Abstract
Cardiac fibrosis is a hallmark of cardiovascular diseases. Several studies have indicated that microRNAs (miRs) are associated with the development of cardiac fibrosis. However, to date, the underlying molecular mechanisms of miR-489 in cardiac fibrosis have not been studied. The present study investigated the biological function of miR-489 in isoproterenol (ISO)-induced cardiac fibrosis. It was observed that miR-489 was downregulated in the heart tissue and cardiac fibroblasts (CFs) obtained from rats with ISO-induced cardiac fibrosis, as compared with the levels in the control group. By contrast, the expression levels of histone deacetylase 2 (HDAC2), collagen I (Col1A1) and α-smooth muscle actin (α-SMA) were increased in the heart tissue and CFs obtained from ISO-treated rats compared with the control group. Furthermore, ISO-treated CFs were transfected with a miR-489 mimic, which resulted in decreased viability and differentiation of CFs compared with the control group. Bioinformatics analysis and a dual-luciferase reporter assay further revealed that HDAC2 is a downstream target of miR-489. Subsequently, a loss-of-function experiment demonstrated that depletion of HDAC2 decreased the expression levels of Col1A1 and α-SMA in CFs. Taken together, the results obtained in the present study revealed that the miR-489/HDAC2 signaling pathway may serve as a novel regulatory mechanism in ISO-induced cardiac fibrosis and may increase the understanding on cardiac fibrosis.
Keywords: cardiac fibrosis, cardiac fibroblasts, isoproterenol, microRNA-489, histone deacetylase 2
Introduction
Cardiac fibrosis is a risk factor for the development of various cardiovascular diseases, including myocardial infarction, arrhythmia and heart failure (1,2). The main pathological features of cardiac fibrosis include abnormal proliferation of cardiac fibroblasts (CFs) and excessive deposition of extracellular matrix in the interstitium and perivascular region (3–6). In response to pathological stimuli, CFs can differentiate into myofibroblasts and increase the secretion of extracellular matrix proteins, which consequently leads to cardiac fibrosis (7–9). Despite advancements in the diagnosis and treatment of cardiac fibrosis, the exact pathogenesis remains unclear.
MicroRNAs (miRNAs) are a family of endogenous non-coding RNAs, 22–25 nucleotides in length, which serve as transcriptional regulators of genes (10). A number of studies have revealed that miRNAs are associated with cellular processes, including cell growth, proliferation and differentiation (11–13). Furthermore, miRNAs have been demonstrated to be key regulators of cardiovascular disease, including cardiac fibrosis (14–17). For instance, overexpression of let-7i attenuated angiotensin II-induced cardiac fibrosis by regulating the expression levels of interleukin-6 and collagen (18). Furthermore, it has been reported that increased miR-489 expression decreased pulmonary fibrosis by targeting MYD88 innate immune signal transduction adaptor (19). Transgenic overexpression of miR-489 was also reported to inhibit cardiac fibrosis following treatment with angiotensin II treatment (20). However, to the best of our knowledge, the underlying mechanism of miR-489 in attenuating the development of cardiac fibrosis has not been previously reported.
Histone deacetylases (HDACs) include 18 isoforms and are subdivided into four classes. HDAC2, a member of HDAC class II, is involved in a number of diseases, including tumorigenesis and cardiovascular disease (21). It has been reported that HDAC inhibitors inhibit fibrosis in a number of organs, such as the lungs and liver (22,23). In addition, overexpression of HDAC2 promotes cardiac hypertrophy (24). As miRNAs regulate the expression of downstream target genes, miR-489 may suppress cardiac fibrosis by downregulating the expression of HDAC2.
The present study revealed that miR-489 inhibited isoproterenol (ISO)-induced cardiac fibrosis in Sprague-Dawley (SD) rats by downregulating the expression of HDAC2. The results may guide the development of novel therapeutic agents for cardiac fibrosis.
Materials and methods
Animal experiments
A total of 30 SD rats (8 weeks old) were obtained from the Laboratory Animal Center of Soochow University. The mice were maintained under 22°C, 50% relative humidity, with a 12-h light/dark cycle and received food and water ad libitum. All animal experiments were approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University (Changzhou, China). The rats were randomly divided into two groups, as follows: i) Cardiac fibrosis group, which consisted of animals subcutaneously injected with 5 mg/kg/day ISO (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 10 days; and ii) control group, which consisted of animals injected with an equivalent volume of saline. Animal health and behavior, such as body temperature, weight loss, behavioral changes and pathological changes, were monitored every day. On day 11, the rats were sacrificed by cervical dislocation following anesthesia with sodium pentobarbital (50 mg/kg), following which their hearts were immediately isolated. Part of the cardiac tissue specimens were cut into 5-µm thick sections and fixed in 4% formaldehyde solution and stained with hematoxylin and eosin (H&E) or Masson solution for 5–10 min at room temperature to detect pathological changes in the cardiac tissues. The remaining section was used for RNA analysis.
Isolation of rat CFs and ISO treatment
Heart tissues were collected from SD rats, homogenized into 1-mm3 sections, placed in D-Hank's solution and subsequently digested using a mixed enzyme solution (trypsin: Collagenase ratio, 2:1). The cells were centrifuged at room temperature and 800 × g for 10 min and cultured in Dulbecco's modified Eagle medium (Corning, Inc., Corning, NY, USA) containing 10% fetal bovine serum (Corning, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C for 2 h. Adherent cells were obtained after discarding the non-adherent cells and were identified as CFs by their characteristic spindle shaped appearance using an inverted phase contrast microscope and immunohistochemical staining for vimentin (25,26). CFs at passage 3 were used in subsequent experiments. The cells were divided into two groups, and treated with 10 µM ISO or saline for 24 h, respectively.
Transfection with small interfering RNA (siRNA), mimics and inhibitor
siRNAs targeting HDAC2 (siHDAC2), siRNA-negative control (siNC), miR-489 mimic, miR-NC, miR-489 inhibitor and NC inhibitor were synthesized by GenePharma Co., Ltd. (Shanghai, China). For HDAC2 overexpression, full length HDAC2 cDNA was amplified by PCR reaction from the cDNA library of HeLa cells (cat. no. R71407; Invitrogen; Thermo Fisher Scientific, Inc.) and subcloned into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.), with empty pcDNA3.1 serving as control, and then transfected into cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Transfection with siHDAC2 or siNC (10 nM), miR-489 or NC mimics (10 nM), miR-489 inhibitor or NC inhibitor (10 nM) and co-transfection with miR-489 mimics and HDAC2 were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cells were subsequently split 36 h after transfection and treated with ISO for 24 h. The transfection sequences were as follows: siHDAC2, 5′-GUAUCAUCAGAGAGUCUUATT-3′; siNC, 5′-UUUGUACUACACAAAAGUACUG-3′; miR-489 mimic, 5′-GUGACAUCACAUAUACGGCAGC-3′; NC mimics, 5′-UUCUCCGAACGUGUCACGUUU-3′; miR-489 inhibitor, 5′-GCUGCCGUAUAUGUGAUGUCAC-3′; NC inhibitor, 5′-CAGUCCUUUUGUGUAGUACAA-3′.
Bioinformatics prediction and dual-luciferase reporter assay
The bioinformatics prediction software TargetScan version 7.2 (www.targetscan.org) was used to predict the potential target genes of miR-489. Next, a dual-luciferase reporter assay was conducted to validate the predicted targets. Wild-type (WT) and mutant (MUT) HDAC2 were cloned into a pGL3 plasmid to construct pGL3-HDAC2-WT and pGL3-HDAC2-MUT vectors, respectively. Subsequently, miR-489 mimics and vectors were co-transfected into 293T cells (American Type Culture Collection) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), and luciferase activity was measured using the Dual-Luciferase Reporter System (Promega Corporation, Madison, WI, USA) at 48 h after transfection.
Determination of serum myocardial injury markers
The rats were anaesthetized and sacrificed by cervical dislocation following anesthesia with sodium pentobarbital (50 mg/kg). Then, the blood samples were collected from the carotid artery of rats and centrifuged at 3,000 × g for 10 min at room temperature. The activities of creatine kinase (CK) and CK isozyme (CK-MB) were measured using the MD-100 multifunctional automatic biochemistry analyzer (Sanhe Medical Equipment Co., Ltd., Sanhe, China). Additionally, the concentration of cardiac troponin I (cTnI) was measured using the VITROS Immuno Diagnostic kit (Ortho-Clinical Diagnostics, Inc., Raritan, NJ, USA). All assays were performed according to the manufacturer's protocol.
Cell viability assay
Cell viability was detected using an MTT assay (KeyGen Biotech Co., Ltd.) according to the manufacturer's protocol. In brief, CFs were seeded into 96-well plates (3,000 cells/well) and transfected with the miR-489 mimic, miR-489 inhibitor or corresponding NC. After 36 h transfection, the cells were treated with ISO for 24 h and total of 50 µl MTT solution was added in each sample well, which were subsequently incubated for 4 h at 37°C. Next, 200 µl dimethyl sulfoxide was added into each well, and the optical density was measured at a wavelength of 570 nm.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cardiac tissues and CFs using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA concentrations were measured using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). For miRNA expression, RT reactions were performed using a One Step PrimeScript miRNA cDNA Synthesis kit (Takara Biotechnology Co., Ltd.) at 37°C for 30 min according to the manufacturer's protocols, followed by qPCR with SYBR® Premix Ex Taq (Takara Biotechnology Co., Ltd.). For mRNA expression, cDNA was synthesized from total RNA using a PrimeScript™ RT Reagent kit (Takara Biotechnology Co., Ltd.). qPCR amplifications were then performed using SYBR® Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). For miR-489 detection, the following thermocycling conditions were used: An initial denaturation step at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 10 sec and annealing at 60°C for 1 min. For the detection of mRNA, the thermocycling conditions were as follows: Initial denaturation at 95°C for 15 sec, denaturation at 94°C for 30 sec, annealing at 60°C for 20 sec, and extension at 72°C for 40 sec for 40 cycles. Fold changes in expression levels were calculated using the 2−ΔΔCq method (27). The primer sequences used in qPCR were as follows: miR-489 forward, 5′-ACACTCCAGCTGGGGTGACATCACATA-3′, and reverse, 5′-TGGTGTCGTGGAGTCG-3′; HDAC2 forward, 5′-GCTATTCCAGAAGATGCTGTTC-3′, and reverse, 5′-GTTGCTGAGCTGTTCTGATTTG-3′; collagen I (Col1A1) forward, 5′-CAGAGCACGATGTCCTGAGA-3′, and reverse, 5′-GCAAATGTGAGCTTCTGTGC-3′; α-smooth muscle actin (α-SMA) forward, 5′-GGAGTGATGGTTGGAATGG-3′, and reverse, 5′-ATGATGCCGTGTTCTATCG-3′; GAPDH forward, 5′-CAAGCTCATTTCCTGGTATGAC-3′, and reverse 5′-CAGTGAGGGTCTCTCTCTTCCT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACATATACTA-3′, and reverse, 5′-ACGAATTTGCGTGTCATCCTTGCG-3′. HDAC2, collagen I and α-SMA mRNA levels were normalized to the internal reference gene GAPDH, while miR-489 levels were normalized to U6.
Statistical analysis
Statistical analyses were performed using SPSS software (version 18.0; SPSS, Inc.). Comparisons of parameters between two groups were performed using a paired Student's t-test. Comparisons among multiple groups were performed by one-way analysis of variance, followed by Tukey's test. Data are presented as the mean ± standard deviation of at least three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
Pathological changes and levels of fibrosis-associated mRNAs in vivo and in vitro
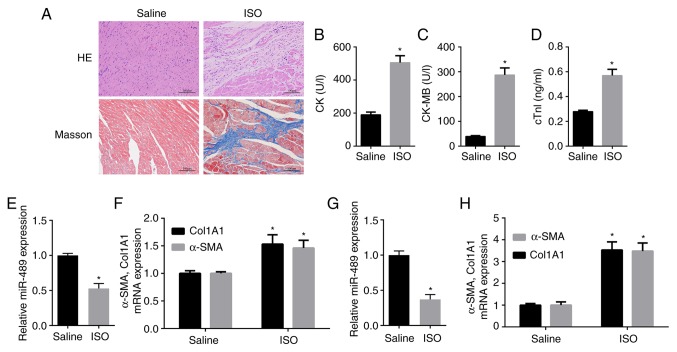
H&E and Masson staining were performed to confirm that the model of ISO-induced cardiac fibrosis was successfully constructed in the rats (Fig. 1A). Furthermore, the serum levels of CK, CK-MB and cTnI (28,29), which serve as diagnostic markers of myocardial damage, were measured. The data revealed that the levels of CK, CK-MB and cTnI were significantly increased in the serum of ISO-treated rats compared with those in the control rats (Fig. 1B-D). Furthermore, RT-qPCR indicated that miR-489 expression was significantly decreased in heart tissues obtained from the ISO-treated rats as compared with the control group (Fig. 1E). By contrast, the mRNA expression levels of Col1A1 and α-SMA were markedly increased in heart tissues obtained from ISO-treated rats compared with the control group (Fig. 1F). Consistent with these in vivo experimental results, miR-489 expression was found to be decreased in ISO-treated CFs compared with the control group, while the expression levels of Col1A1 and α-SMA were significantly increased in the ISO-treated CFs (Fig. 1G and H).
Figure 1.
Pathological changes in myocardial tissue and expression of fibrosis-associated markers in ISO-treated rat heart tissues and CFs. (A) H&E and Masson staining revealed myocardial collagen deposition in myocardial tissues subsequent to ISO injection (scale bar, 100 µm). (B) CK activity, (C) CK-MB activity and (D) cTnI concentration were measured in the serum of ISO-treated and normal control rats. (E) miR-489 expression, and (F) Col1A1 and α-SMA mRNA levels in heart tissues obtained from the control and ISO-treated rats, as well as (G) miR-489, and (H) Col1A1 and α-SMA levels in the control and ISO-treated CFs were determined by reverse transcription-quantitative polymerase chain reaction. The data are presented as the mean ± standard deviation. *P<0.05 vs. saline group. ISO, isoproterenol; CFs, cardiac fibroblasts; CK, creatine kinase; CK-MB, CK isozyme; cTnI, troponin I; miR, microRNA; Col1A1, collagen I; α-SMA, α-smooth muscle actin.
miR-489 inhibits the viability and differentiation of CFs
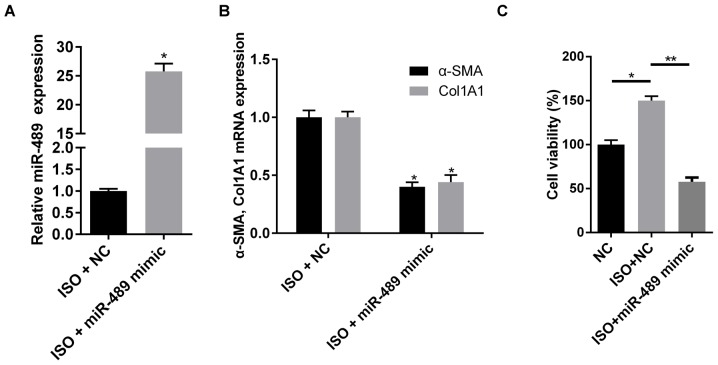
To investigate the effect of miR-489 on cardiac fibrosis, miR-489 mimics were transfected into rat CFs to induce miRNA overexpression (Fig. 2A). As shown in Fig. 2B, overexpression of miR-489 decreased the mRNA expression levels of Col1A1 and α-SMA. In addition, overexpression of miR-489 significantly inhibited the cell viability induced by ISO (Fig. 2C). These data demonstrated that miR-489 inhibited the viability and differentiation of ISO-treated CFs.
Figure 2.
miR-489 overexpression inhibited the viability and differentiation of CFs. (A) miR-489 expression, and (B) Col1A1 and α-SMA mRNA levels in CFs transfected with NC and miR-489 mimic were determined by reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. NC group. (C) An MTT assay was used to investigate the viability of CFs transfected with NC and miR-489 mimic treated with ISO. The data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01. miR, microRNA; CFs, cardiac fibroblasts; NC, negative control; Col1A1, collagen I; α-SMA, α-smooth muscle actin.
HDAC2 is a direct target of miR-489
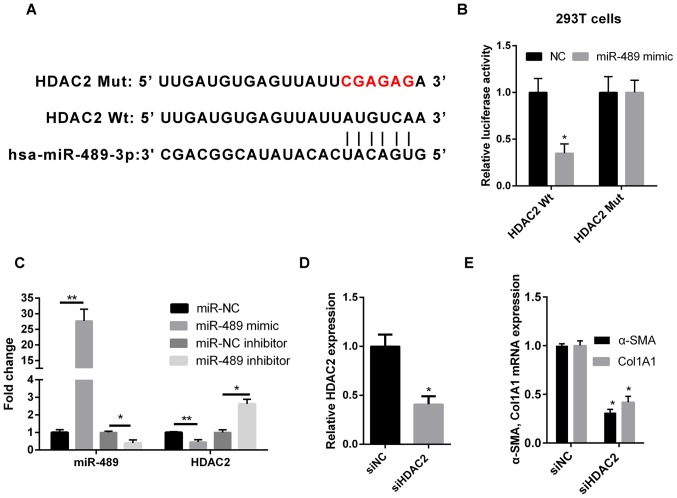
Computational predictions of miR-489 target genes were performed using TargetScan software. As shown in Fig. 3A, the 3′-untranslated region (UTR) of HDAC2 was complementary to miR-489, suggesting that HDAC2 may be a direct downstream target of miR-489. In order to validate this result, WT or MUT HDAC2 sequences were cloned into the 3′-UTR of the firefly luciferase gene. The results revealed that the miR-489 mimic transfection reduced the luciferase activity in 293T cells that were co-transfected with WT HDAC2, but not with MUT HDAC2 (Fig. 3B). Furthermore, RT-qPCR was used to determine the expression level of HDAC2 in ISO-treated CFs transfected with miR-489 mimic, miR-489 inhibitor or the corresponding NC. Compared with the NC group, HDAC2 expression was downregulated in miR-489 mimic-transfected cells and upregulated in miR-489 inhibitor-transfected cells (Fig. 3C).
Figure 3.
miR-489 directly targets HDAC2. (A) Bioinformatics analysis suggested that the 3′-untranslated region of HDAC2 was complementary to miR-489. (B) The luciferase reporter assay revealed that miR-489 binds to WT HDAC2, but not MUT HDAC2 in 293T cells. (C) RT-qPCR was used to determine the expression levels of miR-489 and HDAC2 mRNA in isoproterenol-treated CFs transfected with NC, miR-489 mimic or miR-489 inhibitor. (D) HDAC2 expression, and (E) Col1A1 and α-SMA mRNA levels in CFs transfected with siNC and siHDAC2 were determined by RT-qPCR. The data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01, vs. corresponding NC group. miR, microRNA; HDAC2, histone deacetylase 2; WT, wild-type; MUT, mutant; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CFs, cardiac fibroblasts; NC, negative control; Col1A1, collagen I; α-SMA, α-smooth muscle actin; si, small interfering RNA.
Silencing of HDAC2 decreases the expression levels of Col1A1 and α-SMA in CFs
To further investigate the role of HDAC2 in cardiac fibrosis, HDAC2 expression was knocked down using siRNA followed by treatment with ISO. The transfection efficiency was confirmed by RT-qPCR. Relative HDAC2 expression was significantly reduced in cells transfected with siHDAC2. (Fig. 3D). As shown in Fig. 3E, the expression levels of Col1A1 and α-SMA were markedly reduced in the siHDAC2-transfected group, as compared with those in the siNC group.
miR-489 suppresses cardiac fibrosis via HDAC2
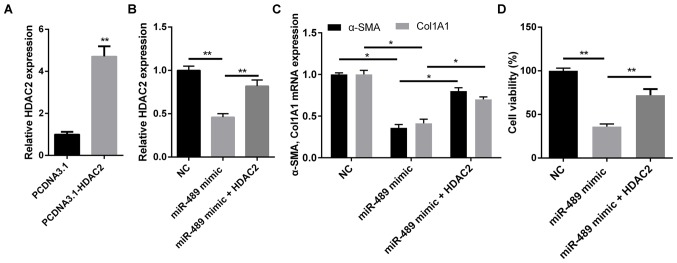
The present study earlier revealed that HDAC2 is a downstream target of miR-489 and was involved in the expression of fibrosis-associated markers. In order to determine whether the biological function of miR-489 in fibrogenesis was mediated by HDAC2, ISO-treated CFs were transfected with NC or miR-489 mimic, or co-transfected with HDAC2 overexpression plasmid and miR-489 mimic. RTqPCR demonstrated that the level of HDAC2 was notably increased in CFs transfected with the HDAC2 overexpression plasmid (Fig. 4A), while HDAC2 markedly reversed the inhibitory effect of miR-489 on HDAC2 expression in co-transfected CFs (Fig. 4B). As shown in Fig. 4C and D, cells co-transfected with HDAC2 overexpression plasmid and miR-489 mimic exhibited significantly increased expression levels of α-SMA and Col1A1, as well as enhanced cell viability, compared with the cells transfected with the miR-489 mimic alone. These results further suggested that HDAC2 serves a role in attenuating the viability and differentiation of ISO-treated CFs.
Figure 4.
miR-489 inhibited the isoproterenol-induced cardiac fibrosis induced via HDAC2. (A) RT-qPCR was used to determine the expression level of HDAC2 in CFs transfected with pcDNA3.1 and pcDNA3.1-HDAC2. (B) HDAC2, and (C) Col1A1 and α-SMA expression levels in CFs transfected with NC, miR-489 mimic or miR-489 mimic plus HDAC2 were determined by RT-qPCR. (D) An MTT assay was used to assess the viability of CFs transfected with NC, miR-489 mimic or miR-489 mimic plus HDAC2. The data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01. miR, microRNA; HDAC2, histone deacetylase 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CFs, cardiac fibroblasts; Col1A1, collagen I; α-SMA, α-smooth muscle actin; NC negative control.
Discussion
The present study revealed that miR-489 served an important role in ISO-induced cardiac fibrosis. Specifically, miR-489 attenuated cardiac fibrosis and decreased the expression levels of Col1A1 and α-SMA by downregulating HDAC2 expression.
Emerging evidence suggests that miRNAs are strongly associated with the development of cardiac fibrosis. For instance, Zhang et al (30) reported that miR-29b inhibited angiotensin II-induced cardiac fibrosis by targeting transforming growth factor (TGF)-β1, thereby inhibiting the TGF-β/SMAD3 signaling pathway. Wang et al (20) further revealed that cardiac hypertrophy-related factor, a long non-coding RNA, regulated cardiac hypertrophy by serving as an endogenous sponge for miR-489, thereby decreasing its expression level. However, the underlying mechanism of miR-489 in cardiac fibrosis has not been fully elucidated. In the present study, Sprague-Dawley rats were treated with ISO to induce cardiac fibrosis, and H&E and Masson staining were used to confirm that the model of cardiac fibrosis was successfully established. Additionally, miR-489 expression was found to be significantly downregulated in ISO-treated cardiac tissues and CFs, whereas the mRNA expression levels of Col1A1 and α-SMA were increased in ISO-treated cardiac tissues and CFs. miR-489 overexpression significantly reduced cell viability and Col1A1 and α-SMA mRNA expression in ISO-treated CFs. Taken together, these findings demonstrated that miR-489 mimic suppressed cell viability and differentiation of ISO-treated CFs.
HDAC inhibitors have been demonstrated to inhibit fibrosis following injury in a number of organs (31,32). Furthermore, the involvement of HDAC2 in heart disease has been reported (24). Trivedi et al (33) also indicated that HDAC2 regulated the cardiac hypertrophy response by modulating glycogen synthase kinase 3β activity. The present study revealed that miR-489 expression was decreased following ISO treatment in vivo or in vitro. Bioinformatics analysis and dual-luciferase reporter assay demonstrated that HDAC2 mRNA directly interacted with miR-489. RT-qPCR analysis found that the overexpression of miR-489 inhibited HDAC2 expression Subsequently, it was observed that HDAC2 knockdown decreased Col1A1 and α-SMA expression levels in CFs, whereas HDAC2 overexpression reversed the inhibitory effects of miR-489b on ISO-treated CFs. Therefore, the data suggested that miR-489 suppressed ISO-induced cardiac fibrosis by downregulating HDAC2. However, other possible targets of miR-489 may exist, whilst HDAC2 could also be subject to the regulation by other miRNAs. Therefore, further experiments of miR-489 on CFs are required to elucidate the mechanism in cardiac fibrosis further and the application of miR-489 for the treatment of cardiovascular diseases.
In conclusion, the results obtained in the present study indicated that miR-489 served an important role in the development of ISO-induced cardiac fibrosis by regulating HDAC2. The present study provided new insight into the mechanisms underlying cardiac fibrogenesis and suggested that miR-489 may serve as a potential therapeutic target for the treatment of cardiac fibrosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XY and TY designed the study. TY and SZ analyzed the data and prepared the figures. XY drafted the manuscript. All authors approved this manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University (Changzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cell Mol Life Sci. 2014;71:549–574. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dzeshka MS, Lip GY, Snezhitskiy V, Shantsila E. Cardiac fibrosis in patients with atrial fibrillation: Mechanisms and clinical implications. J Am Coll Cardiol. 2015;66:943–959. doi: 10.1016/j.jacc.2015.06.1313. [DOI] [PubMed] [Google Scholar]
- 3.Segura AM, Frazier OH, Buja LM. Fibrosis and heart failure. Heart Fail Rev. 2014;19:173–185. doi: 10.1007/s10741-012-9365-4. [DOI] [PubMed] [Google Scholar]
- 4.Edgley AJ, Krum H, Kelly DJ. Targeting fibrosis for the treatment of heart failure: A role for transforming growth factor-β. Cardiovasc Ther. 2012;30:e30–e40. doi: 10.1111/j.1755-5922.2010.00228.x. [DOI] [PubMed] [Google Scholar]
- 5.Espira L, Czubryt MP. Emerging concepts in cardiac matrix biology. Can J Physiol Pharmacol. 2009;87:996–1008. doi: 10.1139/Y09-105. [DOI] [PubMed] [Google Scholar]
- 6.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes JL, Gorin Y. Myofibroblast differentiation during fibrosis: Role of NAD(P)H oxidases. Kidney Int. 2011;79:944–956. doi: 10.1038/ki.2010.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber KT, Sun Y, Tyagi SC, Cleutjens JP. Collagen network of the myocardium: Function, structural remodeling and regulatory mechanisms. J Mol Cell Cardiol. 1994;26:279–292. doi: 10.1006/jmcc.1994.1036. [DOI] [PubMed] [Google Scholar]
- 9.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79:215–262. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Feng H, Wang Y, Su J, Liang H, Zhang CY, Chen X, Yao W. MicroRNA-148a suppresses the proliferation and migration of pancreatic cancer cells by down-regulating ErbB3. Pancreas. 2016;45:1263–1271. doi: 10.1097/MPA.0000000000000677. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Jiaqi C, Zhaoying C, Huimin C. MicroRNA-506-3p regulates neural stem cell proliferation and differentiation through targeting TCF3. Gene. 2016;593:193–200. doi: 10.1016/j.gene.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 13.Vienberg S, Geiger J, Madsen S, Dalgaard LT. MicroRNAs in metabolism. Acta Physiol (Oxf) 2017;219:346–361. doi: 10.1111/apha.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thum T, Catalucci D, Bauersachs J. MicroRNAs: Novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 15.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei C, Kim IK, Kumar S, Jayasinghe S, Hong N, Castoldi G, Catalucci D, Jones WK, Gupta S. NF-κB mediated miR-26a regulation in cardiac fibrosis. J Cell Physiol. 2013;228:1433–1442. doi: 10.1002/jcp.24296. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. MicroRNAs and the regulation of fibrosis. FEBS J. 2010;277:2015–2021. doi: 10.1111/j.1742-4658.2010.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Wang HX, Li YL, Zhang CC, Zhou CY, Wang L, Xia YL, Du J, Li HH. MicroRNA Let-7i negatively regulates cardiac inflammation and fibrosis. Hypertension. 2015;66:776–785. doi: 10.1161/HYPERTENSIONAHA.115.05548. [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Han L, Yan W, Ji X, Han R, Yang J, Yuan J, Ni C. miR-489 inhibits silica-induced pulmonary fibrosis by targeting MyD88 and Smad3 and is negatively regulated by lncRNA CHRF. Sci Rep. 2016;6:30921. doi: 10.1038/srep30921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 21.Yoon S, Eom GH. HDAC and HDAC inhibitor: From cancer to cardiovascular diseases. Chonnam Med J. 2016;52:1–11. doi: 10.4068/cmj.2016.52.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Zheng Y, Yuan H, Liu Y, Wen X. Effects of dynamic changes in histone acetylation and deacetylase activity on pulmonary fibrosis. Int Immunopharmacol. 2017;52:272–280. doi: 10.1016/j.intimp.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Wu XQ, Xu T, Li XF, Yang Y, Li WX, Huang C, Meng XM, Li J. Role of histone deacetylases(HDACs) in progression and reversal of liver fibrosis. Toxicol Appl Pharmacol. 2016;306:58–68. doi: 10.1016/j.taap.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Eom GH, Cho YK, Ko JH, Shin S, Choe N, Kim Y, Joung H, Kim HS, Nam KI, Kee HJ, Kook H. Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011;123:2392–2403. doi: 10.1161/CIRCULATIONAHA.110.003665. [DOI] [PubMed] [Google Scholar]
- 25.Katwa LC, Guarda E, Weber KT. Endothelin receptors in cultured adult rat cardiac fibroblasts. Cardiovascular research. 1993;27:2125–2129. doi: 10.1093/cvr/27.12.2125. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Wang B, Zhou C, Bi Y. Matrine induces apoptosis in angiotensin II-stimulated hyperplasia of cardiac fibroblasts: Effects on Bcl-2/Bax expression and caspase-3 activation. Basic Clin Pharmacol Toxicol. 2007;101:1–8. doi: 10.1111/j.1742-7843.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Zaitone SA, Abo-Gresha NM. Rosuvastatin promotes angiogenesis and reverses isoproterenol-induced acute myocardial infarction in rats: Role of iNOS and VEGF. Eur J Pharmacol. 2012;691:134–142. doi: 10.1016/j.ejphar.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Jiang Y, Jing W, Sun B, Miao C, Ren L. Quercetin provides greater cardioprotective effect than its glycoside derivative rutin on isoproterenol-induced cardiac fibrosis in the rat. Can J Physiol Pharmacol. 2013;91:951–959. doi: 10.1139/cjpp-2012-0432. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Huang XR, Wei LH, Chung AC, Yu CM, Lan HY. miR-29b as a therapeutic agent for angiotensin II-induced cardiac fibrosis by targeting TGF-β/Smad3 signaling. Mol Ther. 2014;22:974–985. doi: 10.1038/mt.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 32.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.