Abstract
Somatic cell nuclear transfer (SCNT) has shown a wide application in the generation of transgenic animals, protection of endangered animals, and therapeutic cloning. However, the efficiency of SCNT remains very low due to some poorly characterized key factors. Compared with fertilized embryos, somatic donor cells lack some important components of sperm, such as sperm small noncoding RNA (sncRNA) and proteins. Loss of these factors is considered an important reason for the abnormal development of SCNT embryo. This study focused on recent advances of SCNT and the roles of sperm in development. Sperm-derived factors play an important role in nucleus reprogramming and cytoskeleton remodeling during SCNT embryo development. Hence, considering the role of sperm may provide a new strategy for improving cloning efficiency.
Keywords: Cytoskeleton remodeling, Nucleus reprogramming, Sperm, Somatic cell nuclear transfer, Animal cloning
Introduction
Somatic cell nuclear transfer (SCNT) has shown great advantages and application prospects in the generation of transgenic animals, protection of endangered animals, and stem cell therapy [1]. With the rapid development of gene-editing biotechnology, the combination of gene editing and SCNT has produced more than 300 genetically modified animals, including pigs, cattle, sheep, and goats. It has great potential in the genetic engineering of livestock [2]. Also, SCNT is probably one of the ways to save endangered species, but studies on the use of SCNT (both intra- and interspecies SCNT) in wildlife species are scarce [3]. The utmost relevance of SCNT in regenerative medicine remains unquestionable due to the fact that it is not only free of somatic epigenetic memory but also more similar to conventional embryonic stem cells (ESCs) derived from in vitro fertilized embryos in their transcriptomic and epigenomic signatures [4]. In SCNT, a somatic cell nucleus from the patient is injected into an enucleated oocyte, and the resulting ESCs are then isolated from cloned blastocysts. These ESCs are capable of differentiating into all cell types, which is a potential approach to treat Parkinson’s disease, lateral amyotrophic sclerosis, and other disorders [5]. However, the use of this technology in clinical treatment may take a long time due to its low efficiency, unknown mechanisms, and other ethical issues.
Since the birth of the first cloning sheep “Dolly” in 1996, various species have been successfully cloned [6–23] (Table 1). Although SCNT has shown great progress over the past two decades, the breakthrough is primarily in different species, and the efficiency is still very low [24]. Common concerns include inferior developmental competence of SCNT embryos, low pregnancy rate, fetal placental abnormalities, large offspring syndrome, immunodeficiency, and early death in the pups [25]. Still many aspects of SCNT remain unknown, and more in-depth studies are urgently needed. Currently, the reasons for the low efficiency of SCNT are roughly classified into the following two aspects: (1) Operations, such as enucleation, electrofusion, and embryo culture, can cause endoplasmic reticulum stress and damage. Changes in the osmotic pressure and accumulation of toxic metabolites in the culture medium can lead to SCNT embryo apoptosis [26]. (2) SCNT embryos manifest abnormal epigenetic and expression patterns, which are currently considered to be the main barrier for reprogramming [24].
Table 1.
Summary of the cloned mammalian species
| Species | Published year | Donor cell type | Born number | Transferred embryos number | Reference |
|---|---|---|---|---|---|
| Sheep | 1996 | Epithelium | 1 | 29 | [6] |
| Fibroblast | 2 | 34 | |||
| Bovine | 1998 | Fetal fibroblast | 3 | 28 | [7] |
| Mouse | 1998 | Cumulus cell | 31 | 1385 | [8] |
| Sertoli cell | 1 | 59 | |||
| Neuron | 1 | 46 | |||
| Goats | 1999 | Fetal fibroblast | 1 | 47 | [9] |
| Pig | 2000 | Granulosa cell | 5 | 72 | [10] |
| Mouflon | 2001 | Granulosa cell | 1 | 7 | [11] |
| Rabbit | 2002 | Cumulus cell | 6 | 371 | [12] |
| Cat | 2002 | Fibroblast | 1 | 81 | [13] |
| Cumulus cell | |||||
| Horse | 2003 | Fibroblast | 1 | 17 | [14] |
| Mule | 2003 | Fibroblast | 3 | 305 | [15] |
| Rat | 2003 | Fibroblast | 3 | 129 | [16] |
| Dog | 2005 | Fibroblast | 2 | 1095 | [17] |
| Ferrets | 2006 | Fibroblast | 1 | 104 | [18] |
| Cumulus cell | 3 | 193 | |||
| Wolf | 2007 | Fibroblast | 2 | 251 | [20] |
| Buffalos | 2007 | Granulosa cell | 3 | 42 | [21] |
| Fibroblast | |||||
| Red deer | 2007 | Periosteum | 8 | 84 | [19] |
| Bone cell | |||||
| Fat cell | |||||
| Camel | 2010 | Cumulus cells | 1 | 139 | [22] |
| Fibroblast | |||||
| Monkey | 2018 | Fibroblast | 2 | 79 | [23] |
Abnormal nuclear reprogramming of SCNT embryos
The DNA sequence of most cells in an individual is exactly the same, but the gene expression patterns in different cell types vary widely. This disparity, which is also regulated by epigenetics, leads to diverse morphology and functions. Histone modification, DNA methylation, genomic imprinting, and noncoding RNA regulation are the main forms of epigenetics. The original epigenetic modifications in donor cell genome are incompletely erased and abnormally reprogramed by the enucleated oocyte cytoplasm.
Histone modification
In fertilized embryos, protamine-encapsulated DNA in sperm chromatins are exchanged by maternal histones, while donor cell histones are rapidly replaced by maternal histones in SCNT embryos [27]. Histone modification plays an important role in regulating gene expression of the reprogramming process. The level of histone acetylation in SCNT embryos is low, and histone deacetylase inhibitors (HDACis), such as trichostatin A (TSA), Scriptaid, and Oxamflatin, are used to increase the level of histone acetylation, significantly improving the SCNT efficiency in mice, bovine, pigs, monkeys, and other mammals [23]. Similar to the adjustment of histone acetylation level, an abnormally high level of H3K9me3 modifications in the heterochromatin region of SCNT embryos can be adjusted by the overexpression of demethylase (KDM4A or KDM4D) or downregulation of methyltransferase (Suv39h1 and Suv39h2) [28]. H3K27me3 is also considered an epigenetic barrier for reprogramming because of its association with the loss of imprinting. The amelioration of abnormal H3K27me3 and H3K4me3 levels in SCNT embryos is another mechanism to improve SCNT efficiency [29].
DNA methylation
Methylation at the 5-position of cytosine (5mC) is an important form of epigenetic modification during mammalian embryo development, which is established and maintained by DNA methyltransferase (Dnmts) and demethylated ten-eleven translocation (TET) protein. DNA methylation of the donor cell genome is a relatively stable marker, and the level of 5mC is significantly higher in SCNT embryos than in fertilized embryos during preimplantation stage [28]. The unusual DNA methylation pattern mostly exists on the satellite sequence, long terminal repeat sequence, long dispersed elements (LINEs), and imprinted control region (ICR). Trophoblast cells in SCNT blastocysts are hypermethylated in intergenic differentially methylated regions, which are associated with subsequent abnormal placental development [30]. Treatment with a methyltransferase inhibitor and interference of Dnmts have been used to reduce the higher DNA methylation level, and this strategy effectively improves the SCNT efficiency [28].
Imprinting
Maternal and paternal genomes contain indispensable components for both embryonic development, and they also play complementary roles in development. Within imprinted genes, only one working copy exists because either maternal copy or paternal copy is silenced. In mammals, most of the epigenetic tags are reprogramed after fertilization, while only imprinted genes keep their epigenetic tags [31]. Improper imprinting can result in two active copies or two inactive copies, leading to severe developmental abnormalities, cancer, and other problems [32]. During the process of injecting a donor nucleus from a nonreproductive cell into an enucleated oocyte in SCNT, imprinting of sperm and oocyte does not exist. Loss of H3K27me3 imprinting in SCNT embryos disrupts post-implantation development [33]. A large number of studies provided the evidence of the abnormal expression of imprinted genes in SCNT animal tissues with abnormalities. Compared with normal reproductive cattle, H19 was highly expressed in the bladder, brain, heart, and lung of dead cloned cattle. In the meantime, IGF2 was highly expressed in the heart, kidney, lung, and spleen, and IGF2R was highly expressed in the bladder and brain of dead cloned cattle [34]. The abnormal expression of imprinted genes was also found in pig, mouse, and other cloned animals [35, 36]. Ameliorating the abnormal imprinting pattern of SCNT embryo might be a promising strategy to improve the SCNT efficiency.
X-chromosome inactivation
During mammalian development, X chromosome inactivation (XCI) is observed in the preimplantation stage of embryos and is inherited to the placental lineage to balance the gene expression on the X chromosome. XCI is triggered by Xist, which is both an imprinted gene and a noncoding RNA [37]. Xist expression was abnormally high in male or female SCNT-derived mouse embryos, leading to the inactivation of almost all X chromosomes [38]. Deletion or downregulation of Xist resulted in a tenfold increase in the birth rates of cloned offspring in mice [38]. Also, other studies have corroborated that the rectification of Xist in preimplantation SCNT embryos has long-term effects on their development and postnatal growth.
Abnormal cytoskeleton remodeling of SCNT embryo
Significant differences in cytoskeletal organization and post-transcriptional modification of cytoskeleton were observed in SCNT and IVF embryos. The abnormal cytoskeleton remodeling was considered to be responsible for the abnormal development of SCNT embryos, in particular abnormal pronuclear structures, faster development kinetics, and higher ratio of blastomere fragments [39].
Cytoskeleton remodeling
The cytoskeleton is composed mainly of microtubules, intermediate filaments (IFs), and microfilaments. The functions of cytoskeleton include regulating intracellular material transport, organelle positioning, maintaining cell shape, regulating cell movement and cell division, and maintaining genomic stability. Vimentin is a type III intermediate filament expressed in fibroblasts, leukocytes, and blood vessel endothelial cells. Studies have found that vimentin persists around the nucleus and damages the DNA of SCNT embryos [40]. Studies on rhesus monkeys have shown that the nuclear mitotic protein (NUMA) cannot recombine in SCNT embryos, causing aneuploidy [41]. The spindle–chromosome complex (SCC) in SCNT embryos showed a higher abnormality rate [42]. Removal of the oocyte SCC can cause an irreversible loss or damage of some cytoplasmic proteins, leading to mitotic abnormalities and aneuploidy in early SCNT embryos.
After a donor cell nucleus is injected into the enucleated oocyte, the M phase–promoting factor (MPF) in oocyte triggers nuclear envelope breakdown (NEBD) and premature chromosome condensation (PCC). In normal fertilized eggs, the sperm-derived protein PLCZ1 activates zygotes by Ca2+ oscillation and decomposition of MPF. Due to the lack of activating factors in somatic cells, artificial activation is needed in SCNT embryos to initiate development. In fertilized eggs, the two nuclei from sperm and oocytes are known as male pronucleus and female pronucleus, while the pronucleus in the SCNT embryo is called pseudo-pronucleus. Abnormality can often be observed in the formation of NEBD, PCC, and pseudo-nucleus of SCNT embryos during the first cell cycle [43]. Abnormal centrosomes and failed “pronuclear” migration are related to errors in spindle morphology, chromosome alignment, and cytokinesis in SCNT embryos, which can lead to chromosome instability, disorder of transcriptional regulation and DNA replication, and DNA damage [44]. The complete deletion of sperm regulators in SCNT embryos may be an important reason for the abnormal PCC and pronucleus formation [39].
Post-translational modification (PTM) of cytoskeleton
There are many forms of PTM, such as phosphorylation, glycosylation, ubiquitination, tyrosinylation, glutamination, glycosylation, acetylation, and SUMOylation. These modifications enable cytoskeletal proteins to perform multiple functions in different organelles and physiological states. Acetylation of lysine at the end of α-tubulin is crucial to cytoskeletal stability [45]. Tyrosinization of microtubules regulates the separation of chromosomes in the late stage of cell division via monitoring the activity of depolymerization kinesin [46]. Polyglutamate modification can induce microtubule shear, regulate the length of mitotic spindles, and control the timely division of cytoplasm. Phosphorylation occurs most frequently on serine, threonine, and tyrosine residues of the cytoskeleton. It participates in various cell cycle pathways [47]. Glycosylation of cytoskeleton involves protein folding, distribution, stability, and activity [48]. SUMOylation of cytoskeleton is involved in cell cycle regulation and DNA damage responses.
PTM of cytoskeleton plays a vital role in fertilization, symmetric and asymmetric cell divisions, morula and blastocyst formation, stem cell maintenance, and differentiation. Particularly in the period before the zygotic genome activation (ZGA) stage, the embryonic genes are silenced, and PTM-mediated cytoskeletal remodeling dramatically changes, which is a rapid and efficient way to regulate embryonic developmental events [49]. PTM mediation by sperm was thought to be critically important for cytoskeletal remodeling, but the knowledge of these events is still sparse. α-Tubulin in microtubules of mouse embryos is acetylated in a specific spatial and temporal sequence during the preimplantation of embryo development [50]. Tyrosinated α-tubulin redistributes toward the apex of cells during the 8-cell and 16-cell stages, while acetylated α-tubulin accumulates near the intercellular contact zone in the basal part of the cell [49]. During the morula stage, acetylated α-tubulin is detected only in a subpopulation located predominantly at the cell cortices [49]. During the blastocyst stage, the cells inside the developing embryos contain more acetylated microtubules compared with the outside cells [49]. The drastic PTM-mediated cytoskeletal remodeling undoubtedly plays an important role during embryo development. A recent study found a different α-tubulin acetylation pattern between SCNT and fertilized embryos [39].
Roles of sperm in reprogramming
During the process of fertilization, the paternal and maternal genomes undergo genome-wide epigenetic modifications or reprogramming and transform into an early embryonic genome model with developmental pluripotency, thus starting a new life course. This process is the period during which the epigenetic modification of the genome is the most frequent and the development events are most concentrated. It is traditionally believed that oocyte modulators are the main factors affecting epigenetic reprogramming and embryonic development, while the sperm contributes only to paternal chromosomes and activates fertilized embryos. The latest studies showed that sperm proteins [51], protein modification [52, 53], DNA methylation [54, 55], and sperm small noncoding RNA (sncRNA) [56–58] were all important carriers for regulating epigenetics. These epigenetic factors can escape the reprogramming of early embryos and regulate genes at the transcriptional, post-transcriptional, translational, or post-translational levels, thus contributing to the embryo, and even offspring, development (Fig. 1), [51–58]. It is also worth mentioning that seminal plasma has an important role on offspring development and health [59, 60].
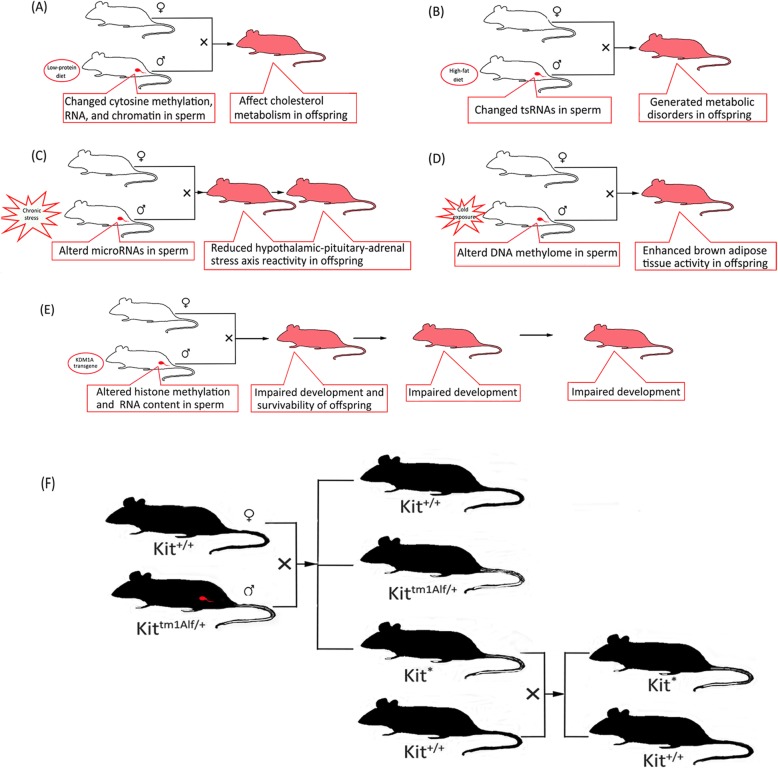
Fig. 1.
Intergenerational and transgenerational epigenetic inheritance via the sperms in mice. a Cytosine methylation, RNA, and chromatin of sperm in male fed a low-protein diet changed, and the offspring exhibited elevated hepatic expression of many genes involved in lipid and cholesterol biosynthesis and decreased levels of cholesterol esters. b Sperm tsRNAs in male fed a high-fat diet exhibited changes in expression profiles and RNA modifications, and this treatment generated metabolic disorders in the F1 offspring. c Chronic paternal stress altered the profiling of sperm microRNAs, which reduced hypothalamic–pituitary–adrenal (HPA) stress axis reactivity in the offspring. d Seasonal or experimental cold exposure induced the epigenetic programming of the sperm such that the offspring harbored hyperactive brown adipose tissue and an improved adaptation to overnutrition and hypothermia. e Disruption of histone methylation in developing sperm by exposure to the KDM1A transgene severely impaired development and survivability of offspring, and the defects occurred in nontransgenic descendants in the absence of KDM1A germline expression. f The genotype of male heterozygous mice Kittm1Alf/+ showed a white tail tip and white feet, and the genotype of mice Kit+/+ showed a full color. The male mice Kittm1Alf/+ were mated with the female mice Kit+/+, and the offspring were of three kinds: offspring (Kit+/+) showed a full color, offspring (Kittm1Alf/+) showed a white tail tip and white feet, and offspring (Kit*) showed a white tail tip and white feet. The genotype of Kit+/+ and Kit* was the same, but the phenotype was different because Kit messenger RNA decreased in the offspring (Kit*). Male mice (Kit*) were mated with female mice (Kit+/+), the offspring still had a white tail tip and white feet, and the degree of this phenomenon reduced
Sperm sncRNA
Sperm contains both coding and noncoding RNAs, including mRNA, miRNA, piRNA, endo-siRNA, and tRNA. Sperm-borne RNA plays an important role in embryonic development following fertilization. Sperm small RNA could regulate the epigenetic state and cytoskeleton remodeling in SCNT embryos (Fig. 2), [39]. Many studies have been performed on individual sample types, such as oocytes, sperm, or donor cells. However, a few studies included all three cell types. In the study by Yang et al., the deep sequencing method was used to measure the expression profiles of sncRNA in sperm, oocytes, and embryos. They used the expression profiles of CD4 + T cell (CD4) as a somatic cell control for illustrating the categories and length distributions of small RNAs [61]. In this review, the expression data of sperm, oocytes, and CD4 in a study by Yang et al. were selected to investigate the potential function of microRNA in sperm. A total of 21 microRNAs were highly expressed in sperm, and 65 microRNAs were only expressed in sperm. The microRNAs only expressed or highly expressed in sperm were compared with conservative microRNA among four species (mice, rats, humans, and rabbits) screened by Yuan et al. [62]. Nine microRNAs (miR-34c-3p, miR-99a-5p, miR-10b-5p, miR-135a-5p, miR-34c-5p, miR-200b-3p, miR-125b-5p, miR-34b-3p, and miR-199a-3p) were found in both sets. Target genes of these nine microRNAs were predicted using microDB (http://mirdb.org/mining.html). Then GO analysis and KEGG pathway analysis were conducted (Table 2). The top enriched biological processes of target genes were related to the regulation of transcription, indicating that sperm microRNAs might have an important role in the regulation of embryonic transcription. The enriched cellular component comprised mainly of cytoplasm, membrane, and nucleus, and the enriched molecular function included protein binding, metal ion binding, DNA binding, nucleotide binding, and transferase activity. These target genes were involved in important signaling pathways related to embryonic development, such as MAPK, cAMP, AMPK, regulation of actin cytoskeleton, and regulating pluripotency of stem cells.
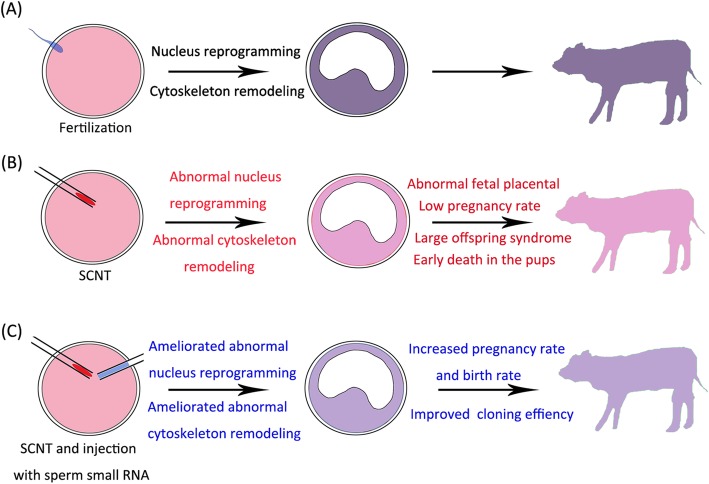
Fig. 2.
Diagram showing the development of fertilized and SCNT embryos; SCNT embryos had sperm small RNA in cattle. a Sperm enters oocytes, fertilizes, and initiates nucleus reprogramming and cytoskeleton remodeling. b Somatic cells were injected into enucleated oocytes, and SCNT embryos manifest abnormal nucleus reprogramming and cytoskeleton remodeling. c Sperm small RNAs were injected into SCNT embryos, and the treatment ameliorated abnormal nucleus reprogramming and cytoskeleton remodeling in SCNT embryos in cattle
Table 2.
The five most-enriched GO categories and KEGG pathways for the target genes of conserved and high sperm miRNAs (P < 0.05, FDR < 0.05)
| ID | Term | Count | P value | FDR |
|---|---|---|---|---|
| Biological process | ||||
| GO:0006351 | Transcription, DNA-templated | 254 | 2.65E−20 | 4.99E−17 |
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 155 | 7.38E−18 | 1.39E−14 |
| GO:0006355 | Regulation of transcription, DNA-templated | 283 | 1.17E−17 | 2.21E−14 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 95 | 1.89E−12 | 3.56E−09 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 111 | 4.19E−12 | 7.88E−09 |
| Cellular component | ||||
| GO:0005737 | Cytoplasm | 668 | 1.71E−22 | 2.56E−19 |
| GO:0016020 | Membrane | 634 | 4.87E−10 | 7.31E−07 |
| GO:0005634 | Nucleus | 588 | 1.92E−15 | 2.83E−12 |
| GO:0005654 | Nucleoplasm | 208 | 4.08E−08 | 6.12E−05 |
| GO:0005829 | Cytosol | 194 | 5.85E−08 | 8.78E−05 |
| Molecular function | ||||
| GO:0005515 | Protein binding | 473 | 8.29E−24 | 1.34E−20 |
| GO:0046872 | Metal ion binding | 353 | 9.70E−11 | 1.57E−07 |
| GO:0003677 | DNA binding | 227 | 5.16E−13 | 8.37E−10 |
| GO:0000166 | Nucleotide binding | 198 | 2.71E−05 | 0.043909 |
| GO:0016740 | Transferase activity | 161 | 4.83E−06 | 0.007827 |
| KEGG pathway | ||||
| mmu04151 | PI3K-Akt signaling pathway | 58 | 3.05E−08 | 3.97E-05 |
| mmu04010 | MAPK signaling pathway | 46 | 4.87E−08 | 6.34E−05 |
| mmu04810 | Regulation of actin cytoskeleton | 38 | 1.74E−06 | 0.002264 |
| mmu04152 | AMPK signaling pathway | 27 | 3.35E−06 | 0.004361 |
| mmu04550 | Signaling pathways regulating pluripotency of stem cells | 27 | 1.40E−05 | 0.018295 |
The expression profiles of microRNA were different between sperms with a high fertilization rate and those with a low fertilization rate. Germline-specific Dicer and Drosha conditional knockout mice produced sperm partially deficient in microRNA, which disrupted maternal transcript turnover and failure in ZGA and caused a significant reduction in developmental potential. The findings in recent years showed that sperm-borne microRNA-449b improved epigenetic reprograming and decreased the apoptosis index of early embryos [63]. Also, microRNA-125b was a key epigenetic regulatory factor that promoted nuclear reprogramming via targeting SUV39H1, which played a vital role in heterochromatin organization, chromosome segregation, and mitotic progression [64]. In another study, deep sequencing was used to analyze small RNA in bovine sperm, and microRNA-125b was found to be highly expressed in bovine sperm [65]. Also, the target genes of sperm microRNAs were associated with placental development, and these imprinted genes were partially responsible for abnormal placenta development and large offspring syndrome [66].
piRNA plays an important role in mammalian spermatogenesis. Impairment of Miwi protein, which binds to piRNA, can lead to a higher exchange barrier between histones and protamines, which severely affect the stability of genomic DNA and chromatin remodeling, leading to male sterility. BTBD18, produced by the meiosis of mouse spermatogenic cells, regulates piRNA via promoting the transcriptional elongation of RNA polymerase II. By silencing BTBD18, the piRNA synthesis and spermatogenesis are disordered, resulting in male mouse sterility. piRNA is also involved in maternal mRNA degradation and plays an important role in regulating protein translation, mediating ubiquitination degradation, germ cell development, sex determination, and embryonic development. The latest study found that H3K9me3 in SCNT embryos significantly reduced, and injecting small RNA into SCNT embryos could significantly enhance cloning efficiency [39]. The molecular mechanism of action of sperm small RNA in regulating SCNT embryo reprogramming is still not clear and needs to be explored in subsequent studies.
Sperm proteins
Up to date, a large number of proteomic studies on sperm or oocytes have been performed. Of the total 6871 proteins found in human mature sperm, 560 have been found to be involved in modulating gene expression, DNA methylation, histone modifications, and noncoding RNA biogenesis, which may be critical for regulation after ZGA and epigenetic transmission of altered phenotypes [51]. It is traditionally considered that zygote is affected mainly by proteins accumulated by oocytes before ZGA, while after ZGA, the embryo is regulated mainly by the embryonic proteins. In fact, sperm enters the oocyte carrying a large amount of proteins, and the half-life of these proteins can vary from 30 min to 8 days, indicating that some sperm proteins may play a role in the development of preimplantation stage embryos [67]. Sperm proteins are necessary for fertilization and embryo development. Sperm protein IZUMO1 guides egg–sperm fusion by binding to oocyte protein JNUO [68]. Sperm protein PLCZ1 induces Ca2+ oscillation, initiates pronuclear formation, and contributes to embryogenesis. Deficiency or abnormal positioning of sperm protein EQTN can lead to abnormal fertilization and cause disorders of gamete fusion [69]. Knockout of some sperm proteins can cause abnormal development at the preimplantation stage, death, or sterility.
Roles of sperm in cytoskeleton remodeling
Recent studies on the regulation of early embryo development focused mainly on epigenetic modification, while a few studies investigated the role of cytoskeleton PTMs. In the last decade, efforts were made to improve the efficiency of SCNT transgenic animals with some achievements. Previous studies found that SCNT embryos showed higher abnormal cytoskeleton remodeling, such as multiple pronuclei formation and abnormal cleavage rate or time. They also demonstrated that sperm microRNAs or proteins derived from sperm were involved in cytoskeletal remodeling. Injecting sperm miR-449b into SCNT embryos slowed down the first cleavage time and improved the development potential of SCNT embryos [63]. Injecting bovine sperm small RNA into SCNT embryos ameliorated the acetylation of α-tubulin K40 and pronuclei formation rate, demonstrating that sperm small RNA played an important role in the cytoskeletal remodeling of SCNT embryos [39]. Proteins, which are rich in sperm, are the most direct regulators, and some proteins are important enzymes for embryonic development. Protamine 1 is an important protein in mature sperm, which binds with high affinity to all DNAs. Upon fertilization, paternal chromosomes rapidly lose protamine 1 and regain a nucleosomal organization built on maternal histones. Establishing somatic donor cells, which exogenously expressed protamine 1, could create spermatid-like chromatin and cytoskeleton structures [70]. The spermatid-like somatic cells were conducive to nuclear reprogramming and beneficial for SCNT [70]. Despite all this, a few studies have examined sperm proteins on SCNT embryos, and attempts are being made to study sperm-derived key proteins associated with SCNT development. Also, an in-depth investigation of cytoskeletal protein modification may be important for understanding embryo development networks and improving SCNT efficiency.
Conclusion and future perspectives
Factors derived from sperm play important roles in nuclear reprogramming and cytoskeletal remodeling during embryo development. Loss of sperm-derived regulators in somatic cells may be an important reason for low cloning efficiency. The roles of sperm factors in epigenetic inheritance and fertilized embryo development increasingly attracted researchers’ attention. A few reports are available on the influence of sperm in SCNT embryos and additional studies on the involvement of sperm factors in embryo development are warranted. Sperm-specific and crucial RNA, proteins, and/or other epigenetic factors should be screened, and a regulatory network of these sperm factors on embryo development should be constructed. Furthermore, the involvement of sperm in nuclear reprogramming and cytoskeletal remodeling in fertilized and SCNT embryos should be elucidated. Considering the roles of sperm factors may improve the cloning efficiency and open a new era for SCNT.
Acknowledgements
We thank Dr. Siyu Liu and Dr. Yue Du for the assistance with language polish.
Abbreviations
- SCNT
Somatic cell nuclear transfer
- sncRNA
Small noncoding RNA
- ESCs
Embryonic stem cells
- HDACis
Histone deacetylase inhibitors
- TSA
Trichostatin A
- KDM4A
Lysine demethylase 4A
- KDM4D
Lysine demethylase 4D
- Suv39h1
Suppressor of variegation 3–9 homolog 1
- Suv39h2
Suppressor of variegation 3–9 homolog 2
- H3K27me3
Histone H3 lysine 27 trimethylation
- H3K4me3
Histone H3 lysine 4 trimethylation
- 5mC
5-Position of cytosine
- Dnmts
DNA methyltransferase
- TET
Ten-eleven translocation
- LINEs
Long dispersed elements
- ICR
Imprinted control region
- H19
H19 imprinted maternally expressed transcript
- IGF2
Insulin-like growth factor 2
- IGF2R
Insulin-like growth factor 2 receptor
- XCI
X chromosome inactivation
- IVF
In vitro fertilization
- IFs
Intermediate filaments
- NUMA
Nuclear mitotic protein
- SCC
Spindle–chromosome complex
- MPF
M phase–promoting factor
- NEBD
Nuclear envelope breakdown
- PCC
Premature chromosome condensation
- PLCZ1
Sperm protein PLCzeta
- PTM
Post-translational modification
- ZGA
Zygotic genome activation
- CD4
CD4 + T cell
- MAPK
Mitogen-activated protein kinase
- cAMP
Cyclic adenosine monophosphate
- AMPK
Adenosine 5′-monophosphate activated protein kinase
- BTBD18
BTB domain containing 18
- IZUMO1
Izumo sperm-egg fusion 1
- JNUO
IZUMO1 receptor
- EQTN
Equatorin
Authors’ contributions
PQ and YW wrote the manuscript. CZ and EL edited the manuscript. All authors read and approved the final manuscript.
Funding
This review was supported by the Natural Science Foundation of Shaanxi Province (Grant No. 2014FWPT07), the Wuzhong Innovation and Entrepreneurship Talent Project of Suzhou (Grant No. WC201526), and the National Natural Science Foundation of China (Grant No. 81270348).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Loi P, Iuso D, Czernik M, Ogura A. A new, dynamic era for somatic cell nuclear transfer? Trends Biotechnol. 2016;34:791–797. doi: 10.1016/j.tibtech.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Tan W, Proudfoot C, Lillico SG, Whitelaw CB. Gene targeting, genome editing: from Dolly to editors. Transgenic Res. 2016;25:273–287. doi: 10.1007/s11248-016-9932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mastromonaco GF, King WA. Cloning in companion animal, non-domestic and endangered species: can the technology become a practical reality? Reprod Fertil Dev. 2007;19:748–761. doi: 10.1071/RD07034. [DOI] [PubMed] [Google Scholar]
- 4.Pan G, Wang T, Yao H, Pei D. Somatic cell reprogramming for regenerative medicine: SCNT vs. iPS cells. BioEssays. 2012;34:472–476. doi: 10.1002/bies.201100174. [DOI] [PubMed] [Google Scholar]
- 5.Zhao MT, Chen H, Liu Q, Shao NY, Sayed N, Wo HT, Zhang JZ, Ong SG, Liu C, Kim Y, et al. Molecular and functional resemblance of differentiated cells derived from isogenic human iPSCs and SCNT-derived ESCs. Proc Natl Acad Sci U S A. 2017;114:E11111–E11120. doi: 10.1073/pnas.1708991114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 7.Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce de Leon FA, Robl JM. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280:1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- 8.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 9.Baguisi A, Behboodi E, Melican DT, Pollock JS, Destrempes MM, Cammuso C, Williams JL, Nims SD, Porter CA, Midura P, et al. Production of goats by somatic cell nuclear transfer. Nat Biotechnol. 1999;17:456–461. doi: 10.1038/8632. [DOI] [PubMed] [Google Scholar]
- 10.Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares DL, et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- 11.Loi P, Ptak G, Barboni B, Fulka J, Jr, Cappai P, Clinton M. Genetic rescue of an endangered mammal by cross-species nuclear transfer using post-mortem somatic cells. Nat Biotechnol. 2001;19:962–964. doi: 10.1038/nbt1001-962. [DOI] [PubMed] [Google Scholar]
- 12.Chesne P, Adenot PG, Viglietta C, Baratte M, Boulanger L, Renard JP. Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat Biotechnol. 2002;20:366–369. doi: 10.1038/nbt0402-366. [DOI] [PubMed] [Google Scholar]
- 13.Shin T, Kraemer D, Pryor J, Liu L, Rugila J, Howe L, Buck S, Murphy K, Lyons L, Westhusin M. A cat cloned by nuclear transplantation. Nature. 2002;415:859. doi: 10.1038/nature723. [DOI] [PubMed] [Google Scholar]
- 14.Galli C, Lagutina I, Crotti G, Colleoni S, Turini P, Ponderato N, Duchi R, Lazzari G. Pregnancy: a cloned horse born to its dam twin. Nature. 2003;424:635. doi: 10.1038/424635a. [DOI] [PubMed] [Google Scholar]
- 15.Woods GL, White KL, Vanderwall DK, Li GP, Aston KI, Bunch TD, Meerdo LN, Pate BJ. A mule cloned from fetal cells by nuclear transfer. Science. 2003;301:1063. doi: 10.1126/science.1086743. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q, Renard JP, Le Friec G, Brochard V, Beaujean N, Cherifi Y, Fraichard A, Cozzi J. Generation of fertile cloned rats by regulating oocyte activation. Science. 2003;302:1179. doi: 10.1126/science.1088313. [DOI] [PubMed] [Google Scholar]
- 17.Lee BC, Kim MK, Jang G, Oh HJ, Yuda F, Kim HJ, Hossein MS, Kim JJ, Kang SK, Schatten G, et al. Dogs cloned from adult somatic cells. Nature. 2005;436:641. doi: 10.1038/436641a. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Sun X, Chen J, Liu X, Wisely SM, Zhou Q, Renard JP, Leno GH, Engelhardt JF. Cloned ferrets produced by somatic cell nuclear transfer. Dev Biol. 2006;293:439–448. doi: 10.1016/j.ydbio.2006.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg DK, Li C, Asher G, Wells DN, Oback B. Red deer cloned from antler stem cells and their differentiated progeny. Biol Reprod. 2007;77:384–394. doi: 10.1095/biolreprod.106.058172. [DOI] [PubMed] [Google Scholar]
- 20.Kim MK, Jang G, Oh HJ, Yuda F, Kim HJ, Hwang WS, Hossein MS, Kim JJ, Shin NS, Kang SK, et al. Endangered wolves cloned from adult somatic cells. Cloning Stem Cells. 2007;9:130–137. doi: 10.1089/clo.2006.0034. [DOI] [PubMed] [Google Scholar]
- 21.Shi D, Lu F, Wei Y, Cui K, Yang S, Wei J, Liu Q. Buffalos (Bubalus bubalis) cloned by nuclear transfer of somatic cells. Biol Reprod. 2007;77:285–291. doi: 10.1095/biolreprod.107.060210. [DOI] [PubMed] [Google Scholar]
- 22.Wani NA, Wernery U, Hassan FA, Wernery R, Skidmore JA. Production of the first cloned camel by somatic cell nuclear transfer. Biol Reprod. 2010;82:373–379. doi: 10.1095/biolreprod.109.081083. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M, et al. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 2018;174:245. doi: 10.1016/j.cell.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 24.Matoba S, Zhang Y. Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell. 2018;23:471–485. doi: 10.1016/j.stem.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keefer CL. Artificial cloning of domestic animals. Proc Natl Acad Sci U S A. 2015;112:8874–8878. doi: 10.1073/pnas.1501718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordova A, King WA, Mastromonaco GF. Choosing a culture medium for SCNT and iSCNT reconstructed embryos: from domestic to wildlife species. J Anim Sci Technol. 2017;59:24. doi: 10.1186/s40781-017-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao S, Chung YG, Parseghian MH, King GJ, Adashi EY, Latham KE. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Gao R, Wang C, Gao Y, Xiu W, Chen J, Kou X, Zhao Y, Liao Y, Bai D, Qiao Z, et al. Inhibition of aberrant DNA re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell. 2018;23:426–435. doi: 10.1016/j.stem.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Xie B, Zhang H, Wei R, Li Q, Weng X, Kong Q, Liu Z. Histone H3 lysine 27 trimethylation acts as an epigenetic barrier in porcine nuclear reprogramming. Reproduction. 2016;151:9–16. doi: 10.1530/REP-15-0338. [DOI] [PubMed] [Google Scholar]
- 30.Hirose M, Hada M, Kamimura S, Matoba S, Honda A, Motomura K, Ogonuki N, Shawki HH, Inoue K, Takahashi S, et al. Aberrant imprinting in mouse trophoblast stem cells established from somatic cell nuclear transfer-derived embryos. Epigenetics. 2018;13:693–703. doi: 10.1080/15592294.2018.1507199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond Ser B Biol Sci. 2013;368:20110336. doi: 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Erice Imprinting G. Genomic Imprinting and physiological processes in mammals. Cell. 2019;176:952–965. doi: 10.1016/j.cell.2019.01.043. [DOI] [PubMed] [Google Scholar]
- 33.Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, et al. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell. 2018;23:343–354. doi: 10.1016/j.stem.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong ZJ, Zhou YY, Xu M, Cai Q, Li H, Yan JB, Wang J, Zhang HJ, Fan SY, Yuan Q, et al. Aberrant expression of imprinted genes and their regulatory network in cloned cattle. Theriogenology. 2012;78:858–866. doi: 10.1016/j.theriogenology.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Han DW, Im YB, Do JT, Gupta MK, Uhm SJ, Kim JH, Scholer HR, Lee HT. Methylation status of putative differentially methylated regions of porcine IGF2 and H19. Mol Reprod Dev. 2008;75:777–784. doi: 10.1002/mrd.20802. [DOI] [PubMed] [Google Scholar]
- 36.Okae H, Matoba S, Nagashima T, Mizutani E, Inoue K, Ogonuki N, Chiba H, Funayama R, Tanaka S, Yaegashi N, et al. RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum Mol Genet. 2014;23:992–1001. doi: 10.1093/hmg/ddt495. [DOI] [PubMed] [Google Scholar]
- 37.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 38.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 39.Qu P, Zuo Z, Liu Z, Niu Z, Zhang Y, Du Y, Ma X, Qiao F, Wang M, Zhang Y, et al. Sperm-borne small RNA regulate alpha-tubulin acetylation and epigenetic modification of early bovine somatic cell nuclear transfer embryos. Mol Hum Reprod. 2019;25:471–82. [DOI] [PubMed]
- 40.Gall L, Brochard V, Ruffini S, Laffont L, Fleurot R, Lavin TA, Jouneau A, Beaujean N. Intermediate filaments promote nuclear mechanical constraints during somatic cell nuclear transfer in the mouse. Cell Reprogram. 2012;14:497–504. doi: 10.1089/cell.2012.0027. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Yang SH, Ding CH, He XC, Xie YH, Hildebrandt TB, Mitalipov SM, Tang XH, Wolf DP, Ji WZ. A comparative approach to somatic cell nuclear transfer in the rhesus monkey. Hum Reprod. 2006;21:2564–2571. doi: 10.1093/humrep/del216. [DOI] [PubMed] [Google Scholar]
- 42.Simerly C, Navara C, Hyun SH, Lee BC, Kang SK, Capuano S, Gosman G, Dominko T, Chong KY, Compton D, et al. Embryogenesis and blastocyst development after somatic cell nuclear transfer in nonhuman primates: overcoming defects caused by meiotic spindle extraction. Dev Biol. 2004;276:237–252. doi: 10.1016/j.ydbio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Mitalipov SM, Zhou Q, Byrne JA, Ji WZ, Norgren RB, Wolf DP. Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. Hum Reprod. 2007;22:2232–2242. doi: 10.1093/humrep/dem136. [DOI] [PubMed] [Google Scholar]
- 44.Katayama M, Zhong Z, Lai L, Sutovsky P, Prather RS, Schatten H. Mitochondrial distribution and microtubule organization in fertilized and cloned porcine embryos: implications for developmental potential. Dev Biol. 2006;299:206–220. doi: 10.1016/j.ydbio.2006.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forer A, Johansen KM, Johansen J. Movement of chromosomes with severed kinetochore microtubules. Protoplasma. 2015;252:775–781. doi: 10.1007/s00709-014-0752-7. [DOI] [PubMed] [Google Scholar]
- 47.Ramkumar A, Jong BY, Ori-McKenney KM. ReMAPping the microtubule landscape: how phosphorylation dictates the activities of microtubule-associated proteins. Dev Dyn. 2018;247:138–155. doi: 10.1002/dvdy.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarbet HJ, Dolat L, Smith TJ, Condon BM, O'Brien ET 3rd, Valdivia RH, Boyce M. Site-specific glycosylation regulates the form and function of the intermediate filament cytoskeleton. Elife. 2018;7:e31807. [DOI] [PMC free article] [PubMed]
- 49.Schatten H, Sun QY. Posttranslationally modified tubulins and other cytoskeletal proteins: their role in gametogenesis, oocyte maturation, fertilization and pre-implantation embryo development. Adv Exp Med Biol. 2014;759:57–87. doi: 10.1007/978-1-4939-0817-2_4. [DOI] [PubMed] [Google Scholar]
- 50.Schatten G, Simerly C, Asai DJ, Szoke E, Cooke P, Schatten H. Acetylated alpha-tubulin in microtubules during mouse fertilization and early development. Dev Biol. 1988;130:74–86. doi: 10.1016/0012-1606(88)90415-0. [DOI] [PubMed] [Google Scholar]
- 51.Castillo J, Jodar M, Oliva R. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum Reprod Update. 2018;24:535–555. doi: 10.1093/humupd/dmy017. [DOI] [PubMed] [Google Scholar]
- 52.Lesch BJ, Tothova Z, Morgan EA, Liao Z, Bronson RT, Ebert BL, Page DC. Intergenerational epigenetic inheritance of cancer susceptibility in mammals. Elife. 2019;8:e39380. [DOI] [PMC free article] [PubMed]
- 53.Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016;26:1034–1046. doi: 10.1101/gr.201541.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun W, Dong H, Becker AS, Dapito DH, Modica S, Grandl G, Opitz L, Efthymiou V, Straub LG, Sarker G, et al. Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring. Nat Med. 2018;24:1372–1383. doi: 10.1038/s41591-018-0102-y. [DOI] [PubMed] [Google Scholar]
- 55.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 56.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 57.Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A. 2015;112:13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 59.Morgan HL, Watkins AJ. The influence of seminal plasma on offspring development and health. Semin Cell Dev Biol. 2020;97:131–137. doi: 10.1016/j.semcdb.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 60.Selokar NL, Sharma P, Saini M, Sheoran S, Rajendran R, Kumar D, Sharma RK, Motiani RK, Kumar P, Jerome A, et al. Successful cloning of a superior buffalo bull. Sci Rep. 2019;9:11366. doi: 10.1038/s41598-019-47909-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Q, Lin J, Liu M, Li R, Tian B, Zhang X, Xu B, Liu M, Zhang X, Li Y, et al. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci Adv. 2016;2:e1501482. doi: 10.1126/sciadv.1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuster A, Tang C, Xie Y, Ortogero N, Yuan S, Yan W. SpermBase: a database for sperm-borne RNA contents. Biol Reprod. 2016;95:99. doi: 10.1095/biolreprod.116.142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M, Gao Y, Qu P, Qing S, Qiao F, Zhang Y, Mager J, Wang Y. Sperm-borne miR-449b influences cleavage, epigenetic reprogramming and apoptosis of SCNT embryos in bovine. Sci Rep. 2017;7:13403. doi: 10.1038/s41598-017-13899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Qu P, Zhou C, Liu X, Ma X, Wang M, Wang Y, Su J, Liu J, Zhang Y. MicroRNA-125b is a key epigenetic regulatory factor that promotes nuclear transfer reprogramming. J Biol Chem. 2017;292:15916–15926. doi: 10.1074/jbc.M117.796771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du Y, Wang X, Wang B, Chen W, He R, Zhang L, Xing X, Su J, Wang Y, Zhang Y. Deep sequencing analysis of microRNAs in bovine sperm. Mol Reprod Dev. 2014;81:1042–1052. doi: 10.1002/mrd.22426. [DOI] [PubMed] [Google Scholar]
- 66.Su J, Liu X, Sun H, Wang Y, Wu Y, Guo Z, Zhang Y. Identification of differentially expressed microRNAs in placentas of cloned and normally produced calves by Solexa sequencing. Anim Reprod Sci. 2015;155:64–74. doi: 10.1016/j.anireprosci.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 67.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 68.Bianchi E, Doe B, Goulding D, Wright GJ. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ito C, Yamatoya K, Yoshida K, Fujimura L, Sugiyama H, Suganami A, Tamura Y, Hatano M, Miyado K, Toshimori K. Deletion of Eqtn in mice reduces male fertility and sperm-egg adhesion. Reproduction. 2018;56:579–90. [DOI] [PubMed]
- 70.Czernik M, Iuso D, Toschi P, Khochbin S, Loi P. Remodeling somatic nuclei via exogenous expression of protamine 1 to create spermatid-like structures for somatic nuclear transfer. Nat Protoc. 2016;11:2170–2188. doi: 10.1038/nprot.2016.130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.