Abstract
During evolution, several algae and plants became heterotrophic and lost photosynthesis; however, in most cases, a nonphotosynthetic plastid was maintained. Among these organisms, the colourless alga Polytomella parva is a special case, as its plastid is devoid of any DNA, but is maintained for specific metabolic tasks carried out by nuclear encoded enzymes. This makes P. parva attractive to study molecular events underlying the transition from autotrophic to heterotrophic lifestyle. Here we characterize metabolic adaptation strategies of P. parva in comparison to the closely related photosynthetic alga Chlamydomonas reinhardtii with a focus on the role of plastid‐localized PII signalling protein. Polytomella parva accumulates significantly higher amounts of most TCA cycle intermediates as well as glutamate, aspartate and arginine, the latter being specific for the colourless plastid. Correlating with the altered metabolite status, the carbon/nitrogen sensory PII signalling protein and its regulatory target N‐acetyl‐l‐glutamate‐kinase (NAGK; the controlling enzyme of arginine biosynthesis) show unique features: They have co‐evolved into a stable hetero‐oligomeric complex, irrespective of effector molecules. The PII signalling protein, so far known as a transiently interacting signalling protein, appears as a permanent subunit of the enzyme NAGK. NAGK requires PII to properly sense the feedback inhibitor arginine, and moreover, PII tunes arginine‐inhibition in response to glutamine. No other PII effector molecules interfere, indicating that the PII‐NAGK system in P. parva has lost the ability to estimate the cellular energy and carbon status but has specialized to provide an entirely glutamine‐dependent arginine feedback control, highlighting the evolutionary plasticity of PII signalling system.
Keywords: algal metabolomics, arginine biosynthesis, N‐acetyl‐l‐glutamate kinase, Nonphotosynthetic plastids, PII‐signalling, TCA/GS‐GOGAT cycles
The major target of the PII signalling protein in Archaeplastida is N‐acetyl glutamate kinase (NAGK). PII is known as a transiently interacting signalling protein. In this work, we demonstrate that NAGK co‐evolved with PII signalling protein towards a stable hetero‐oligomeric complex that tunes arginine feedback inhibition (+ve/−ve) in response to glutamine status in nonphotosynthetic alga Polytomella parva. This is likely to be a metabolic adaptation strategy in line with the evolutionary loss of photosynthesis in this species.
Abbreviations
- 2‐OG
2‐oxoglutarate
- Arg
arginine
- CrNAGK
Chlamydomonas reinhardtii NAGK protein
- CrPII
Chlamydomonas reinhardtii PII protein
- Gln
glutamine
- GOGAT
glutamate synthase
- GS
glutamine synthase
- NAGK
N‐acetyl‐l‐glutamate kinase
- OsPII
Oryza sativa PII protein
- PEP
phosphoenolpyruvate
- PpaNAGK
Polytomella parva NAGK protein
- PpaPII
Polytomella parva PII protein
- SEC‐MALS
size exclusion chromatography coupled with multiangle light scattering
- SPR
surface plasmon resonance spectroscopy
- TCA
tricarboxylic acid cycle
Introduction
The loss of photosynthesis is always accompanied by heterotrophic lifestyles and arose in diverse eukaryotic lineages 1. In the course of evolution, many algal species and land plants lost photosynthesis and became heterotrophic 1, 2, 3, 4, 5. Most of these nonphotosynthetic organisms still retain the plastids, which contain a small genome to carry out various nonphotosynthetic metabolic reactions 1, 5, 6. Several of these colourless algae evolved into parasites, such as the Apicomplexa lineage. Polytomella is a genus of colourless, free‐living unicellular nonphotosynthetic green algae, closely related to the photosynthetic green alga Chlamydomonas reinhardtii 1, 7, 8. Recently, the plastids of Polytomella spp. have been identified to be the first algae harbouring‐plastid devoid of any plastid genomes 1, while, the Rafflesia genus was identified to be the first parasitic plant with no recognizable plastid genome 5.
RNA‐seq analysis of Polytomella parva uncovered transcripts for a large set of nuclear encoded, plastid‐targeted enzymes mainly involved in carbohydrate and starch metabolism as well as amino acid and fatty acid biosynthesis 1. This implies that P. parva has maintained a nonphotosynthetic plastid for metabolic purposes as a specialized anabolic organelle 1, 2, 7, 8. Therefore, P. parva is an attractive model system for exploring the evolutionary pressure to maintain plastids in the absence photosynthesis. Up to now, the question how the primary metabolism in Polytomella spp., has adapted to the loss of photosynthesis has not been experimentally approached. Therefore, we started a first characterization concerning the biochemical and metabolic adaptation strategies of P. parva in response to different nitrogen regimes. Notably, P. parva was found to possess nuclear genes predicted to encode a plastid‐targeted PII signalling protein (PpaPII, plastid‐targeted) and the enzymes of the ornithine/arginine biosynthesis pathway, in particular the target of PII regulation, N‐acetyl‐l‐glutamate kinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/8.html) (PpaNAGK, plastid‐targeted), which catalyses the commented step of arginine biosynthesis. These two proteins co‐evolved in the course of endosymbiotic generation of plastids 9 and therefore, represent a prominent test case to address issues of metabolic adaptation strategies.
The PII signalling proteins constitute a large superfamily occurring in all domains of life 10, 11. The PII proteins are trimeric in the structure and are present in almost all bacteria, in nitrogen‐fixing archaea 10, 11 and in the eukaryotic Archaeplastida domain 9, 10, 11, 12. The PII homologues (GlnB and GlnK), which contain the conserved PROSITE motifs (PTM‐site: PS00496 and C‐terminal signature: PS00638) 13, are referred as canonical PII proteins (reviewed in 9, 10, 11, 12). The PII members, which demonstrate the same trimeric architectural principle as GlnB/GlnK proteins but lack their typical PROSITE signature pattern, are termed as the PII‐like proteins 10, 14.
In contrast to the high structural conservation of PII proteins, the PII controlled targets are distinct and versatile in different phylogenetic lineages. In eukaryotes, PII homologues have only been identified and characterized in Chloroplastida (green algae and land plants), where they are nuclear encoded 15, 16, 17 and in Rhodophyta, where they are coded by the plastid genome 12, 18. In both groups of eukaryotic phototrophs, PII is localized in the plastid 15, 16, 17, 18. In cyanobacteria and plants, the PII signalling proteins were found to regulate the activity of NAGK, the controlling enzyme of arginine biosynthesis 16, 17, 18, 19, 20. In green algae and land plants, NAGK activity is controlled by the cellular glutamine (Gln) levels via glutamine‐dependent PII‐NAGK complex formation, which leads to increased enzyme activity 16, 17. In contrast to PII proteins from Chloroplastida, PII of the red alga Porphyra purpurea controls NAGK in a similar way as shown in cyanobacteria: PII‐NAGK complex formation is antagonized 2‐oxoglutarate (2‐OG) but independent of glutamine 18. Through complex formation with PII, NAGK gets relieved from feedback inhibition by arginine (Arg) 16, 17, 18, 19, 20, leading to enhanced activity. It appears that the biochemical features of PII‐NAGK complexes reflect the metabolic adaptations during endosymbiotic evolution 9.
The present study is the first to address metabolic adaptation strategies of the nonphotosynthetic alga P. parva in response to nitrogen limitation in comparison to the closely related photosynthetic alga C. reinhardtii by performing a relative quantification of the intracellular metabolites. To gain mechanistic insights in the metabolic specialization of the P. parva plastid, we studied the PII‐mediated regulation of NAGK activity, which is a key step in the control of arginine biosynthesis. Surprisingly, we found unique features not described for PII‐NAGK complexes so far. PpaPII forms an unusually stable complex with PpaNAGK, irrespective of effector molecules. In this complex, PII tunes arginine feedback inhibition of NAGK specifically in response to varying glutamine levels, whereas the tricarboxylic acid (TCA) cycle intermediate 2‐oxoglutarate (2‐OG) and ATP/ADP nucleotides had no regulatory effect. These data indicate that the PII‐NAGK system in this nonphotosynthetic alga evolved into a hetero‐oligomeric enzyme complex that has lost the ability to estimate the current energy and carbon status of the cells but specifically responds with high sensitivity to the arginine/glutamine level of the cells.
Results
Metabolomic analysis
To investigate the impact of the nonphotosynthetic lifestyle of P. parva on its metabolomic landscape, we applied an untargeted LC‐MS metabolomics approach to characterize the changes in the metabolomic pool sizes of P. parva cells under different nitrogen regimes (Fig. 1A,B) in comparison to the closely related alga C. reinhardtii grown under optimal mixotrophic conditions 21. We were able to identify 11 metabolites of the central carbon (C) and nitrogen (N) metabolism, mainly of TCA and GS‐GOGAT cycles (Fig. 1 and Table S1), which were significantly different between C. reinhardtii and P. parva and changed upon shift from high to low nitrogen.
Figure 1.
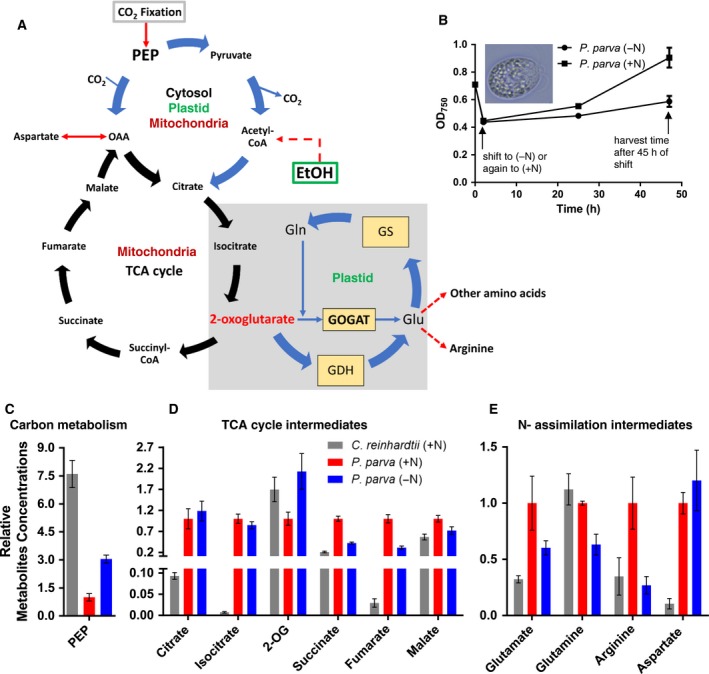
Central C‐ and N‐ metabolism in nonphotosynthetic alga Polytomella parva and in photosynthetic alga Chlamydomonas reinhardtii. (A) Inferred metabolic pathways in non‐ and photosynthetic algae P. parva and C. reinhardtii, respectively, with special reference to the TCA‐ and GS/GOGAT‐cycles. The scheme of metabolic pathway is compartmentalized in terms of mitochondrion, plastid and cytosol, according to 21. Polytomella parva uses ethanol as a carbon source for, while C. reinhardtii fixes CO 2 or/and uses acetate as external carbon. (B) Growth of P. parva (inset; scale 10 μm) under N‐limited and N‐rich conditions. The arrow at the right end shows the time point (45 h) of harvesting P. parva for metabolite analysis. The experiment was started with an exponentially growing culture of P. parva under nitrogen‐rich conditions, which was collected and shifted to N‐limiting (0.375‐mm NH 4 +) conditions or back again to the N‐rich conditions (7.5‐mm NH 4 +) (arrow left). Significant metabolic alterations of (C) PEP (C‐metabolism), (D) TCA‐cycle intermediates and (E) the major amino acids of N‐assimilation reactions and GS/GOGAT‐cycle intermediates within the nonphotosynthetic algae P. parva cells after shift from rich‐ to low‐nitrogen conditions in comparison to the photosynthetic algae C. reinhardtii under rich nitrogen (7.5‐mm NH 4 +) condition. The metabolite concentrations are relative to P. parva cells under high‐nitrogen supply (normalized to 1.0, red bars) for three independent replicates, and the standard deviation (SD) is indicated by error bars.
Remarkably, the pools of most tricarboxylic acid (TCA) cycle intermediates (citrate, isocitrate, succinate and fumarate), except malate and 2‐OG were much higher in P. parva cells (Fig. 1). In striking contrast to most TCA intermediates, the levels of 2‐OG were lower in P. parva than in C. reinhardtii. This suggests an efficient nitrogen assimilatory system in P. parva that constantly keeps the 2‐OG levels relatively low, as compared to other TCA cycle intermediates. During nitrogen deprivation, the 2‐OG level increases in P. parva, as expected 22, since the consumption of 2‐OG through nitrogen assimilatory reactions is reduced. Intriguingly, the levels of phosphoenolpyruvate (PEP) show the inverse pattern than most TCA intermediates. Of note, PEP is synthesized in phototrophes from the CO2 fixation product 3‐phosphoglycerate (3PGA) through a few glycolytic reactions 23.
Under nitrogen‐rich conditions, P. parva cells accumulate around 2.9‐fold more arginine, 9.4‐fold more aspartate and 3.1‐fold more glutamate than the C. reinhardtii cells. This suggests again, in agreement with lower levels of 2‐OG, an efficient nitrogen assimilatory system. Nitrogen assimilation and arginine synthesis appears to take place in the colourless plastid 1, as the corresponding C. reinhardtii homologous enzymes, glutamine synthase (GS) and glutamate synthase (GOGAT) as well as arginine biosynthesis enzymes are plastid localized 1, 21. In contrast to the elevated levels of Glu, Asp and Arg in P. parva, the Gln‐levels were relatively low, which suggests a high activity of GOGAT that constantly consumes glutamine and 2‐OG to produce glutamate. The high levels of Glu correlate with high Arg levels, indicating that the controlling enzyme of the ornithine/arginine pathway, PpaNAGK, should be adapted to the specific metabolic alterations in P. parva.
Upon shift of P. parva cells from nitrogen‐rich to ‐poor conditions, marked changes were mainly observed for metabolites of the TCA and GS/GOGAT cycles. The arginine, succinate and fumarate pools dropped by more than 50% (Fig. 1), whereas the malate, glutamate and glutamine (the primary nitrogen assimilation product) pools dropped by 30% to 40% (Fig. 1). The amount of aspartate increased slightly, which can be explained by diminished aspartate consumption for arginine synthesis through the argininosuccinate synthase reaction. As expected, the central TCA product 2‐OG showed a more than twofold increase upon shift to low nitrogen condition, whereas the 2‐OG precursors citrate and isocitrate did not show marked changes. The threefold increase of PEP levels under N‐limitation reflects the shift in the C:N ratio during external N‐limitation. Due to the limitation of nitrogen assimilation reactions under N‐poor conditions, the decreased utilization of glycolytic intermediates for various amino acid biosynthesis reactions could lead to increased levels of the glycolytic metabolite PEP. Overall, these metabolic changes reflect the limitation of nitrogen availability, which goes along with a slightly reduced growth of P. parva cells under these nitrogen‐poor conditions (Fig. 1B).
Together, the main metabolic difference between the photosynthetic alga C. reinhardtii and its heterotrophic relative P. parva concerns major metabolites of the TCA cycle, and nitrogen assimilation products glutamate, aspartate as well as arginine as a final nitrogen‐storage molecule 24. The higher levels of TCA intermediates agree with the dominance of mitochondrial metabolism in P. parva. The high levels of nitrogen assimilation products, in particular the nitrogen‐storage amino acid arginine, which is produced in the colourless plastid, indicates a prominent metabolic role of this organelle. To get mechanistic insights into the high Arg levels, we decided to study the interaction of the nitrogen regulatory PII protein with the key enzyme of arginine synthesis, NAGK in P. parva, which proved crucial in the activation of the committed step of arginine biosynthesis in plants, photosynthetic algae and cyanobacteria 9, 10, 11, 12, 16, 17, 18, 19, 20, 24, 25, 26.
PpaPII is a canonical plant PII protein
The predicted full‐length PpaPII polypeptide encoded by the P. parva GLB1 gene consists of 209 amino acids with a calculated molecular weight of 22 745 Da and contains predicted plastid transit peptide using ChloroP 1.1 Server (amino acid residues 1‐49). As expected, the mature PpaPII demonstrated the highest degree of identity with C. reinhardtii PII (61.78%). We performed primary sequence alignment of PII from P. parva with canonical PII proteins from other Archaeplastida and bacteria. The alignment of PpaPII indicates extremely high local identities over two signature patterns that have been defined at the PROSITE (PS00496 and PS00638) in all canonical PII proteins (Fig. 2) 13, 27. Moreover, similar to PII homologues of Chloroplastida, PpaPII protein contains the unique C‐terminal segment including the Q‐loop, which is responsible for glutamine sensing 16, 17. The alignment also showed a high degree of conservation of the functional important regions of PII proteins, including the T‐loop residues, which are involved in NAGK interactions 17, 24, 25, 26. The only noticeable variable in a contact site to NAGK concerns the tip to the T‐loop, with a Gly residue in PpaPII (corresponding to Arg47 in bacterial PII proteins). The NAG1 gene of P. parva encodes the full‐length NAGK polypeptide (PpaNAGK) consisting of 329 amino acids with a calculated molecular weight of 34 819 Da comprising a putative N‐terminal plastid transit peptide. The PpaNAGK sequence exhibits the N‐terminal signature pattern of arginine‐sensitive NAGK enzymes and the allosteric arginine‐binding site appears conserved (Fig. 3) 28. The calculated molecular weight of the predicted mature PpaNAGK polypeptide is 32 587 Da.
Figure 2.
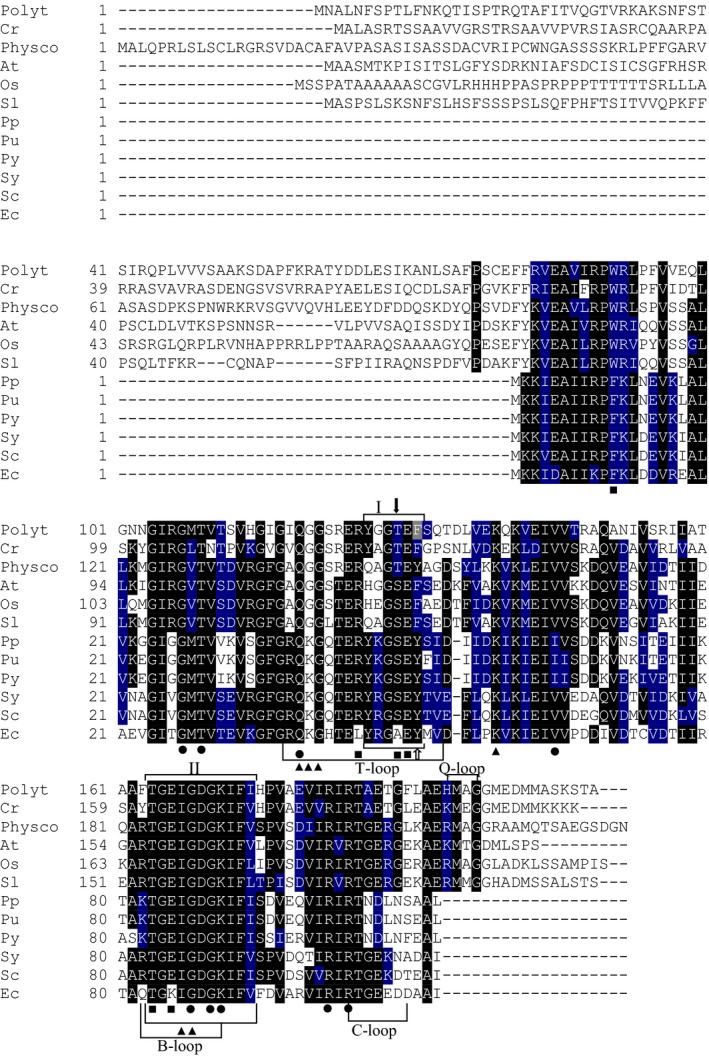
Multiple amino acid sequence alignment of PII proteins. The protein sequences were derived from NCBI database. The sequences are derived from PII polypeptides of the nonphotosynthetic alga Polytomella parva (Polyt), green photosynthetic alga Chlamydomonas reinhardtii (Cr; XP_001703658.1), land plants Physcomitrella patens (Physco; BAF36548.1), Arabidopsis thaliana (At; NP_192099.1), Oryza sativa Japonica (Os; Os05g0133100) and Solanum lycopersicum (Sl; AAR14689.1), red algae Porphyra purpurea (Pp; NP_053864.1), Porphyra umbilicalis (Pu; AFC39923.1) and Pyropia yezoensis (Py; AGH27579.1), cyanobacteria Synechococcus elongatus PCC 7942 (Sy; P0A3F4.1), Synechocystis sp. PCC 6803 (Sc; CAA66127.1) and Escherichia coli (Ec; CAQ32926.1). All the indicated regions and residues have been characterized in previous work 17, 24, 25, 26, 27. The regions referring to T‐, B‐, C‐ and Q‐loops are indicated 17. Highlighted residues in black are invariant in at least 55% of aligned PIIs proteins. Amino acids in blue represent similar residues. Boxs I and II indicate PII signature patterns. The positions of known PIIs post‐translational modification sites: the phosphorylation site in cyanobacterial S. elongatus PII (S49) and the uridylation site in E. coli PII (Y51) are indicated by solid black and white arrows, respectively. The amino acid residues involved in binding of ATP (●), NAGK (■) and 2‐OG (▲) are indicated 24, 25, 26, 27. The alignment was done using the ClustalW program and manually refined.
Figure 3.
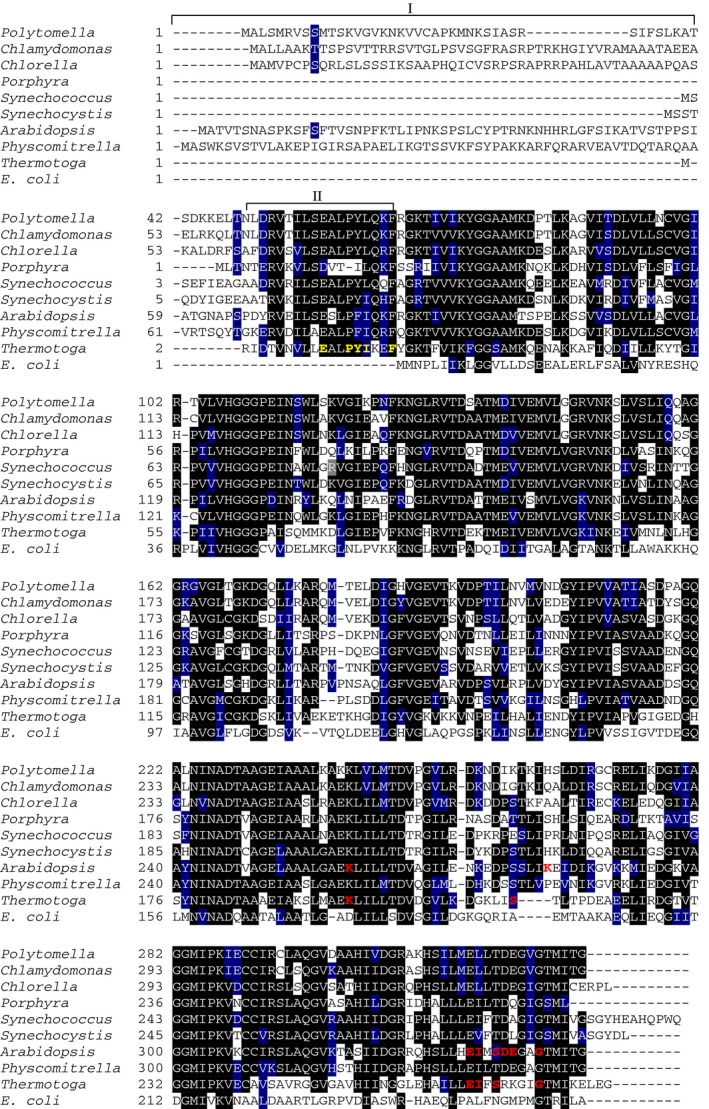
Multiple amino acid sequence alignment of NAGK proteins. The NAGK protein sequences were derived from UniProt database. The sequences are derived from NAGK polypeptides from nonphotosynthetic alga Polytomella parva (A6XGV3), green photosynthetic alga Chlamydomonas reinhardtii (A8HPI1) and Chlorella variabilis (E1ZQ49), land plants Physcomitrella patens (A0JC02) and Arabidopsis thaliana (Q9SCL7), red algae Porphyra purpurea (P69365), cyanobacteria Synechococcus elongatus PCC 7942 (Q6V1L5) and Synechocystis sp. PCC 6803 (P73326), and bacteria Thermotoga maritima (Q9X2A4) and Escherichia coli (P0A6C8). Highlighted residues in black are invariant in at least 55% of aligned NAGK proteins. Amino acids in blue represent similar residues. Box I refers to plastid‐targeting signal peptides sequence (ChloP server). Box II indicates an N‐terminal signature extension of Arg‐sensitive NAGK proteins, which is absent in Arg‐insensitive E. coli NAGK 28. In Box II, the previously identified signature sequence of Arg‐sensitive NAGK from Thermotoga maritima is highlighted in yellow, which is involved in forming the allosteric Arg binding site 28. Amino acid residues directly involved in allosteric Arg binding are highlighted in red and are deduced from known structures of NAGK: Arg complexes from Thermotoga maritima NAGK (PDB: http://www.rcsb.org/pdb/search/structidSearch.do?structureId=2BTY) 28 and Arabidopsis thaliana (PDB: http://www.rcsb.org/pdb/search/structidSearch.do?structureId=2RD5) 26. The alignment was done using the ClustalW program and manually refined.
To gain further insights into biochemical properties of PpaPII and PpaNAGK proteins and their mode of interaction, we prepared respective recombinant proteins. Therefore, a recombinant N‐terminal His‐tagged variant of the predicted mature PpaNAGK protein without the plastid transit peptide (amino acid residues 1‐40) (the theoretical molecular mass of monomeric recombinant PpaNAGK protein is 32.6 kDa), and a recombinant C‐terminal strep‐tagged version of the mature PpaPII protein without the plastid transit peptide (the theoretical molecular mass of monomeric recombinant PpaPII protein is 19.4 kDa) were overexpressed in Escherichia coli and affinity‐purified.
PpaNAGK catalytic efficiency in the absence of arginine is not influenced by PII
The kinetic constants of the purified recombinant PpaNAGK enzyme in the absence of the feedback inhibitor arginine exhibited an apparent K m value for NAG of 2.35 ± 0.22 mm and a v max of 58.1 ± 3.0 U·mg−1 (corresponding to a k cat of 211.5 ± 4.1 s−1) (Fig. 4A). In the presence of PpaPII, the apparent K m for NAG and the specific activity substantially increased to 3.99 ± 0.62 mm and 99.1 ± 1.2 U·mg−1 (k cat of 385.4 ± 16.2 s−1), respectively. The PpaPII‐triggered changes in the kinetic parameters of PpaNAGK indicate that, as in photosynthetic alga, PpaPII interacts with NAGK in P. parva. However, the overall catalytic efficiency (k cat/K m) was very similar for free (90 × 103) or PpaPII‐complexed (96.5 × 103 s−1·m −1) PpaNAGK. Strikingly, addition of Gln did not cause any increase in k cat/K m catalytic efficiency (96.9 × 103 s−1·m −1), in stark contrast to the situation in C. reinhardtii 17, and therefore, the overall PpaNAGK catalytic efficiency was not affected by PpaPII, neither in the presence nor in the absence of Gln.
Figure 4.
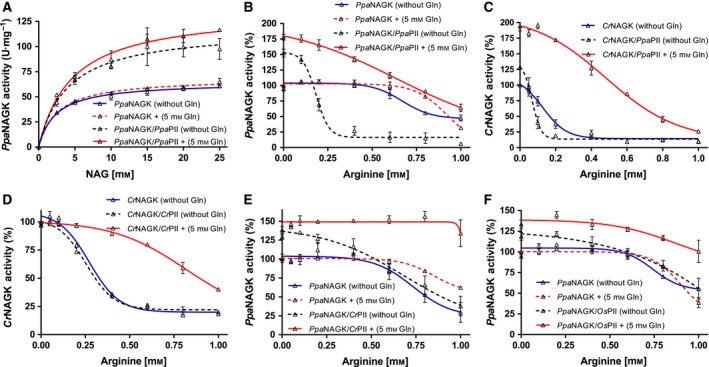
Characterization of PIIs modulated NAGK activity. (A) Catalytic activity of Ppa NAGK in presence or absence of Ppa PII and of 5‐mm Gln, as indicated. NAG was used as a variable substrate, as indicated. (B–F) Arginine feedback inhibition of NAGK enzymes in presence or absence of PII proteins, with or without 5‐mm glutamine, as indicated. (B) Ppa NAGK with Ppa PII; (C) Cr NAGK with Ppa PII; (D) Cr NAGK with Cr PII; (E) Ppa NAGK with Cr PII and (F) Ppa NAGK with Os PII. The Arg‐IC 50 in (D) for free Cr NAGK (0.27 ± 0.02 mm), Cr PII‐Cr NAGK in absence of Gln (0.25 ± 0.01) and Cr PII‐Cr NAGK in presence of Gln (0.82 ± 0.09 mm) were comparable to the previously published data 17. All data were fitted using GraphPad prism program. The arginine feedback inhibition data were fitted according to a sigmoidal dose‐response curve, yielding an IC 50 for arginine. SD as indicated by error bars, represents independent triplicate measurements.
Arginine sensitivity of PpaNAGK activity is enhanced by PII and modulated by glutamine
As the relief from arginine inhibition by PII‐NAGK complex formation is crucial for metabolic control of arginine biosynthesis in Cyanobacteria and Chloroplastida 17, 18, 19, 20, we asked if the presence of PpaPII could change the arginine inhibition profile of PpaNAGK. In the absence of PpaPII, feedback inhibition by arginine of PpaNAGK occurred with a half maximal inhibitory concentration (IC50) of 0.67 ± 0.04 mm (Fig. 4B). Strikingly, addition of PpaPII protein to PpaNAGK enhanced arginine sensitivity of NAGK by dropping the IC50 for arginine by 3.7‐fold to 0.18 ± 0.01 mm. By contrast, in the presence of glutamine (5 mm), PpaPII strongly relieved PpaNAGK from arginine feedback inhibition. However, in the absence of PpaPII, glutamine alone had no remarkable influence on NAGK activity, indicating that PpaPII either enhances or reduces the arginine‐sensitivity of PpaNAGK, depending on the presence of glutamine (Fig. 4B).
To investigate further whether the increased sensitivity of the PpaPII‐PpaNAGK complex towards Arg, as compared to PpaNAGK alone, is due to properties of PpaPII or of PpaNAGK, we performed heterologous enzymatic assays using the respective C. reinhardtii proteins (CrNAGK and CrPII). Of note, the CrNAGK protein is inherently more sensitive towards arginine than PpaNAGK 17. Furthermore, we tested the PII protein from rice plant Oryza sativa (OsPII) over PpaNAGK.
Strikingly, the addition of PpaPII protein to CrNAGK further increased the arginine‐sensitivity of CrNAGK: The IC50 for arginine dropped from 0.12 ± 0.03 in the absence of PpaPII to 0.07 ± 0.01 in presence of PpaPII (Fig. 4C). However, when 5‐mm glutamine was added to the assay, PpaPII behaved as shown previously for CrPII (Fig. 4D) 17, strongly relieving arginine feedback inhibition, as evidenced by the fourfold increase of the IC50 for arginine to 0.48 ± 0.04 mm (Fig. 4C). In contrast to PpaPII, the CrPII and OsPII proteins did not raise the arginine sensitivity of the PpaNAGK in the absence of Gln (Fig. 4E,F). But contrary, the CrPII and OsPII proteins slightly enhanced PpaNAGK activity at low concentrations of Arg (up to 0.5 mm). At high arginine concentrations (in the absence of Gln), PpaNAGK activity dropped in the absence (IC50 of 0.67 mm) or presence of the heterologous PII proteins (corresponding to IC50 values of 0.68 ± 0.15 with CrPII and 1.5 mm with OsPII) (Fig. 4E,F). In the presence of 5‐mm glutamine, CrPII and OsPII proteins relieved arginine feedback inhibition of PpaNAGK, as expected 17 with IC50 values of 1.2 and 2.3 mm, respectively. Together, these results showed that the PpaPII‐mediated enhancement of arginine‐sensitivity of NAGKs is an intrinsic property of PpaPII, which it can deploy in heterologous assays with other NAGK enzymes.
Because glutamine increases the activity of the PpaPII‐NAGKs complex in the presence of arginine (Fig. 4B,C), we next tested the activation of arginine‐inhibited PpaPII‐PpaNAGK complex (with 0.5‐mm Arg) by glutamine in a concentration‐dependent manner (Fig. 5A). The half‐maximal effective concentration (EC50) of glutamine for activation of the PpaPII‐PpaNAGK complex was determined to be 1.8 mm. A similar value was obtained for glutamine‐dependent activation of CrNAGK by PpaPII, with a glutamine EC50 of 1.1 mm (Fig. 5A). By comparison, the EC50 of glutamine for stimulation of CrNAGK activity by Chlamydomonas CrPII or Chlorella variabilis PII (CvPII) proteins were 2.4 ± 0.8 mm 17 or 6.5 ± 1.1 mm 16, respectively. Moreover, the microalga Myrmecia incisa PII (MiPII) also required high concentrations of Gln (3‐12 mm) to relive arginine feedback inhibited MiNAGK 29 and the activation of Arabidopsis thaliana NAGK (AtNAGK) by Physcomitrella patens PII or rice OsPII required also high concentrations of Gln with EC50 of 6.6 mm and 9.2 mm, respectively 17. This suggests that PpaPII has evolved to sense lower glutamine concentrations than the other so‐far studied plant PII proteins.
Figure 5.
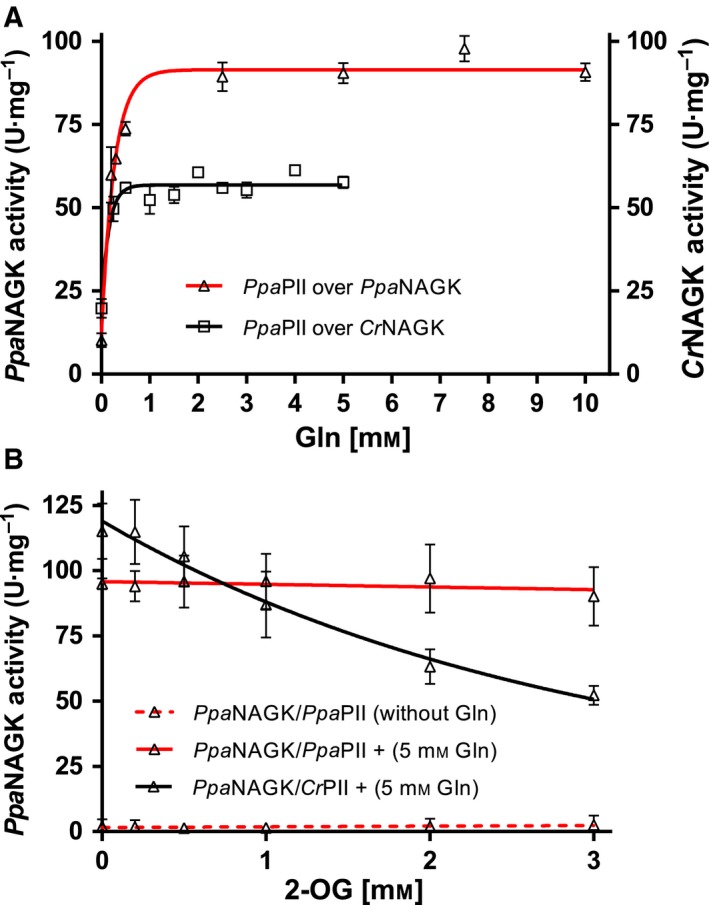
Effect of glutamine and 2‐OG on PII‐mediated NAGKs activation. (A) Glutamine‐dependent activation of arginine‐inhibited Ppa NAGK or arginine‐inhibited Cr NAGK by Ppa PII, as indicated. (B) Effect of 2‐OG on Ppa PII and Cr PII proteins in the presence of 5‐mm glutamine on activation of arginine‐inhibited Ppa NAGK, as indicated. The assays were performed in presence of 0.5‐mm arginine for Ppa NAGK or 0.12‐mm arginine for Cr NAGK. Data were fitted using a graphpad prism, yielding an EC 50 for Gln and an IC 50 for 2‐OG. SD as indicated by error bars, represents triplicate independent measurements.
PpaPII protein lacks the response to 2‐oxoglutarate
Most PII proteins were found to sense 2‐OG as the principle effector molecule in a synergistic binding reaction with ATP 10, 18, 22, 27. Recent studies have identified PII proteins from some Chloroplastida that lack 2‐OG responses 18. Therefore, we assessed the effect of 2‐OG on the modulation of PpaNAGK activity by PpaPII. As shown in Fig. 5B, addition of 2‐OG to a reaction mixture containing PpaPII‐PpaNAGK complex together with 5‐mm Gln and 0.5‐mm Arg did not lead to inhibition of PpaNAGK activity, which would be expected if the complex would dissociate. As a control, the expected response towards 2‐OG was obtained for the heterologous assay with CrPII, which senses 2‐OG 17, 18 with an IC50 of 1.99 mm. Together, it appears that the PpaPII protein does not respond to 2‐OG, unlike CrPII 17, 18, but it complexes with NAGK to tune its response towards the feedback inhibitor arginine in a glutamine‐dependent manner.
Glutamine‐independent PpaPII‐PpaNAGK complex formation
The above described enzyme tests suggested that PpaPII‐PpaNAGK complex formation must be different from all previously tested cases 16, 17, 18, 19, 20, 24, 25, 26, since PpaPII enhances the arginine sensitivity of NAGK in the absence of glutamine. Of note, in the absence of Gln, the PII proteins from representative Chloroplastida were not able to effectively form a complex with NAGK, even in the presence Mg2+‐ATP 17. PpaPII contains the C‐terminal Q‐loop responsible for glutamine binding (Fig. 2), that was shown previously to promote glutamine‐dependent complex formation of CrPII‐CrNAGK or of other plant PII proteins except Arabidopsis 17. To monitor any changes in molecular weight due to complex formation, we characterized PII‐NAGK complexes in the presence or absence of Gln using analytical size exclusion chromatography (SEC) coupled to multiangle light scattering (MALS). First, we determined the oligomerization state of PpaPII and PpaNAGK proteins. As expected, the PpaPII protein eluted as a trimer and PpaNAGK as a hexamer (Fig. 6A) 17, 20, 24, 25, 26. When an excess of PpaPII was mixed with PpaNAGK (4 : 1 monomeric concentrations), a PpaPII‐PpaNAGK complex was detected with a clearly detectable peak shift for the PpaNAGK hexamer. In agreement with the enzymatic characterization, glutamine was not required for complex formation, nor did it induce a remarkable shift in the size of the complex (Fig. 6A).
Figure 6.
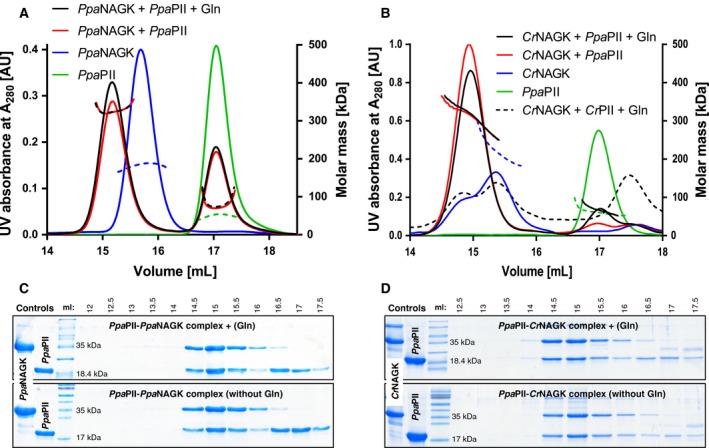
Complex formation of Ppa PII‐NAGKs analysed by SEC‐MALS. Gel filtration of PII‐NAGK complexes was carried out as described in Methods. SEC‐MALS profiles for (A) Ppa NAGK in presence or absence of Ppa PII and 5‐mm glutamine and (B) Cr NAGK in presence or absence of Ppa PII or of Cr PII with or without glutamine, as indicated. The mass of the eluted particles was determined via MALS and plotted on the right y‐axis. The protein elution profile was monitored using UV signal at 280 nm and plotted on the y‐left. (C and D) The eluted protein fractions between 12.5 to 17.5 mL corresponding to Ppa PII‐Ppa NAGK complexes as shown in (A) or for Ppa PII‐Cr NAGK complexes as shown in (B) were collected and subjected to Glycine‐SDS/PAGE, and revealed the presence of Ppa PII and NAGK proteins after Coomassie blue stain.
To confirm that the PpaPII protein is responsible for glutamine‐independent complex formation with NAGK, we investigated complex formation with the CrNAGK protein. The CrNAGK eluted as a hexamer like previously reported 17. Independent of the absence or presence of Gln, the PpaPII protein was able to form a stable complex with CrNAGK and both proteins co‐eluted together (Fig. 6B). In agreement, the SDS/PAGE analysis of the collected complexes’ peaks showed the presence of both PpaPII with PpaNAGK or with CrNAGK (Fig. 6C,D). Together, the results demonstrated that the PpaPII protein forms complexes with NAGKs independent of glutamine.
Influence of different effector molecules on PII‐NAGK complex formation
To further confirm that the direct interaction between PpaPII and PpaNAGK is glutamine‐independent and to test the influence of the other known PII effectors molecules (ATP, ADP, 2‐OG and Gln) on the PII‐NAGK complex formation, we assessed the complex formation using surface plasmon resonance (SPR) spectroscopy. In SPR experiments, the His‐tagged NAGK protein was immobilized on a Ni‐NTA sensor chip and the strep‐tagged PII protein was injected together with or without different combinations of effectors molecules to monitor the difference in the response unites (ΔRU) due to the PII‐NAGK complex formation.
We showed previously that formation of the CrPII‐CrNAGK complex from C. reinhardtii was strictly Mg2+‐ATP and glutamine‐dependent and was not supported by ADP (Fig. 7A) 17. By contrast, PpaPII was able to form a strong complex with NAGK on the SPR surface, independent of presence or absence of ADP, ATP, 2‐OG and Gln (Fig. 7B). Remarkably, the PpaPII‐PpaNAGK complex was extraordinary stable and dissociated very slowly in the course of the assay with an estimated K d value of 93.8 ± 29.9 nm (Fig. 7C,D). The percent of PpaPII‐PpaNAGK complex dissociation from the sensor chip at 330 and 660 sec after the end of the injection phase was 60.7% and 41.9%, respectively (RU at 110 sec was taken as 100%) (Fig. 7D), indicating the stability of the complex. By contrast, in the case of CrPII‐CrNAGK, the complex dissociated spontaneously at the end of the injection as soon as it encountered a buffer devoid of Mg2+‐ATP and Gln (compare Fig. 7A,C).
Figure 7.
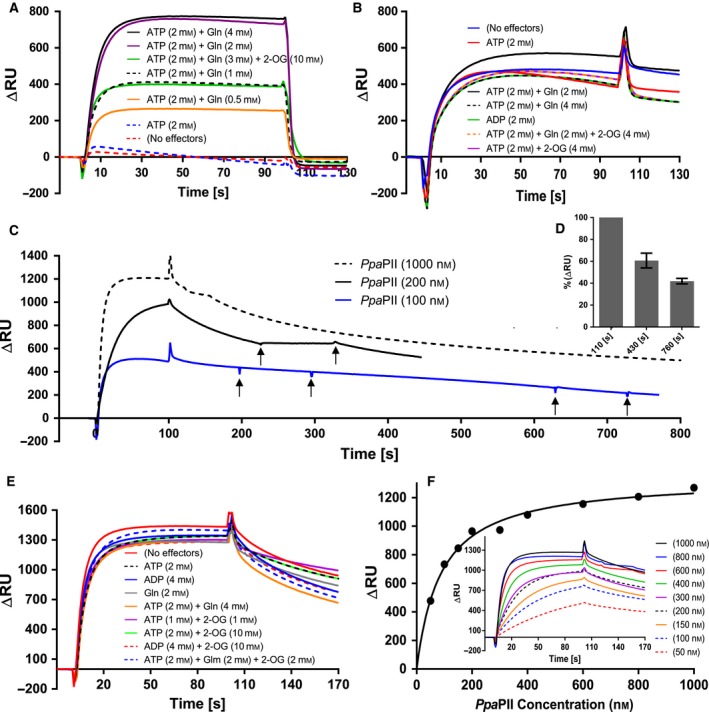
Surface plasmon resonance spectroscopy analysis of PII‐NAGK complex formation. Cr PII or Ppa PII were injected to FC2‐immobilized Cr NAGK or Ppa NAGK. (A) Strict Mg2+‐ATP/Gln dependency of 1000‐nm Cr PII binding to Cr NAGK. (B–F) Binding of 1000‐nm Ppa PII to various NAGK enzymes under various conditions. (B): Binding of Ppa PII to Ppa NAGK, as indicated. (C) Stability of the Ppa PII‐Ppa NAGK complex formed by injection of 100‐, 200‐ or 1000‐nm Ppa PII, as indicated, in absence of any effector molecules during SPR dissociation. The arrows indicate the injection of 2‐mm ADP, which did not affect complex stability/dissociation. (D) Dissociation of Ppa PII‐Ppa NAGK complex; shows the average of the response signals shown in (C) in form of % at t:430s and at t:760s (330s and 660s after the end of the injection, respectively). The signals at t:110s (10s after the end of the injection) were normalized to 100%. SD as indicated by error bars, represents triplicate independent measurements. (E) Binding of 1000‐nm Ppa PII to Cr NAGK, as indicated. (F) K d value for binding Ppa PII to NAGK calculated from ∆RU at t:100s. The inset in (F) shows the Ppa PII titration (from 50 to 1000 nm) to NAGK in absence of effectors molecules, as indicated.
Furthermore, we reported previously that ADP and 2‐OG negatively affected cyanobacterial PII‐NAGK interaction by promoting the dissociation of the complex, and further, the injection of 1 mm of ADP caused immediate dissociation of PII‐NAGK complexes 30. Remarkably, the PpaPII‐PpaNAGK complex was resistant against the injection of ADP, indicating that PpaPII is unable to sense ADP (Fig. 7B,C). The 2‐OG effector also showed no antagonistic effect on PpaPII‐PpaNAGK complex formation (Fig. 7B) in agreement with the inability of 2‐OG to inhibit the NAGK enzymatic activity (Fig. 5B). This result resembled a previous result on CrPII‐CrNAGK interaction, where 2‐OG had no influence on the CrPII‐CrNAGK complex formation, while the 2‐OG mediated inhibition of CrNAGK activity in complex with CrPII‐complex appeared to occur postbinding 17.
To gain further insights in the Gln‐independent complex formation of PpaPII, we repeated the previous SPR experiments using CrNAGK as a binding partner. Regardless of the effector molecules added to the assay mixture (using 1‐μm PpaPII protein), the PpaPII protein formed a strong complex with CrNAGK in an Mg2+‐ATP and Gln‐independent manner, and moreover, neither ADP nor 2‐OG had influence on PpaPII‐CrNAGK complex formation (Fig. 7E). As before, PpaPII was able to bind to CrNAGK without any effector molecules (K d value of 86.3 ± 9.4 nm, Fig. 7F). Together, these experiments suggest that the Gln‐independent formation of the PpaPII‐NAGKs complex is a unique feature of PpaPII in comparison to other plant PII proteins that possess a functional Q‐loop, the latter requiring Gln for NAGK interaction, as shown for PII from green algae (C. reinhardtii and Chlorella variabilis) 16, 17, microalga (Myrmecia incisa) 29, or higher plants (Oryza sativa and Physcomitrella patens) 17.
Finally, we asked whether the PpaNAGK protein may also provide features to the glutamine‐independence of PpaPII‐NAGKs complex formation. Therefore, we tested the ability of CrPII and OsPII proteins to form heterologous complexes with PpaNAGK in the absence or presence of effector molecules Mg2+‐ATP and Gln using SPR. As already mentioned, the formation of the CrPII‐CrNAGK complex is strictly dependent on Mg2+‐ATP and Gln (Fig. 7A), whereas Mg2+‐ADP did not support complex formation 17. Remarkably, CrPII and OsPII proteins were able to bind to PpaNAGK without any effector molecules (Fig. 8), indicating that PpaNAGK attracts the heterologous PII proteins in a glutamine‐independent manner. Nevertheless, with CrPII, the presence of Mg2+‐ATP alone or in combination Gln moderately or strongly enhanced the binding to PpaNAGK, respectively. Interestingly, in the presence of Mg2+‐ADP, CrPII was still able to form a weak complex with PpaNAGK, similar to the absence of effector molecules (Fig. 8A), indicating that CrPII lost the ability to sense ADP, confirming our previous reports 17, 18. The addition of Gln in presence of Mg2+‐ADP enhanced the CrPII‐PpaNAGK complex formation (Fig 8B), however, Gln in combination with Mg2+‐ATP stimulated much stronger complex CrPII‐PpaNAGK formation (compare Fig 8A,B). As for the homologue CrPII‐CrNAGK complex 17, 2‐OG did not show any influence on the CrPII‐PpaNAGK complex (Fig. 8C). Moreover, OsPII interacted with PpaNAGK independent of any effector molecules (Fig. 8D). These results indicate that PpaNAGK strongly influences the binding properties of various PII proteins and implies a role in the sensory properties of the entire PII‐NAGK complex.
Figure 8.
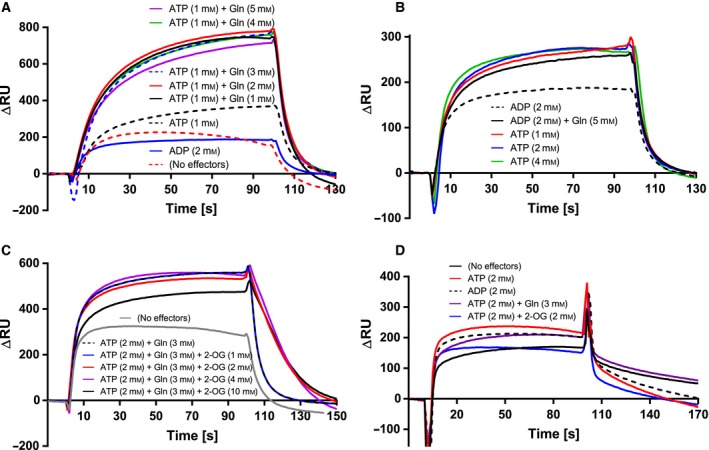
Ability of Ppa NAGK to bind Cr PII or Os PII in effector molecule‐independent manner. Interaction between Ppa NAGK and Cr PII or Os PII was analysed by SPR; 1000 nm of Cr PII or Os PII were injected to FC2‐immobilized Ppa NAGK. (A and B) Binding of Cr PII to Ppa NAGK, as indicated. (C) 2‐OG independent binding of Cr PII to Ppa NAGK, as indicated. (D) Binding of Os PII to Ppa NAGK, as indicated; shows no negative influence of ADP or 2‐OG on Os PII‐Ppa NAGK complex.
Discussion
Many species of Chlorophyceae, to which the Polytomella spp. lineages belong, including P. parva, contain a strongly reduced mitochondrial genome and more strikingly, the Polytomella spp. are the first discovered plastid‐bearing algae devoid of a plastid genome 1. Apparently, the P. parva plastid seems to carry out essential anabolic functions including amino acid, fatty acid, carbohydrate and lipid biosynthesis, which have not been re‐located to the cytoplasm during evolution. In P. parva, the genes encoding for NAGK and PII proteins were found among the plastid‐targeted/nuclear encoded genes. We hypothesized that P. parva must possess strong metabolic adaptations to cope with the evolutionary loss of photosynthesis.
As a consequence of the lifestyle switch towards a purely heterotrophic metabolism, the mitochondria in P. parva are the primary energy‐generating organelles through their respiratory activity, whereas in C. reinhardtii, their respiratory function is mainly limited to dark periods. In the organotrophic lifestyle of P. parva, it uses ethanol as a carbon source and oxidatively metabolizes it by mitochondrial activity for energy release. In agreement with the prominent role of mitochondrial metabolism, the levels of most of the TCA cycle intermediates in P. parva are strongly increased as compared to C. reinhardtii. By contrast, in C. reinhardtii, the elevated PEP pool (as compared to P. parva) agrees with a flow of carbon from CO2 fixation into lower glycolysis 31, to provide the cells with precursors for most anabolic pathways. Whereas, in the heterotrophic P. parva, PEP has to be synthesized via gluconeogenetic reactions starting from the carbon source ethanol, which can explain the 7.6‐fold decreased level of PEP in P. parva (Fig. 1).
Under conditions of nitrogen excess, the levels of the amino acids arginine, aspartate and glutamate are clearly elevated in P. parva compared with C. reinhardtii (Fig. 1), which suggests fast nitrogen‐assimilation reactions in P. parva. The GS/GOGAT cycle is the primary route for nitrogen assimilation, suggesting that this reaction cycle should be highly active in P. parva, in agreement with the localization of GS and GOGAT enzymes in the P. parva plastid 1 and the fast growth rate of P. parva with generation time of ~ 4.7 h at 25 °C 32. As compared to C. reinhardtii, the level of 2‐OG is relatively the lowest of all TCA cycle metabolites in P. parva. This agrees with an efficient GOGAT reaction, constantly depleting the 2‐OG pool. Since this reaction also consumes glutamine, we also find relatively lower levels of glutamine than of glutamate, aspartate or arginine. The highly active nitrogen assimilation activity results not only in elevated glutamate levels but also in high arginine levels. The controlling enzyme of the arginine synthesis pathway, NAGK, therefore, needs to be highly active. As shown here, the control of NAGK by the PII signalling protein shows unique features, which probably result from the evolutionary pressure of a nonphotosynthetic environment with a restrained adenylate energy charge to maintain NAGK at high activity.
The Arg sensitivity profile of free PpaNAGK (Arg‐IC50 of 0.67 mm) is intermediate between the more sensitive Chlamydomonas CrNAGK (Arg‐IC50 of 0.11 mm) 17, and the low‐sensitive NAGKs from Arabidopsis AtNAGK (Arg‐IC50 of 1.0 mm) 19 or Chlorella CvNAGK (Arg‐IC50 of 1.2 mm) 16. Thus, the higher levels of arginine required to inhibit PpaNAGK in comparison to CrNAGK is in good agreement with the observed higher levels of Arg production in P. parva (Fig. 1). Multiple sequence alignment (Fig. 3) of NAGK sequences shows that all residues participating in the allosteric Arg binding site in AtNAGK (PDB: http://www.rcsb.org/pdb/search/structidSearch.do?structureId=2RD5) 26 are perfectly conserved in the other plant NAGKs, providing no clue to the different sensitivity towards Arg. Structural analysis of the PpaPII‐PpaNAGK complex in the absence or presence of arginine is required for a mechanistic explanation. In any case, in complex with its cognate PII protein, Arg‐sensitivity of PpaNAGK (Arg‐IC50 of 0.18 mm) is similar to that of free CrNAGK 17.
Unlike all other PII‐NAGK couples from cyanobacteria and plants investigated so far, the PpaPII‐PpaNAGK complex associates in an almost irreversible manner. The effector molecules ADP or Mg2+‐ATP/2‐OG, which cause efficient dissociation of the complex, are ineffective in the case of the P. parva proteins. Even more strikingly, the complex forms in a completely glutamine‐independent manner, although the glutamine‐sensing C‐terminal extension, the Q‐loop 17 is perfectly conserved in PpaPII (Fig. 2). Since the amino acid sequences of PII and NAGK are highly conserved between P. parva and C. reinhardtii (with 61.7% and 84.5% identity, respectively), these unique features were unexpected. A few amino acid substitutions in PII may be sufficient to change the transient PII‐NAGK complex into a stable hetero‐oligomeric enzyme complex. In this respect, one residue at the tip of the T‐loop may be of particular importance: This residue, which corresponds to R47 in bacterial PII proteins, has been shown to be of key function for PII‐AmtB interaction 33. Moreover, mutation of the R47 residue in a cyanobacterial PII protein to Ala strongly reduced the affinity of Synechococcus PII to NAGK 25. In contrast to cyanobacteria and red algae, where this Arg residue is highly conserved, it is replaced by Ala or Glu, in many PII proteins from Chlorophyta, or in the case of Arabidopsis and P. parva, it is replaced by Gly. Conspicuously, both, A. thaliana and P. parva, bind NAGK independent of Gln, with the difference that P. parva nevertheless senses Gln, but A. thaliana PII does not, due to a truncation in its Q‐loop segment 17.
In P. parva, the PII protein has turned into a stably attached regulatory subunit of NAGK. The very high in vitro stability of the complex suggests that these proteins are co‐evolved towards the formation of a stable hetero‐oligomeric complex in an effector molecule‐independent manner, and probably are always complexed in vivo. In this complex, the Gln binding site resides in the PII subunit and NAGK exhibits the allosteric Arg site. Importantly, the entire complex is required for sensitive reaction of NAGK towards Arg. This requires that both PpaPII and PpaNAGK proteins are constantly expressed and co‐localized in the nonphotosynthetic plastid, which still needs to be experimentally proven. However, in support of this assumption, both proteins possess a plastid signal peptide at their N‐termini (amino acid residues 1‐49 for PpaPII and 1‐21 for PpaNAGK) (Fig. 3). Furthermore, in C. reinhardtii and in the microalga Myrmecia incisa, NAGK and PII proteins were already found to be plastid localized 16, 29, 34.
The regulatory effects of the effector molecules Gln and Arg occur in the PpaPII‐PpaNAGK complex at the postbinding stage. At low Gln‐levels (corresponding to N‐poor conditions), the complex is highly sensitive towards arginine. This indicates that in complex with PpaPII, PpaNAGK adopts a conformation that has high affinity for the allosteric feedback inhibitor arginine. By contrast, when Gln binds to the complex (under N‐rich conditions), PpaPII strongly relieves PpaNAGK from arginine feedback inhibition, indicating that glutamine, through binding to the C‐terminal Q‐loop, imposes a conformational change on the entire complex that counteracts the feedback inhibitory effect of arginine, like previously shown for PII‐NAGK complexes from oxygenic phototrophs 16, 17, 29. The half‐maximal effective concentration (EC50) of glutamine to stimulate PpaPII‐NAGKs activity in presence of arginine is ~ 50% lower than the EC50 for CrPII, showing that the PpaPII protein has evolved to allow enhanced PpaNAGK activity at lower glutamine concentrations. Therefore, we speculate that the default mode of the PII‐NAGK system in P. parva is a high arginine production through the extremely active PpaPII‐PpaNAGK complex, unless there is a severe nitrogen (Gln) limitation.
Analysis of the heterologous complexes formed between P. parva proteins and plant PII or CrNAGK proteins allowed us to conclude that in P. parva, both partner proteins have co‐evolved towards a stable complex, with both proteins contributing to the enhanced complex stability. PpaNAGK showed avid binding of CrPII regardless of glutamine and of effector molecules that usually dissociate PII‐NAGK complexes (Mg2+‐ATP/2‐OG or ADP). Conversely, PpaPII is prone to bind to NAGK proteins irrespective of effector molecules, as demonstrated by the effector molecule‐independent binding of PpaPII to CrNAGK (which usually only accepts glutamine‐ligated CrPII as partner). This shows that the PpaPII protein has evolved to exclusively sense the glutamine level in a very sensitive manner. The loss of sensing the ATP/ADP ratio and of 2‐OG might be attributed to the loss of photosynthetic activity in the plastid with consequent metabolic changes. Recently we found that the PII protein from the moss Physcomitrella patens 18 has also lost the ability to sense ADP and 2‐OG. This suggests that the detailed sensing properties of the PII proteins can easily be adjusted to the regulatory need of the respective metabolic situation in an organism.
Collectively, our finding extends the knowledge of PII signalling in plants. Apparently, it seems that during the evolution of Chlorophyta, the PII proteins diverged in their properties, becoming very heterogeneous with respect to 2‐OG and to ADP binding and towards complex formation with NAGK. P. parva is an extreme case, where the PII protein specialized its function towards a glutamine‐regulated subunit of the key enzyme of the arginine pathway NAGK. Possibly, other targets of PII regulation might have been lost during the reductive evolution of the nonphotosynthetic organelle, allowing PII to exclusively focus on NAGK regulation. It would be interesting in future to investigate PII‐NAGK systems in other secondary nonphotosynthetic organisms, to reveal if the unique feature of the PII‐NAGK complex in P. parva is related to the loss of photosynthesis during evolution.
Materials and methods
Strains and cultivation conditions
The whole cloning procedure was performed in E. coli NEB 10‐beta, while protein expression and purification were done using E. coli LEMO‐21(DE3) and PII‐deficient E. coli RB9060 35 in LB medium. The Polytomella parva SAG 63‐3 culture was obtained kindly from the algal culture collection (SAG‐Göttingen University, Germany) as an environmental nonaxenic culture. The culture was excessively treated with antibiotics until we were able to isolate a clean axenic culture of P. parva SAG 63‐3 (Fig. 1B). Polytomella parva was cultivated in REP media containing 40‐mm EtOH as a carbon source and 7.5‐mm NH4Cl as a nitrogen source, pH 4.0 36 at 22 °C under day/night cycles. The wild‐type Chlamydomonas reinhardtii CC‐125 mt+ [137c] was kindly obtained from Erik Schäffer lab. (ZMBP, Tübingen University), and cultivated in tris‐acetate‐phosphate (TAP) medium containing 7.5‐mm NH4Cl 37 under day/night cycles at 22 °C.
To induce nitrogen deprivation, an exponentially growing culture of P. parva under nitrogen‐rich condition (7.5‐mm NH4Cl) was harvested, washed twice in nitrogen‐free media, then suspended in fresh media, and divided into two subcultures. One subculture was re‐inoculated again into nitrogen‐rich (7.5‐mm NH4Cl) condition, while the other half was re‐inoculated into nitrogen‐limiting (0.375‐mm NH4Cl) condition. After 45 h, the P. parva cultures were harvested to determine the intracellular metabolites using LC‐MS, in comparison to standard growing culture of C. reinhardtii under nitrogen‐rich condition (7.5‐mm NH4Cl).
Metabolite extraction and quantification
For quantification the intracellular metabolites of 50 mL of exponentially growing cells under the day cycle of P. parva under different nitrogen regimes (excess nitrogen of 7.5‐mm NH4Cl or poor nitrogen of 0.375‐mm NH4Cl), and of C. reinhardtii under nitrogen‐rich condition (7.5‐mm NH4Cl) were shock‐cooled in ice for 5 min, then rapidly harvested by centrifugation at 4 °C. After discarding of liquid media, the cell pellets were immediately frozen in liquid nitrogen. Metabolite extraction and quantification was done according to 38. Briefly, the cells were lyophilized followed by an extraction of the metabolites using a Retsch ball mill (two cycles, 30 s each). Extraction was done twice using 400 μL of 80% methanol containing 0.1% formic acid followed by a second extraction step with 400 μL of 20% methanol also containing 0.1% formic acid. The extracted metabolites were combined and concentrated in a Speed‐Vac, then dissolved in 150 μL of 20% methanol containing 0.1% formic acid (HPLC‐grade). LC/MS‐analyses were done on a Waters UPLC‐SynaptG2 LC/MS system. Chromatography was carried out on a 2.1 × 100 mm, 1.8‐μm Waters Acquity HSST3 column. For separation, a 10‐min gradient from 99% water to 99% methanol (both solvents with 0.1% formic acid) was used. The mass spectrometer was operated in ESI negative and positive mode and scanned from 50 to 2000 m/z with a scan rate of 0.5 s. For the determination of peak areas, extracted ion chromatograms were generated and integrated. The quantification of the intracellular metabolites was normalized to cell‐dry weight.
Cloning of PpaPII and PpaNAGK‐like proteins
The sequences for PpaNAGK homologue and PpaPII were derived from iMicrobe database under project ID MMETSP0052 (https://www.imicrobe.us/#/projects/104) with sequence ID: for PpaNAGK (MMETSP0052_2‐20121109|9957_1) and for PpaPII (MMETSP0052_2‐20121109|12411_1). Gene Blocks, with optimized codon usage for cloning and expression into E. coli, encoding for amino acid sequences of mature PpaNAGK and PpaPII genes without plastid signal peptides, were synthesized by IDT, USA. The first Gene Block fragment for the amino acid sequence of the PpaNAGK was derived from a DNA sequence starting with the 41st amino acid (TSDKK); the gene was amplified using the forward primer 5′‐TCATCATCATCACAGCAGCGGCCTGGTGCCGCGCGGCAGC‐3′ and the reverse primer 5′‐TATGCTCGAGGATCCGGCTGCTAACAAAGCCCGAAAGGAA‐3′. The second Gene Block for the DNA sequence of PpaPII was derived from the amino acid sequence starting with the 50th amino acid (SAAKS) and was amplified with the forward primer 5′‐AATAGTTCGACAAAAATCTAGATAACGAGGGCAAAAAATG‐3′ and the reverse primer 5′‐CTGCAGGGGGACCATGGTCTCAGCGCTTGGAGCCACCCGC‐3′. Using Gibson assembly, the Gene Blocks for PpaNAGK and PpaPII were cloned directly into NdeI‐digested pET15b vector (Novagen, Darmstadt, Germany) and BsaI‐digested pASK‐IBA3 vector (IBA, Munich, Germany), respectively, as described previously 39. The plastid signal peptides were determined using a ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/) 40, 41.
Expression and purification of PpaNAGK, CrNAGK, PpaPII, CrPII and OsPII proteins
The overexpression of the recombinant N‐terminal fused His6‐tagged PpaNAGK and CrNAGK was performed in E. coli LEMO‐21(DE3) and the proteins were affinity purified on a Ni‐NTA columns according to 18, 42. Overexpression of the recombinant C‐terminal fused strep‐tagged PII proteins (PpaPII, CrPII and OsPII) were performed in PII‐deficient E. coli RB9060 35 and the proteins were affinity purified on a Strep‐Tactin II column according to 14, 20.
Coupled NAGK activity assay
The activity of NAGK was assessed using a coupled enzyme assay in which the production of ADP after the consumption of ATP for phosphorylation of NAG was associated with the oxidation of NADH by pyruvate kinase and lactate dehydrogenase as described previously 18, 19. The standard reaction mixture consisted of 50‐mm imidazole pH 7.5, 50‐mm KCl, 20‐mm MgCl2, 0.4‐mm NADH, 1‐mm phosphoenolpyruvate, 5‐mm ATP, 0.5‐mm DTT, 11‐U lactate dehydrogenase, 15‐U pyruvate kinase and 50‐mm NAG and the reaction was started by the addition of 1.5‐μg NAGK. When necessary, PII protein was added to the reaction mix in equimolar concentration. When needed, the effector molecules 2‐OG, Gln and Arg were added to the reaction mixtures at concentrations as indicated. The oxidation of NADH was measured at 340 nm for 10 min with a SPECORD‐spectrophotometer (model‐210 PLUS, Analytik Jena AG). One molecule oxidation of NADH is proportional to one molecule phosphorylation of NAG. One unit of NAGK catalyses the conversion of 1 μmol of NAG min−1, calculated with the molar absorption coefficient of NADH of 6178 L mol−1·cm−1 at 340 nm. Means of triplicate experimental determinations are shown with a standard deviation of less than 5%. The enzymatic constants K m, k cat, IC50 and EC50 were calculated from the velocity slopes using the graphpad prism software program (GraphPad Software, San Diego, CA, USA).
Surface plasmon resonance spectroscopy analysis (SPR spectroscopy)
SPR experiments were done at 25 °C using a BIAcore‐X biosensor system (Biacore AB, Uppsala, Sweden) in HBS buffer (10‐mm HEPES, 150‐mm NaCl, 2‐mm MgCl2 and 0.005% NP‐40, pH 7.5) with a flowrate of 15‐μL·min−1, as described previously 17, 42. The recombinant His6‐tagged NAGKs (PpaNAGK and CrNAGK) proteins were immobilized on the flow cell (FC2) of the Ni2+‐loaded NTA‐biosensor chip. NAGKs in HBS buffer were injected (50 μL) until a saturation of NTA‐biosensor chip by a signal of ~ 3000‐4000 resonance units (RU), which corresponds to a surface concentration change of 3‐4 (ng·mm−2). To evaluate the effect of the effector molecules on the PII‐NAGK complex formation for the binding of PII (PpaPII, CrPII and OsPII) proteins to the immobilized His6‐tagged NAGKs, the strep‐tagged PII proteins (100‐1000 nm) as indicated in HBS buffer, were incubated in ice for 5 min with or without different combinations of the effector molecules (as indicated). PII proteins (25 μL) were injected as an analyte into both FC1 (control for unspecific binding of PII to the sensor chip) and FC2 (immobilized NAGKs) on the sensor chip. The specific binding of PIIs to NAGKs was recorded as the difference in the response signal of FC2‐FC1 (∆RU). CrPII protein dissociates immediately after the end of the injection, making immobilized NAGKs ready for the next injection. By contrast, the PpaPII protein formed a strong complex with NAGK and dissociates very slowly over the time. Therefore, to refresh the NTA sensor chip for another assay, 25 μL of 1‐M imidazole pH 7.0 was injected to remove the immobilized NAGKs. To regenerate the NTA sensor chip, 50 μL of 0.4‐M EDTA pH 7.5 was injected and subsequently, the sensor chip was reloaded with Ni2+ and fresh NAGKs as described. The regeneration procedure was done when the response of PII binding to the immobilized NAGK started to decrease.
Size exclusion chromatography and multiangle light scattering analysis
Analytical size exclusion chromatography was carried out as described previously 43, 44 on a Micro‐Äkta purifier system equipped with Superose 6 Increase 10/300 GL column (GE Healthcare, Freiburg, Germany). The Superose column was coupled to a triple‐angle light scattering (MALS) detector (MiniDAWN™ TREOS® system; Wyatt Technology Corp., CA, USA) and a refractometer (Optilab T‐rEX, Wyatt). The column was calibrated using standard proteins: thyroglobulin (670 kDa), ferritin (440 kDa), globulin (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), RNase (13.7 kDa) (Bio‐Rad gel filtration standard, GE Healthcare LMW gel filtration calibration kit). Bovine serum albumin (BSA) was used to calibrate and validate the MALS analysis. The running buffer consisted of 10‐mm Tris pH 7.8, 300‐mm NaCl, 2‐mm MgCl2, 0.02% NaN3 and 2% glycerol. The samples were centrifuged for 5 min at 18 407 g, and 100 μL of the supernatant were injected for analysis with a flow rate 0.5 mL·min−1. The resulting data were analysed with ASTRA program (Wyatt Technology, Dernbach, Germany). The elution volume was plotted against the UV signal and molecular weight profiles. The apparent molecular weights were derived from MALS data. The chromatographic elution profiles were collected (0.5‐mL fractions) and analysed by Glycine‐SDS/PAGE.
Conflicts of interest
The authors declare no conflict of interests.
Author contributions
EE and KF conceived and initiated the project. KAS and KF designed the experiments. KAS and TL performed experiments. KAS interpreted the results and wrote the manuscript. KAS, EE and KF commented and edited on the manuscript. All authors analysed the results and approved the final version of the manuscript.
Supporting information
Table S1. List of identified metabolites by LC‐MS normalized to 1 mg of algal cell dry weight including standard deviation (SD) of three biological replicates for Polytomella parva (under nitrogen excess and limiting conditions) and Chlamydomonas reinhardtii (under nitrogen‐rich conditions).
Acknowledgements
This work was supported by Saint‐Petersburg State University (Grant No. 1.65.38.2017) to EE, and DFG to KF (Fo195/9‐2, Fo195/13‐1). The authors are grateful to Jörg Scholl and Heinz Grenzendorf (IMIT, Tübingen University) for the technical assistance, the staff of the analytics unite (ZMBP, Tübingen University), and Christine Kiefer (Schäffer lab, ZMBP, Tübingen University) for providing C. reinhardtii strain.
Contributor Information
Karl Forchhammer, Email: karl.forchhammer@uni-tuebingen.de.
Elena Ermilova, Email: e.ermilova@spbu.ru.
References
- 1. Smith DR & Lee RW (2014) A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella . Plant Physiol 164, 1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asmail SR & Smith DR (2016) Retention, erosion, and loss of the carotenoid biosynthetic pathway in the nonphotosynthetic green algal genus Polytomella . New Phytol 209, 899–903. [DOI] [PubMed] [Google Scholar]
- 3. Keeling PJ (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365, 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wicke S, Muller KF, dePamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS & Schneeweiss GM (2013) Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25, 3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA et al (2014) Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol Biol Evol 31, 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pombert JF, Blouin NA, Lane C, Boucias D & Keeling PJ (2014) A lack of parasitic reduction in the obligate parasitic green alga Helicosporidium . PLoS Genet 10, e1004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith DR & Asmail SR (2014) Next‐generation sequencing data suggest that certain nonphotosynthetic green plants have lost their plastid genomes. New Phytol 204, 7–11. [DOI] [PubMed] [Google Scholar]
- 8. Smith DR, Hua J, Archibald JM & Lee RW (2013) Palindromic genes in the linear mitochondrial genome of the nonphotosynthetic green alga Polytomella magna . Genome Biol Evol 5, 1661–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chellamuthu VR, Alva V & Forchhammer K (2013) From cyanobacteria to plants: conservation of PII functions during plastid evolution. Planta 237, 451–462. [DOI] [PubMed] [Google Scholar]
- 10. Forchhammer K & Lüddecke J (2016) Sensory properties of the PII signalling protein family. FEBS J 283, 425–437. [DOI] [PubMed] [Google Scholar]
- 11. Huergo LF, Chandra G & Merrick M (2013) P(II) signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol Rev 37, 251–283. [DOI] [PubMed] [Google Scholar]
- 12. Uhrig RG, Ng KK & Moorhead GB (2009) PII in higher plants: a modern role for an ancient protein. Trends Plant Sci 14, 505–11. [DOI] [PubMed] [Google Scholar]
- 13. Sant'Anna FH, Trentini DB, de Souto Weber S, Cecagno R, da Silva SC & Schrank IS (2009) The PII superfamily revised: a novel group and evolutionary insights. J Mol Evol 68, 322–336. [DOI] [PubMed] [Google Scholar]
- 14. Selim KA, Haase F, Hartmann MD, Hagemann M & Forchhammer K (2018) PII‐like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response. Proc Natl Acad Sci USA 115, E4861–E4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ermilova E, Lapina T, Zalutskaya Z, Minaeva E, Fokina O & Forchhammer K (2013) PII signal transduction protein in Chlamydomonas reinhardtii: localization and expression pattern. Protist 164, 49–59. [DOI] [PubMed] [Google Scholar]
- 16. Minaeva E, Forchhammer K & Ermilova E (2015) Glutamine assimilation and feedback regulation of L‐acetyl‐N‐glutamate kinase activity in Chlorella variabilis NC64A results in changes in arginine pools. Protist 166, 493–505. [DOI] [PubMed] [Google Scholar]
- 17. Chellamuthu VR, Ermilova E, Lapina T, Lüddecke J, Minaeva E, Herrmann C, Hartmann MD & Forchhammer K (2014) A widespread glutamine‐sensing mechanism in the plant kingdom. Cell 159, 1188–1199. [DOI] [PubMed] [Google Scholar]
- 18. Lapina T, Selim KA, Forchhammer K & Ermilova E (2018) The PII signaling protein from red algae represents an evolutionary link between cyanobacterial and Chloroplastida PII proteins. Sci Rep 8, 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beez S, Fokina O, Herrmann C & Forchhammer K (2009) N‐acetyl‐L‐glutamate kinase (NAGK) from oxygenic phototrophs: P(II) signal transduction across domains of life reveals novel insights in NAGK control. J Mol Biol 389, 748–758. [DOI] [PubMed] [Google Scholar]
- 20. Heinrich A, Maheswaran M, Ruppert U & Forchhammer K (2004) The Synechococcus elongatus P signal transduction protein controls arginine synthesis by complex formation with N‐acetyl‐L‐glutamate kinase. Mol Microbiol 52, 1303–1314. [DOI] [PubMed] [Google Scholar]
- 21. Johnson X & Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12, 776–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huergo LF & Dixon R (2015) The emergence of 2‐Oxoglutarate as a master regulator metabolite. Microbiol Mol Biol Rev 79, 419–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwarz D, Nodop A, Hüge J, Purfürst S, Forchhammer K, Michel KP, Bauwe H, Kopka J & Hagemann M (2011) Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol 155, 1640–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Llácer JL, Fita I & Rubio V (2008) Arginine and nitrogen storage. Curr Opin Struct Biol 18, 673–81. [DOI] [PubMed] [Google Scholar]
- 25. Llácer JL, Contreras A, Forchhammer K, Marco‐Marín C, Gil‐Ortiz F, Maldonado R, Fita I & Rubio V (2007) The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc Natl Acad Sci USA 104, 17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mizuno Y, Moorhead GB & Ng KK (2007) Structural basis for the regulation of N‐acetylglutamate kinase by PII in Arabidopsis thaliana . J Biol Chem 282, 35733–35740. [DOI] [PubMed] [Google Scholar]
- 27. Fokina O, Chellamuthu VR, Forchhammer K & Zeth K (2010) Mechanism of 2‐oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc Natl Acad Sci USA 107, 19760–19765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramón‐Maiques S, Fernández‐Murga ML, Gil‐Ortiz F, Vagin A, Fita I & Rubio V (2006) Structural bases of feed‐back control of arginine biosynthesis, revealed by the structures of two hexameric N‐acetylglutamate kinases, from Thermotoga maritima and Pseudomonas aeruginosa . J Mol Biol 356, 695–713. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Liu W, Sun LP & Zhou ZG (2017) Evidence for PII with NAGK interaction that regulates Arg synthesis in the microalga Myrmecia incisa in response to nitrogen starvation. Sci Rep 7, 16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fokina O, Herrmann C & Forchhammer K (2011) Signal‐transduction protein P(II) from Synechococcus elongatus PCC 7942 senses low adenylate energy charge in vitro. Biochem J 440, 147–156. [DOI] [PubMed] [Google Scholar]
- 31. Court SJ, Waclaw B & Allen RJ (2015) Lower glycolysis carries a higher flux than any biochemically possible alternative. Nat Commun 6, 8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith DR & Lee RW (2011) Nucleotide diversity of the colorless green alga Polytomella parva (Chlorophyceae, Chlorophyta): high for the mitochondrial telomeres, surprisingly low everywhere Els. J Eukaryot Microbiol 58, 471–473. [DOI] [PubMed] [Google Scholar]
- 33. Forcada‐Nadal A, Llácer JL, Contreras A, Marco‐Marín C & Rubio V (2018) The PII‐NAGK‐PipX‐NtcA regulatory axis of cyanobacteria: a tale of changing partners, allosteric effectors and non‐covalent interactions. Front Mol Biosci 5, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terashima M, Specht M & Hippler M (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57, 151–68. [DOI] [PubMed] [Google Scholar]
- 35. Bueno R, Pahel G & Magasanik B (1985) Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli . J Bacteriol 164, 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Atteia A, van Lis R, Ramírez J & González‐Halphen D (2000) Polytomella spp. growth on ethanol. Extracellular pH affects the accumulation of mitochondrial cytochrome c550. Eur J Biochem 267, 2850–2858. [DOI] [PubMed] [Google Scholar]
- 37. Sager R & Granick S (1954) Nutritional control of sexuality in Chlamydomonas reinhardii . J Gen Physiol 37, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watzer B, Engelbrecht A, Hauf W, Stahl M, Maldener I & Forchhammer K (2015) Metabolic pathway engineering using the central signal processor PII. Microb Cell Fact 14, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd & Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- 40. Emanuelsson O, Brunak S, von Heijne G & Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2, 953–971. [DOI] [PubMed] [Google Scholar]
- 41. Emanuelsson O, Nielsen H & von Heijne G (1999) ChloroP, a neural network‐based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maheswaran M, Urbanke C & Forchhammer K (2004) Complex formation and catalytic activation by the PII signaling protein of N‐acetyl‐L‐glutamate kinase from Synechococcus elongatus strain PCC 7942. J Biol Chem 279, 55202–55210. [DOI] [PubMed] [Google Scholar]
- 43. Hauf K, Kayumov A, Gloge F & Forchhammer K (2016) The molecular basis of TnrA control by glutamine synthetase in Bacillus subtilis . J Biol Chem 291, 3483–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walter J, Selim KA, Leganés F, Fernández‐Piñas F, Vothknecht UC, Forchhammer K, Aro EM & Gollan PJ (2019) A novel Ca2+‐binding protein influences photosynthetic electron transport in Anabaena sp. PCC 7120. Biochim Biophys Acta Bioenerg 1860, 519–532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of identified metabolites by LC‐MS normalized to 1 mg of algal cell dry weight including standard deviation (SD) of three biological replicates for Polytomella parva (under nitrogen excess and limiting conditions) and Chlamydomonas reinhardtii (under nitrogen‐rich conditions).


