Abstract
Objectives
To give an overview of the genetic and structural abnormalities occurring in fetuses with nuchal translucency (NT) measurement exceeding the 95th percentile at first‐trimester screening and to investigate which of these abnormalities would be missed if cell‐free fetal DNA (cfDNA) were used as a first‐tier screening test for chromosomal abnormalities.
Methods
This is a national study including 1901 pregnancies with NT≥95th percentile referred to seven university hospitals in the Netherlands between 1 January 2010 and 1 January 2016. All cases with unknown pregnancy outcome were excluded. Results of detailed ultrasound examinations, karyotyping, genotyping, pregnancy and neonatal outcomes, investigation by a clinical geneticist and post‐mortem investigations were collected.
Results
In total, 821 (43%) pregnancies had at least one abnormality. The rate of abnormalities was 21% for fetuses with NT between 95th and 99th percentile and 62% for fetuses with NT≥99th percentile. Prevalence of single‐gene disorders, submicroscopic, chromosomal and structural abnormalities was 2%, 2%, 30% and 9%, respectively.
Conclusion
Although cfDNA is superior to the combined test, especially for the detection of trisomy 21, 34% of the congenital abnormalities occurring in fetuses with increased NT may remain undetected in the first trimester of pregnancy, unless cfDNA is used in combination with fetal sonographic assessment, including NT measurement.
Short abstract
What's already known about this topic?
Nuchal translucency is associated with a wide range of chromosomal and structural abnormalities.
What does this study add?
If cell‐free DNA were used as the only first trimester screening test, 34% of fetal congenital abnormalities would be missed in the first trimester of pregnancy.
In high‐risk pregnancies with increased nuchal translucency (NT≥95th percentile), 23% of abnormalities are found in fetuses with NT between 95th and 99th percentile.
What's already known about this topic?
Nuchal translucency is associated with a wide range of chromosomal and structural abnormalities.
What does this study add?
If cell‐free DNA were used as the only first trimester screening test, 34% of fetal congenital abnormalities would be missed in the first trimester of pregnancy.
In high‐risk pregnancies with increased nuchal translucency (NT≥95th percentile), 23% of abnormalities are found in fetuses with NT between 95th and 99th percentile.
1. INTRODUCTION
Nuchal translucency (NT), defined as the subcutaneous accumulation of fluid behind the fetal neck, can effectively be measured by an ultrasound investigation between 11 and 13+6 weeks of gestation. Nuchal translucency was first described in 1992 by Nicolaides 1 as a marker for fetal chromosomal abnormalities and especially Down syndrome. Since its first appearance in the prenatal screening paradigm, many authors have studied the value of NT measurement for the detection of fetal congenital abnormalities and it is now well established that an increased NT (defined as NT≥3.5 mm – corresponding to the 99th centile) is not only associated with chromosomal abnormalities but also with a wide range of structural defects2, 3 and genetic aberrations.2, 4, 5 In the Netherlands, NT measurement is offered since 2007 as part of the first trimester combined test (CT). Since 2017, genome‐wide cell‐free fetal DNA (cfDNA) has become the preferred first tier screening test for fetal aneuploidies, next to the CT. Women are informed on the test characteristics of the two tests and, owing to the better performance6 of cfDNA, the lower false positive rates7, 8, 9, 10, 11 and the possibility to detect large fetal chromosomal aberrations,12 the proportion of women choosing for the CT has dramatically dropped.13 Now the CT is mainly performed in di‐chorionic multiple pregnancies. Women choosing for cfDNA generally undergo a dating scan at around 10 weeks' gestation. This means that, although an enlarged NT (≥99th percentile) and fetal abnormalities are considered exclusion criteria for cfDNA, the lack of a systematic scan at 12‐13 weeks prevents the application of these exclusion criteria. Before endorsing this change in first trimester screening policy it is important to assess, once more, the role of the NT measurement as marker for congenital abnormalities that cannot be detected by cfDNA. In this study we aim at doing so on a national dataset obtained before the introduction of cfDNA, when the CT was still being offered as the only screening test during the first trimester of pregnancy.
2. METHODS
2.1. Population
This is a retrospective cohort study including 1901 pregnancies with a nuchal translucency measurement exceeding the 95th percentile, measured at 11‐13+6 weeks' gestation, between 1 January 2010 and 1 January 2016 and referred to one of seven university hospitals in The Netherlands (University Medical Center Groningen, Maastricht University Medical Center, University Medical Centers Amsterdam location VU and AMC, Leiden University Medical Center, Erasmus Medical Center Rotterdam and Radboud University Medical Center Nijmegen). In the Netherlands first trimester CT and NT measurements are performed by sonographers accredited for the performance of the scan14 and working in ultrasound clinics or in fetal medicine units (FMU). Increased NT was defined as a measurement ≥95th percentile by CRL‐adjusted percentiles (CRL range 45‐84 mm), in accordance to the standards of the FMF.15 According to the Dutch screening protocol, a NT≥3.5mm (corresponding to the 99th percentile) is an indication for referral to a tertiary‐care center for further investigation in the form of additional ultrasound scans and genetic testing, irrespective of the CT risk and of whether parents decline karyotyping. The CT risk cut‐off used in the Netherlands is 1:200. Fetuses with NT 95th‐99th percentile and CT risk lower than 1:200 are not referred for karyotyping, except in case of suspicion of fetal abnormalities. In our study, pregnancies with NT 95th‐99th percentile therefore included women referred after the CT either in view of an increased CT risk or because of suspicion of congenital abnormalities or women undergoing a first trimester scan at one of the FMUs because of increased a‐priori risk for congenital abnormalities. The dataset of one of the seven university centers only included patients with NT≥3.5 mm (n=67) referred from other ultrasound clinics in the region, as the center did not perform first trimester screening.
All NT measurement ≥95th percentile by CRL‐adjusted percentiles recorded in the study period were retrieved from local databases. Only cases with known pre‐ and postnatal information, with emphasis on results of detailed ultrasound examination, karyotyping by amniocentesis or chorionic villous sampling (CVS), chromosomal microarrays (CMA), delivery reports, post‐mortem examination and neonatal physical examination by a clinical geneticist of babies with visible abnormalities at birth, were included and analyzed. Genetic testing, in the form of QF‐PCR or karyotyping was offered to all mothers with an increased risk at the CT defined as equal to or more than 1:200. Chromosomal microarray (CMA) investigation was offered in case of NT≥3.5mm and/or fetal structural abnormalities at the ultrasound investigation. All centers used a cut‐off of 5MB for CMA, except for one center using a cut‐off of 0.15MB. As the study covered six year the availability and indications for CMA analysis changed in the course of the study. Moreover, not all the centers used the same protocols for offering CMA investigation, especially when the increased NT was isolated.
2.2. Data analysis
Maternal and clinical data locally stored in the clinical databases (Astraia software gmbh and Mosos Clinical record) of the academic hospitals were collected and transferred into a single electronic database. SPSS Statistics Version 23.0 (IBM Corporation, NY, USA) was used to perform descriptive and comparative statistics.
2.3. Ethical statement
This study has been approved by the Medical Ethical Committee of the University Medical Center of Groningen.
3. RESULTS
In the study period, a total of 23494 NT measurements were recorded in the databases of the seven medical centers. After exclusion of all cases with NT<95th percentile (n=21486, 91.5%), 2008 (8.5%) fetuses with NT≥95th percentile were included in the analysis. One‐hundred (5%) cases were excluded because of missing follow‐up and 7 cases were excluded because of lack of a final diagnosis. In total 1901 pregnancies with a NT≥95th percentile (Figure 1) were analyzed. Mean maternal age at the time of NT measurement was 34 years (range 18‐48) and the median NT was 3.6 mm (IQR: 2.8 – 5.1 mm). Of all fetuses with NT≥95th percentile, 894 (47%) had a NT between 95th and 99th percentile and 1007 (53%) had a NT≥95th percentile (Table 1).
Figure 1.
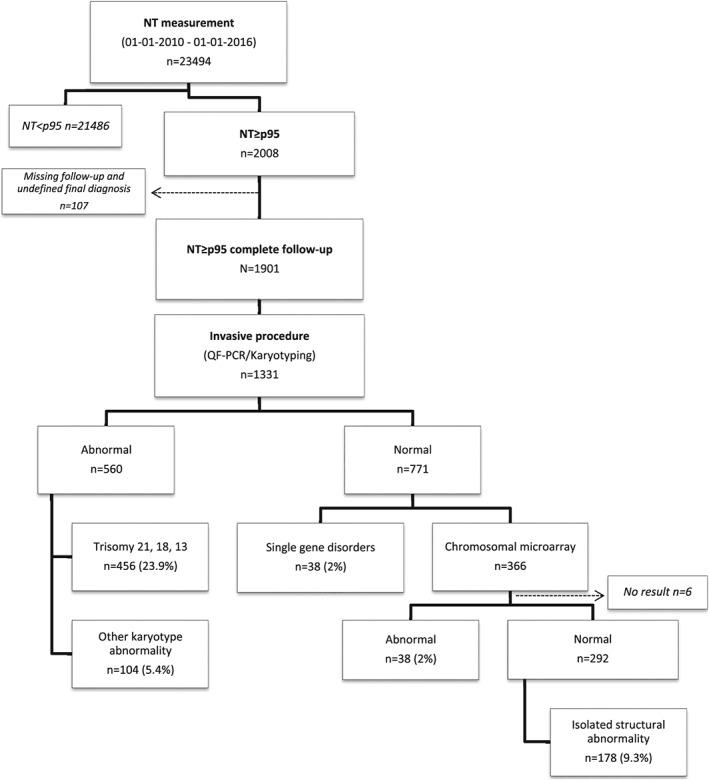
Flowchart patient population
Table 1.
Congenital abnormalities associated with increased NT
| NT (mm) | All fetuses | Congenital abnormality n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| All abnormal fetuses | Detected genetic abnormality (n=636, 33.3%) | Structural (n=178, 9.3%) | ||||||
| Chromosomal (n=560, 29.4%) | Submicroscopic ‡ | Single‐gene disorders § | ||||||
| Total | T21‐18‐13 * | Other ¶ | ||||||
| p95‐p99 | 894 (47) | 190 (21.3) | 124 (13.8) | 112 (12.5) | 12 (1.3) | 8 (0.9) | 5 (0.6) | 53 (5.9) |
| ≥p99 | 1007 (53) | 624 (62) | 436 (43.2) | 344 (34) | 92 (9.1) | 30 (3) | 33 (3.3) | 125 (12.4) |
| 3.5‐4.9 | 492 (26) | 213 (43.3) | 138 (28) | 122 (24.7) | 16 (3.2) | 16 (3.2) | 6 (1.2) | 53 (10.8) |
| 5.0‐6.4 | 199 (10.5) | 153 (76.8) | 113 (56.8) | 87 (43.5) | 26 (13) | 7 (3.5) | 11 (5.5) | 22 (11) |
| 6.5‐7.9 | 155 (8.2) | 129 (83.2) | 93 (60) | 79 (50.6) | 14 (9) | 5 (3.2) | 4 (2.6) | 27 (17.3) |
| ≥8.0 | 162 (8.5) | 129 (79.6) | 92 (56.7) | 56 (34.4) | 36 (22.1) | 2 (1.2) | 12 (7.4) | 23 (14.1) |
| Total | 1901 | 814 (43) | 560 (29.4) | 456 (23.9) | 104 (5.4) | 38 (2.0) | 38 (2.0) | 178 (9.3) |
Trisomy 21 (n=272), trisomy 18 (n=134), trisomy 13 (n=50).
Other chromosomal abnormalities detected by classic karyotyping (Table 4).
Submicroscopic aberrations <5Mb detectable only by chromosomal microarrays.
DNA sequence variations causing single‐gene disorders detectable by sequencing.
3.1. Congenital abnormalities
In total, 814 (43%) pregnancies had at least one abnormality. Abnormalities were observed in 21.3% (n=190) of fetuses with a NT between 95th and 99th percentile and in 62% (n=624) of cases with a NT≥99th percentile. Among fetuses with NT≥99th percentile, the percentage of congenital abnormalities exponentially increased from 43.3% (n=213) in cases with a NT between 3.5 and 4.9 mm to 79.6% (n=129) when NT was ≥8.0 mm (Table 1).
3.2. Genetic abnormalities
Of all fetuses, 33.3% (n=636) had genetic abnormalities (Table 1). Chromosomal abnormalities were diagnosed in 29.4% (n=560) of cases. Single‐gene disorders (Table 2) in 2% (n=38) and submicroscopic genetic aberrations smaller than 5 Mb also in 2% (n=38) of fetuses (Table 3). Among chromosomal abnormalities, trisomy 21, 18 and 13 were observed in 272 (45.5%), 134 (22.4%) and 50 (8.4%) cases, respectively . In the remaining cases, other aneuploidies and karyotype abnormalities (17.4%, n=104) were found (Table 4). Other genetic disorders were especially present at higher degrees of NT enlargement. Of the 38 single‐gene disorders, 20 (52%) were RASopathies (Table 5).
Table 2.
Single‐gene disorders in the study population
| Monogenic disease (gene) | Total | P95‐p99 | ≥p99 |
|---|---|---|---|
| Rasopathies ‐ total | 20 | ||
| ‐Noonan syndrome ‐ total | 18 | 4 | 14 |
| Noonan syndrome (SOS1) | 5 | 2 | 3 |
| Noonan syndrome (PTPN11) | 8 | 2 | 6 |
| Noonan syndrome (LZTR1) | 1 | 1 | |
| Noonan syndrome (RIT1) | 3 | 3 | |
| Noonan syndrome (BRAF) | 1 | 1 | |
| ‐Leopard syndrome (PTPN11) | 1 | 1 | |
| ‐Cardio‐facio‐cutaneous syndrome (MAP 2K1) | 1 | 1 | |
| KAT6A Syndrome (KAT6A) | 1 | 1 | |
| Pena‐Shokeir syndrome (NEB) | 1 | 1 | |
| Cornelia de Lange syndrome (NIPBL) | 2 | 1 | 1 |
| Roberts syndrome (ESCO2) | 2 | 2 | |
| Congenital abnormalities of the kidney and urinary tract (KIF14) | 1 | 1 | |
| Beals syndrome (FBN2) | 1 | 1 | |
| Spinal muscular atrophy type 1 (SMN1) | 1 | 1 | |
| Alpha‐thalassemia x‐linked intellectual disability syndrome (ATRX) | 1 | 1 | |
| Kabuki syndrome (KDMA6) | 1 | 1 | |
| Zellweger syndrome (PEX1) | 1 | 1 | |
| Donnai‐Barrow syndrome (LRP2) | 1 | 1 | |
| APERT syndrome (FGFR2) | 1 | 1 | |
| Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke (MELA) | 1 | 1 | |
| Multiple pterygium syndrome (CHRNG) | 1 | 1 | |
| 38 | 8 | 30 |
Table 3.
Submicroscopic aberrations detected by CMA in the study population
| Microscopic chromosomal arrays | N (total) | n (p95‐p99) | n (≥p99) |
|---|---|---|---|
| ‐ Del 2q13 ‐Pierre Robin sequence | 1 | 1 | |
| ‐ Dup 10q21.3 | 1 | 1 | |
| ‐ Del 3p26.3 | 1 | 1 | |
| ‐ Dup 7q21.12q21.13 | 1 | 1 | |
| ‐ Del 4q28.3 and dup 7p22.3 | 1 | 1 | |
| ‐ Dup 49 kb at 11p14.1 | 1 | 1 | |
| ‐ Interstitial 6q del | 1 | 1 | |
| ‐ Del 15q11.2 ‐ Prader/Angelman Syndrome | 1 | 1 | |
| ‐ 8p23.1 microdeletion syndrome | 1 | 1 | |
|
‐ 22q11 deletion – Di George syndrome ‐ Dup 22q11 ‐ Del sub telomere region in 18p |
5 1 1 |
5 1 1 |
|
| ‐ Dup of 4.1 Mb in 2p25.3 and del of 6.2 Mb in8p23.3p23.1 | 1 | 1 | |
| ‐ Del of 715 kb in 9q34.3 | 1 | 1 | |
| ‐ Dup of 263 kb at 10p12.31 and dup of 85 kb at 17p13.2 | 1 | 1 | |
| ‐ Unbalanced translocation chrom 11 and chrom 14* | 1 | 1 | |
| ‐ 9p del syndrome | 1 | 1 | |
| ‐ Dup of ~435 kb in 11q13.4 | 1 | 1 | |
| ‐ Unbalanced translocation chrom 4 and chrom 18* | 1 | 1 | |
| ‐ Dup 6p25.3 (6p25.3(1,519,929‐1,708,856)x3 pat) | 1 | 1 | |
| ‐ XY translocation, Yp11.2p11.31 translocated in Xp22.33 | 1 | 1 | |
| ‐ 47,XY,+i(12)(p10)de novo | 1 | 1 | |
| ‐ 17q21.31 microdeletion syndrome | 1 | 1 | |
|
‐ 46,XY der(11)t(2;11)(p11.2;q2.4)pat ‐ Del 13q13.3 q33.3 |
1 1 |
1 1 |
|
| ‐ Unbalanced translocation partial monosomy 13p, partial trisomy 16p* | 1 | 1 | |
| ‐ Del 732kb 5p* | 1 | 1 | |
| ‐ Del 15q 26.2 | 1 | 1 | |
| ‐ Del 4p16.2 | 1 | 1 | |
| ‐ 918 kb dup 10q11.21 and 110 kb del 22q12.3 | 1 | 1 | |
| ‐ Dup in 13q12.11 and 245 kb dup in Xp22.33 | 1 | 1 | |
| ‐ 183 kb del in 7p15.3 | 1 | 1 | |
| ‐ Dup 11p15.4(4,041,195‐4,239,042)x4mat | 1 | 1 | |
| ‐ 46,XY, der(11)t(2;11) (p11.2;q2.4) | 1 | 1 | |
| 38 | 8 | 30 |
Breakpoints for these cases could not be retrieved.
Table 4.
Other karyotype abnormalities in the study population
| Karyotype abnormality | N (total) | n (p95‐p99) | n (≥p99) |
|---|---|---|---|
| Aneuploidy | 101 | 12 | 89 |
| 45,X | 74 | 4 | 70 |
| Triploidy | 8 | 2 | 6 |
| 47,XXY | 7 | 3 | 4 |
| Trisomy 22 | 4 | 1 | 3 |
| Trisomy 15 | 2 | 2 | |
| Trisomy 16 | 2 | 1 | 1 |
| Trisomy 7 | 1 | 1 | |
| Trisomy 11 | 1 | 1 | |
| Trisomy 19 | 1 | 1 | |
| Tetrasomy 9 | 1 | 1 | 0 |
| Balanced translocations | 3 | 3 | |
| 46,XX,t(5;6) | 1 | 1 | |
| 46,XX,t(1:9)(q32;q13.3) | 1 | 1 | |
| 46,XY,inv(1)(p11;q21) | 1 | 1 | |
| 104 | 12 | 92 |
Table 5.
NT measurement in fetuses with RASopathies in the study population
| RASopathy | n | NT mm (median, range) |
|---|---|---|
| Noonan syndrome | 18 | 5.9 (3.1–14.3) |
| Leopard syndrome | 1 | 12.0 |
| Cardio‐facio‐cutaneous syndrome | 1 | 16.7 |
| All | 20 | 6.6 (3.1–16.7) |
3.3. Fetal structural abnormalities after genetic anomalies are ruled out
Structural abnormalities were diagnosed in 178 (9.3%) chromosomally normal fetuses. Of the isolated structural abnormalities, cardiac defects were the most common ones (n=74, 3.9%), followed by abnormalities of the urogenital tract (n=20, 1%) and of the central nervous system. Body stalk anomaly was diagnosed in 11 fetuses (0.6%) (Table 6). Multiple congenital abnormalities were diagnosed in 29 fetuses (1.6%). Almost half of the structural defects (n=79, 44.4%) were diagnosed at the time of the NT scan, between 11 and 13+6 weeks of gestation, and another 20.2% (n=36) at the moment the pregnancy was referred to a fetal medicine unit (FMU) for fetal karyotyping. This was in all cases before 18 weeks' gestation. Especially abdominal wall defects were amenable to early diagnosis (detection rate (DR) at the NT scan: 7/9, 77.8%). Of the cardiac abnormalities 21/74 (28.4%) had already been diagnosed at the early scan and another 20/74 (27%) at referral to a tertiary center before 18 weeks' gestation. Finally 26/74 (35%) were diagnosed after 18 weeks. Overall, 64.6% (n=115) of all structural abnormalities were diagnosed before 18 weeks' gestation. Table 6 shows the mean age at termination of pregnancy (TOP) depending on the moment of diagnosis of the abnormalities. The majority of abnormalities (n=79) were detected at NT measurement and the largest number of TOP (n=45) occurred at a mean gestational age of 15 weeks +3 days (Table 6).
Table 6.
Structural abnormalities in the study population divided by organ system and moment of diagnosis
| Structural anomaly | Referral | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total n (%) | At NT measurement n (%) | Time TOP# Mean (n) | <18 weeks n (%) | Time TOP# Mean (n) | >18 weeks n (%) | Time TOP§ Mean (n) | After birth n (%) | Unknown n (%) | |
| Cardiac | 74 (3.9) | 21 (28.4) | 15+1 (9) | 20 (27) | 17+3 (7) | 26 (35.1) | 22+6 (2) | 5 (6.8) | 2 (6.8) |
| Urogenital tract | 20 (1.04) | 4 (20) | 14+3 (2) | 2 (10) | 19+3 (1) | 12 (60) | ‐ | 2 (10) | 0 |
| Body stalk & CNS | 11 (0.6) | 4 (36.4) | 15+1 (3) | 2 (18.2) | 18+3 (1) | 2 (18.2) | ‐ | 1 (9.1) | 2 (18.2) |
| Skeletal | 13 (0.7) | 9 (69.2) | 15+3 (7) | 4 (30.8) | 17+5 (2) | 0 | 0 | 0 | |
| Abdominal wall | 9 (0.5) | 7 (77.8) | 15+3 (3) | 1 (11.1) | ‐ | 1 (11.1) | ‐ | 0 | 0 |
| Pulmonary | 7 (0.4) | 1 (14.3) | 18+3 (1) | 2 (28.6) | ‐ | 2 (28.6) | ‐ | 1 (14.3) | 1 (14.3) |
| Digestive tract | 5 (0.5) | 0 | 0 | 1 (20.0) | ‐ | 3 (60.0) | 1 (20.0) | ||
| Facial | 1 (0.3) | 0 | 1 (100) | ‐ | 0 | 0 | 0 | ||
| Other* | 9 (0.5) | 7 (77.8) | 12+6 (2) | 0 | 0 | 0 | 2 | ||
| MCA¶ | 29 (1.6) | 25 (86.2) | 13+6 (18) | 4 (13.4) | 17+2 (3) | ‐ | ‐ | ‐ | |
| Total | 178 | 79 (44.4) | 15+3 (45) | 36 (20.2) | 18+2 (14) | 44 (24.7) | 22+6 (2) | 12 (6.7) | 8 (4.5) |
Others: 1 case siamese twins, 1 case limb body wall anomaly, 1 case hydrops with AVSD and multiple heart anomalies, 1 case with heterotaxia, 1 case with teratoma, 1 with case severe hydrothorax, 3 cases with hydrops because of unspecified heart anomalies.
Multiple congenital anomalies: (parents declined genetic testing in 8 fetuses. No anomaly was found by karyotyping, QF‐PCR and MCA in 18 cases. 2 fetuses with neuromuscular disorders and 1 case with normal karyotype and failed MCA investigation which was not repeated).
Mean timing of termination of pregnancy (TOP) in weeks.
3.4. NT cut‐off: 95th‐99th percentile
Receiver operating characteristic (ROC) analysis was firstly used to determine the NT cut‐off with the highest sensitivity and specificity for the prediction of congenital abnormalities and, secondly, for the prediction of adverse pregnancy outcome (termination of pregnancy or pregnancy loss). The best cut‐off was at a NT measurement of 3.55 mm; this gave an Area Under the Curve (AUC) of 0.791 with a sensitivity of 74% and specificity of 71% (Figure 2). The best cut‐off for the prediction of adverse pregnancy outcome was at a NT measurement of 3.6 mm, with an AUC of 0.779 with a sensitivity of 70% and specificity of 75% (Figure 3).
Figure 2.
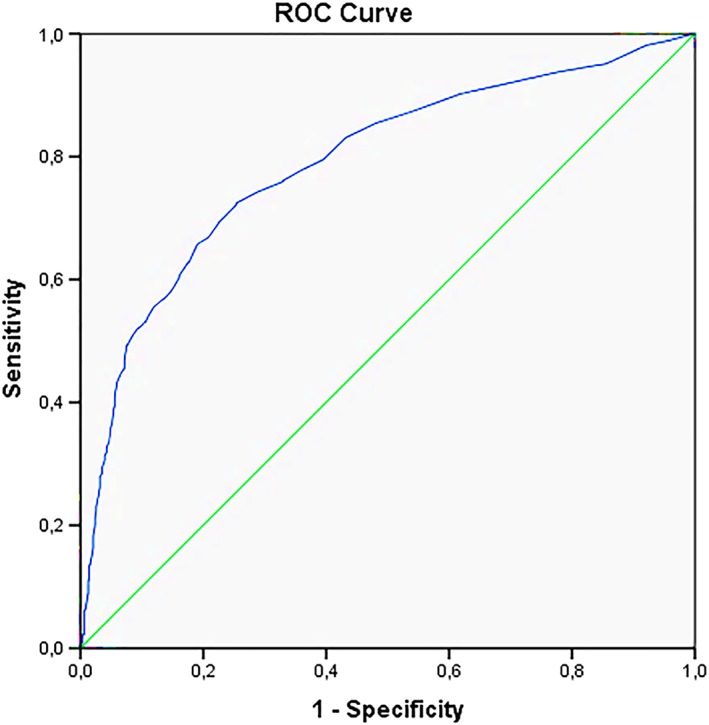
ROC curve all congenital abnormalities [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
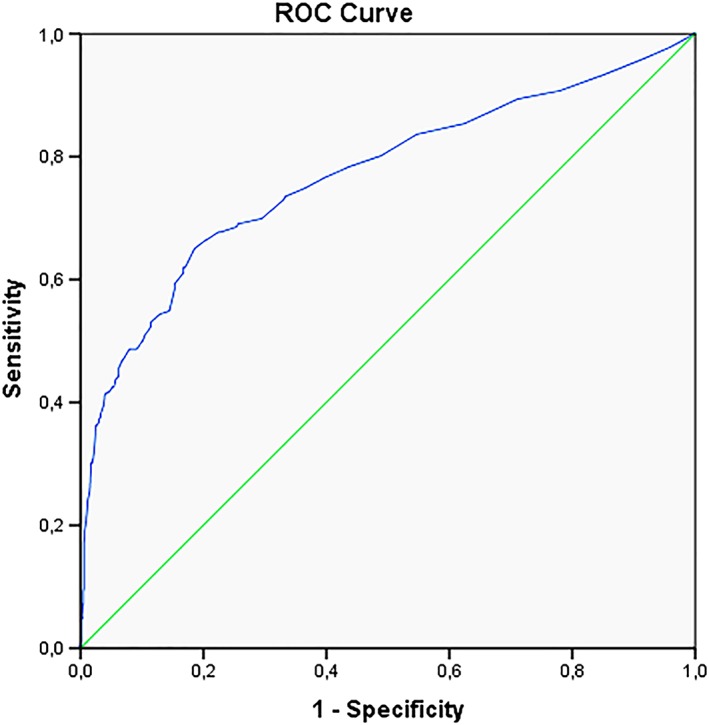
ROC curve pregnancy outcome [Colour figure can be viewed at http://wileyonlinelibrary.com]
4. DISCUSSION
This study shows that 43% of fetuses with NT≥95th percentile had either genetic or structural abnormalities, with rates increasing proportionally to the degree of NT enlargement. Sixty‐two percent of fetuses with NT≥99th percentile had an abnormality and the percentage increased from 43.3% (n=213) for NT between 3.5 and 4.9 mm to 79.6% (n=129) for NT≥8.0 mm.
Of the fetuses with an NT between 95th and 99th percentile one in five had a congenital abnormality. The high rate of abnormalities in this group is most likely due to the fact that especially cases with a NT>95th percentile and with an increased CT risk were referred to a FMU. This selection bias may result in an overestimation of the true prevalence of abnormalities in this group, limiting the extrapolation of the data to the whole group of fetuses with NT between 95th and 99th percentile. Nevertheless, our results are in keeping with previous studies confirming the strong relationship between degree of NT thickness and congenital abnormalities.1, 16, 17, 18
Chromosomal abnormalities (n=560) were diagnosed in 29.4% of the fetuses with NT≥95th percentile. Of these, 456 (81%) were common trisomies. Other chromosomal unbalances were detected in 104 (5.4%) fetuses and 38 single‐gene disorders in an additional 2% of fetuses.
4.1. Chromosomally abnormal fetuses
The rate of chromosomal abnormalities in fetuses with NT≥3.5mm (43%) was higher in this study than that reported by Srebniak and colleagues19 (38%). We confirm that, after trisomy 21, 18 and 13, monosomy X (3.8%) is the next most common numerical chromosomal aberration. Even though this abnormality is usually amenable to first trimester diagnosis when accompanied by a very large nuchal fluid accumulation, this implies that a scan needs to be performed. Since in The Netherlands cfDNA is offered as first tier screening test without sex chromosomal analysis, and because a scan is usually performed to date the pregnancy at a very early gestational age (9‐10 weeks), it can be speculated that of the 74 cases of monosomy X in this study, the majority may have remained undetected, had the current screening paradigm been used. The same would apply to the 8 cases of triploidy in the cohort.
Chromosomal microarray analysis revealed submicroscopic aberrations <5Mb in 3% of the pregnancies with NT≥3.5mm. The frequency of copy number variants (CNVs) in these cases varies considerably in the literature from 0–15%, according to the unselected or selected nature of the pregnant population.20, 21, 22, 23 In our study, CMA was not consistently performed in all cases with enlarged NT, but primarily in cases with ultrasound abnormalities and more systematically in the last years of the study. This may explain our lower rate of pathogenic CNVs, in comparison with the meta‐analysis by Grande and colleagues, reporting 6%‐9% CNVs detected by CMA in fetuses with increased nuchal translucency and with ultrasound abnormalities.24 However our rate of CNV is similar to the 1.6% pathogenic structural unbalanced chromosome aberration plus 0.8 % susceptibility locus for neurodevelopmental disorders reported by Srebniak.19 With advances in laboratory techniques, cfDNA can now detect a wider range of chromosomal abnormalities than only the common trisomies for which it was initially offered.7 These include deletions and duplications >10Mb and maternal sex chromosome abnormalities.25 If in our cohort genome‐wide cfDNA had been performed instead of the CT, we estimate that 4 (10.5%) of the 38 atypical chromosomal aberrations could have potentially been detected. However, as detection rates reported to date are very variable this may be an optimistic assessment.
The use of genome‐wide cfDNA panels impacts the usually extremely low false positive rate of the test, due to cases of confined placental mosaicism (CPM).12 CPM is especially common in sex chromosomes, particularly monosomy X,26 and rare autosomal trisomies such as trisomy 7, 16, 8 and 20. This also negatively affects the positive predictive value of the screening test, which inevitably decreases.12, 26, 27 The net result is that genome‐wide cfDNA undermines the main advantage of this screening test above the CT, which is to reduce the need for invasive testing. Equally concerning is that parents must face a stressful time of insecurity for conditions of uncertain and often limited clinical significance.28 Finally, although there is a trend towards decreasing costs of cfDNA, these are still high. The use of extended panels will inevitably also add to the costs related to the higher screen positive rate requiring, besides more invasive procedures, also extensive genetic counseling. This can be extremely challenging since many incidental findings are rare and their natural history is not yet understood and cannot be predicted.25 Alternatively to universal screening by cfDNA, some authors have suggested a strategy of cfDNA contingent to first‐line screening by CT as more cost effective.29 In this case, women with high risk at the CT are referred for invasive testing and women with intermediate risk (cut‐off depending on financial means) are offered cfDNA.29, 30, 31, 32 This results in higher sensitivity and specificity at considerably lower costs.
4.2. Chromosomally normal fetuses
Of all fetuses with NT>95th percentile and normal karyotype, 9.3% had structural abnormalities. This study confirms that the prevalence of structural abnormalities ranges between 3% and 50%,18 depending on the NT cut‐off and on the study population. The most common abnormalities are cardiac,33, 34 followed by pulmonary, gastrointestinal, genitourinary and musculoskeletal2, 3 abnormalities. Since about half of all structural abnormalities are amenable to early diagnosis,35, 36, 37, 38 ultrasound investigation is essential, regardless of the chosen screening paradigm. In our study, 44.4% of structural abnormalities were diagnosed at the time of NT measurement and an additional 20.2% at referral and in any case before the 20‐week scan. Thus, 64.6% of structural abnormalities would have potentially been missed in the first trimester if cfDNA had been used as first trimester screening test without an early ultrasound scan.
Overall, of the 821 congenital abnormalities, 34% would have remained undiagnosed in the first trimester if cfDNA had been offered as the only screening test. These include sex chromosome abnormalities (n=81), triploidy (n=7) single gene disorders (n=38), submicroscopic aberrations <5Mb (n=38) and structural abnormalities diagnosed in the first trimester (n=115).
Our study confirms the strong association between nuchal fluid accumulation and RASopathies, of which Noonan syndrome is the most common.39 This group represented 52% of the single gene disorders diagnosed in the cohort. Of note is that all the fetuses with RASopathies had all NT larger than 3.1 mm. in a recent study RASopathy analysis is recommended in all cases of NT ≥5mm, even in the absence of additional markers.39
4.3. NT cut‐off
The most widely used NT cut‐off as indication for additional ultrasound and genetic investigations is 3.5mm, corresponding to the 99th centile.40 Our results support that this cut‐off has the highest sensitivity and specificity to detect congenital abnormalities. However, 23.4% of abnormalities were found in pregnancies with a NT measurement between 95th and 99th percentile. Recently, a cut‐off of 3.0mm has been recommended as indication for CMA.41 Although using lower cut‐offs may be less cost‐effective, the choice of undergoing invasive testing at lower NT measurements should be left to parents' discretion and the indication for CMA be primarily determined by the presence of structural abnormalities.
In conclusion, although cfDNA has proven to be superior to the CT for the detection of common trisomies, chromosomal abnormalities are less frequent than structural abnormalities in the general population. Considering the fact that structural abnormalities, and especially many lethal ones, can already be diagnosed in the first trimester of pregnancy, an ultrasound scan remains an essential part of the screening paradigm. In a holistic first trimester screening approach genotyping should be coupled to phenotyping. Finally, new improvements in cfDNA panel coverage are promising for the future, but their clinical utility is still unclear and needs further investigation.
CONFLICT OF INTEREST STATEMENT
All authors declare they have no conflicts of interest.
FUNDING STATEMENT
No funding was available for this study.
Bardi F, Bosschieter PFN, Verheij JBGM, et al. Is there still a role for nuchal translucency measurement in the changing paradigm of first trimester screening?. Prenatal Diagnosis. 2020;40:197–205. 10.1002/pd.5590
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.
REFERENCES
- 1. Nicolaides KH, Azar G, Byrne D, Mansur C, Marks K. Fetal nuchal translucency: ultrasound screening for chromosomal defects in first trimester of pregnancy. BMJ. 1992;304(6831):867‐869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bilardo CM, Muller MA, Pajkrt E, Clur SA, Van Zalen MM, Bijlsma EK. Increased nuchal translucency thickness and normal karyotype: time for parental reassurance. Ultrasound Obstet Gynecol. 2007;30(1):11‐18. [DOI] [PubMed] [Google Scholar]
- 3. Baer RJ, Norton ME, Shaw GM, et al. Risk of selected structural abnormalities in infants after increased nuchal translucency measurement. Am J Obstet Gynecol. 2014;211(6):675.e1‐675.e19. [DOI] [PubMed] [Google Scholar]
- 4. Pilu G, Nicolaides K, Ximenes R JP: The 18‐23 week scan. https://fetalmedicine.org/var/uploads/18-23_Weeks_Scan.pdf [9 June 2018].
- 5. The Fetal Medicine Foundation : Certificates of competence, Training and Certification. https://fetalmedicine.org/training-n-certification/certificates-of-competence [12 June 2018].
- 6. Lichtenbelt KD, Diemel BDM, Koster MPH, et al. Detection of fetal chromosomal anomalies: does nuchal translucency measurement have added value in the era of non‐invasive prenatal testing? Prenat Diagn. 2015;35(7):663‐668. [DOI] [PubMed] [Google Scholar]
- 7. Gil MM, Quezada MS, Bregant B, Ferraro M, Nicolaides KH. Implementation of maternal blood cell‐free DNA testing in early screening for aneuploidies. Ultrasound Obstet Gynecol. 2013;42(1):34‐40. [DOI] [PubMed] [Google Scholar]
- 8. Palomaki GE, Kloza EM, Lambert‐Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011. Nov;13(11):913‐920. [DOI] [PubMed] [Google Scholar]
- 9. Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14(3):296‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204(3):205.e1‐205.e11. [DOI] [PubMed] [Google Scholar]
- 11. Norton ME, Brar H, Weiss J, et al. Non‐Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207(2):137.e1‐137.e8. [DOI] [PubMed] [Google Scholar]
- 12. Grati FR. Implications of fetoplacental mosaicism on cell‐free DNA testing: a review of a common biological phenomenon. Ultrasound Obstet Gynecol. 2016;48(4):415‐423. [DOI] [PubMed] [Google Scholar]
- 13. Van Schendel RV, Page‐Christiaens GCML, Beulen L, et al. Women's Experience with Non‐Invasive Prenatal Testing and Emotional Well‐being and Satisfaction after Test‐Results. J Genet Couns. 2017;26(6):1348‐1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The Fetal Medicine Foundation . Nuchal translucency scan. Certificates of competence. Training & Certification. https://fetalmedicine.org/nuchal-translucency-scan [16 October 2018].
- 15. Wright D, Kagan KO, Molina FS, Gazzoni A, Nicolaides KH. A mixture model of nuchal translucency thickness in screening for chromosomal defects. Ultrasound Obstet Gynecol. 2008;31(4):376‐383. [DOI] [PubMed] [Google Scholar]
- 16. Souka AP, Snijders RJM, Novakov A, Soares W, Nicolaides KH. Defects and syndromes in chromosomally normal fetuses with increased nuchal translucency thickness at 10‐14 weeks of gestation. Ultrasound Obstet Gynecol. 1998;11(6):391‐400. [DOI] [PubMed] [Google Scholar]
- 17. Kagan KO, Avgidou K, Molina FS, Gajewska K, Nicolaides KH. Relation Between Increased Fetal Nuchal Translucency Thickness and Chromosomal Defects. Obstet Gynecol. 2006;107(1):6‐10. [DOI] [PubMed] [Google Scholar]
- 18. Souka AP, Von Kaisenberg CS, Hyett JA, Sonek JD, Nicolaides KH. Increased nuchal translucency with normal karyotype. Am J Obstet Gynecol. 2005;192(4):1005‐1021. [DOI] [PubMed] [Google Scholar]
- 19. Srebniak MI, de Wit MC, Diderich KEM, et al. Enlarged NT (≥3.5 mm) in the first trimester – not all chromosome aberrations can be detected by NIPT. Mol Cytogenet. 2016;9(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaffer LG, Rosenfeld JA, Dabell MP, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn. 2012;32(10):986‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scott F, Murphy K, Carey L, et al. Prenatal diagnosis using combined quantitative fluorescent polymerase chain reaction and array comparative genomic hybridization analysis as a first‐line test: results from over 1000 consecutive cases. Ultrasound Obstet Gynecol. 2013;41(5):500‐507. [DOI] [PubMed] [Google Scholar]
- 22. Hillman SC, McMullan DJ, Hall G, et al. Use of prenatal chromosomal microarray: Prospective cohort study and systematic review and meta‐analysis. Ultrasound Obstet Gynecol. 2013;41(6):610‐620. [DOI] [PubMed] [Google Scholar]
- 23. Huang J, Poon LC, Akolekar R, Choy KW, Leung TY, Nicolaides KH. Is high fetal nuchal translucency associated with submicroscopic chromosomal abnormalities on array CGH? Ultrasound Obstet Gynecol. 2014;43(6):620‐624. [DOI] [PubMed] [Google Scholar]
- 24. Grande M, Jansen FAR, Blumenfeld YJ, et al. Genomic microarray in fetuses with increased nuchal translucency and normal karyotype: a systematic review and meta‐analysis. Ultrasound Obstet Gynecol. 2015;46(6):650‐658. [DOI] [PubMed] [Google Scholar]
- 25. Di Renzo GC, Bartha JL, Bilardo CM. Expanding the indications for cell‐free DNA in the maternal circulation: clinical considerations and implications. Am J Obstet Gynecol. 2019; 220(6):537‐542. [DOI] [PubMed] [Google Scholar]
- 26. Grati FR, Bajaj K, Zanatta V, et al. Implications of fetoplacental mosaicism on cell‐free DNA testing for sex chromosome aneuploidies. Prenat Diagn. 2017;37(10):1017‐1027. [DOI] [PubMed] [Google Scholar]
- 27. Yao H, Zhang L, Zhang H, et al. Noninvasive prenatal genetic testing for fetal aneuploidy detects maternal trisomy X. Prenat Diagn. 2012;32(11):1114‐1116. [DOI] [PubMed] [Google Scholar]
- 28. Benn P, Grati FR. Genome‐wide non‐invasive prenatal screening for all cytogenetically visible imbalances. Ultrasound Obstet Gynecol. 2018;51(4):429‐433. [DOI] [PubMed] [Google Scholar]
- 29. Miltoft CB, Rode L, Ekelund CK, et al. Contingent first‐trimester screening for aneuploidies with cell‐free DNA in a Danish clinical setting. Ultrasound Obstet Gynecol. 2018;51(4):470‐479. [DOI] [PubMed] [Google Scholar]
- 30. Chitty LS, Wright D, Hill M, et al. Uptake, outcomes, and costs of implementing non‐invasive prenatal testing for Down's syndrome into NHS maternity care: prospective cohort study in eight diverse maternity units. BMJ. 2016;354:i3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gil MM, Revello R, Poon LC, Akolekar R, Nicolaides KH. Clinical implementation of routine screening for fetal trisomies in the UKNHS: cell‐free DNA test contingent on results from first‐trimester combined test. Ultrasound Obstet Gynecol. 2016;47(1):45‐52. [DOI] [PubMed] [Google Scholar]
- 32. Persico N, Boito S, Ischia B, et al. Cell‐free DNA testing in the maternal blood in high‐risk pregnancies after first‐trimester combined screening. Prenat Diagn. 2016;36(3):232‐236. [DOI] [PubMed] [Google Scholar]
- 33. Axt‐Fliedner R, Gembruch U. Nuchal translucency and fetal cardiac malformations. Ultraschall Med. 2010;31(2):144‐150. [DOI] [PubMed] [Google Scholar]
- 34. Clur SA, Mathijssen IB, Pajkrt E, et al. Structural heart defects associated with an increased nuchal translucency: 9 years experience in a referral centre. Prenat Diagn. 2008;28(4):347‐354. [DOI] [PubMed] [Google Scholar]
- 35. Grande M, Arigita M, Borobio V, Jimenez JM, Fernandez S, Borrell A. First‐trimester detection of structural abnormalities and the role of aneuploidy markers. Ultrasound Obstet Gynecol. 2012;39(2):157‐163. [DOI] [PubMed] [Google Scholar]
- 36. Syngelaki A, Chelemen T, Dagklis T, Allan L, Nicolaides KH. Challenges in the diagnosis of fetal non‐chromosomal abnormalities at 11‐13 weeks. Prenat Diagn. 2011;31(1):90‐102. [DOI] [PubMed] [Google Scholar]
- 37. Kenkhuis MJA, Bakker M, Bardi F, et al. Effectiveness of a 12‐13 week scan for the early diagnosis of fetal congenital anomalies in the cell‐free DNA era. Ultrasound Obstet Gynecol. 2017;51:463‐469. [DOI] [PubMed] [Google Scholar]
- 38. Karim JN, Roberts NW, Salomon LJ, Papageorghiou AT. Systematic review of first trimester ultrasound screening in detecting fetal structural anomalies and factors affecting screening performance. Ultrasound Obstet Gynecol. 2016;50:429‐441. [DOI] [PubMed] [Google Scholar]
- 39. Stuurman KE, Joosten M, van der Burgt I, et al. Prenatal ultrasound findings of rasopathies in a cohort of 424 fetuses: update on genetic testing in the NGS era. J Med Genet. 2019;56(10):654‐661. doi:10.1136 [DOI] [PubMed] [Google Scholar]
- 40. Snijders RJM, Noble P, Sebire N, Souka A, Nicolaides KH. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal‐translucency thickness at 10‐14 weeks of gestation. Lancet. 1998;352(9125):343‐346. [DOI] [PubMed] [Google Scholar]
- 41. Maya I, Yacobson S, Kahana S, et al. Cut‐off value of nuchal translucency as indication for chromosomal microarray analysis. Ultrasound Obstet Gynecol. 2017;50(3):332‐335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.


