKB1 loss and gene expression data to allow calculation of functional LKB1-loss signature. All cell lines, mutation phenotypes, and LKB1-loss scores are listed in Supplementary Tables 1–3. As previously demonstrated, LKB1 mutant NSCLC cell lines express higher functional loss scores compared to wild-type lines (P = 3.7E-14; Student’s T-test; Fig. 1A). Moreover, here we show that LKB1 mutant non-NSCLC cell lines across diverse histological types also exhibit increased signature expression compared to wild-type counterparts (P = 1.0E-16; Student’s T-test; Fig. 1B). Using the discriminatory score cutoff of 0.2 reported in our previous analysis, 73% of non-NSCLC LKB1-deficient cell lines were classified as LKB1 loss, compared to 19% of wild-type lines (P = 9.0E-13 by Fisher’s exact test). Among NSCLC cell lines 92% of mutant lines are correctly classified as LKB1 loss, compared to 19% of wild-type lines (P = 2.0E-15 by Fisher’s exact test). The coherence of expression of these genes is higher in NSCLC than among other cell lines, suggesting that some downstream consequences of LKB1 loss may be influenced by additional factors across the histologically diverse set (Supplementary Fig. 1). Nevertheless, the strong statistical significance demonstrates commonalities in the effects of LKB1 loss on gene expression across multiple tumor types.
Figure 1. LKB1 loss signature is expressed by LKB1 mutant cell lines of NSCLC and other histologic subtypes.
A, LKB1-loss signature scores among LKB1 mutant and LKB1 wild-type NSCLC cell lines. B, LKB1-loss signature scores among LKB1 mutant and LKB1 wild-type non-NSCLC cell lines. Box plots represent 25th, 50th, and 75th percentiles. P-values from Student’s T-test. C, Comparison of NanoString LKB1 loss score with score derived from RNA sequencing expression data for 24 resected lung adenocarcinomas. LKB1 mutant tumors are shown as indicated. P-value from linear regression. D, Comparison of NanoString LKB1-loss score with score derived from publicly available microarray data for the 23 cell lines depicted in panel. P-value from linear regression.
Transcriptional phenotype allows determination of LKB1-loss in clinical specimens and cell lines.
The LKB1-loss transcriptional signature may be a useful adjunct to the detection of somatic mutations, as other mechanisms of tumor suppressor loss account for approximately half of the cases of LKB1 loss (2). We developed an assay to quantify this validated signature using a commercial platform (NanoString, Seattle WA) to measure the expression of the 16 genes comprising the signature as well as housekeeping ‘control’ genes. This was applied to a cohort of 24 resected lung adenocarcinomas that was further characterized by RNAseq gene expression analysis. The LKB1-loss score calculated based on RNAseq expression correlated well with that obtained from the NanoString assay (Fig. 1C; R squared = 0.70; P = 3.3E-07 by linear regression). Similarly, across a 23 cell line panel, the NanoString LKB1 signature score correlated highly with that calculated from previously published microarray datasets (Fig. 1D; R-squared = 0.83, P = 2.1E-10 by linear regression).
Discovery analysis identifies MEK inhibition as a drug candidate for LKB1-deficient tumors.
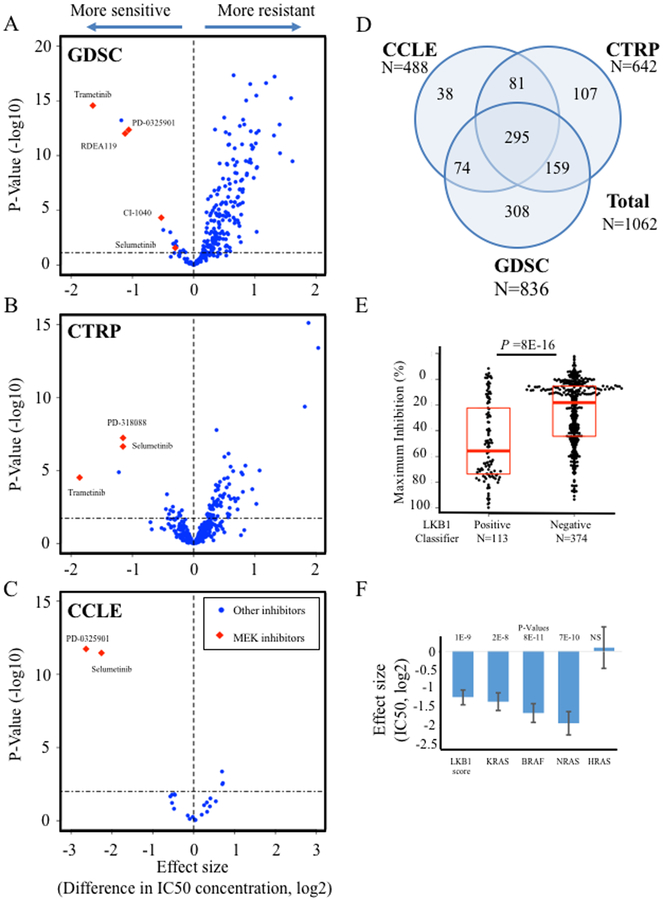
The transcriptional state represented by this signature may reflect differences in signaling pathway activity. Thus, we tested whether expression was associated with susceptibility to targeted pathway inhibitors using data from three large systematic analyses of drug sensitivity phenotypes (15–18). The GDSC comprises response data on 253 inhibitors tested over 987 cell lines (16,17). The CTRP tests 481 inhibitors over 682 cell lines (18). The CCLE tests 24 cell lines over 504 cell lines (15). Using linear models we calculated association between reported IC50 values and LKB1 loss signature across each of these studies (Fig. 2A–C; Supplementary Table 4). A total of 1062 cell lines with both drug sensitivity and gene expression data were used in our analysis (Fig. 2D). Inhibitors of the MEK pathway showed the strongest association to the LKB1 signature in each of the three studies. Every MEK inhibitor tested was significantly associated. The CCLE data included maximum inhibition phenotypes, which was also strongly associated with LKB1 signature expression (Fig. 2E). The top ten associations showing increased sensitivity among cell lines expressing the LKB1-loss signature are highlighted (Table 1). Because MEK inhibitors are being actively investigated in clinical trials and showed consistently strong association across independent studies and multiple compounds, we selected this target for further study.
Figure 2. Drug sensitivity associations with LKB1-loss signature.
A, ‘Volcano plot’ displaying association between LKB1-loss signature and 254 pharmacologic inhibitors from the GDSC study. B, Associations between LKB1-loss signature and 481 inhibitors in the CTRP study. C, Associations between LKB1-loss signature and 24 inhibitors in the CCLE study. For A-C, horizontal axis shows effect size, denoted as fold change in IC50 concentration associated with 1.0 increase in signature expression; vertical axis shows P-values from linear regression. Horizontal dashed lines show cutoff for 10% false discovery rate. D, Number of cell lines from each study that had expression data and were used for our analyses, and overlap between studies. E, Maximum inhibitory effect of selumetinib is shown for cell lines with high expression of the LKB1 classifier (score >0.2) compared to those with low expression in the CCLE dataset. D, Multivariate linear regression coefficients associated with mutations in RAS/RAF family members or the LKB1 signature are shown for trametinib. Values correspond to differences in trametinib IC50 (log2) in the GDSC study. Bars indicate standard error of regression estimates.
Table 1. Inhibitors with increased sensitivity associated with LKB1-loss signature.
Results for top 10 compounds exhibiting increased sensitivity associated with signature expression in the GDSC and CTRP datasets. Effect size represents fold change in IC50 concentration associated with 1.0 increase in signature expression; P-values from linear regression.
| Inhibitor (CTRP) | Reported target | Effect size | P-value |
|---|---|---|---|
| PD318088 | MEK | −1.15 | 5.79E-08 |
| selumetinib | MEK | −1.15 | 2.24E-07 |
| austocystin D | −1.22 | 1.30E-05 | |
| trametinib | MEK | −1.86 | 3.01E-05 |
| MI-2 | MEN1 | −0.44 | 0.00042 |
| VAF-347 | AHR | −0.34 | 0.003 |
| erlotinib | EGFR | −0.45 | 0.0044 |
| GW-405833 | CNR2 | −0.19 | 0.005 |
| GDC-0879 | BRAF | −0.38 | 0.005 |
| fluorouracil | TYMS | −0.34 | 0.006 |
| Inhibitor (GDSC) | Reported target | Effect size | P-value |
| Trametinib | MEK | −1.65 | 2.67E-15 |
| 17-AAG | HSP90 | −1.18 | 5.98E-14 |
| PD-0325901 | MEK | −1.12 | 9.61E-13 |
| RDEA119 | MEK | −1.09 | 8.98E-12 |
| CI-1040 | MEK | −0.53 | 4.71E-05 |
| BMS-754807 | IGF1R | −0.5 | 0.00062 |
| rTRAIL | TR10A/B | −0.38 | 0.0010 |
| FH535 | unknown | −0.28 | 0.0068 |
| OSI-906 | IGF1R | −0.32 | 0.011 |
| Afatinib | EGFR | −0.34 | 0.011 |
MEK sensitivity associated with LKB1 loss signature is independent of mutations in the RAS/RAF pathway.
To account for potential confounding variables, we next studied MEK sensitivity IC50 data from each study cohort using a general linear model with the LKB1-loss signature and mutations in KRAS, NRAS, HRAS, and BRAF. The association between trametinib IC50 concentration and the LKB1-loss signature was independent of these mutations in the GDSC study, and the effect size similar to that seen for mutations in the RAS/RAF pathway (Fig. 2F; P = 1E-9). Similar independence was seen for selumetinib in the CCLE and for PD-318088 in CTRP (data not shown). The association between selumetinib sensitivity and LKB1-loss expression is significant among KRAS mutant, BRAF mutant and RAS/RAF wild-type cell lines (Supplemental Fig. 2A–C). LKB1 mutant cell lines with signature expression greater than classifier cutoff score of 0.2 were significantly more sensitive to selumetinib (Supplemental Fig. 2D).
NSCLC cell lines with LKB1 loss are more sensitive to MEK inhibition and display attenuated AKT/FOXO3 activation.
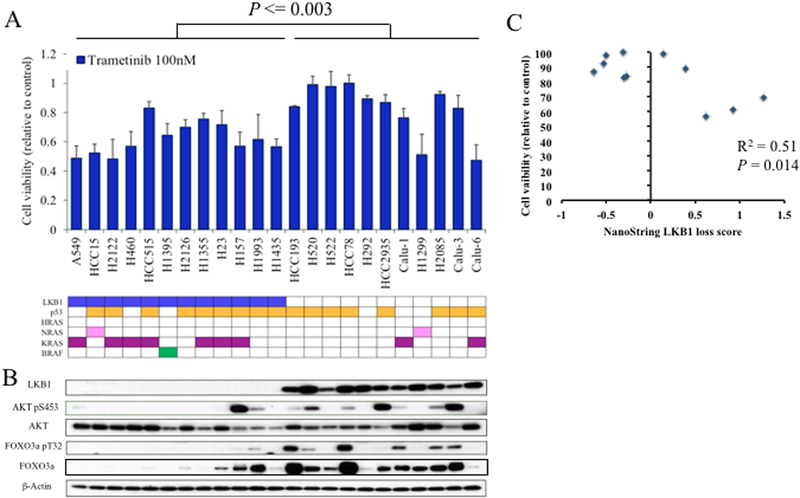
We measured in vitro trametinib sensitivity and AKT/FOXO3 protein expression across twenty-three NSCLC cell lines, including 12 LKB1 mutants. LKB1-deficient cell lines were significantly more sensitive to trametinib (Fig. 3A; P = 0.003 by Student’s T-test). LKB1 deficient lines expressed decreased phosphorylated FOXO3 and a trend toward decreased phosphorylated AKT (Fig. 3B; P = 0.027 for pFOXO3, P = 0.083 for pAKT, Wilcoxon rank sum test of densitometric measurements). Trametinib sensitivity was associated with RAS/RAF mutations (P = 0.00097; Student’s t-test), and pFOXO3 (P = 0.014; linear regression), with a non-significant trend observed for pAKT (P = 0.08; linear regression). Furthermore, the LKB1 loss signature was significantly associated with trametinib sensitivity among RAS/RAF wild-type cell lines (P = 0.014; linear regression; Fig. 3C). These results support the findings from the discovery analysis that LKB1 loss is associated with trametinib sensitivity independent of previously identified drivers of susceptibility.
Figure 3. LKB1 loss in NSCLC is associated with trametinib sensitivity and decreased AKT/FOXO3 phosphorylation.
A, Alamar Blue colorometric assay was measured after 72 hours of treatment and trametinib (100nM) treated cells reported relative to vehicle treated control for 23 NSCLC cell lines. Error bars represent standard deviation of three measurements. Mutations status of LKB1, p53, HRAS, NRAS, KRAS, and BRAF are shown. P-value from student’s T-test. B, Results from western blots with the noted antibodies are shown for respective cell lines. C, Cell viability after treatment with trametinib 100nM, relative to vehicle treated control after 72 hours, plotted against NanoString LKB1-loss score among RAS/RAF wild-type NSCLC cell lines. P-value from linear regression.
LKB1 induces resistance to MEK inhibition in LKB1 mutant cell lines.
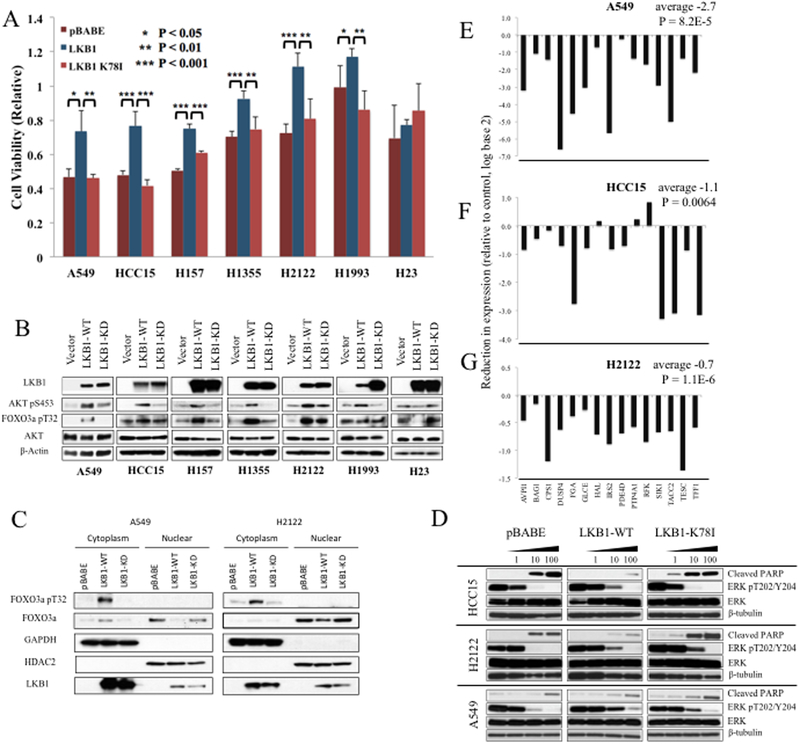
To test direct effects of LKB1 on MEK sensitivity we stably expressed wild-type LKB1 in mutant cell lines. Seven LKB1 mutant cell lines – A549, HCC15, H157, H1355, H2122, H1993, and H23 – were stably selected for expression of wild-type LKB1, kinase deficient K78I mutant LKB1, or empty vector using pBABE retrovirus and puromycin selection. Growth assays in the presence of trametinib (100nM) demonstrated LKB1-mediated resistance in six of the seven cell lines tested (Fig. 4A). A549 and H2122 were similarly tested with temsirolimus, BEZ-235, MK2206, dasatinib, and paclitaxel, which showed no resistance (Supplementary Fig. 3).
Figure 4. Wild-type LKB1 induces MEK resistance and alterations in AKT and FOXO3 signaling.
A, Alamar Blue colorometric assay was measured for indicated cell lines after 72 hours of treatment with 100nM trametinib. Mean values are shown relative to vehicle treated cells; error bars represent standard deviation. P-values from Student’s t-test. B, Immunoblots shown for indicated antibodies and cell lines, stably transfected with wild-type LKB1 or either vector or LKB1 K78I control. C, Nuclear and cytoplasmic fractions were purified from derivatives of A549 and H2122 cell lines using NE-PER, and immunoblots performed using the indicated antibodies. GAPDH and HDAC2 were used as cytoplasmic and nuclear markers. D, Immunoblots from HCC15, H2122, and A549 stably transduced with the indicated constructs are shown after inhibition with trametinib for 48 hours at the indicated concentrations. E-G, Fold reduction after expression of wild-type LKB1 relative to vector control, for fifteen target genes of LKB1-loss signature, assessed by NanoString assay for A549, HCC15, and H2122. Average (log2 transformed) reduction in LKB1-loss expression is shown across the genes shown. P-value from paired Student’s t-test comparing gene expression in vector vs wild-type LKB1 cells across the 15 genes.
LKB1 also affected the expression of MEK resistance pathways. Wild-type LKB1 induced pAKT in six of the seven cell lines, and increase in pFOXO3 in five of seven (Fig. 4B). No change in AKT/FOXO3 phosphorylation was seen in H23, which also had no change in MEK sensitivity. Nuclear cytoplasmic fractionation of A549 and H2122 lysates demonstrated FOXO3 cellular localization. Phosphorylated FOXO3 was detected only in the cytoplasm, as expected. LKB1 resulted in reduction in nuclear FOXO3 consistent with phosphorylation-dependent shift in localization (Fig. 4C).
Cleaved PARP and phosphorylated ERK were measured from cell lysates from A549, HCC15, and H2122 after 48 hours of trametinib treatment (Fig. 4D). Cleaved PARP was reduced in both the HCC15 and H2122 cell lines, though not in A549. Reduction in phosphorylated ERK was similar to controls. The effects of restoring LKB1 on gene expression signature were studied in A549, HCC15, and H2122 using the NanoString assay, demonstrating significant reduction in the expression of this signature in each cell line (Fig. 4E, 4F, and 4G).
Loss of LKB1 induces MEK sensitivity in wild-type cell lines through alterations in FOXO3 signaling.
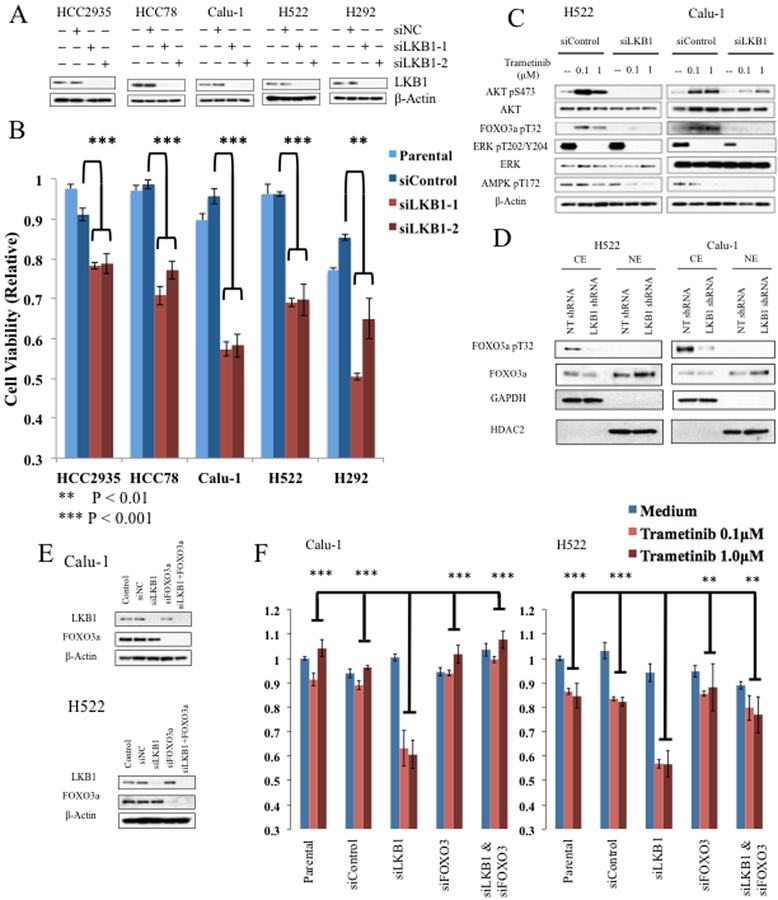
RNA interference was used to inhibit LKB1 expression in wild-type cell lines. HCC2935, HCC78, Calu-1, H522, and H292 were transfected with siRNA targeting LKB1 or control constructs (Fig. 5A). Resulting cells were then treated with trametinib (100nM) for 48 hours. In each of the five cell lines tested, trametinib sensitivity was induced by LKB1-knockdown relative to controls (Fig. 5B).
Figure 5. LKB1 knockdown induces MEK sensitivity mediated by FOXO3 activation.
A, LKB1 wild-type cell lines, HCC2935, HCC78, Calu-1, H522, and H292 were transfected with control siRNA (siNC) or two siRNA constructs targeting LKB1. Lysates from parental cells and each of the three siRNA transfections were collected 48 hours after transfection, and immunoblots performed with the indicated antibodies. B, Alamar Blue colormetric assay was measured after 48 hours of treatment with trametinib at indicated concentrations and shown relative to vehicle treated parental cell lines for the same cell line derivatives. Error bars represent standard deviation. P-values from Student’s t-test. C, H522 or Calu-1 cell lines were transfected with LKB1 targeted siRNA or control. Western blots are shown for indicated antibodies after treatment with trametinib at indicated concentrations for 48 hours. D, Nuclear and cytoplasmic fractions were purified from siRNA derivatives of Calu-1 and H522 cell lines using NE-PER, and immunoblots were performed on resulting lysates using the indicated antibodies. GAPDH and HDAC2 were used as cytoplasmic and nuclear markers. E, H522 or Calu-1 cell lines were transfected as indicated with control siRNA or siRNA targeting LKB1 or FOXO3a. Cell lysates were subjected to western blot with the indicated antibodies. F, Alamar Blue colormetric assay was measured for indicated cell lines after 48 hours of treatment with trametinib at indicated concentrations. Mean values relative to vehicle treated parental cell lines are depicted; error bars represent standard deviation. P-value from Student’s t-test comparing trametinib treated LKB1 knockdown cells to trametinib treated parental, siNC, siFOXO3, or combined siFOXO3/siLKB1 cells, as indicated.
The effects of LKB1 loss on AKT and FOXO3 activation were tested in H522 and Calu-1 cells. Cells were treated with siRNA as above and protein lysates harvested after treatment with trametinib or vehicle for 48 hours. Both control and LKB1 knockdown cell lines showed no ERK phosphorylation after trametinib. However, trametinib induced AKT and FOXO3 phosphorylation in control cells that was reduced or absent in the LKB1 knockdown cells (Fig. 5C). Fractionation of nuclear and cytoplasmic components showed decreased phosphorylation to be associated with increased FOXO3 nuclear localization (Fig. 5D).
This experiment was then performed in cells treated with FOXO3 targeting siRNA or control. In both Calu-1 and H522, sensitivity to trametinib induced by LKB1 knockdown was reversed in the double FOXO3/LKB1 knockdowns, consistent with a direct mechanistic role for this pro-apoptotic transcription factor (Fig. 5E and 5F).
Loss of LKB1 confers sensitivity to single agent MEK inhibition in vivo.
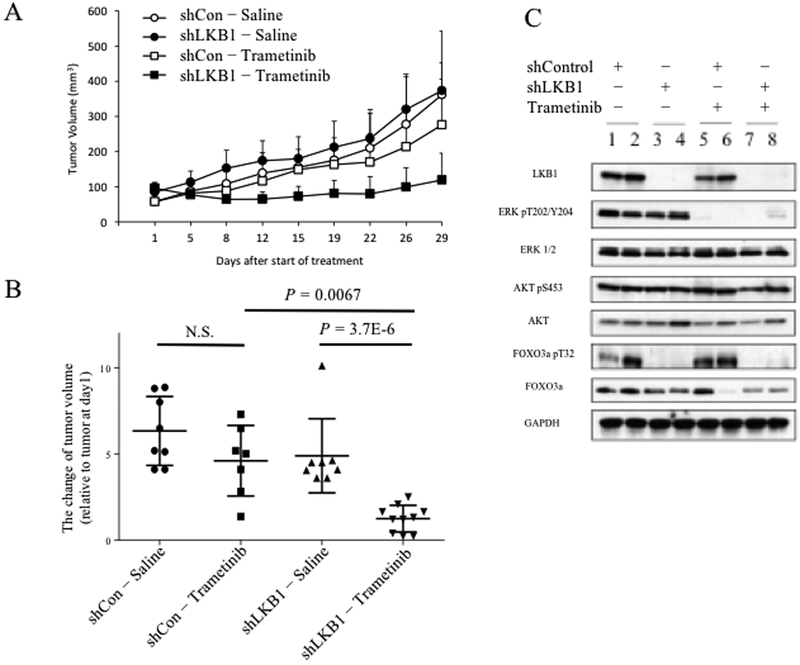
Calu-1 was used to generate derivative lines with stable shRNA-mediated knockdown of LKB1 or non-targeted control. Knockdown of LKB1 induced MEK sensitivity (Supplementary Fig. 4). These derivatives were injected subcutaneously to form orthotopic xenografts in the flanks of female NOD-SCID mice. When tumor volumes reached 50 to 100 mm3 mice were treated with trametinib or saline control. With saline treatment, Calu-1 control and shLKB1 tumors grew at comparable rates. However, trametinib significantly inhibited the growth of shLKB1 xenografts (Fig. 6A and 6B; P = 3.7e-6; Student’s t-test), while there was no significant effect on growth of shRNA control xenografts.
Figure 6. LKB1 knockdown confers sensitivity to trametinib in vivo.
A, The flank of each mouse was subcutaneously injected with 5×106 Calu-1 cells stably expressing LKB1 targeting shRNA or control shRNA. Mice were treated with intraperitoneal injections of trametinib at dose of 1 mg/kg or equal volume of normal saline. Tumor volumes were assessed twice a week by caliper and are reported as mean volumes for eight mice per group; error bars represent standard deviation of measurements. B, Volumes after thirty days of treatment are indicated for individual tumors, relative to volume on day one. Mean and standard deviation are indicated. P-values from Student’s t-test. C, Mice were sacrificed thirty days after start of treatment. Protein lysates were harvested from two tumors representing each group, as indicated, and western blots performed using indicated antibodies.
Mice were sacrificed after 30 days of treatment and protein lysates were harvested from resulting tumors. LKB1, ERK, phosphorylated ERK, AKT, phosphorylated AKT, FOXO3, and phosphorylated FOXO3 were measured by western blot (Fig. 6C). Trametinib induced equivalent attenuation of phosphorylated ERK in LKB1 knockdown and control tumors. Phosphorylated AKT showed no difference in LKB1 knockdown cells. However, phosphorylation of FOXO3 was reduced in both untreated and treated cells that lacked LKB1, consistent with our in vitro results.
Discussion:
Identifying therapeutic strategies for subsets of tumors with tumor suppressor loss is a challenging yet pressing need in clinical oncology. This is especially true for LKB1, where loss is seen in approximately 30% of lung adenocarcinomas, which are almost exclusively EGFR wild-type (2) and have been shown to exhibit low expression of immunomodulatory targets such as PDL-1 and PDL-2 (9,10). Our results join previous studies that work to define the therapeutic vulnerabilities of LKB1 deficient and other genetic subsets of tumors.
Here we use a novel approach that takes advantage of the consistent effect of LKB1 loss on transcriptional phenotype to identify robust and reproducible associations with candidate drugs not apparent through analysis of LKB1 mutations alone. Prior to this work the only report of association between LKB1-loss and MEK sensitivity was based on a very small observational study of ten cell lines (26). The drug sensitivity studies from which we perform our analyses reported a modest association between LKB1 mutations and MEK inhibition, that fell below the level of significance after correcting for multiple hypothesis testing. The difference in results could be due in part to incomplete characterization of LKB1 mutations and other mechanisms of LKB1 loss. It may also relate to phenotypic information captured by the gene signature. Although the functional state that drives signature expression has not been defined, it may denote differences in relative LKB1-AMPK pathway activation, with variation seen across LKB1 mutant and wild-type groups that is regulated by multiple pathways. Alternatively, the signature may be directly related to MEK responsiveness. Lending support to this notion, LKB1 mutant cell lines with low signature expression are significantly less sensitive to selumetinib. Given the complexity of the issue, additional work is required to fully address these questions.
The association between our gene signature and LKB1 loss is clearly established in NSCLC, where our previous validation demonstrates that this signature is expressed by 97% of LKB1-mutant lung adenocarcinomas in the TCGA cohort, and identifies many additional LKB1 deficient tumors that are not LKB1 mutated by sequencing. LKB1 is a large gene with difficult-to-sequence regions, and many commercial sequencing platforms do not have complete coding exon coverage; furthermore, intragenic deletions, non-exonic mutations or other regulatory mechanisms can cause LKB1 functional loss. Our signature is a direct assessment of functional loss. We describe an assay to measure these genes using the commercially available NanoString platform to allow us to study variability of this signature in response to drug or genetic perturbations. This assay works on small FFPE samples and thus would be readily applied to clinical samples. A similar NanoString based assay has been recently applied to detection of LKB1 loss in lung cancer specimens and improves detection compared to sequencing efforts alone (27). Ultimately we expect that an approach that combines mutational sequencing with signature analysis will facilitate a better understanding of LKB1 biology, yield improved classification of the LKB1-loss phenotype in patient specimens, and will be useful in performing and interpreting clinical trials of candidate treatments for tumors with LKB1 loss.
As noted above, the statistical association observed between the LKB1-loss signature and MEK sensitivity is open to multiple interpretations. Thus, we tested the hypothesis that LKB1 loss directly affects MEK sensitivity using an isogenic cell line derivative system, and show consistent effects across many cell lines and in an in vivo xenograft model. Such systems have been used in multiple previous studies to explore the effects of LKB1 on drug sensitivity (3,6,7,10,14). Genetically engineered murine models represent an alternative system to test LKB1-associated effects and show the opposite effect with regard to MEK sensitivity, with LKB1/KRAS mutated tumors exhibiting less response to trametinib in comparison to p53/KRAS tumors (13). This difference could be related to the observation that gene expression patterns from murine models do not exhibit the characteristic features observed in human tumors, suggesting underlying differences in signaling and transcriptional programs (2,10). In contrast, LKB1-deficient cell lines exhibit gene expression patterns similar to resected human tumors, and experimental manipulation of LKB1 results in alteration of these expression patterns (2). It will be difficult to determine which model system better reflects patient phenotypes without objective clinical data.
Mechanistically, we link LKB1-deficient MEK sensitivity to decreased AKT activity and increased FOXO3 activation, an association we have described in resected LKB1-deficient lung adenocarcinomas (2). These proteins regulate apoptosis and MEK sensitivity (28,29). However, the mechanism by which LKB1 influences this signaling is less clear. It is possible that mTOR activation resulting from LKB1 loss could result in feedback inhibition of PI3K/AKT signaling (30), similar to effects in tumors with TSC2 loss (31,32). LKB1 interacts with AKT (33), and can facilitate AKT activation and enhance its anti-apoptotic effects (33, 34). LKB1 and AMPK also exhibit cross talk directly with RAS/RAF/MEK signaling pathways (35,36), in which MEK phosphorylates LKB1 leading to LKB1 and AMPK attenuation, whereas AMPK activation potentiates RAS/RAF/MEK signaling (37). Thus, in LKB1 wild-type cells, metformin and MEK inhibition demonstrate synergistic anti-cancer effects (37). These various pathways exhibit complex feedback and autoregulation, and further studies of these signaling networks will inform understanding of drug sensitivity phenotypes and improve treatment, e.g. through the rational design of combinatorial approaches to overcome resistance mechanisms.
Our study furthers such translational aims, specifically highlighting the importance of LKB1 loss as a novel determinant of MEK sensitivity. MEK inhibitors have shown efficacy in the treatment of BRAF mutant melanoma (38) and promising results have been presented in NSCLC (39), including a phase II clinical trial in KRAS mutant advanced stage NSCLC (40). Unfortunately, tumor biopsy samples from these trials are extremely limited or do not exist. Thus, analysis of LKB1 loss and the expression of the associated gene signature in prospective clinical trials of MEK inhibition is warranted to conclude whether these phenotypes predict patient outcome and could therefore be used to guide treatment decisions.
Supplementary Material
Acknowledgements.
We gratefully acknowledge the work of the Vanderbilt High Throughput Sequencing Core Facility and Travis Clark, PhD and Yan Guo, PhD for their assistance in generating RNAseq gene expression data, and the OSUCCC Genomic Shared Resource for work developing nanoString assay. Proof reading and revisions from Dwight Kaufman, MD, PhD are much appreciated.
Financial support: This work was supported by NIH grants P50 CA90949, U10- CA180950, and U01-CA114771.
Footnotes
Conflict of interest statement: The authors declare no conflicts of interest.
References
- 1.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448:807–10. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, Carbone DP. LKB1 loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. J Thoracic Oncol 2014;9:794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A 2004;101:3329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 2004;116:457–66. [DOI] [PubMed] [Google Scholar]
- 5.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature Rev Cancer 2009;9:563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004;6:91–9. [DOI] [PubMed] [Google Scholar]
- 7.Momcilovic M, McMickle R, Abt E, Seki A, Simko SA, Magyar C, et al. Heightening energetic stress selectively targets LKB1-deficient non–small cell lung cancers. Cancer Res 2015;75:4910–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2015;Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS ONE 2012;7:e36530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell 2010;17:547–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012;483:613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 2013;23:143–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A landscape of pharmacogenomic interactions in cancer. Cell 2016; S0092–8674(16) 30746–2 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483:570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov 2015;5:1210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 2013; 155:552–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucl Acids Res 2010;39:D945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform 2014;15:879–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Zhao S, Sheng Q, Ye F, Li J, Lehmann B, et al. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics 2014;103:323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: Multiperspective Approach for RNAseq Differential Expression Analysis and Quality Control. BioMed Res Int 2014:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahoney CL, Choudhury B, Davies H, Edkins S, Greenman C, Haaften GV, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signaling inhibition. Br J Cancer 2009;100:370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Engel BE, Welsh EA, Yoder SJ, Brantley SG, Chen DT, et al. A sensitive NanoString-based assay to score STK11 (LKB1) pathway disruption in lung adenocarcinoma. J Thoracic Oncol 2016;11:838–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopal YNV, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res 2010;70:8736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng J, Fang B, Liao Y, Chresta CM, Smith PD, Roth JA. Apoptosis induction by MEK inhibition in human lung cancer cells is mediated by Bim. PLoS ONE 2010;5:e13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 2011;1:248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 2010;18:472–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Feedback inhibition of AKT signaling limits the growth of tumors lacking Tsc2. Genes Dev 2005;19:1773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez-López N, Varela-Rey M, Fernández-Ramos D, Woodhoo A, Vázquez-Chantada M, Embade N, et al. Activation of LKB1-AKT pathway independent of phosphoinositide 3-kinase plays a critical role in the proliferation of hepatocellular carcinoma from nonalcoholic steatohepatitis. Hepatology 2010;52:1621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong D, Liu X, Khuri FR, Sun SY, Vertino PM, Zhou W. LKB1 is necessary for AKT-mediated phosphorylation of proapoptotic proteins. Cancer Res 2008;68:7270–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esteve-Puig R, Canals F, Colomé N, Merlino G, Recio JÁ. Uncoupling of the LKB1-AMPKα energy sensor pathway by growth factors and oncogenic BRAF. PLoS ONE 2009;4:e4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng B, Jeong JH, Asara JM, Yuan Y-Y, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell 2009;33:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Della Corte CM, Ciaramella V, Di Mauro C, Castellone MD, Papaccio F, Fasano M, et al. Metformin increases antitumor activity of MEK inhibitors through GLI1 downregulation in LKB1 positive human NSCLC cancer cells. Oncotarget 2016;7:4265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stinchcombe TE, Johnson GL. MEK inhibition in non-small cell lung cancer. Lung Cancer 2014;86:121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jänne PA, Shaw AT, Pereira JR, Jeannin G. Selumetinib plus docetaxel for KRAS mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo controlled, phase 2 study. Lancet Oncol 2013;14:38–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.