Abstract
Colorectal cancer (CRC) is one of the most common causes of cancer death worldwide. While standard chemotherapy and new targeted therapy have been improved recently, problems such as multidrug resistance (MDR) and severe side effects remain unresolved. RNAs are essential to all biological processes including cell proliferation and differentiation, cell cycle, apoptosis, activation of tumor suppressor genes, suppression of oncogenes. Therefore, there are various potential approaches to address genetic disease like CRC at the RNA level. In contrast to conventional treatments, RNA-based therapeutics such as RNA interference, antisense oligonucleotides, RNA aptamer, ribozymes, have the advantages of high specificity, high potency and low toxicity. It has gained more and more attention due to the flexibility in modulating a wide range of targets. Here, we highlight recent advances and clinical studies involving RNA-based therapeutics and CRC. We also discuss their advantages and limitations that remain to be overcome for the treatment of human CRC.
Keywords: RNA-based therapeutics, Colorectal Cancer, Chemical modifications, RNA interference, Gene Delivery, Clinical trials
Graphical abstract
1. Introduction
Colorectal cancer (CRC) is the third commonly diagnosed and second leading cause of cancer death worldwide in 2018 [1]. Currently, the most common treatment for patients with stage I and II colorectal cancer is surgical resection. The patients with stage III disease may receive surgery and adjuvant chemotherapy to lower the risk of recurrence. The main treatment for stage IV metastatic colorectal cancer (mCRC) is chemotherapy. The use of targeted therapies is another option to treat stage IV colorectal cancer.
The treatment for early stage disease has the best effect, however, about half of colorectal cancer patients are diagnosed with metastatic disease or will develop advanced-stage disease subsequently or have cancer recurrence in the following months [2]. For patients with inoperable metastatic disease, the prognosis is poor, with a 5-year survival rate around 14% [3].
Chemotherapy drugs for CRC patients include fluorouracil (5-FU) plus leucovorin (folinic acid), capecitabine, irinotecan, oxaliplatin, and trifluridine plus tipiracil [4]. In most cases, two or more of these chemotherapy drugs are combined during treatment to achieve a better clinical outcome. For advanced colorectal cancer, chemotherapy improves life quality and prolongs survival rate [5]. However, chemotherapy has its limitations. The patients who receive chemotherapy may suffer from hair loss, bone marrow suppression, nausea, vomiting, diarrhea, increased risk of infections, and neuropathy [6]. Another limitation of chemotherapy is multidrug resistance (MDR), which may cause cancer relapse and metastasis [7]. Drug resistance is responsible for treatment failure in 90% of patients with metastatic cancers [8]. Currently, 5-FU remains the most effective chemotherapeutic agent for the treatment of colorectal cancer, however, nearly half of the mCRC patients do not respond to it [9].
“One fits all” has been the principle of cancer therapeutics for a long time and chemotherapy is based on this principle. However, when CRC patients at the same disease stage receive the same treatment, the clinical outcomes for different individuals are quite different [10]. Advances in research and technologies have shown that cancer is a disease with high heterogeneity [11–13]. The CRC progression involves a variety of molecular biological changes such as genetic mutations or epigenetic abnormalities [14][15], the key property for cancer cells to gain drug resistance and evade the immune system of hosts. Studies have shown that correcting the abnormalities in gene expression can be detrimental to the cancer cells [16][17]. Therefore, new therapeutic strategies tailoring to the genetic changes and aiming at several cancer hallmarks simultaneously are in urgent need.
Ribonucleic acid (RNA)-based therapeutics with high specificity, high potency and low toxicity hold enormous potential as a new therapeutic approach [18]. The options for RNA therapeutics include RNA interference (RNAi), microRNA mimics and inhibitors, antisense oligonucleotides (ASO), aptamers, ribozymes, long non-coding RNA (lncRNA), and a variety of other RNAs. This wide range of choices offers flexible approaches to target disease-causing proteins at the RNA level. In contrast to conventional treatments, RNA-based therapeutics offer competitive advantages [19]. The “undruggable” proteins for conventional medicines can be selectively modulated by RNAs. They can even target proteins that have multiple homologous family members. In addition, RNAs can be designed for the targeted genes with a known sequence and easily be synthesized.
In this review, we summarize the recent researches on the biological actions of different RNAs and their role in regulating genes that are involved in CRC progression, which include effects on CRC cell growth, apoptosis, invasion, and metastasis. We also summarize clinical studies of RNA-based therapeutic strategies and discuss their advantages and limitations in CRC treatment.
2. RNA-based Therapeutics in Colorectal Cancer
RNA therapeutics refer to the use of RNAs as therapeutic agents. These consist different classes of RNAs, each of them regulates gene expression or translation through different mechanisms of action. Small interfering RNAs (or short silencing RNAs, siRNA), microRNAs (miRNA), and antisense oligonucleotides (ASO) are the most commonly used RNA therapeutics for silencing gene expression. Examples of RNA-based therapeutics in CRC treatment are summarized in Table 1.
Table 1.
RNA-based therapeutics used in the targeted treatment of CRC
| Agent | Targeted cells | Targets | Functions | References |
|---|---|---|---|---|
| B7-H4 siRNA | LOVO | CXCL12/CXCR4 and JAK2/STAT3 signaling | Inhibits proliferation, invasion, and migration | 22 |
| Antibody-siRNA complexes | HCT116, LoVo, SW480, HCT15, DLD1 | KRAS | Deactivates ERK and the MAPK pathway; inhibits tumor growth | 23 |
| siRNA | SW620 | LSD1 | Suppresses proliferation, migration and invasion of CRC cells | 24 |
| siRNA-PEG and SN-38 | LS174T | VEGF | Inhibits tumor growth, and enhances the antitumor effect of the chemotherapeutic drug | 25 |
| siRNA and Dox | HCT116 | Snail | Changes EMT genes, induces cell apoptosis, inhibit migration | 26 |
| siRNAs | HCT116 | KRAS and PK3CA | A synergistic decrease in proliferation and an increase in apoptosis | 27 |
| miR-204–5p | LoVo and HCT116 | RAB22A | Inhibits migration and invasion, promotes the sensitivity to chemotherapy | 45 |
| miR 217 | RKO and SW480 | MAPK1 | Inhibits tumor growth and enhances apoptosis in CRC | 46 |
| miR-143 | SW480, Lovo | KRAS | Suppresses CRC cell growth | 48 |
| SW480, 228 | DNMT3A | Decreases tumor cell growth | 49 | |
| SW620 | MACC1 | Inhibits cell growth, migration and invasion | 50 | |
| ASO | HT29 | EGFR | Reduces cell proliferation | 54 |
| ASO | HCT116 | miR-21 | Inhibits cell proliferation, reduces invasion and migration | 55 |
| ASO | LS174T and DLD-1 | MDM2 | Anti-tumor activity in vitro and in vivo | 57 |
| ASO AZD4785 | SW480 | KRAS mRNA | Inhibits the proliferation of tumor cells | 58 |
| S-1 aptamer | 119X | protein DHX9 | Binds to DHX9 and localizes to tumors in vivo | 62 |
| Aptamer | LS174T, LoVo, SW480 | CEA | Inhibits homotypic aggregation, migration, and invasion | 64 |
| Aptamer | HT29 | EpCAM | Delivers siRNA to cancer stem cells in vivo | 68 |
| Hammerhead ribozyme | SW480 | KRAS | Induces growth suppression, apoptosis and alters angiogenic factor expression | 73 |
| Ribozyme | HT-29 | hTERT | Targets and treats the tumor | 74 |
| Hammerhead ribozyme | HCT-8DDPA | γ-GCS | Suppresses multidrug resistance-associated protein | 75 |
| Ribozyme (Angiozyme) | KM12 | VEGF-1 | Reduces the number of metastases | 76 |
| smRNA | HCT116, HT29 | P21 | Inhibits cell proliferation an’ induces apoptosis | 97 |
CXCL12: C-X-C Motif Chemokine Ligand 12, JAK2: Janus Kinase 2, STAT3: Signal Transducer and Activator of Transcription 3, LSD1: Lysine-specific Histone Demethylase 1, PEG: Polyethylene Glycol, VEGF: Vascular Endothelial Growth Factor, MAPK1: Mitogen-Activated Protein Kinase 1, DHX9: DExH-Box Helicase 9, CEA: Carcinoembryonic Antigen, EpCAM: Epithelial Cell Adhesion Molecule, hTERT: human Telomerase Reverse Transcriptase, γ-GCS: gamma-glutamylcysteine synthetase, VEGF-1: vascular endothelial growth factor-1.
2.1. RNA interference (RNAi)
RNAi is a naturally biological process used by cells to inhibit gene expression through targeting the mRNA molecules. Fire and Mello revealed the mechanism of action in the nematode worm Caenorhabditis in 1998 and won the 2006 Nobel Prize for their work [20]. In many types of cancer, including colorectal cancer, RNAi opens up a new avenue for the treatment. It has been used for silencing oncogenes or proto-oncogenes that are up-regulated due to gene mutations or overexpression. SiRNA and miRNA are the most well-known RNA interference.
2.1.1. siRNA
SiRNAs are a class of double-stranded RNAs (dsRNAs), usually 21~25 nucleotides in length with 3’ overhanging nucleotides at each end [21]. SiRNAs can be either produced from long dsRNAs and hairpin looped RNA catalyzed by a RNase III enzyme Dicer, or artificially synthesized and introduced into the cells by transfection. They are incorporated into other proteins to form RNA-induced silencing complex (RISC), and unwound to single strand siRNAs subsequently. The single strand siRNAs which remain part of RISC find and bind to complementary mRNAs, and induce the mRNAs cleavage.
SiRNA can be used for a variety of purposes, including the suppression of CRC cell proliferation and induction of cell apoptosis, overcoming multi-drug resistance, and prevention of CRC metastasis. Peng et al. have reported that B7-H4 siRNA is able to inhibit proliferation, invasion, and migration of colorectal cancer cell line LOVO effectively through targeting CXCL12/CXCR4 and JAK2/STAT3 signaling [22]. In another study, the siRNA against KRAS has been applied to inhibit the proliferation of KRAS mutated CRC cells and to slow the tumor growth in a xenograft mouse model [23]. Ding et al. knocked down a histone demethylase called lysine-specific demethylase 1 (LSD1) with siRNA, resulting in suppressing of proliferation, migration and invasion of CRC in vitro [24]. Due to the complexity of cancer, combination therapy has gained more attention to overcome drug resistance or to active cancer cell apoptosis. Many studies have reported combination approaches for siRNA with small molecule anticancer drugs to achieve synergistic anticancer effects by combining different mechanisms of action. Lee et al. have shown that the synergistic effect to suppress tumor growth by using VEGF siRNA and SN-38 (7-ethyl-10- hydroxycamptothecin) in a LS174T tumor-bearing mouse xenograft model [25]. A study conducted by Sadreddini et al. used the combination of doxorubicin and snail siRNA. Snail is an important mediator of epithelial mesenchymal transition (EMT), a process by which epithelial cells gain migratory and invasive properties. The combination regimen inhibited proliferation, induced apoptosis, and reduced migration in human CRC cell line HCT-116 [26].
Combined siRNA treatments have also been studied. Valentino et al. applied a co-targeting strategy that targets mutated PI3K/AKT/mTOR and RAS pathways by siRNAs in CRC cell lines with PIK3CA and KRAS mutations. The combined therapy of siRNA (PIK3CA + KRAS or Akt2 + KRAS) provided a synergistic inhibition of CRC cell proliferation and an increase in apoptosis [27].
For siRNA to be effective to treat CRC, there are mainly three steps. Firstly, the targeted gene that plays an important role in the CRC progression need to be identified; secondly, the siRNAs for targeted gene silencing will be designed and synthesized; thirdly, the siRNAs need to be delivered into the CRC cells. Each step comes along with different challenges, which include unintended off-target effects [28], length of effect [29], short half-life [30], requirement of a delivery approach [31], and risk of immune system activation [32]. There are hypothesized theories for off-target effects. 1) siRNAs are able to bind to non-targeted mRNAs via imperfect complementarity and silence the non-targeted gene expression [33]. 2) siRNAs may enter endogenous miRNA systems that could imperfectly bind to non-targeted mRNAs and silence those targets [34]. To be clinically applicable, siRNAs should exert effects over a certain period time since siRNA will be degraded quickly after introduction. This may require repeated siRNA delivery or chemical modification of the siRNA to increase stability [35]. In order for siRNA entered the target cells, certain delivery strategies need to be taken due to the clearance by nucleases in the bloodstream. In addition, when the cells are introduced with siRNAs, they may consider it as a by-product of viral and activate innate immune responses [32].
2.1.2. microRNA (miRNA)
MiRNAs are short non-coding RNAs of 20~25 nucleotides in length that regulate gene expression post-transcriptionally by imperfect base pairing to the complementary sequences in the 3’ untranslated region (UTR) of mRNAs. MiRNAs are able to regulate cellular functions including cell development, proliferation, invasion, and apoptosis [36]. It has been found aberrant expressions of miRNAs are associated with the initiation, progression and metastasis of CRC [37–39]. MiRNA biogenesis is controlled by various enzymes such as Drosha, Dicer, DGCR8, thus abnormal expression of these proteins due to gene mutations or dysregulated epigenetic changes such as DNA hypomethylation [40], DNA hypermethylation [41], and histone deacetylation [42]. In CRC, some miRNAs whose expression are increased have been identified as oncogene miRNAs (or oncomiRs). OncomiRs regulate cancer development by inhibiting tumor suppressor genes, genes involved in cell differentiation or apoptosis. For example, Asangani et al. have reported that miR-21 induces invasion, intravasation and metastasis in colorectal cancer cell lines by negatively regulating tumor suppressor Pdcd4 at the post-transcriptional level [43]. Fang et al. have found that miR-17–5p induces drug resistance by targeting PTEN, a tumor suppressor that dominates the PTEN/AKT/PI3K pathway [44].
Other miRNAs whose expression is decreased in CRC cells act like tumor suppressors. Yin et al. have demonstrated that miR-204–5p is a tumor suppressor in CRC by inhibiting RAB22A, a member of the RAS oncogene family [45]. Tang et al. have shown that miR-93 suppresses CRC development by down regulating Wnt/β-catenin [46]. Zhang et al. have reported that miR-217 inhibits tumor growth and enhances cell apoptosis in CRC via down regulating MAPK signaling [47].
A single miRNA can regulate a variety of disease-associated targets simultaneously. It is a major advantage for the treatment of a disease like cancer where multiple genes are mutated or dysregulated. It has been reported that miR-143 suppresses CRC cell growth by regulating the oncogene KRAS expression [48]. Another study has demonstrated that miR-143 exerts its tumor suppressor role in CRC by repressing DNMT3A [49]. Furthermore, miRNA-143 has been found to inhibit CRC cell invasion and migration by targeting metastasis-associated in colon cancer-1 (MACC1), a CRC tumorigenesis and metastasis related gene [50].
Although miRNA therapy possesses great potential for the treatment of CRC, several obstacles remain to be overcome including an efficient miRNA delivery system, and the safety issues related to side effects.
2.2. Antisense Oligonucleotides (ASOs)
ASOs are synthesized nucleic acids and their base sequences are complementary to the targeted RNAs in the nucleus (e.g., pre-mRNA) or cytoplasm (e.g., miRNA, mRNA) via Watson-Crick hybridization [51]. Among all the RNA therapeutics, ASOs have a relatively longer track record, first being introduced by Paterson et al. to inhibit translation in 1977 [52]. Since then different modifications in the structure (e.g., backbone, sugar moiety) have been applied to optimize their therapeutic potential. ASOs have been tested for the treatment of cancer in more than 50 clinical trials (https://clinicaltrials.gov).
Targets of ASOs for treatment of CRC include genes that play an essential role in cell proliferation, cell differentiation, cell cycle, and cell death. Growth factors such as epidermal growth factor receptor (EGFR) play an important role in CRC cellular proliferation [53]. Najar et al. established EGFR ASO encapsulated within polyamidoamine (PAMAM) nanoparticles. The nanoparticles reduced the proliferation of human colon cancer cell line HT29 [54].
In addition, genes that are important for proliferation-independent processes in CRC progression such as adhesion, invasion, and migration are also potential targets. For example, Tao et al. constructed a eukaryotic expression vector encoding ASOs against an oncomiR miR-21. The down- regulation of miR- 21 by ASOs significantly reduced the invasion and migration of CRC cells [55].
Regulation of proto-oncogenes such as c-MYC by ASOs has been explored. Abaza et al. have reported that c-MYC ASOs inhibited the growth of CRC cells and increase the sensitivity of CRC cells to several chemotherapeutic drugs including paclitaxel, 5-FU, doxorubicin, and vinblastine [56]. Oncogene MDM2 is a negative regulator of the tumor suppressor p53. Wang et al. inhibited MDM2 expression with an ASO and found MDM2 ASO had both in vitro and in vivo antitumor activity, indicating its potential as a CRC cancer therapeutic [57]. It has been reported that mutations of the RAS oncogenes result in persistent activation of the downstream pathways, leading to carcinogenesis in several human cancers including CRC. Ross et al. evaluated a high-affinity constrained ethyl-containing therapeutic ASO AZD4785 targeting KRAS mRNA and found AZD4785 effectively inhibited proliferation in KRAS mutant cells [58].
2.3. RNA Aptamer
RNA aptamers are single-strand RNA oligonucleotides with various shapes that can bind to targets such as proteins, peptides, and small molecules. RNA aptamers are identified through several rounds of selection called systematic evolution of ligands by exponential enrichment (SELEX) [59]. Due to their stable three-dimensional shape, the binding to targets has high affinity and specificity [60].
Varieties of RNA aptamers have been identified to bind different targets and have shown great potential as diagnostic, prognostic and therapeutic tools in CRC. Mi et al. have screened a large number of nuclease-resistant RNA oligonucleotides in a tumor-bearing mouse model to identify RNA aptamers that can localize to hepatic colon cancer metastases. They have found an RNA aptamer that binds to p68, an RNA helicase that is upregulated in CRC [61]. RNA aptamers are isolated against not only cell surface markers, but also intracellular key components. Mi et al. have identified another aptamer that binds to the protein DHX9, another RNA helicase that is upregulated in CRC. The aptamer is selectively localized to the nucleus of cancer cells in vivo, indicating that it may facilitate targeted delivery to the nucleus [62].
In addition to their targeting capability, RNA aptamers can act like antagonists to inhibit the interactions between extracellular targets and their ligands. The overexpression of carcinoembryonic antigen (CEA) in CRC cells induces cell adhesion, enhances the resistant to anoikis, and promotes hepatic metastasis [63]. Lee et al. have identified an RNA aptamer against the domain of CEA required for metastasis with high affinity and specificity using SELEX. They have demonstrated that the RNA aptamer could inhibit CEA interactions with heterogeneous nuclear ribonucleoprotein M4 and death receptor 5, resulting a suppression of hepatic metastasis of colon cancer cells in mice [64].
Aptamers are called “chemical antibodies”, which have several competitive advantages comparing to their protein counterpart. Cell-free assembly allows aptamer production to be cost effective, rapid, and reproducible in large scale [65]. Aptamers induce less immunogenicity after chemical modifications [66], and they have an enhanced tissue penetration due to their smaller size [67]. Another significant advantage of aptamers is being able to introduce other functional groups to the backbone. For example, aptamers can form a chimeric structure with siRNA to deliver siRNA to targeted cell population [68], conjugated to chemo drugs such as doxorubicin [69], or combined with three-way junction RNA to construct multifunctional RNA nanoparticle to target metastatic CRC cells [70].
2.4. Ribozymes
Ribozymes are RNA molecules that function as enzymes to catalyze biochemical reactions, thus they have great potential for gene therapy. The most common activities of natural ribozymes are to catalyze RNA cleavage and ligation reactions [71]. Gene-tailored ribozymes have been designed, synthesized and delivered to specific cells to regulate the expression of targeted genes [72].
Ribozymes have been reported to target a variety of oncogenes and the drug resistance genes in CRC. A hammerhead ribozyme has been designed to preferentially cleave oncogene KRAS mRNA in human colon cancer cell lines with KRAS mutations, resulting in tumor growth suppression and alteration of angiogenic gene expression [73]. Jeong et al. have shown that systemic delivery of adenovirus harboring trans-splicing ribozyme can recognize cancer-specific transcripts and significantly reduce tumor burden in colon cancer mouse model [74]. Glutamylcysteine synthetase (γ-GCS) plays an important role in both cisplatin and multidrug resistance. Nagata et al. have designed a hammerhead ribozyme against γ-GCS mRNA to specifically down-regulated γ-GCS gene expression in the HCT-8DDP human colon cancer cell line. The downregulation of γ-GCS expression leads to reversal of resistance to cisplatin, doxorubicin and etoposide [75]. RPI.4610 (Angiozyme) is a ribozyme that targets vascular endothelial growth factor receptor 1 (VEGF-1). It has been reported that Angiozyme can inhibit metastases in a human KM12 colorectal cancer xenograft model and increased survival in a murine 4T1 tumor model [76].
2.5. Long Noncoding RNAs (IncRNAs)
LncRNAs are a class of RNAs with a length over 200 nucleotides that are not translated to proteins. The role of lncRNA in cancer development has attracted more and more attention [77]. Although the research on lncRNAs are still developing and at a preliminary stage, it is believed that lncRNAs are important regulators for gene expression.
lncRNAs have complex secondary and tertiary structures due to their large sizes. The structure complexity enables their regulatory abilities via binding to protein, DNA, or RNA (see Table 2 for details). Accumulating evidence has indicated that lncRNAs are a powerful factor in the CRC progression. LncRNAs can epigenetic and transcriptional regulate gene expressions. For example, a lncRNA called CCAT1-L is transcribed specifically in human CRC from a locus 515 kb upstream of MYC. CCAT1-L is important for MYC transcriptional regulation and promotes long-range chromatin looping. CCAT1-L knockdown have reduced long-range interactions between the MYC promoter and its enhancers [78]. CRC patients have higher HOTAIR expression levels in CRC tissue than adjacent normal tissues [79]. It has been demonstrated that lncRNA HOTAIR acts as a scaffold to assemble polycomb repressive complex 2 (PRC2) and lysine specific demethylase 1 (LSD1) complexes, leading to epigenetic silencing of cancer-related genes [80].
Table 2.
Examples of lncRNAs serve as important regulators in CRC progression
| Category of lncRNA mechanisms | Examples | Functions | References |
|---|---|---|---|
| Chromatin interactions | CCAT1 | Promotes long-range chromatin looping at the MYC locus | 78 |
| Protein interactions | HOTAIR | Acts as a scaffold to assemble PRC2 and LSD1 complexes | 80 |
| miRNA interactions | CRNDE | Represses miR-181a-5p expression | 85 |
| UCA1 | Inhibits a tumor suppressive miRNA miR-204–5p | 82 | |
| HNF1A-AS1 | Regulates SIRT1 by competitively binding miR-34a | 89 | |
| mRNA interactions | SNHG5 | Binds target mRNAs to block STAU1 mediated degradation | 81 |
| UCA1 | Interacts with mRNA 3-UTRs | 83 |
CCAT: Colon Cancer Associated Transcript 1, HOTAIR: HOX Transcript Antisense RNA, PRC2: Polycomb Repressive Complex 2, LSD1: Lysine-specific Histone Demethylase 1, CRNDE: Colorectal Neoplasia Differentially Expressed, UCA1: Urothelial Cancer Associated 1, HNF1A-AS1: HNF1A antisense RNA 1, SIRT1: Sirtuin 1, SNHG5: Small Nucleolar RNA Host Gene 5, STAU1: Staufen Double-Stranded RNA Binding Protein 1.
Besides the transcriptional and epigenetic regulations, it has been found that lncRNAs play important roles in posttranscriptional regulation. Damas et al. have demonstrated that lncRNA SNHG5 promotes tumor cell survival in CRC by binding to target mRNAs to block double-stranded RNA-binding protein staufen homolog 1 (STAU1) mediated degradation [81]. Bian et al. have demonstrated that lncRNA UCA1 exerts its regulatory function by sponging and inhibiting a tumor suppressive miRNA miR-204–5p [82]. Another study has shown that lncRNA UCA1 can also control cancer-related pathways by interacting with mRNA 3’-UTRs, preventing them from miRNA-mediated degradation [83].
Recent breakthroughs have provided many examples of lncRNAs’ regulatory capacities. LncRNAs associated with CRC development include AFAP1-AS1 [84], CCAT1 [78], CRNDE [85], DUSP [86], DANCR [87], GHET1 [88], HOTAIR-G [79], HNF1A-AS1 [89], MALAT1 [90], NEAT1 [91], SNHG12 [92], UCA1 [83] and others.
2.6. Small Activating RNAs
Small activating RNAs (saRNAs) are small double-stranded RNAs, which work through a distinct mechanism called RNA activation [93][94]. Several studies have demonstrated that saRNAs are able to up-regulate protein that are important for tumor progression by targeting gene promoters to induce transcriptional gene activation [95] [96]. Wang et al. have demonstrated that saRNA p21-saRNA-322 can inhibit CRC growth by stimulating the expression of p21, a downstream protein of P53, in vivo [97].
3. Improving the Effectiveness of RNA therapeutics for the treatment of Colorectal Cancer
Although RNA-based therapeutics have great potential for the treatment of CRC, problems such as degradation by nuclease, delivery to the CRC cells, and potential immunogenicity remain to be addressed [98]. Many studies have been reported that these issues can be overcome with the chemical modification of the RNAs [99]. In addition, advances in nanotechnology have improved systemic delivery of RNA therapeutics.
3.1. Chemical Structure Modifications
RNAs are unstable and rapidly cleaved in vivo due to a large amount of ribonucleases in serum and cells. RNAs, coding and non-coding, can be modified to affect RNA structure and function. In order to improve the stability of RNAs without reducing their activities in biological fluids, various chemical modifications to the base, sugar or phosphate moieties of nucleotides have been performed.
Chemical modifications to RNAs may also increase specificity and reduce off-target effects [100], enhance activity [101], improve pharmacokinetic (PK) and pharmacodynamic (PD) properties [102], and decrease immunological activation [103].
The chemical modifications for short synthesized RNA oligonucleotides include phosphorothioate (PS) backbone modification; 2’-fluoro (2’-F), 2’-O-methyl (2’-OMe), 2’-O-methoxyethyl (2’- MOE), locked nucleic acid (LNA) sugar substitutions; and conjugation with cholesterol or polyethylene glycol (PEG). The PS backbone modification replaces the non-bridging phosphate oxygen by a sulfur atom. This simple substitution increases resistance to nuclease degradation and improved affinity to plasma proteins with less renal clearance [104]. Substitutions at the 2’ position of the sugar ring with 2’-F, 2’-OMe, 2’-MOE or LNS group improve the potency, stability, and overall PK and PD properties [105].
Non-nucleotide chemical modifiers can also be applied. Kitade et al. added aromatic benzene-pyridine (BP-type) analogs to the 3’-overhang region of the miR-143, resulting in greater activity and increased resistance to nuclease. The modified miR-143 showed a significant tumor-suppressive effect on xenografted tumor of CRC DLD-1 cells [106].
3.2. Targeted Nanoparticles for RNA Delivery to Colorectal Cancer Cells
RNAs are negatively charged and have large molecular weight, they cannot pass into the CRC cells. This obstacle can be overcome with delivery systems. There are various delivery systems currently being applied to improve the efficacy of RNA therapeutics. The two major types are viral and non-viral delivery vector.
Viral vectors such as retroviral, lentiviral, adenovirus, adeno-associated virus have high transfection efficiency, however the shortcomings of potential immunogenicity, cytotoxicity and insertional mutagenesis limit their applications [107]. Nanoparticle-based non-viral vectors such as liposomes, polymersomes, dendrimers, and inorganic nanoparticles have the advantages of low cost, ease of production in large amount, and lower pathogenicity, which offer a promising alternative for RNA therapeutics delivery [108]. Nanoparticles can reach the tumor tissue passively due to their sizes. In addition, the enhanced permeation and retention (EPR) effect allows the accumulation within the cancer tissue longer to improve the therapeutic efficacy [109].
Tanggudu et al. [110] have reported that oral delivery of c-Myc conjugated polyethyleneimine (PEI)-macromolecule polyglycidal methacrylate (PGMA) nanoparticles suppressed tumor growth efficiently and increased animal survival in a colon cancer model. Sureban et al. [111] encapsulated siDCAMKL-1 in Poly(lactide-co-glycolide)-based nanoparticles (NP-siDCAMKL-1) and treated mice carrying CRC tumor with NP-siDCAMKL-1, resulting in CRC tumor xenograft growth arrest.
Targeting is essential for the treatment of CRC. The most commonly used biomarkers in CRC include carcinoembryonic antigen (CEA) [112], death receptor 5 (DR5, also known as TRAIL receptor 2) [113], epithelial growth factor receptor (EGFR) [114], tumor-associated glycoprotein (TAG)-72 [115], and folate receptor-α (FRα). The conjugation of ligands on the surface of nanocarriers for the targeted delivery of RNA therapeutics to CRC cells has been widely used. Pi et al. have demonstrated that folate-displaying extracellular vesicles (EVs) can specifically deliver survivin siRNA to CRC cells and efficiently inhibit colorectal cancer growth in patient-derived colorectal cancer xenograft mouse model [117]. Kim et al. have developed anti-TAG-72 PEG-immunoliposomes (PILs) to target TAG-72 overexpressing CRC cells. Intravenous administration of anti-TAG-72 PILs efficiently accumulated in the tumor tissues, indicating that the immunoliposomes have great potential as gene delivery to human CRC cells.
However, nanocarriers still have several limitations such as immunogenicity and toxicity, which limit the therapeutic applications. Thus, more studies on the nanocarrier delivery in CRC therapy are needed.
4. Clinical Application of RNA-based Therapy in Colon Cancer
To date, the U.S. Food and Drug Administration (FDA) has approved only a limited number of RNA-based therapeutics: five ASO drugs (fomivirsen, mipomersen, eteplirsen, nusinersen, inotersen), one aptamer drug (pegaptanib), and one siRNA drug (patisiran). These successful clinical translations show great potential of utilizing of RNA-based therapy for tackling various diseases through gene regulation. In August 2018, the FDA approved the first targeted RNA-based therapy siRNA for treat a rare disease polyneuropathy, which fully demonstrates siRNAs as a new drug class with great potential. Although currently no RNA therapeutics is approved by the FDA for CRC treatment, many clinical investigations are ongoing.
An example of this is Angiozyme, the first synthetic ribozyme tested in clinical trial. Angiozyme targets the mRNA of vascular endothelial growth factor receptor-1 (VEGFR-1) to inhibit angiogenesis and tumor growth. A Phase I clinical trial has demonstrated that Angiozyme have minimal toxicities and good bioavailability for 31 patients with refractory solid tumors, including colorectal cancer [118].
Several ASOs have reached clinical trials for the treatment of CRC. ASO LY2275796 targets an oncogene eukaryotic translation initiation factor 4E (eIF-4E) in solid tumors, including colon cancer. In a phase 1 dose escalation, pharmacokinetic and pharmacodynamics study of LY2275796, tumor elF-4E expression was decreased but no tumor responses observed, suggesting LY2275796 should combine with other treatment modalities [119]. ISIS 183750 is another ASO that inhibits the production of eIF4E. In preclinical study, the EIF4e ASO demonstrated its abilities to reduce EIF4e expression and inhibit CRC cell proliferation. In addition, it had enhanced activity in combination with irinotecan (Iri). The researchers conducted a phase I/II clinical trial to test the combination therapy activity. The study has shown that the ASO can penetrate tumor cells, however its activity has been compromised by extensive stromal binding. The combination of ISIS183750 and Iri did not show objective responses. Future studies may need combination with other chemotherapies or treatment for patient with less advanced disease [120].
RNAi is a highly targeted therapeutics for gene silencing, however, only a few of them have progressed into clinical trials. Atu027 is a RNA interference therapeutic that consists of liposomal particles and chemically stabilized siRNA that silences the expression of protein kinase N3 (PKN3) in the vascular endothelium. A phase 1 study of Atu027 was tested in patients with advanced solid tumors including colon cancer [121]. The study demonstrated that Atu027 was well tolerated in the patients. Based on the mode of action, future studies that examine efficiency of combination with traditional cytotoxic drugs are recommended. CALAA-01 is a cyclodextrin-based polymeric nanoparticle including a siRNA that reduces expression of the M2 subunit of ribonucleotide reductase (R2) [122]. A phase I study reported that the delivery system can provide a targeted delivery of siRNA and CALAA-01 was well tolerated during the phase Ia study [123].
5. Advantages and Limitations in developing RNA-based therapeutics
RNA-based therapeutics modulate the internal machinery of cells, changing the expression of targeted genes. It offers a broad range of potential applications and provides a high degree of flexibility, making it possible to manipulate previously “undruggable” targets. A variety of different approaches are available for researchers to either up-regulate or down-regulate certain genes within a cell. In addition, depending on the approach being used, a specific protein or a broader range of proteins can be targeted at the same time. In contrast, DNA-based therapies or gene editing technologies such as CRISPR aims to repair a dysfunctional gene or introduce a correct version into the cells. They have the high risk to permanently alter the genome, which limits a wide application in human.
Although RNA-based therapeutics offer advantages as mentioned above, there are challenges remain to be overcome. Besides stability and delivery issues, one challenge for RNAi is the “off-target” effect [124]. Due to the tolerance for mismatches with targets, those small RNAs may have a large number of potential targets in the genome and could affect many mRNAs that are not their intended targets [125]. In addition, the off-target effects may affect the phenotype of cells much more extensively than we originally anticipated [28].
Another challenge is tissue distribution. The majority of the systematically administered siRNAs and miRNAs tend to accumulate in organs that are parts of monoculear phagocyte system such as spleen, liver and lung [126]. ASOs have been shown to have a high distribution in liver and kidney, while the distribution to tumors is less extensive [127]. The identification of an appropriate target is another issue. To apply RNA-based therapeutics, we need to understand the importance of target gene for the progress of CRC, the degree of gene suppression needed, and the mechanisms that other genes may compensate to the loss of function of targeted gene.
6. Conclusion
RNA-based therapeutics have gained increased attention, mainly due to the ability to target disease genes that are previously unmanipulatable and the flexibility in modulating a large range of targets. However, because of challenges in stability and delivery, it may take time for clinical practice becoming a reality. Advances in medicine chemistry and nanotechnology will help to solve those issues and RNA based therapies will become more widely adopted. To be a new class of treatment modality alongside medicines such as small molecules or antibodies, RNA modalities need to have superior outcomes for patients. Currently pharmaceutical scientists equipped with new biotechnology are working on different RNA-based mechanisms to reveal the fullest potential for RNA-based therapeutics.
Figure 1.
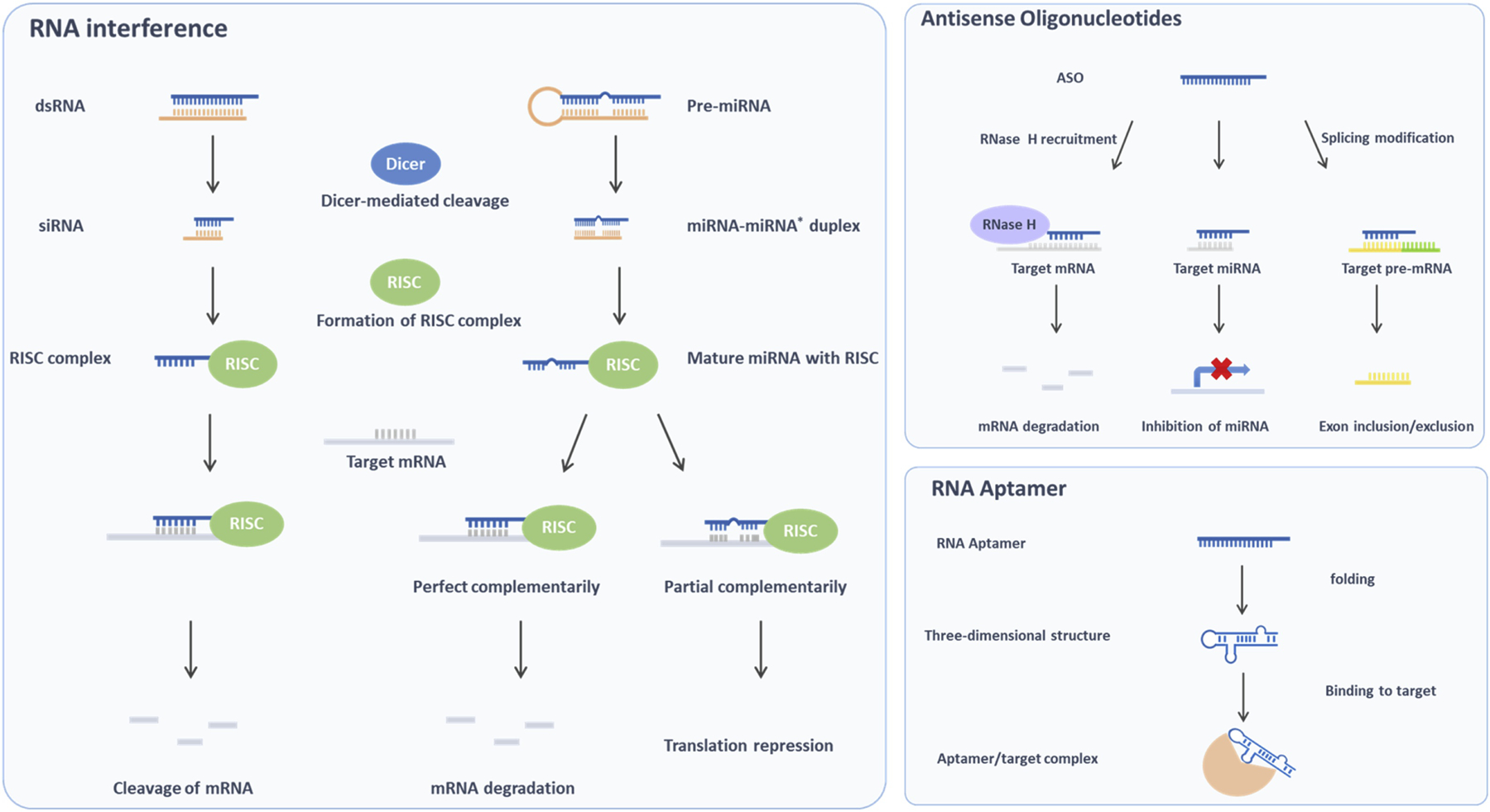
Common RNA-based therapeutics (RNA interference, ASO, amd RNA aptamer) mechanism of action.
Acknowledgment
This work was supported in part by NIH Grant CA186100.
Abbreviations
- CRC
colorectal cancer
- lncRNA
long non-coding RNA
- ASO
antisense oligonucleotides
- RNAi
RNA interference
- RISC
RNA-induced silencing complex
- miRNA
microRNA
- saRNA
Small activating RNA
- PK
pharmacokinetic
- EPR
enhanced permeation and retention
- EMT
epithelial-mesenchymal transition
- PD
pharmacodynamics
- MDR
multidrug resistance
- CXCL12
C-X-C motif chemokine ligand 12
- JAK2
janus kinase 2
- STAT3
signal transducer and activator of transcription 3
- LSD1
lysine-specific histone demethylase 1
- PEG
polyethylene glycol
- VEGF
vascular endothelial growth factor
- MAPK1
mitogen-activated protein kinase 1
- DHX9
DExH-box helicase 9
- CEA
carcinoembryonic antigen
- EpCAM
epithelial cell adhesion molecule
- hTERT
human telomerase reverse transcriptase
- γ-GCS
gamma-glutamylcysteine synthetase
- VEGF-1
vascular endothelial growth factor-1
- UTR
3’ untranslated region
- PAMAM
polyamidoamine
- EGFR
epidermal growth factor receptor
- CEA
carcinoembryonic antigen
- SELEX
systematic evolution of ligands by exponential enrichment
- CCAT
colon cancer associated transcript 1
- HOTAIR
HOX transcript antisense RNA
- PRC2
polycomb repressive complex 2
- CRNDE
colorectal neoplasia differentially expressed
- UCA1
urothelial cancer associated 1
- HNF1A-AS1
HNF1A antisense RNA 1
- SIRT1
sirtuin 1
- SNHG5
small nucleolar RNA host gene 5
- PS
phosphorothioate
- 2’-F
2’-fluoro
- 2’-OMe
2’-O-methyl
- 2’- MOE
2’-O-methoxyethyl
- LNA
locked nucleic acid
- PEI
polyethyleneimine
- PGMA
polyglycidal methacrylate
- DR5
death receptor 5
- FRα
folate receptor-a
- EVs
extracellular vesicles
- eIF-4E
eukaryotic translation initiation factor 4E
- PKN3
protein kinase N3
Chemical compounds studied in this article
Capecitabine (PubChem CID: 60953); Doxorubicin (PubChem CID: 31703); Etoposide (PubChem CID: 36462); 5-Fluorouracil (PubChem CID: 3385); Irinotecan (PubChem CID: 60838); Leucovorin (PubChem CID: 135403648); Paclitaxel (PubChem CID: 36314); Tipiracil (PubChem CID: 6323266); Trifluridine (PubChem CID: 6256); Vinblastine (PubChem CID: 241903)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018. November;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Primrose JN. Treatment of colorectal metastases: surgery, cryotherapy, or radiofrequency ablation. Gut. 2002. January 1;50(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RG, Barzi A et al. , Colorectal cancer statistics, 2017. CA: a cancer journal for clinicians. 2017. May 6;67(3):177–93. [DOI] [PubMed] [Google Scholar]
- [4].Engstrom PF, Arnoletti JP, Benson AB, Chen YJ, Choti MA, Cooper HS et al. , Colon cancer. Journal of the National Comprehensive Cancer Network. 2009. September 1;7(8):778–831. [DOI] [PubMed] [Google Scholar]
- [5].Neugut AI, Matasar M, Wang X, McBride R, Jacobson JS, Tsai WY et al. , Duration of adjuvant chemotherapy for colon cancer and survival among the elderly. J Clin Oncol. 2006. May 20;24(15):2368–75. [DOI] [PubMed] [Google Scholar]
- [6].Mols F, Beijers T, Lemmens V, van den Hurk CJ, Vreugdenhil G, van de Poll-Franse LV. Chemotherapy-induced neuropathy and its association with quality of life among 2-to 11-year colorectal cancer survivors: results from the population-based PROFILES registry. J Clin Oncol. 2013. July 20;31(21):2699–707. [DOI] [PubMed] [Google Scholar]
- [7].Van der Jeught K, Xu HC, Li YJ, Lu XB, Ji G. Drug resistance and new therapies in colorectal cancer. World journal of gastroenterology. 2018. September 14;24(34):3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Advanced pharmaceutical bulletin. 2017. September;7(3):339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wilson PM, El-Khoueiry A, Iqbal S, Fazzone W, LaBonte MJ, Groshen S et al. , A phase I/II trial of vorinostat in combination with 5-fluorouracil in patients with metastatic colorectal cancer who previously failed 5-FU-based chemotherapy. Cancer chemotherapy and pharmacology. 2010. April 1;65(5):979–88. [DOI] [PubMed] [Google Scholar]
- [10].Tanaka T, Tanaka M, Tanaka T, Ishigamori R. Biomarkers for colorectal cancer. International journal of molecular sciences. 2010. September;11(9):3209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013. September;501(7467):338–45. [DOI] [PubMed] [Google Scholar]
- [12].Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. science. 2013. March 29;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013. July;499(7457):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Willett CG, Chang DT, Czito BG, Meyer J, Wo J. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012.(5). International Journal of Radiation Oncology Biology Physics. 2013. May 1;86(1). [Google Scholar]
- [15].Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO et al. , Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. The Journal of pathology. 2013. September;231(1):63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and nononcogene addiction. Cell. 2009. March 6;136(5):823–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H et al. , Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015. June 18;161(7): 1539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nature chemical biology. 2006. December;2(12):711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. nature. 2001. May;411(6836):494. [DOI] [PubMed] [Google Scholar]
- [20].Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. nature. 1998. February;391(6669):806. [DOI] [PubMed] [Google Scholar]
- [21].GroBhans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008. January 23;451(7177):414. [DOI] [PubMed] [Google Scholar]
- [22].Peng HX, Wu WQ, Yang DM, Jing R, Li J, Zhou FL et al. , Role of B7-H4 siRNA in proliferation, migration, and invasion of LOVO colorectal carcinoma cell line. BioMed research international. 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baumer S, Baumer N, Appel N, Terheyden L, Fremerey J, Schelhaas S et al. , Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clinical Cancer Research. 2015. March 15;21(6): 1383–94. [DOI] [PubMed] [Google Scholar]
- [24].Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S et al. , LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. British journal of cancer. 2013. August;109(4):994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee SY, Yang CY, Peng CL, Wei MF, Chen KC, Yao CJ et al. , A theranostic micelleplex co-delivering SN-38 and VEGF siRNA for colorectal cancer therapy. Biomaterials. 2016. April 1;86:92–105. [DOI] [PubMed] [Google Scholar]
- [26].Sadreddini S, Safaralizadeh R, Baradaran B, Aghebati-Maleki L, Hosseinpour-Feizi MA, Shanehbandi D et al. , Chitosan nanoparticles as a dual drug/siRNA delivery system for treatment of colorectal cancer. Immunology letters. 2017. January 1;181:79–86. [DOI] [PubMed] [Google Scholar]
- [27].Valentino JD, Li J, Song J, Rychahou P, Weiss H, Evers M. Novel SiRNA Cotargeting Strategy as Treatment for Colorectal Cancer. Journal of Surgical Research. 2012. February 1;172(2):305–6. [Google Scholar]
- [28].Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K et al. , Off-target effects by siRNA can induce toxic phenotype. Rna. 2006. July 1;12(7): 118896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xue HY, Wong HL. Tailoring nanostructured solid-lipid carriers for time-controlled intracellular siRNA kinetics to sustain RNAi-mediated chemosensitization. Biomaterials. 2011. April 1;32(10):2662–72. [DOI] [PubMed] [Google Scholar]
- [30].Hong J, Huang Y, Li J, Yi F, Zheng J, Huang H et al. , Comprehensive analysis of sequence-specific stability of siRNA. The FASEB Journal. 2010. December;24(12):4844–55. [DOI] [PubMed] [Google Scholar]
- [31].Xu CF, Wang J. Delivery systems for siRNA drug development in cancer therapy. Asian Journal of Pharmaceutical Sciences. 2015. February 1;10(1):1–2. [Google Scholar]
- [32].Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature biotechnology. 2005. April;23(4):457. [DOI] [PubMed] [Google Scholar]
- [33].Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L et al. , Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. Rna. 2006. July 1;12(7): 1179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Petri S, Meister G. siRNA design principles and off-target effects InTarget Identification and Validation in Drug Discovery 2013. (pp. 59–71). Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- [35].Larsson E, Sander C, Marks D. mRNA turnover rate limits siRNA and microRNA efficacy. Molecular systems biology. 2010. January 1;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. Journal of physiology and biochemistry. 2011. March 1;67(1):129–39. [DOI] [PubMed] [Google Scholar]
- [37].Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M et al. , Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5–6):397–402. [DOI] [PubMed] [Google Scholar]
- [38].Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S et al. , MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008. April;27(15):2128–36. [DOI] [PubMed] [Google Scholar]
- [39].Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer?. Cancer Epidemiology and Prevention Biomarkers. 2011. July 1;20(7):1272–86. [DOI] [PubMed] [Google Scholar]
- [40].Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J et al. , MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013. September 1;62(9): 1315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y et al. , Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer research. 2008. June 1;68(11):4123–32. [DOI] [PubMed] [Google Scholar]
- [42].Humphreys KJ, Cobiac L, Le Leu RK, Van der Hoek MB, Michael MZ. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR - 17 – 92 cluster. Molecular Carcinogenesis. 2013. June 1;52(6):459–74. [DOI] [PubMed] [Google Scholar]
- [43].Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S et al. , MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008. April;27(15):2128–36. [DOI] [PubMed] [Google Scholar]
- [44].Fang L, Li H, Wang L, Hu J, Jin T, Wang J et al. , MicroRNA-17–5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014. May;5(10):2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J et al. , miR-204–5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clinical cancer research. 2014. December 1;20(23):6187–99. [DOI] [PubMed] [Google Scholar]
- [46].Tang Q, Zou Z, Zou C, Zhang Q, Huang R, Guan X et al. , MicroRNA-93 suppress colorectal cancer development via Wnt/p-catenin pathway downregulating. Tumor Biology. 2015. March 1;36(3):1701–10. [DOI] [PubMed] [Google Scholar]
- [47].Zhang N, Lu C, Chen L. miR-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancer. Oncology letters. 2016. December 1;12(6):4589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y et al. , Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009. March;28(10):1385. [DOI] [PubMed] [Google Scholar]
- [49].Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ et al. , MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. British journal of cancer. 2009. August;101(4):699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang Y, Wang Z, Chen M, Peng L, Wang X, Ma Q et al. , MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Molecular cancer. 2012. December;11(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dean NM, Bennett CF. Antisense oligonucleotide-based therapeutics for cancer. Oncogene. 2003. December;22(56):9087. [DOI] [PubMed] [Google Scholar]
- [52].Paterson BM, Roberts BE, Kuff EL. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proceedings of the National Academy of Sciences. 1977. October 1;74(10):4370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F et al. , Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. The lancet oncology. 2005. May 1;6(5):279–86. [DOI] [PubMed] [Google Scholar]
- [54].Najar AG, Pashaei-Asl R, Omidi Y, Farajnia S, Nourazarian AR. EGFR antisense oligonucleotides encapsulated with nanoparticles decrease EGFR, MAPK1 and STAT5 expression in a human colon cancer cell line. Asian Pacific Journal of Cancer Prevention. 2013;14(1):495–8. [DOI] [PubMed] [Google Scholar]
- [55].Tao YJ, Li YJ, Zheng W, Zhao JJ, Guo MM, Zhou Y et al. , Antisense oligonucleotides against microRNA-21 reduced the proliferation and migration of human colon carcinoma cells. Cancer cell international. 2015. December;15(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Abaza MS, Al-Saffar A, Al-Sawan S, Al-Attiyah R. c-Myc antisense oligonucleotides sensitize human colorectal cancer cells to chemotherapeutic drugs. Tumor Biology. 2008;29(5):287–303. [DOI] [PubMed] [Google Scholar]
- [57].Wang H, Nan L, Yu D, Lindsey JR, Agrawal S, Zhang R. Anti-tumor efficacy of a novel antisense anti-MDM2 mixed-backbone oligonucleotide in human colon cancer models: p53-dependent and p53-independent mechanisms. Molecular Medicine. 2002. April 1;8(4):185–99. [PMC free article] [PubMed] [Google Scholar]
- [58].Ross SJ, Revenko AS, Hanson LL, Ellston R, Staniszewska A, Whalley N et al. , Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Science translational medicine. 2017. June 14;9(394):eaal5253. [DOI] [PubMed] [Google Scholar]
- [59].Gopinath SC. Methods developed for SELEX. Analytical and bioanalytical chemistry. 2007. January 1;387(1):171–82. [DOI] [PubMed] [Google Scholar]
- [60].Patel DJ, Suri AK, Jiang F, Jiang L, Fan P, Kumar RA et al. , Structure, recognition and adaptive binding in RNA aptamer complexes. Journal of molecular biology. 1997. October 10;272(5):645–64. [DOI] [PubMed] [Google Scholar]
- [61].Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA et al. , In vivo selection of tumor-targeting RNA motifs. Nature chemical biology. 2010. January;6(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mi J, Ray P, Liu J, Kuan CT, Xu J, Hsu D et al. , In vivo selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. Molecular Therapy-Nucleic Acids. 2016. January 1;5:e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wirth T, Soeth E, Czubayko F, Juhl H. Inhibition of endogenous carcinoembryonic antigen (CEA) increases the apoptotic rate of colon cancer cells and inhibits metastatic tumor growth. Clinical & experimental metastasis. 2002. March 1;19(2): 155–60. [DOI] [PubMed] [Google Scholar]
- [64].Lee YJ, Han SR, Kim NY, Lee SH, Jeong JS, Lee SW. An RNA aptamer that binds carcinoembryonic antigen inhibits hepatic metastasis of colon cancer cells in mice. Gastroenterology. 2012. July 1;143(1):155–65. [DOI] [PubMed] [Google Scholar]
- [65].Sun H, Zu Y. Aptamers and their applications in nanomedicine. Small. 2015. May;11(20):2352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nature reviews Drug discovery. 2010. July;9(7):537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xiang D, Zheng C, Zhou SF, Qiao S, Tran PH, Pu C et al. , Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics. 2015;5(10):1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].AlShamaileh H, Wang T, Xiang D, Yin W, Tran PH, Barrero RA et al. , Aptamer-mediated survivin RNAi enables 5-fluorouracil to eliminate colorectal cancer stem cells. Scientific reports. 2017. July 19;7(1):5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer - doxorubicin physical conjugate as a novel targeted drug - delivery platform. Angewandte chemie international edition. 2006. December 11;45(48):8149–52. [DOI] [PubMed] [Google Scholar]
- [70].Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY et al. , Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS nano. 2015. February 10;9(2):1108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002. July;418(6894):222. [DOI] [PubMed] [Google Scholar]
- [72].Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Current opinion in pharmacology. 2010. October 1;10(5):551–6. [DOI] [PubMed] [Google Scholar]
- [73].Tokunaga T, Tsuchida T, Kijima H, Okamoto K, Oshika Y, Sawa N et al. , Ribozyme-mediated inactivation of mutant K-ras oncogene in a colon cancer cell line. British journal of cancer. 2000. September;83(6):833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jeong JS, Lee SW, Hong SH, Lee YJ, Jung HI, Cho KS et al. , Antitumor effects of systemically delivered adenovirus harboring trans-splicing ribozyme in intrahepatic colon cancer mouse model. Clinical Cancer Research. 2008. January 1;14(1):281–90. [DOI] [PubMed] [Google Scholar]
- [75].Nagata J, Kijima H, Hatanaka H, Asai S, Miyachi H, Takagi A et al. , Reversal of cisplatin and multidrug resistance by ribozyme-mediated glutathione suppression. Biochemical and biophysical research communications. 2001. August 17;286(2):406–13. [DOI] [PubMed] [Google Scholar]
- [76].Kobayashi H, Eckhardt SG, Lockridge JA, Rothenberg ML, Sandler AB, O’Bryant CL et al. , Safety and pharmacokinetic study of RPI. 4610 (ANGIOZYME), an anti-VEGFR-1 ribozyme, in combination with carboplatin and paclitaxel in patients with advanced solid tumors. Cancer chemotherapy and pharmacology. 2005. October 1;56(4):329–36. [DOI] [PubMed] [Google Scholar]
- [77].Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2014. November 1;1839(11):1097–109. [DOI] [PubMed] [Google Scholar]
- [78].Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z et al. , Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014. May;24(5):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H et al. , Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2014. November 27;30(2):303–10. [DOI] [PubMed] [Google Scholar]
- [80].Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. , Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010. August 6;329(5992):689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Damas ND, Marcatti M, Come C, Christensen LL, Nielsen MM, Baumgartner R et al. , SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nature communications. 2016. December 22;7:13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y et al. , LncRNA—UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204–5p. Scientific reports. 2016. April 5;6:23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A et al. , LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Molecular Therapy-Nucleic Acids. 2018. September 7;12:229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomedicine & pharmacotherapy. 2016. July 1;81:152–9. [DOI] [PubMed] [Google Scholar]
- [85].Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY et al. , The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/p-catenin signaling. Molecular cancer. 2017. December;16(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Forrest ME, Saiakhova A, Beard L, Buchner DA, Scacheri PC, LaFramboise T et al. , Colon cancer-upregulated long non-coding RNA lincDUSP regulates cell cycle genes and potentiates resistance to apoptosis. Scientific reports. 2018. May 9;8(1):7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. International journal of clinical and experimental pathology. 2015;8(9):11480. [PMC free article] [PubMed] [Google Scholar]
- [88].Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2016. May 2;23(6):303–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B et al. , Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer letters. 2017. December 1;410:50–62. [DOI] [PubMed] [Google Scholar]
- [90].Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P et al. , High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. International journal of clinical and experimental pathology. 2014;7(6):3174. [PMC free article] [PubMed] [Google Scholar]
- [91].Peng W, Wang Z, Fan H. LncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of Akt signaling. Pathology & Oncology Research. 2017. July 1;23(3):651–6. [DOI] [PubMed] [Google Scholar]
- [92].Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Brazilian Journal of Medical and Biological Research. 2017;50(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nature chemical biology. 2007. March;3(3):166. [DOI] [PubMed] [Google Scholar]
- [94].Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S et al. , Small dsRNAs induce transcriptional activation in human cells. Proceedings of the National Academy of Sciences. 2006. November 14;103(46):17337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hu J, Chen Z, Xia D, Wu J, Xu H, Ye ZQ. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochemical Journal. 2012. November 1;447(3):407–16. [DOI] [PubMed] [Google Scholar]
- [96].Yang K, Shen J, Xie YQ, Lin YW, Qin J, Mao QQ et al. , Promoter-targeted double-stranded small RNAs activate PAWR gene expression in human cancer cells. The international journal of biochemistry & cell biology. 2013. July 1;45(7):1338–46. [DOI] [PubMed] [Google Scholar]
- [97].Wang LL, Guo HH, Zhan Y, Feng CL, Huang S, Han YX et al. , Specific up-regulation of p21 by a small active RNA sequence suppresses human colorectal cancer growth. Oncotarget. 2017. April 11;8(15):25055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nature biotechnology. 2017. March;35(3):222. [DOI] [PubMed] [Google Scholar]
- [99].Shukla S, Sumaria CS, Pradeepkumar PI. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. ChemMedChem. 2010. March 1;5(3):328–49. [DOI] [PubMed] [Google Scholar]
- [100].Fluiter K, Mook OR, Baas F. The therapeutic potential of LNA-modified siRNAs: reduction of off-target effects by chemical modification of the siRNA sequence InsiRNA and miRNA Gene Silencing 2009. (pp. 1–15). Humana Press. [DOI] [PubMed] [Google Scholar]
- [101].Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL et al. , Uniformly modified 2’-deoxy-2’-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. Journal of medicinal chemistry. 1993. April;36(7):831–41. [DOI] [PubMed] [Google Scholar]
- [102].Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB et al. , Pharmacokinetic properties of 2’-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. Journal of Pharmacology and Experimental Therapeutics. 2001. March 1;296(3):890–7. [PubMed] [Google Scholar]
- [103].Sioud M, Furset G, Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochemical and biophysical research communications. 2007. September 14;361(1): 122–6. [DOI] [PubMed] [Google Scholar]
- [104].Geary RS. Antisense oligonucleotide pharmacokinetics and metabolism. Expert opinion on drug metabolism & toxicology. 2009. April 1;5(4):381–91. [DOI] [PubMed] [Google Scholar]
- [105].Gao S, Dagnaes-Hansen F, Nielsen EJ, Wengel J, Besenbacher F, Howard KA et al. , The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Molecular therapy. 2009. July 1;17(7):1225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kitade Y, Akao Y. MicroRNAs and their therapeutic potential for human diseases: microRNAs, miR-143 and-145, function as anti-oncomirs and the application of chemically modified miR-143 as an anti-cancer drug. Journal of pharmacological sciences. 2010:1010080465-. [DOI] [PubMed] [Google Scholar]
- [107].Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nature Reviews Genetics. 2003. May;4(5):346. [DOI] [PubMed] [Google Scholar]
- [108].Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chemical reviews. 2008. December 3;109(2):259–302. [DOI] [PubMed] [Google Scholar]
- [109].Fang J, Nakamura H, Maeda H. The EPR effect: unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Advanced drug delivery reviews. 2011. March 18;63(3):136–51. [DOI] [PubMed] [Google Scholar]
- [110].Tangudu NK, Verma VK, Clemons TD, Beevi SS, Hay T, Mahidhara G et al. , RNA Interference Using c-Myc-Conjugated Nanoparticles Suppresses Breast and Colorectal Cancer Models. Molecular cancer therapeutics. 2015. May 1;14(5):1259–69. [DOI] [PubMed] [Google Scholar]
- [111].Sureban SM, May R, Mondalek FG, Qu D, Ponnurangam S, Pantazis P et al. , Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. Journal of nanobiotechnology. 2011. December;9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Tiernan JP, Perry SL, Verghese ET, West NP, Yeluri S, Jayne DG et al. , Carcinoembryonic antigen is the preferred biomarker for in vivo colorectal cancer targeting. British journal of cancer. 2013. February;108(3):662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Perraud A, Akil H, Nouaille M, Petit D, Labrousse F, Jauberteau MO et al. , Expression of p53 and DR5 in normal and malignant tissues of colorectal cancer: correlation with advanced stages. Oncology reports. 2011. November 1;26(5):1091–7. [DOI] [PubMed] [Google Scholar]
- [114].Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R et al. , Impact of EGFR expression on colorectal cancer patient prognosis and survival. Annals of Oncology. 2005. January 1;16(1):102–8. [DOI] [PubMed] [Google Scholar]
- [115].Kim KS, Lee YK, Kim JS, Koo KH, Hong HJ, Park YS. Targeted gene therapy of LS174 T human colon carcinoma by anti-TAG-72 immunoliposomes. Cancer gene therapy. 2008. May;15(5):331. [DOI] [PubMed] [Google Scholar]
- [116].Shia J, Klimstra DS, Nitzkorski JR, Low PS, Gonen M, Landmann R et al. , Immunohistochemical expression of folate receptor a in colorectal carcinoma: patterns and biological significance. Human pathology. 2008. April 1;39(4):498–505. [DOI] [PubMed] [Google Scholar]
- [117].Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P et al. , Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nature nanotechnology. 2018. January;13(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Weng DE, Masci PA, Radka SF, Jackson TE, Weiss PA, Ganapathi R et al. , A phase I clinical trial of a ribozyme-based angiogenesis inhibitor targeting vascular endothelial growth factor receptor-1 for patients with refractory solid tumors. Molecular cancer therapeutics. 2005. June 1;4(6):948–55. [DOI] [PubMed] [Google Scholar]
- [119].Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L et al. , A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clinical Cancer Research. 2011. October 15;17(20):6582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Duffy AG, Makarova - Rusher OV, Ulahannan SV, Rahma OE, Fioravanti S, Walker M et al. , Modulation of tumor eIF4E by antisense inhibition: A phase I/II translational clinical trial of ISIS 183750—an antisense oligonucleotide against eIF4E—in combination with irinotecan in solid tumors and irinotecan - refractory colorectal cancer. International journal of cancer. 2016. October 1;139(7): 1648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O et al. , First-inhuman phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. Journal of clinical oncology. 2014. November 17;32(36):4141–8. [DOI] [PubMed] [Google Scholar]
- [122].US National Institutes of Health. ClinicalTrials. gov: Safety Study of CALAA-01 to Treat Solid Tumor Cancers. [Google Scholar]
- [123].Zuckerman JE, Gritli I, Tolcher A, Heidel JD, Lim D, Morgan R et al. , Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proceedings of the National Academy of Sciences. 2014. August 5;111(31):11449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006. September;443(7109):359. [DOI] [PubMed] [Google Scholar]
- [125].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell. 2005. January 14; 120(1): 15–20. [DOI] [PubMed] [Google Scholar]
- [126].Park J, Park J, Pei Y, Xu J, Yeo Y. Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Advanced drug delivery reviews. 2016. September 1; 104:93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Geary RS, Norris D, Yu R, Bennett CF. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Advanced drug delivery reviews. 2015. June 29;87:46–51. [DOI] [PubMed] [Google Scholar]


