Summary
Determining where species diversify (cradles) and persist (museums) over evolutionary time is fundamental to understanding the distribution of biodiversity and for conservation prioritization. Here, we identify cradles and museums of angiosperm generic diversity across tropical Africa, one of the most biodiverse regions on Earth.
Regions containing nonrandom concentrations of young (neo‐) and old (paleo‐) endemic taxa were identified using distribution data of 1719 genera combined with a newly generated time‐calibrated mega‐phylogenetic tree. We then compared the identified regions with the current network of African protected areas (PAs).
At the generic level, phylogenetic diversity and endemism are mainly concentrated in the biogeographically complex region of Eastern Africa. We show that mountainous areas are centres of both neo‐ and paleo‐endemism. By contrast, the Guineo‐Congolian lowland rain forest region is characterized by widespread and old lineages. We found that the overlap between centres of phylogenetic endemism and PAs is high (> 85%).
We show the vital role played by mountains acting simultaneously as cradles and museums of tropical African plant biodiversity. By contrast, lowland rainforests act mainly as museums for generic diversity. Our study shows that incorporating large‐scale taxonomically verified distribution datasets and mega‐phylogenies lead to an improved understanding of tropical plant biodiversity evolution.
Keywords: angiosperms, CANAPE, East Africa, endemism, mountains, phylogenetic diversity, protected areas
Introduction
Understanding the ecological and evolutionary processes that shape biodiversity is crucial to establishing conservation priorities (McNeely et al., 1990; Myers et al., 2000). Endemic taxa are defined as being geographically restricted and their spatial concentration often highlights evolutionary processes. Traditional methods have mainly focused on using taxonomic diversity (i.e. taxon richness) to depict biodiversity patterns. However, adding the evolutionary history allows a more detailed picture of such patterns (Faith, 1992; Forest et al., 2007). Combining data from phylogenetic trees and taxon diversity enables the calculation of phylogenetic diversity (PD), that is, the total length of branches of a phylogenetic tree connecting taxa in a particular area (Faith, 1992). Regions with high PD contain important levels of evolutionary history (Sechrest et al., 2002; Forest et al., 2007). Phylogenetic endemism (PE) is the spatial restriction of phylogenetic lineages, that is, the degree to which a part of the evolutionary history is range‐restricted (Rosauer et al., 2009). In this context, two types of endemic taxa can be distinguished (Kruckeberg & Rabinowitz, 1985; Nekola, 1999; Ferreira & Boldrini, 2011; Mishler et al., 2014). Neo‐endemics are taxa that diverged recently (i.e. sustained by short phylogenetic branches) and whose narrow range is generally linked to a lack of time to disperse and expand. By contrast, paleo‐endemics are taxa that diverged much earlier and which are also range‐restricted, either because of range reduction or because of a lack of adaptation or dispersal opportunities. Spatial concentrations of neo‐ or paleo‐endemic taxa are referred to as centres of neo‐endemism or paleo‐endemism, respectively (Fig. 1a,b), and concentrations of both neo‐ and paleo‐endemic taxa are called centres of mixed‐endemism (Fig. 1c) (Mishler et al., 2014). Centres of neo‐endemism can be viewed as ‘cradles of diversity', that is, regions with high speciation rates, and are centres of diversification, whereas centres of paleo‐endemism can be viewed as ‘museums of diversity', where taxa have persisted over time. By studying the distribution of endemic taxa together with their evolutionary history, we can generate hypotheses about historical processes that shaped biodiversity (Mishler et al., 2014; Thornhill et al., 2016, 2017; Scherson et al., 2017). Such knowledge is crucial for the improvement of conservation management in the context of an unprecedented biodiversity decline (Humphreys et al., 2019).
Figure 1.
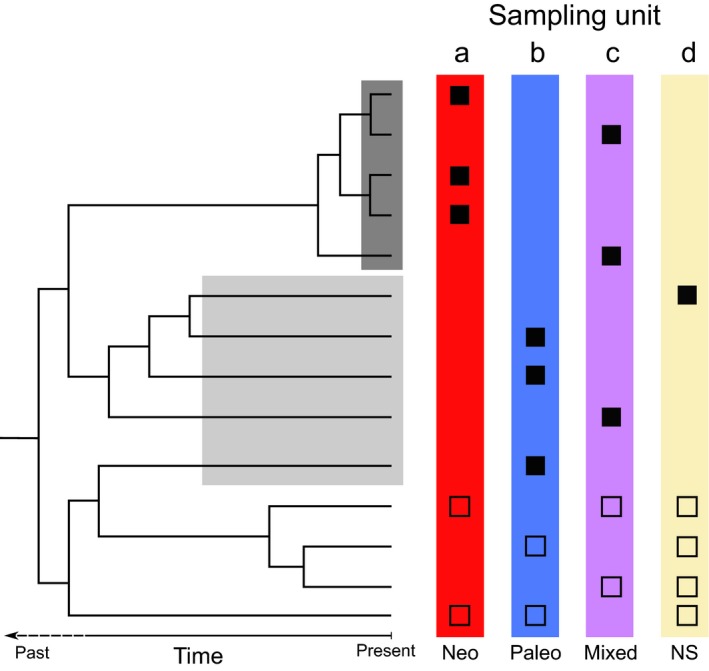
Theoretical framework for identifying the four categories of phylogenetic endemism based on a phylogenetic tree with branch lengths calibrated in units of time and the distribution of 14 taxa across four sites of equal taxonomic richness. Neo‐endemic, endemic taxon sustained by a relatively short branch (highlighted in the dark grey box); paleo‐endemic, endemic taxon sustained by a relatively long branch (highlighted in the light grey box). Sampling units are as follows: a, centre of neo‐endemism with an over‐representation of neo‐endemics; b, centre of paleo‐endemism with an over‐representation of paleo‐endemics; c, centre of mixed‐endemism with an over‐representation of both neo‐ and paleo‐endemics; d, nonsignificant site. Black filled squares, endemic taxon (i.e. present in a relatively low number of sites); unfilled squares, nonendemic taxon (i.e. present in a relatively high number of sites).
Tropical Africa contains some of the most biologically diverse regions of the world (Myers et al., 2000; Linder, 2001; Küper et al., 2004). Although it has lower species diversity when compared with the Neotropics and Southeast Asia, tropical Africa still harbours an estimated 30 000–35 000 species of plants (Couvreur, 2015). High species richness and centres of taxonomic endemism are found in Cameroon, Gabon, eastern Democratic Republic of the Congo (DRC), and eastern Tanzania (Linder, 2001; Küper et al., 2004; Sosef et al., 2017). Furthermore, Africa contains the second largest continuous tropical rainforest block after the Amazon basin (Malhi et al., 2013). Heterogeneous topography and climate across the continent gave rise to intricate biogeographical patterns forming numerous bioregions and transition zones (White, 1983; Linder et al., 2012; Fayolle et al., 2014; Droissart et al., 2018). Tropical African plant diversity patterns have been relatively well explored by taxonomic approaches (Brenan, 1978; White, 1983; Lovett et al., 2000; Linder, 2001; Küper et al., 2004; Klopper et al., 2007; Droissart et al., 2018). However, in an evolutionary context our understanding about where diversity originated and where it is maintained is limited.
The lowland rainforests of the Guineo‐Congolian region have been suggested to be Pleistocene cradles of diversity (Table 1) with glacial forest refugia triggering speciation (Diamond & Hamilton, 1980; Maley, 1996; Robbrecht, 1996; Sosef, 1996; Huntley & Voelker, 2016). However, molecular dating estimates in several plant and animal clades have shown that many species originated before the start of the Pleistocene, suggesting that forest refugia patterns reflect mainly Pliocene climatic fluctuation (Plana et al., 2004; Voelker et al., 2010). This led to the idea that lowland rainforests are museums of diversity (Table 1, Murienne et al., 2013). In other studies, these lowland regions have been found to be concentrations of old and widespread lineages, especially in birds (Table 1; Fjeldså, 1994; Fjeldså & Lovett, 1997; Fjeldså & Bowie, 2008; Fjeldså et al., 2012).
Table 1.
Hypotheses and categorical analysis of neo‐ and paleo‐endemism (CANAPE) predictions tested across the tropical African angiosperm flora.
| Hypotheses | CANAPE predictions | References |
|---|---|---|
| Lowland rainforests are museums of diversity | Significant concentration of old and range‐restricted (paleo‐endemics) or old and widespread taxa in lowland rainforest SUs | Fjeldså (1994); Fjeldså & Lovett (1997); Fjeldså & Bowie (2008); Fjeldså et al. (2012) |
| Lowland rainforests are cradles of diversity (past refugia as ‘species pump') | Significant concentration of young and range‐restricted taxa (neo‐endemic) in lowland rainforest SUs | Maley (1996); Sosef (1996); Huntley & Voelker (2016) |
| Montane regions are centres of diversification (‘montane speciation model'), i.e. cradles of diversity | Significant concentrations of neo‐endemic taxa (centres of neo‐endemism) in montane SUs | Roy (1997); Dimitrov et al. (2012); Schwery et al. (2014) |
| Montane regions are centres of persistence of lineages, i.e. museums of diversity | Significant concentration of paleo‐endemic taxa (centres of paleo‐endemism) in montane SUs | Fjeldså & Lovett (1997); López‐Pujol et al. (2011); Tolley et al. (2011) |
| Montane regions are centres of both diversification and persistence of lineages | Significant concentration of both paleo‐ and neo‐endemic taxa (centres of mixed‐endemism) in montane SUs | Wasser & Lovett (1993); Fjeldså & Lovett (1997); López‐Pujol et al. (2011) |
SU, sampling unit (100 km2).
Cradles of diversity have also been suggested in montane regions of Africa (Table 1), such as the Albertine Rift and the Eastern Arc Mountains (Fjeldså & Lovett, 1997; Roy, 1997; Dimitrov et al., 2012), thereby supporting a ‘montane speciation model' (Roy, 1997). Indeed, mountains are important centres of global diversification that represent heterogeneous and dynamic landscapes, as well as isolation and reconnection processes that are linked with past climate changes (Peterson et al., 1997; Fjeldså et al., 2012; Hoorn et al., 2013; Antonelli et al., 2018; Muellner‐Riehl, 2019). This diversification is the consequence of different mechanisms, for example, fragmented habitats promoting allopatric divergence by isolation of small populations (Hughes & Eastwood, 2006; Schwery et al., 2014; Merckx et al., 2015; see Wen et al., 2014 for a review of all the mechanisms). Mountains have also been acknowledged as museums of diversity as their complex topography supports relatively stable habitats by buffering climatic fluctuations and by allowing the movement of habitats over relatively short altitudinal distances to respond to temperature shifts, and thereby allowing their persistence over long periods of time (Peterson et al., 1997; Fjeldså & Lovett, 1997; Hewitt, 2000; Tzedakis et al., 2002; Loarie et al., 2009; López‐Pujol et al., 2011; Tolley et al., 2011). However, studies focusing on bird and few plant clade distributions have identified co‐occurrence of neo‐ and paleo‐endemic taxa in African mountain areas (Wasser & Lovett, 1993; Fjeldså & Lovett, 1997; López‐Pujol et al., 2011), suggesting that mountains are both cradles and museums of diversity (Fjeldså & Lovett, 1997) (Table 1).
For plants, these hypotheses have not yet been tested using both large distribution datasets and dated molecular phylogenies. Botanical datasets recording African plant distribution information are becoming increasingly comprehensive (Stropp et al., 2016; Dauby et al., 2016) and are significantly improving our understanding of biodiversity and phytogeographical patterns across tropical Africa (Sosef et al., 2017; Droissart et al., 2018). In parallel, the incorporation of evolutionary history information is now possible thanks to the generation of large phylogenies using existing data available on GenBank (Hinchliff & Smith, 2014; Smith & Brown, 2018). Together, this allows us to test the described hypotheses at new levels of precision.
Here, we evaluate five main hypotheses (Table 1) related to the evolutionary and historical processes shaping tropical African biodiversity. We identify significant centres of neo‐ and paleo‐endemism across the entire tropical African angiosperm flora at generic level based on a taxonomically verified distribution dataset and a newly generated mega‐phylogeny of angiosperms. At the generic level, we identified areas of significant PD, PE and centres of neo‐, paleo‐ or mixed‐endemism across tropical Africa to answer the following questions: do previously identified areas of high taxon diversity display more PD and PE than expected by chance; are African mountains centres of diversification, centres of lineage persistence, or both; are African lowland rainforests museums or cradles of diversity? Finally, as centres of PE are of high conservation interest, we explored the overlap between the protected area network across tropical Africa and the different centres of endemism identified.
Materials and Methods
Study area
Here, tropical Africa is defined by the ecoregions characterized by Olson et al. (2001) with the ‘south Saharan steppe and woodlands' ecoregion and Mauritania as northern limit and the ‘Drakensberg montane grassland', ‘Highveld grassland', ‘Kalahari Acacia Baikiaea woodland', ‘Kalahari xeric savannah' and ‘Namibian savannah woodlands' ecoregions as southern limits (Sosef et al., 2017; Droissart et al., 2018). This study includes continental Africa and the Guinean Gulf islands (Bioko, Sao Tomé and Principe), but excludes Madagascar (Fig. 2).
Figure 2.
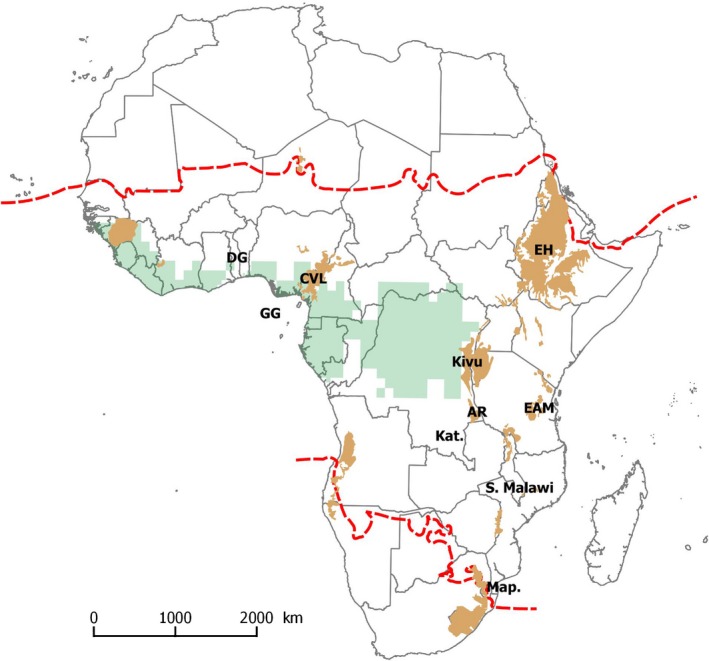
Map of Africa. The red dotted line delineates the study zone. Green shading delineates the Guineo‐Congolian bioregion as defined by Droissart et al. (2018). Brown shading delineates ‘montane regions' (GMBA mountain inventory_V1.2; Körner et al., 2017). Locations cited in the main text: AR, Albertine Rift; CVL, Cameroon Volcanic Line; DG, Dahomey Gap; EAM, Eastern Arc Mountains; EH, Ethiopian Highlands; GG, Guinean Gulf; Kat., Katanga; Map., Maputaland; S. Malawi, South Malawi. The map is projected in the Africa Albers Equal Area Conic coordinate reference system (ESRI:102022).
Taxonomic level
We conducted this study on tropical African plants at the genus level. This taxonomic level presents advantages and drawbacks. Conceptually, genera should represent clear morphological differences of the flora and older evolutionary events than species, which is the main focus of this study. From a methodological point of view, generic names are comparatively stable taxonomic entities, which circumvent biases as a result of taxonomic uncertainties and potential misidentifications of species (but see Discussion). Moreover, mega‐phylogenies are better resolved at the genus than at the species level (Hinchliff & Smith, 2014; Smith & Brown, 2018). Finally, genus‐level studies are useful for addressing broad‐scale biodiversity patterns (Forest et al., 2007; Thornhill et al., 2016; Scherson et al., 2017). A drawback is that really recent species cradles will be missed by this approach (see Discussion).
Datasets
Plant occurrence data
We used the RAINBIO dataset (Dauby et al., 2016), a database of georeferenced occurrences of African vascular plants with comparatively high taxonomic quality. It represents a compilation of 614 022 botanical records collected from 1782 to 2015 providing distribution information for 25 356 species and 3158 genera within 273 families. This represents c. 89% of all known tropical African plant species.
Species occurrence data were projected using the Africa Albers Equal Area Conic coordinate reference system (ESRI:102022, http://spatialreference.org/). For that, we transformed coordinates from decimal degrees into metres, the base unit of this coordinate reference system. The study area was divided into 100 × 100 km equal‐area square sampling units (SUs).
Using the RAINBIO dataset, we applied three filters, leading to the RAINBIOfiltered dataset we used for all our analyses:
Angiosperm filter. As our study and phylogeny focused on Angiospermae, Pteridophyta and Gymnospermae were excluded.
Edge effect filter. When considering patterns of relative endemism in a delimited zone, bias towards over‐representation of artificial endemic taxa at the boundaries, is frequent. This artefact is a result of the rough cut within the geographic distribution of the taxa occurring both in and outside the study area, thereby artificially leading to the identification of these taxa as endemics although some of them may occur on both sides of the boundary. In order to avoid this edge effect, we kept only genera with > 90% of their occurrences falling within our study area.
Occurrence filter. Specimen density in RAINBIO is heterogeneous across tropical Africa. Poorly sampled SUs occur in poorly known regions and thus are more likely to include taxa that were collected only once (Sosef et al., 2017). This can artificially increase the endemism of these SUs. To avoid potential bias caused by poorly sampled SUs, we included in our analyses only SUs in which at least 100 recorded occurrences are present. A threshold of at least 100 occurrences per SU was selected after testing different threshold values (results not presented here).
Phylogenetic data
We used a newly generated dated angiosperm phylogenetic tree (see Janssens et al., 2019 for details). Briefly, phylogenetic inference was conducted based on a sampling of two plastid markers (matK and rbcL) retrieved from GenBank for 36 234 plant species distributed across 8357 genera. The alignment was conducted with mafft (Katoh et al., 2002) and refined with geneious 7.0 (Auckland, New Zealand). The final topology results from maximum likelihood tree inference with raxml 7.4.2 (Stamatakis, 2006), constrained at the family level, under the general time‐reversible substitution model with gamma rate heterogeneity. Divergence time was estimated using 52 fossil calibration points scattered among the angiosperms and dated using the penalized likelihood algorithm implemented in treePL (Smith & O'Meara, 2012). The hard maximum and minimum age constraints of the angiosperms were set at 220 and 140 Myr, respectively (Soltis et al., 2002; Bell et al., 2005; Moore et al., 2007; Bell et al., 2010; Magallón et al., 2015). Finally, generic names in the RAINBIO dataset and in the phylogenetic tree were standardized using the Taxonomic Name Resolution Service tool (Boyle et al., 2013).
Data analyses
Biodiversity analyses
To explore the diversity, distribution and evolutionary history of the angiosperm in tropical Africa, we used the RAINBIOfiltered dataset and the resulting dated phylogenetic tree to calculate a set of taxonomic and phylogenetic indices for each SU across our study area. All metrics were calculated using biodiverse v.2.0 (Laffan et al., 2010). We used the ‘Biodiverse pipeline' to run biodiverse directly from R (https://github.com/NunzioKnerr/biodiverse_pipeline).
Taxonomic indices
Genus richness (GR) is the number of distinct genera (g) present in each SU:
Weighted endemism (WE) is the sum of the inverse of each genus's (g) geographical range (Rg). Rg is measured as the number of SUs in which the genus occurs (Crisp et al., 2001):
Phylogenetic indices
Phylogenetic diversity (PD) is the sum of branch lengths connecting the root of the phylogenetic tree to all genera (tips of the phylogeny) within each SU (Faith, 1992). In the following formula, is the total number of branches connecting all genera within a SU, is a branch (a single segment between two nodes, representing a clade), and the branch length:
Relative PD (RPD) is the ratio of the PD measured on the original phylogenetic tree (PDorig) divided by the PD measured on a theoretical tree (PDtheor). The theoretical tree has the same topology as the original tree but all branches are of equal length. The RPD index thus measures the relative branch length within a SU: high RPD indicates an over‐representation of long‐branched genera whereas low RPD indicates over‐representation of short‐branched genera (Mishler et al., 2014):
Phylogenetic endemism (PE) is the sum of branch lengths, weighted by the inverse of the branch's range (Rc), for each branch (c) connecting the roots of the phylogenetic tree to the genera (tips of the phylogeny) within a SU (Rosauer et al., 2009). Rc is measured as the number of SUs in which the branch c occurs:
Relative PE (RPE) is the ratio of the PE measured on the original phylogenetic tree (PEorig) divided by the PE measured on a theoretical tree (PEtheor). The theoretical tree has the same topology as the original but all branches are of equal length (Mishler et al., 2014). This index is the base for the categorical analyses of neo‐ and paleo‐endemism (see later):
Null model – randomizations
To test which SUs had significantly higher or lower observed values than expected given the genus richness of the SU and the geographical range of all genera, we ran 999 randomizations using biodiverse v.2.0 (Laffan et al., 2010). For every run, the algorithm randomly reassigns all the genera to each SU without replacement. In order to respect both genus richness patterns and the geographical range of taxa, we constrained the procedure to keep the original number of genera in each SU and to keep the original number of SUs in which each genus occurs (‘rand_struct' option in biodiverse).
Then, for each metric, the observed value is compared with the 999 randomization values. Significantly greater or lower than expected was defined as being > 97.5% or < 2.5% of the random values, respectively (two‐tailed test, α = 0.05). High degrees of significance were established for observed values > 99% or < 1% of the random values.
This randomization test was carried out for WE, PD, PE and RPD, resulting in the assignment of a significance class for each of these metrics for each SU: significantly very low (‘< 0.01'), significantly low (‘< 0.025'), ‘not significant', significantly high (‘> 0.975') or significantly very high (‘> 0.99').
Categorical analysis of neo‐ and paleo‐Endemism (CANAPE)
The categorical analysis of neo‐ and paleo‐endemism (CANAPE) discriminates SUs with significantly high PE in neo‐ or paleo‐endemism based on taxon occurrences and the dated phylogenetic tree (Mishler et al., 2014). First, we calculate the PE based on the original phylogenetic tree (PEorig) and the PE based on the theoretical tree (PEtheor, branches of equal length) for each randomization of genera composition and subsequently compare those with the observed values. This allows us to select SUs that are centres of high PE, that is, showing either significantly high PEorig or PEtheor, or both (observed values of PEorig and PEtheor > 95% of the random values; one‐tailed test, α = 0.05). Second, significant SUs are categorized into four nonoverlapping categories. If the RPE ratio (PEorig/PEtheor) is higher than expected (two‐tailed test, α = 0.05), the SU contains significantly more endemic genera on long branches and is identified as a centre of ‘paleo‐endemism'. If the RPE ratio is lower than expected (two‐tailed test, α = 0.05), the SU contains significantly more endemic genera on short branches and is identified as a centre of ‘neo‐endemism'. If the RPE is not significantly high or low, but both PEorig and PEtheor are significantly high at the level of α = 0.05, SU is tagged as centre of ‘mixed‐endemism' (i.e. a mix of endemic genera with both long and short branches). Finally, if a ‘mixed‐endemism' SU presents both high PEorig and PEtheor at the level of α = 0.01, the SU is tagged as a centre of ‘super‐endemism' (i.e. highly significant concentration of endemic long and short branches).
As CANAPE results might be sensitive to the SU size, CANAPE was also carried out with four different SU sizes across the study zone: 50, 75, 200 and 300 km square. The SUs contain at least 50, 75, 200 and 300 occurrences, respectively.
Overlap with montane regions
A ‘montane' region can be defined according to differences in elevation, relief or steepness. We adopted the montane definition of Körner et al. (2011) based on the concept of steepness, the basic and consistent feature that reflects landscape heterogeneity found in mountains. Indeed, a definition based solely on elevation would consider highland plateaus as mountains (e.g. Tibetan Plateau, Altiplano) even if they are relatively flat landscapes. For continental‐scale purposes, steepness is estimated with the ruggedness, that is, the elevation range between a grid cell and the eight adjacent cells in the grid (Körner et al., 2011). A cell is then considered as a ‘mountain' if its ruggedness exceeds 200 m. Global mountain biodiversity assessment (GMBA) provides ruggedness data for 2.5‐arcminute (c. 4.6 × 4.6 km at the equator) plots across the globe (GMBA mountain definition_V1.0 database; Körner et al., 2011) and shapefiles delineating ‘montane areas' (GMBA mountain inventory_V1.2; Körner et al., 2017). For each SU, we calculated the mean ruggedness, the percentage of montane plots (i.e. percentage of 4.6 × 4.6 km plots whose ruggedness exceeds 200 m) and the percentage of overlap with ‘montane areas' defined by GMBA.
By contrast, we defined ‘lowlands' on the elevation feature to avoid considering a highland plateau as a lowland area. Elevation data were retrieved from the digital elevation modelling data produced by the NASA Shuttle Radar Topographic Mission (SRTM; http://srtm.csi.cgiar.org/). We downloaded the 250 m resolution (SRTM v.4.1) data in GeoTiff raster format, and extracted a raster file of the tropical Africa region using qgis 2.18. From this raster file, we calculated the mean elevation for each SU.
We compared the distribution of the mean ruggedness, the percentage of montane plots and the percentage of overlap with ‘montane areas' of the SUs across:
the three categories of the PD results (significantly high or very high PD; not significant; significantly low or very low PD);
the five categories of the CANAPE results (neo‐endemism; paleo‐endemism; mixed‐endemism; super‐endemism, not significant).
We compared the distribution of the mean elevation of the SUs across the three categories of the RPD results (significantly high or very high RPD; not significant; significantly low or very low RPD).
As normality of the residuals (one of the fundamental assumptions for ANOVAs) was not fulfilled for the three comparisons (results not shown), we used a nonparametric Kruskal–Wallis test. If at least one of the distributions was significantly different from the others (P < 0.05), then Wilcoxon pairwise comparison (two‐tailed tests with Holm's correction) was performed to disentangle which categories were significantly different from each other (P < 0.05).
CANAPE and protected areas network overlap
CANAPE results were overlapped with the African protected areas network retrieved from the World Database on Protected Areas (WDPA, https://protectedplanet.net/, accessed June 2018). Protected areas exclusively related to marine or faunal protection (e.g. ‘bird reserve', ‘hunting reserve', ‘faunal reserve') were excluded. The number of SUs that contained protected areas (PA) were counted for any PA and for the most restrictive PAs in terms of conservation (reported as International Union for Conservation of Nature (IUCN) categories Ia, Ib and II; PAs that have a main focus on biodiversity) (IUCN, 2008).
Results
Datasets
Plant occurrence
The RAINBIOfiltered dataset contained 547 273 occurrences of 2345 genera distributed across 638 equal‐area square SUs. GR is unevenly distributed across tropical Africa, ranging from 39 to 853 genera per SU (Fig. 3). The most diverse SUs are found around the Guinean Gulf, particularly in Gabon, western Cameroon, in the Dahomey Gap, the Eastern Arc Mountains and the Albertine Rift region, and to a lesser extent isolated SUs in the DRC and the Ethiopian Highlands.
Figure 3.
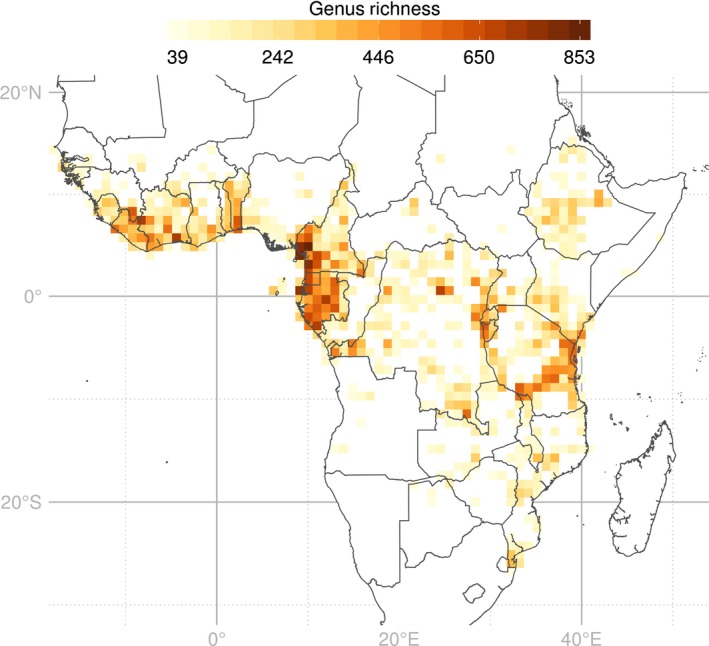
Genus richness map of the tropical African angiosperms, based on the distribution of 547 273 occurrences of 2345 genera across 638 equal area (100 × 100 km) sampling units. White regions contain < 100 records.
Phylogeny
The unfiltered phylogenetic tree contained 36 234 species and 8357 genera. Based on the RAINBIOfiltered dataset, this phylogenetic tree was pruned and included a final total of 1719 genera. Of these, 36% (618/1719) were not monophyletic in the unfiltered phylogenetic tree. In this case, we randomly sampled one of the branches of the nonmonophyletic genera. The tropical African flora at generic level represents 113 687 Myr of cumulative evolutionary history (i.e. the cumulative age of the taxa, estimated by the sum of all branch lengths on the tree).
Biodiversity analyses
Weighted endemism
Raw values of WE (Fig. 4a) followed a similar pattern to GR (Fig. 3) (R 2 = 0.764; Supporting Information Fig. S1). Significantly high WE SUs (Fig. 4b) are located in the eastern parts of tropical Africa, in Ethiopia, Kenya‐Tanzania, Katanga (southeast DRC), Maputaland and scattered in southeast Africa. This indicated that these regions harbour more endemic genera than expected from random. By contrast, regions in Central and West Africa (i.e. the Guineo‐Congolian region) are characterized by a significantly less WE than expected at random (Fig. 4b).
Figure 4.
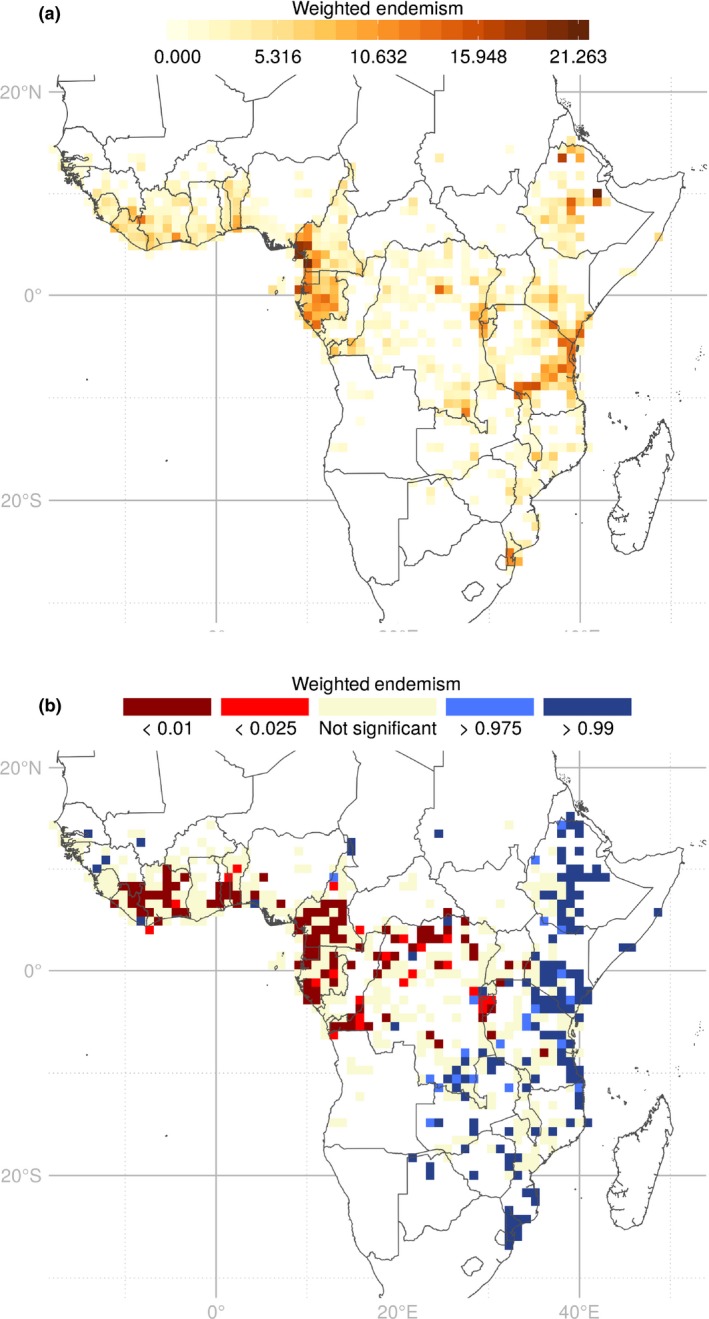
Weighted endemism (WE) maps of 2345 tropical African angiosperm genera. (a) Observed values of WE; (b) significant WE results from the randomization test. Sampling units (SUs): 100 × 100 km. Red SUs contain less WE than expected; blue SUs contain more WE than expected; beige SUs are not significant; white regions contain < 100 records and were not included in the analyses.
Randomization tests for PD
Calculations of phylogenetic metrics were conducted on the 1719 genera found both in the phylogenetic tree and in the RAINBIOfiltered dataset. PD is highly correlated with genus richness (R 2 = 0.992; Figs S2, S3). Higher PD than expected was found in 22 SUs (Fig. 5a), located in mountainous areas (Fig. S4; Table S1) around Lake Victoria, near the Eastern Arc Mountains, in northern Kivu, in Central Ethiopia and in the Cameroon Volcanic Line (Fig. 5a). Conversely, 230 SUs presented lower PD than expected. These are distributed broadly across the study zone, with the main centres occurring in the Dahomey Gap and to the west, in northern Cameroon, central Gabon and coastal Congo, and across eastern Africa (Fig. 5a).
Figure 5.
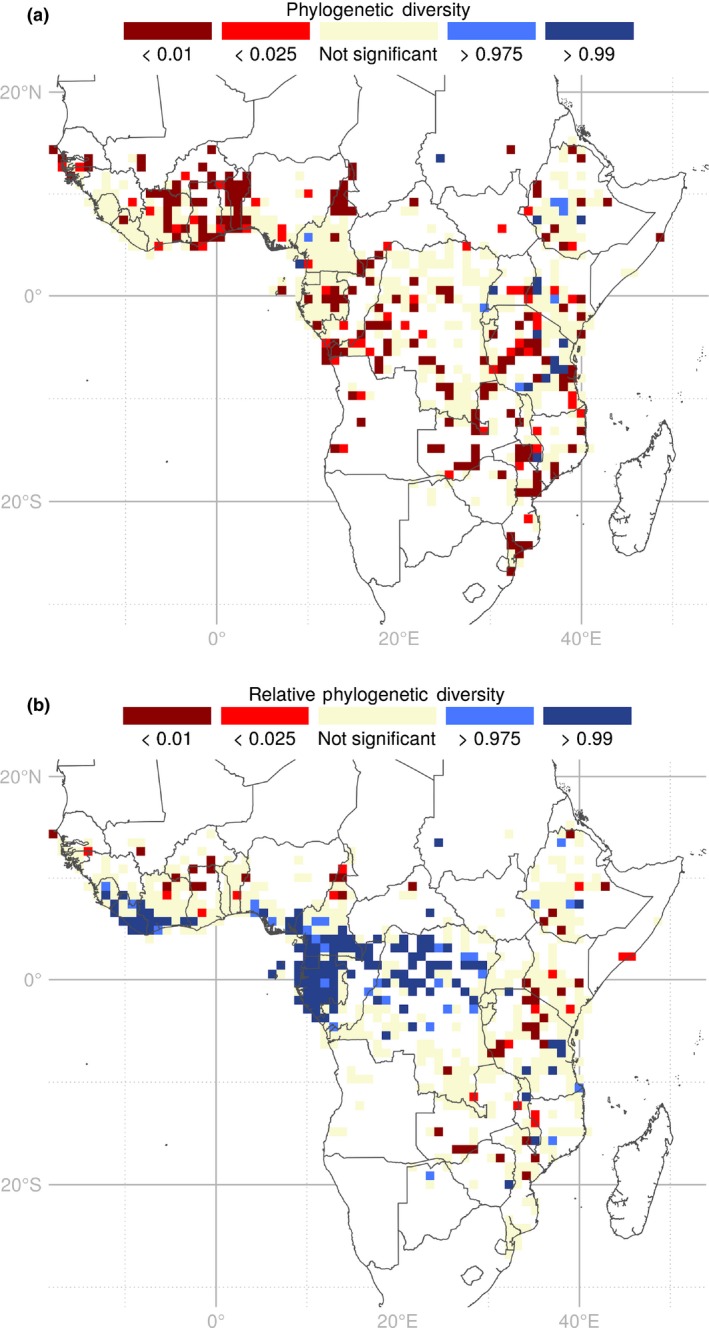
Maps of significant (a) phylogenetic diversity (PD) and (b) relative phylogenetic (RPD) resulting from the randomization tests for 1719 tropical African angiosperm genera. Sampling units (SUs) are 100 × 100 km squares. Red SUs contain less PD or RPD than expected; blue SUs contain more PD or RPD than expected; beige SUs are not significant; white regions contain < 100 records and were not included in the analyses.
SUs of significantly high RPD, representing concentrations of long‐branched (i.e. old) genera, are clustered in the Guineo‐Congolian region, western African forests and scattered in eastern Africa (Fig. 5b). SUs of significant low RPD, representing concentrations of short‐branched (i.e. young) genera, are scattered in the north of the Guineo‐Congolian region and in eastern Africa (Fig. 5b).
Categorical Analyses of neo‐ and paleo‐endemism
As for PD and GR, PE is highly correlated with WE (R 2 = 0.985; Fig. S5) and presents a similar pattern of significance as the WEs (Fig. S6). A total of 155 SUs were revealed by the CANAPE analyses as containing significantly more PE than expected (red, blue and purple SUs in Fig. 6). These are concentrated mainly in the eastern part of Africa. Areas of mixed‐ and super‐endemism are the most common significant CANAPE SUs (58 and 50, respectively). Only 11 SUs are classified as areas of neo‐endemism and these are generally surrounded by areas of mixed‐, super‐ or paleo‐endemism. As for neo‐endemism, we found the 36 SUs of paleo‐endemism surrounded by mixed‐ and super‐endemism SUs. The Ethiopian Highlands show a concentration of mixed, super and paleo‐endemic SUs. From central Kenya towards northern Tanzania, a similar cluster is found, with some neo‐endemic SUs. Maputaland also concentrates PE (mostly paleo‐). Smaller clusters occurred across eastern Africa, such as in coastal and southwestern Tanzania, Katanga and other scattered areas in southeast Africa. Concentrations of significant PE in West and Central Africa are very low and mainly located in northern Cameroon, Bioko Island (Equatorial Guinea, Guinean Gulf), Guinea and Sierra Leone.
Figure 6.
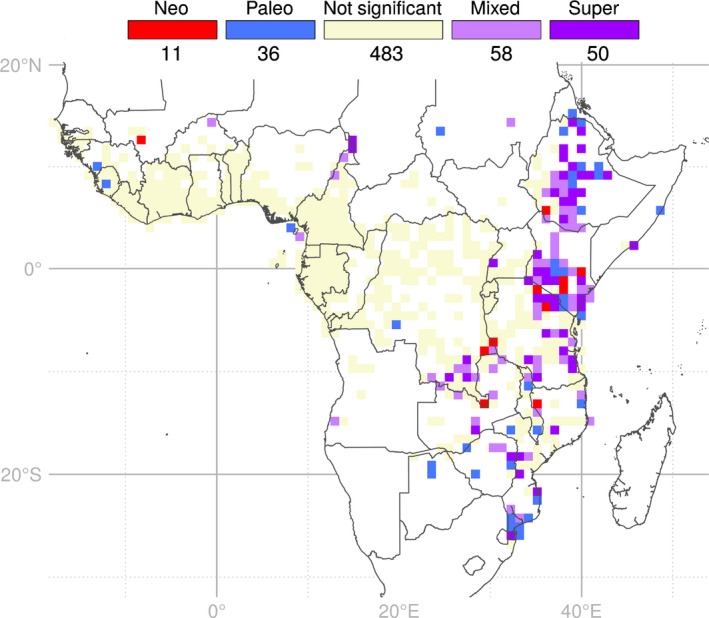
Map of significant phylogenetic endemism (PE) identified by the categorical analysis of neo‐ and paleo‐endemism (CANAPE) for 1719 tropical African angiosperm genera. Sampling units (SUs) are 100 × 100 km squares. Coloured SUs present significantly high PE: red, centres of neo‐endemism; blue, centres of paleo‐endemism; violet, centres of mixed‐endemism (i.e. mix of neo‐ and paleo‐endemism), with darker violet indicating centres of super‐endemism; beige, not significant; white regions contain < 100 records and were not included in the analyses. Figures below the legend indicate the number of SUs included in each category.
Different SU sizes give a similar overall CANAPE results, with higher coverage of the study zone with large SUs (200 and 300 km square) and lower coverage with small SUs (50 and 75 km square) (Fig. S7).
Montane and lowland regions
The mean elevation (Fig. 7) differs significantly between the RPD randomization categories (Kruskal–Wallis test, P < 0.05; Table S2). The high RPD SUs are significantly lower in elevation than nonsignificant and low RPD SUs (Wilcoxon pairwise comparison, P < 0.05; Table S2). Some SUs are outliers of high RPD distribution (solid dots), meaning these few SUs are occurring at high elevation.
Figure 7.
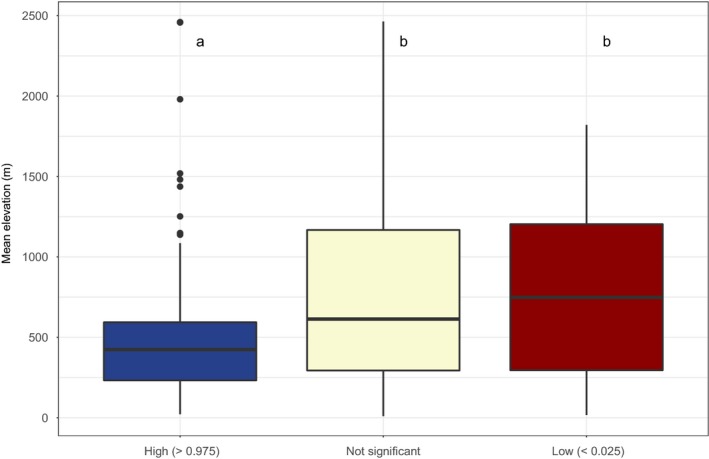
Boxplot of the distribution of the mean elevation of the sampling units (SUs) depending on their relative phylogenetic diversity significance class. Distributions of the mean elevation of the SUs are significantly different (Kruskal–Wallis test, P < 0.05). Different letters indicate significant differences (P < 0.05; pairwise comparison using Wilcoxon test with Holm's correction; see Supporting Information Table S2). For each box, the bold horizontal line corresponds to the median; the lower and upper bounds of the box correspond to first and third quartiles, respectively; the upper vertical line extends from the upper bound of the box to the highest value of the distribution, no further than 1.5 × interquartile range (IQR, or the distance between the first and third quartiles); the lower vertical line extends from the lower bound of the box to the lowest value of the distribution, no further than 1.5 × IQR; black dots are values beyond IQR (‘outlier' values).
The distribution of the mean ruggedness (Fig. 8), the percentage of montane plots (Fig. S8) and the percentage of overlap with ‘montane areas' (Fig. S6) differ significantly between the CANAPE categories (P < 0.05; Tables S3–S5). The mean ruggedness of mixed‐ and super‐endemic SUs is significantly higher than the ruggedness of nonsignificant SUs (Fig. 8) (Wilcoxon pairwise comparison, P < 0.05; Table S3). The mean ruggedness of neo‐ and paleo‐endemic SUs is not different from all the others (Wilcoxon pairwise comparison, P < 0.05; Table S3).
Figure 8.
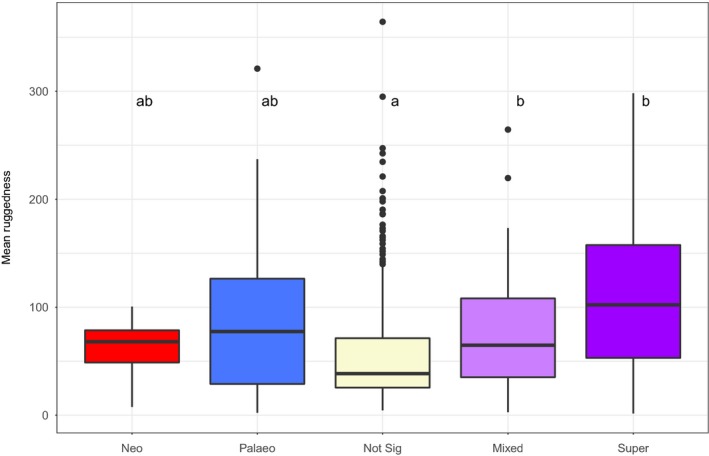
Boxplot of the distribution of the mean ruggedness of the sampling units depending on their categorical analysis of neo‐ and paleo‐endemism (CANAPE) category. Distributions of the mean ruggedness of the sampling units (SUs) are significantly different (Kruskal–Wallis test, P < 0.05). Different letters indicate significant difference (P < 0.05; pairwise comparison using Wilcoxon test with Holm's correction; see Supporting Information Table S3). For each box, the bold horizontal line corresponds to the median; the lower and upper bounds of the box correspond to first and third quartiles, respectively; the upper vertical line extends from the upper bound of the box to the highest value of the distribution, no further than 1.5 × interquartile range (IQR, or the distance between the first and third quartiles); the lower vertical line extends from the lower bound of the box to the lowest value of the distribution, no further than 1.5 × IQR; black dots are values beyond IQR (‘outlier' values).
The distributions of the percentage of montane plots and of the overlap with ‘montane areas' of the SUs show similar differences across the five CANAPE categories, except for paleo‐endemic SUs that are significantly different from nonsignificant SUs (Figs S8, S9; Tables S4, S5).
Protected areas network overlap
The overlap between the African PA network and centres of significant PE detected by CANAPE is partial (Fig. 9). More than 85% of the CANAPE cells contain at least a part of a PA, and > 42% of the CANAPE cells partially overlap with one of the most restrictive PA IUCN categories (Ia, Ib or II). The whole PA network covers 18.7% of the CANAPE cell surface, and 5.9% when considering only IA, Ib and II PA categories (Tables S6, S7).
Figure 9.
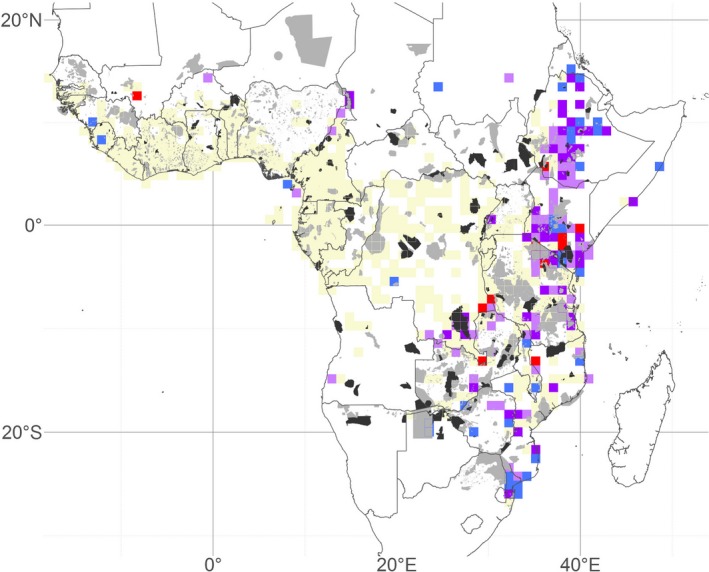
Map of the protected areas of Africa overlaid onto categorical analysis of neo‐ and paleo‐endemism (CANAPE) results. Dark grey areas are the most restrictive protected areas (PAs) (International Union for Conservation of Nature (IUCN) categories Ia, Ib or II); light grey areas are other PAs (IUCN categories III, IV, V or VI, or with no category assigned). Data were retrieved from the World Database on Protected Areas (WDPA, https://protectedplanet.net/, accessed June 2018). Sampling units (SUs) are 100 × 100 km squares. Coloured SUs present significantly high PE: red, centres of neo‐endemism; blue, centres of paleo‐endemism; violet, centres of mixed‐endemism (i.e. mix of neo‐ and paleo‐endemism), with darker violet indicating centres of super‐endemism; beige, not significant; white SUs contain < 100 records.
Discussion
Significant concentrations of evolutionary history and taxonomic endemism
In this study, we combined for the first time at the near continental level a large floristic dataset within a phylogenetic framework to examine more than 113 000 Myr of evolutionary history of tropical African angiosperms. Our GR map (Fig. 3) corroborates previous studies highlighting Cameroon, Gabon, the Albertine Rift, the Eastern Arc Mountains and coastal Tanzania as containing high taxon richness (Linder, 2001; Küper et al., 2004; Lovett et al., 2005; Plumptre et al., 2007; Sosef et al., 2017).
Despite the correlation between GR and PD (Fig. S2), our randomization procedure reveals that in some regions, PD is higher or lower than expected by chance (Fig. 5a). A decoupled pattern between GR and PD was also observed in South Africa (Forest et al., 2007). Across tropical Africa, 230 out of 638 SUs showed significantly lower PD than expected by chance (phylogenetic clustering). These are located in a lowland rainforest in Gabon and central DRC, but also in drier regions such as the Dahomey Gap, northern Cameroon, savannah regions in western DRC and in eastern Africa (Fig. 5a). In savannah (i.e. wooded grassland) regions, phylogenetic clustering is probably caused by the dominance of Poaceae and Fabaceae (Jacobs, 2004). In lowland tropical rainforest regions, phylogenetic clustering suggests evolutionary conservatism in tropical forest adaptations. Analogous phylogenetic clustering has been observed in some tropical forest and arid regions in Borneo, Central America, Australia and Chile (Webb, 2000; Swenson et al., 2007; Thornhill et al., 2016; Scherson et al., 2017). By contrast, only six SUs had significantly higher PD than expected by chance (phylogenetic overdispersion). These are mainly located in montane areas (Fig. S5) of the Cameroon Volcanic Line, Ethiopian Highlands, northern Kivu, central Kenya, the Eastern Arc Mountains and southern Malawi (Fig. 5a). Phylogenetic overdispersion may be related to competitive exclusion of closely related genera whose niches show a large overlap (Webb et al., 2002), extinction (Kissling et al., 2012; Couvreur, 2015), or colonization of phylogenetically distinct lineages. Another reason for this pattern could be linked to the complex topography of montane areas containing heterogeneous habitats and various soil types harbouring distinctly adapted plant lineages (Peterson et al., 1997; Hoorn et al., 2010, 2013; Antonelli et al., 2018). Phylogenetically overdispersed SUs have also been observed in ecotone regions in the Cape region (Forest et al., 2007) and were argued to be of high conservation importance as they represent an important amount of evolutionary history (Faith, 1992; Swenson et al., 2007; Forest et al., 2015).
The regions of high taxonomic diversity mentioned earlier (Cameroon, Gabon, the Albertine Rift, the Eastern Arc Mountains and coastal Tanzania) are also known to contain a high number of endemic species (Linder, 2001; Küper et al., 2004; Plumptre et al., 2007). We indeed found a similar pattern between GR and observed WE (Figs 4a, S1). However, our randomization procedure revealed that these regions present lower amounts of generic endemism than expected (Fig. 4b) – except for southern coastal Tanzania. This does not mean, however, that Cameroon, Gabon, the Albertine Rift and the Eastern Arc Mountains are not rich in endemic genera, but that given the high taxonomic richness, the concentration of endemic genera they harbour is not exceptional. Instead, generic level distribution ranges are wider than expected, indicating an over‐representation of widespread genera in the Guineo‐Congolian region (Fig. 4b). By contrast, East Africa, particularly Ethiopia, Kenya, Tanzania, Katanga (DRC), Maputaland and other SUs in southeast Africa, harbour more endemic genera than expected (high WE; Fig. 4b). These represent important endemic portions of evolutionary history (high PE; Fig. 6), possibly related to the topographic and edaphic complexity of East Africa. Indeed, East Africa has a greater elevation range than West or Central Africa (Guillocheau et al., 2018) and the tectonic activity that occurred in East Africa generated places that differ edaphically from surrounding areas. Edaphic complexity plays a great role in generating endemism (Bruchmann & Hobohm, 2014; Rahbek et al., 2019), and the habitat heterogeneity and fragmentation of montane regions are often associated with range‐restricted taxa (Moritz et al., 2002; Hughes & Eastwood, 2006; Kier et al., 2009; Fjeldså et al., 2012). Moreover, East Africa is acknowledged to be a ‘complex biogeographical mixture' containing distinct taxa adapted to various localized environmental gradients generated by the complex topography and climate (Linder et al., 2012; Droissart et al., 2018). Endemic lineages are thus more likely to occur in greater proportion in the regions that are topographically and edaphically complex than in regions of low habitat heterogeneity (such as lowlands) or low edaphic complexity (such as, inter allia, the Eastern Arc Mountains). Finally, endemism in eastern Africa seems to have been undervalued in previous continental‐scale studies (Fjeldså & Lovett, 1997; Linder, 2001; but see Küper et al., 2004). Our incorporation of evolutionary history and randomization procedures allowed the identification of new regions of high evolutionary and conservation interests (Fig. 6).
Cradles and museums of diversity
In our study, most SUs identified as containing significantly high PE are centres of mixed‐ or super‐endemism, concentrating both neo‐ and paleo‐endemic genera (Fig. 6). These mixed‐endemism and super‐endemism SUs occur significantly more frequently in montane regions than did nonsignificant SUs (Fig. 8). In addition, neo‐endemism SUs also show a nonsignificant tendency to occur mainly in montane areas (Fig. 8). Thus, even though mountains have been suggested to act as cradles of diversity (Roy, 1997; Jetz et al., 2004; Fjeldså & Rahbek, 2006; Schwery et al., 2014; Merckx et al., 2015), our results support the hypothesis that montane regions, particularly in East Africa, are both museums and cradles of diversity (Wasser & Lovett, 1993; Fjeldså & Lovett, 1997; López‐Pujol et al., 2011). Our results also suggest that the montane speciation model, where mountain ‘cradles' are considered to feed surrounding (generally lowland) regions in species (Roy, 1997; Jetz et al., 2004; Fjeldså & Rahbek, 2006; Hoorn et al., 2013), is hard to support at genus level across tropical Africa. This hypothesis appears reductive as mountains can also act as ‘museums', and centres of diversification may also occur elsewhere, such as around river networks or in lowland ecotonal zones (e.g. transitions between forests and savannah) (Fjeldså, 1994; Plana, 2004).
Furthermore, by identifying Ethiopian highlands, Kenya, Tanzania, Katanga, southeast Africa and Maputaland as centres of mixed‐endemism, we demonstrate that within these regions, processes of recent diversification occur together with favourable conditions for lineage persistence leading to a greater accumulation of evolutionary history than expected by chance. Several studies also found areas of mixed‐endemism in other montane regions, such as in South America (Fjeldså, 1994; Fjeldså & Lovett, 1997; Bitencourt & Rapini, 2013), and China (López‐Pujol et al., 2011). In tropical Africa, previous identification of centres of neo‐ and paleo‐endemism in plants focused on only two plant clades a priori assumed to be neo‐ or paleo‐endemics, because of a lack of dated plant phylogenetic trees (Fjeldså & Lovett, 1997). Fjeldså & Lovett (1997) reported co‐occurrences of neo‐ and paleo‐endemics in some mountains, such as the Cameroon Volcanic Line or the Albertine Rift, but they did not detect the same East African regions to act as both museums and cradles, as reported here (Fig. 6). This underlines the importance of integrating randomization and categorization procedures together with a comprehensive phylogeny and angiosperm wide datasets to detect patterns of significant neo‐, paleo‐ and mixed‐endemism (Mishler et al., 2014; Thornhill et al., 2016).
By contrast, most parts of West and Central Africa present nonsignificant CANAPE SUs, indicating that these regions harbour no more endemic genera than expected. Randomization of the RPD revealed concentrations of old genera in lowland rainforests of Guineo‐Congolia (Figs 5b, 7). As mentioned earlier, these genera also appear to be widespread (low WE; Fig. 4b). Several local museums of biodiversity have been proposed to occur within this region (Fjeldså & Lovett, 1997; Murienne et al., 2013), which, according to Linder (2014), contain the oldest flora of tropical Africa, and here appears to be a museum of diversity as a whole.
Our study highlights the importance of incorporating large‐scale taxonomically verified distribution datasets with mega‐phylogenies, which lead to an improved understanding of tropical plant biodiversity evolution. Similar studies in other megadiverse regions such as the Amazon basin will allow comparison of tropical biodiversity origins and maintenance processes across the tropics.
Taxonomic and geographic resolution effects
Biodiversity and phylogenetic analyses at genus level have already demonstrated their relevance in understanding patterns of biodiversity evolution (Forest et al., 2007; Thornhill et al., 2016; Scherson et al., 2017). However, interpretation must not be extrapolated to other taxonomic levels. This is particularly the case for analyses that are focused on species endemism, as different species within a genus may show various geographic distribution patterns. For example, the genus Impatiens (Balsaminaceae) is widespread in the Old World tropics and subtropics, but the majority (c. 80%) of its African species are range‐restricted in regions such as the Eastern Arc Mountains (Grey‐Wilson, 1980; Lovett et al., 2000). Another drawback is that it is harder to detect centres of neo‐endemism when using generic‐level data (Thornhill et al., 2016). This makes the 11 SUs detected as centres of neo‐endemism (Fig. 6) strong cradles of diversity. The completeness of the plant distribution dataset as well as the resolution of the phylogeny may also affect our estimate of PE. Here, even if the RAINBIO dataset represents c. 89% of the known diversity in tropical Africa, it is the best‐quality data that are available to date (Dauby et al., 2016). The relatively high proportion of nonmonophyletic genera (36%) that are present in the unfiltered angiosperm tree is inherent and unavoidable when making use of large‐scale phylogenetic approaches. On one hand, this is caused by the compromise between the amount of markers used and the number of species included in the dataset (see, e.g., Qian & Jin, 2016; Smith & Brown, 2018). On the other hand, there are still some taxonomical uncertainties regarding the generic status of certain angiosperm African genera (e.g. Rubiaceae, Convolvulaceae, Cyperaceae) in which several genera have not been taxonomically revised (Muasya et al., 2009; De Block et al., 2015; Simões & Staples, 2017). Despite this, we are confident that given the broad scale of our analysis, our results will be robust to improved phylogenetic studies at the genus level.
In tropical Africa, documented regions with high degrees of diversity and endemism, and postulated to act as both cradles and museums of species diversity are predominantly mountainous (e.g. Cameroon Volcanic Line, the Albertine Rift and the Eastern Arc Mountains) (Fjeldså & Lovett, 1997; Lovett et al., 2005; Burgess et al., 2007; Plumptre et al., 2007). These mountain blocks are generally small in terms of size (e.g. < 70 × 70 km for the Eastern Arc Mountains; Burgess et al., 2007). Thus, the resolution of our analyses (100 × 100 km SUs) may have diluted the montane endemism effect with the high diversity and potentially widespread genera of adjacent lowlands regions. Using smaller SUs gave a similar overall CANAPE result, but with less coverage of the study zone because of a higher number of poorly sampled SUs that were excluded from the analyses (Fig. S7). Moreover, in mixed‐endemism regions, neo‐endemics and paleo‐endemics may be locally concentrated in different places within a mountain block (Bitencourt & Rapini, 2013).
Conservation implications
Identifying and conserving areas of evolutionary potential, harbouring important processes leading to diversification and/or lineage persistence, are of crucial conservation importance (Fjeldså, 1994; Mace et al., 2003; Faith et al., 2010; Kraft et al., 2010). In eastern Africa, most of the centres of PE are centres of mixed‐endemism (Fig. 6), somewhat bypassing the need to choose between conserving regions of active diversification or persistence (Cowling & Pressey, 2001).
The coverage of the identified centres of PE by the PA network is high, as 85% of the CANAPE SUs contain at least a portion of PA, and 42% contain at least a portion of the most restrictive PAs (Ia, Ib or II types). This means that important regions of diversification and persistence are already represented in the tropical African PA network. In terms of surface, only 18% of the CANAPE SUs' surface is covered by PAs (Fig. 9; Tables S6, S7). This is as expected, as PAs are generally small areas and are unlikely to cover a complete 100 × 100 km SU. Significant CANAPE areas that are not well represented by PAs are mainly situated in South Malawi and, to a lesser extent, in Ethiopia (Fig. 9). Still, it is possible that the actual occurrences of endemics and the PAs within a SU do not coincide, so it might be worthwhile analysing diversity patterns at a fine scale when adapting or adding PAs.
The regions containing significant high PE, particularly regions of mixed‐endemism that are simultaneously ‘cradles' and ‘museums' of diversity, should be considered in future conservation planning or PA extension. Considerable increases in the coverage of multiple facets of biodiversity (taxonomic, phylogenetic and functional) are possible with small expansions of protected areas, if the planning is achieved thoroughly and with an explicit consideration of these multiple facets (Pollock et al., 2015, 2017).
Author contributions
L‐PMJD and TLPC conceived the study; L‐PMJD and SBJ undertook the analyses; L‐PMJD, SBJ, GD, AB‐O, BAM, VD, J‐CS, MSMS, TS, DJH, BS, JJW, OJH and TLPC contributed data and ideas; L‐PMJD and TLPC led the writing; L‐PMJD, SBJ, GD, AB‐O, BAM, VD, J‐CS, MSMS, TS, DJH, BS, JJW, OJH and TLPC read and approved the final version. L‐PMJD and SBJ contributed equally to this work.
Supporting information
Fig. S1 Relationship between weighted endemism (WE) and genus richness (GR).
Fig. S2 Relationship between phylogenetic diversity (PD) and genus richness (GR).
Fig. S3 Map of observed phylogenetic diversity.
Fig. S4 Boxplot of the distributions of the mean ruggedness (in metres) of the SUs depending on their phylogenetic diversity (PD) significance class.
Fig. S5 Relationship between phylogenetic endemism (PE) and weighted endemism (WE).
Fig. S6 Maps of phylogenetic endemism.
Fig. S7 Maps of CANAPE results with different SU sizes.
Fig. S8 Boxplot of the distribution of the percentage of montane plots of the sampling units depending on their CANAPE category.
Fig. S9 Boxplot of the distribution of the percentage of overlap of the SUs with ‘montane areas' depending on their CANAPE category.
Table S1 P‐values resulting from the pairwise comparisons of mean ruggedness per sampling unit in each PD significance class.
Table S2 P‐values resulting from the pairwise comparisons of mean elevation per sampling unit in each RPD significance class.
Table S3 P‐values resulting from the pairwise comparisons of mean ruggedness per sampling unit in each CANAPE significance class.
Table S4 P‐values resulting from the pairwise comparison of the percentage of montane plots of the sampling units depending on their CANAPE category.
Table S5 P‐values resulting from the pairwise comparison of the distribution of the percentage of overlap of the SUs with ‘montane areas' depending on their CANAPE category.
Table S6 Number of significant CANAPE SUs overlaid by at least one protected area.
Table S7 Surface intersection between significant CANAPE SUs and protected areas.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was co‐funded by the French Foundation for Research on Biodiversity (FRB) and the Provence‐Alpes‐Côte d'Azur région (PACA) region through the Centre for Synthesis and Analysis of Biodiversity data (CESAB) programme, as part of the RAINBIO research project (http://rainbio.cesab.org/) and the Agence Nationale de la Recherche (grant number ANR‐15‐CE02‐0002‐01), both to TLPC. JCS considers this work a contribution to his VILLUM Investigator project ‘Biodiversity Dynamics in a Changing World' funded by VILLUM FONDEN (grant 16549) and his TREECHANGE project funded by Independent Research Fund Denmark|Natural Sciences (grant 6108‐00078B). GD was partly funded by the Belgian Fund for Scientific Research (F.R.S.‐FNRS).
References
- Antonelli A, Kissling WD, Flantua SGA, Bermúdez MA, Mulch A, Muellner‐Riehl AN, Kreft H, Linder HP, Badgley C, Fjeldså J et al 2018. Geological and climatic influences on mountain biodiversity. Nature Geoscience 11: 718–725. [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. 2005. The age of the angiosperms: a molecular timescale without a clock. Evolution 59: 1245–1258. [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms re‐revisited. American Journal of Botany 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- Bitencourt C, Rapini A. 2013. Centres of endemism in the espinhaço range: identifying cradles and museums of Asclepiadoideae (Apocynaceae). Systematics and Biodiversity 11: 525–536. [Google Scholar]
- Boyle B, Hopkins N, Lu Z, Raygoza Garay JA, Mozzherin D, Rees T, Matasci N, Narro ML, Piel WH, Mckay SJ et al 2013. The taxonomic name resolution service: an online tool for automated standardization of plant names. BMC Bioinformatics 14: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenan JPM. 1978. Some aspects of the phytogeography of tropical Africa. Annals of the Missouri Botanical Garden 65: 437–478. [Google Scholar]
- Bruchmann I, Hobohm C. 2014. Factors that create and increase endemism In: Hobohm C, ed. Plant and vegetation. Endemism in vascular plants. Dordrecht, the Netherlands: Springer, 51–68. [Google Scholar]
- Burgess ND, Butynski TM, Cordeiro NJ, Doggart NH, Fjeldså J, Howell KM, Kilahama FB, Loader SP, Lovett JC, Mbilinyi B et al 2007. The biological importance of the Eastern Arc Mountains of Tanzania and Kenya. Biological Conservation 134: 209–231. [Google Scholar]
- Couvreur TLP. 2015. Odd man out: why are there fewer plant species in African rain forests? Plant Systematics and Evolution 301: 1299–1313. [Google Scholar]
- Cowling RM, Pressey RL. 2001. Rapid plant diversification: planning for an evolutionary future. Proceedings of the National Academy of Sciences, USA 98: 5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp MD, Laffan S, Linder HP, Monro A. 2001. Endemism in the Australian flora. Journal of Biogeography 28: 183–198. [Google Scholar]
- Dauby G, Zaiss R, Overgaard A‐B, Catarino L, Damen T, Deblauwe V, Dessein S, Dransfield J, Droissart V, Duarte MC et al 2016. RAINBIO: a mega‐database of tropical African vascular plants distributions. PhytoKeys 74: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block P, Razafimandimbison SG, Janssens S, Ochoterena H, Robbrecht E, Bremer B. 2015. Molecular phylogenetics and generic assessment in the tribe Pavetteae. Taxon 64: 79–95. [Google Scholar]
- Diamond AW, Hamilton AC. 1980. The distribution of forest passerine birds and Quaternary climatic change in tropical Africa. Journal of Zoology 191: 379–402. [Google Scholar]
- Dimitrov D, Nogués‐Bravo D, Scharff N. 2012. Why do tropical mountains support exceptionally high biodiversity? The eastern arc mountains and the drivers of Saintpaulia diversity. PLoS ONE 7: e48908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droissart V, Dauby G, Hardy OJ, Deblauwe V, Harris DJ, Janssens S, Mackinder BA, Blach‐Overgaard A, Sonké B, Sosef MSM et al 2018. Beyond trees: Biogeographical regionalization of tropical Africa. Journal of Biogeography 45: 1153–1167. [Google Scholar]
- Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biological Conservation 61: 1–10. [Google Scholar]
- Faith DP, Magallón S, Hendry AP, Conti E, Yahara T, Donoghue MJ. 2010. Evosystem services: an evolutionary perspective on the links between biodiversity and human well‐being. Current Opinion in Environmental Sustainability 2: 66–74. [Google Scholar]
- Fayolle A, Swaine MD, Bastin J‐F, Bourland N, Comiskey JA, Dauby G, Doucet J‐L, Gillet J‐F, Gourlet‐Fleury S, Hardy OJ et al 2014. Patterns of tree species composition across tropical African forests. Journal of Biogeography 41: 2320–2331. [Google Scholar]
- Ferreira PMA, Boldrini II. 2011. Potential reflection of distinct ecological units in plant endemism categories. Conservation Biology 25: 672–679. [DOI] [PubMed] [Google Scholar]
- Fjeldså J. 1994. Geographical patterns for relict and young species of birds in Africa and South America and implications for conservation priorities. Biodiversity & Conservation 3: 207–226. [Google Scholar]
- Fjeldså J, Bowie RCK. 2008. New perspectives on the origin and diversification of Africa's forest avifauna. African Journal of Ecology 46: 235–247. [Google Scholar]
- Fjeldså J, Bowie RCK, Rahbek C. 2012. The role of mountain ranges in the diversification of birds. Annual Review of Ecology, Evolution, and Systematics 43: 249–265. [Google Scholar]
- Fjeldså J, Lovett JC. 1997. Geographical patterns of old and young species in African forest biota: the significance of specific montane areas as evolutionary centres. Biodiversity & Conservation 6: 325–346. [Google Scholar]
- Fjeldså J, Rahbek C. 2006. Diversification of tanagers, a species rich bird group, from lowlands to montane regions of South America. Integrative and Comparative Biology 46: 72–81. [DOI] [PubMed] [Google Scholar]
- Forest F, Crandall KA, Chase MW, Faith DP. 2015. Phylogeny, extinction and conservation: embracing uncertainties in a time of urgency. Philosophical Transactions of the Royal Society B: Biological Sciences 370: 20140002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest F, Grenyer R, Rouget M, Davies TJ, Cowling RM, Faith DP, Balmford A, Manning JC, Procheş Ş, van der Bank M et al 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445: 757–760. [DOI] [PubMed] [Google Scholar]
- Grey‐Wilson C. 1980. Impatiens of Africa. Rotterdam, the Netherlands: A. A. Balkema. [Google Scholar]
- Guillocheau F, Simon B, Baby G, Bessin P, Robin C, Dauteuil O. 2018. Planation surfaces as a record of mantle dynamics: the case example of Africa. Gondwana Research 53: 82–98. [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- Hinchliff CE, Smith SA. 2014. Some limitations of public sequence data for phylogenetic inference (in plants). PLoS ONE 9: e98986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoorn C, Mosbrugger V, Mulch A, Antonelli A. 2013. Biodiversity from mountain building. Nature Geoscience 6: 154. [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez‐Meseguer A, Anderson CL, Figueiredo JP et al 2010. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330: 927–931. [DOI] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. 2006. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences, USA 103: 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys AM, Govaerts R, Ficinski SZ, Nic Lughadha E, Vorontsova MS. 2019. Global dataset shows geography and life form predict modern plant extinction and rediscovery. Nature Ecology & Evolution 3: 1043–1047. [DOI] [PubMed] [Google Scholar]
- Huntley JW, Voelker G. 2016. Cryptic diversity in Afro‐tropical lowland forests: The systematics and biogeography of the avian genus Bleda. Molecular Phylogenetics and Evolution 99: 297–308. [DOI] [PubMed] [Google Scholar]
- IUCN . 2008. Guidelines for applying protected area management categories (Dudley N, ed.). Gland, Switzerland: IUCN. [Google Scholar]
- Jacobs BF. 2004. Palaeobotanical studies from tropical Africa: relevance to the evolution of forest, woodland and savannah biomes. Philosophical Transactions of the Royal Society B: Biological Sciences 359: 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Couvreur TLP, Mertens A, Dauby G, Dagallier L‐P, Abeele SV, Vandelook F, Mascarello M, Beeckman H, Sosef M et al 2019. A large‐scale species level dated angiosperm phylogeny for evolutionary and ecological analyses. bioRxiv 809921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetz W, Rahbek C, Colwell RK. 2004. The coincidence of rarity and richness and the potential signature of history in centres of endemism. Ecology Letters 7: 1180–1191. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kier G, Kreft H, Lee TM, Jetz W, Ibisch PL, Nowicki C, Mutke J, Barthlott W. 2009. A global assessment of endemism and species richness across island and mainland regions. Proceedings of the National Academy of Sciences, USA 106: 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissling WD, Eiserhardt WL, Baker WJ, Borchsenius F, Couvreur TLP, Balslev H, Svenning J‐C. 2012. Cenozoic imprints on the phylogenetic structure of palm species assemblages worldwide. Proceedings of the National Academy of Sciences, USA 109: 7379–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopper RR, Gautier L, Chatelain C, Smith GF, Spichiger R. 2007. Floristics of the angiosperm flora of Sub‐Saharan Africa: an analysis of the African Plant Checklist and Database. Taxon 56: 201–208. [Google Scholar]
- Körner C, Jetz W, Paulsen J, Payne D, Rudmann‐Maurer KM, Spehn E. 2017. A global inventory of mountains for bio‐geographical applications. Alpine Botany 127: 1–15. [Google Scholar]
- Körner C, Paulsen J, Spehn EM. 2011. A definition of mountains and their bioclimatic belts for global comparisons of biodiversity data. Alpine Botany 121: 73. [Google Scholar]
- Kraft NJB, Baldwin BG, Ackerly DD. 2010. Range size, taxon age and hotspots of neoendemism in the California flora: California plant neoendemism. Diversity and Distributions 16: 403–413. [Google Scholar]
- Kruckeberg AR, Rabinowitz D. 1985. Biological aspects of endemism in higher plants. Annual Review of Ecology and Systematics 16: 447–479. [Google Scholar]
- Küper W, Sommer JH, Lovett JC, Mutke J, Linder HP, Beentje HJ, Van Rompaey RSAR, Chatelain C, Sosef M, Barthlott W. 2004. Africa's hotspots of biodiversity redefined. Annals of the Missouri Botanical Garden 91: 525–535. [Google Scholar]
- Laffan SW, Lubarsky E, Rosauer DF. 2010. Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography 33: 643–647. [Google Scholar]
- Linder HP. 2001. Plant diversity and endemism in sub‐Saharan tropical Africa. Journal of Biogeography 28: 169–182. [Google Scholar]
- Linder HP. 2014. The evolution of African plant diversity. Frontiers in Ecology and Evolution 2: 1–14. [Google Scholar]
- Linder HP, de Klerk HM, Born J, Burgess ND, Fjeldså J, Rahbek C. 2012. The partitioning of Africa: statistically defined biogeographical regions in sub‐Saharan Africa: African regionalization. Journal of Biogeography 39: 1189–1205. [Google Scholar]
- Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. 2009. The velocity of climate change. Nature 462: 1052–1055. [DOI] [PubMed] [Google Scholar]
- López‐Pujol J, Zhang F‐M, Sun H‐Q, Ying T‐S, Ge S. 2011. Centres of plant endemism in China: places for survival or for speciation? Journal of Biogeography 38: 1267–1280. [Google Scholar]
- Lovett JC, Marchant R, Taplin J, Küper W. 2005. The oldest rainforests in Africa: Stability or resilience for survival and diversity? In: Purvis A, Gittleman JL, Brooks T, eds. Phylogeny and conservation. Cambridge, UK: Cambridge University Press, 198–229. [Google Scholar]
- Lovett JC, Rudd S, Taplin J. 2000. Patterns of plant diversity in Africa south of the Sahara and their implications for conservation management. Biodiversity and Conservation 9: 37–46. [Google Scholar]
- Mace GM, Gittleman JL, Purvis A. 2003. Preserving the tree of life. Science 300: 1707–1709. [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez‐Acevedo S, Sánchez‐Reyes LL, Hernández‐Hernández T. 2015. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Maley J. 1996. The African rain forest: main characteristics of changes in vegetation and climate from the Upper Cretaceous to the Quaternary (IJ Alexander, MD Swaine, and R Watling, Eds.). Proceedings of the Royal Society of Edinburgh, Section B 104: 31–73. [Google Scholar]
- Malhi Y, Adu‐Bredu S, Asare RA, Lewis SL, Mayaux P. 2013. African rainforests: past, present and future. Philosophical Transactions of the Royal Society B: Biological Sciences 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely JA, Miller KR, Reid W, Mittermeier RA, Werner TB. 1990. Conserving the world's biological diversity (International Union for Conservation of Nature and Natural Resources, Ed.). Gland, Switzerland, and Washington, DC, USA: International Union for Conservation of Nature and Natural Resources; World Resources Institute; Conservation International; World Wildlife Fund‐US; World Bank. [Google Scholar]
- Merckx VSFT, Hendriks KP, Beentjes KK, Mennes CB, Becking LE, Peijnenburg KTCA, Afendy A, Arumugam N, de Boer H, Biun A et al 2015. Evolution of endemism on a young tropical mountain. Nature 524: 347–350. [DOI] [PubMed] [Google Scholar]
- Mishler BD, Knerr N, González‐Orozco CE, Thornhill AH, Laffan SW, Miller JT. 2014. Phylogenetic measures of biodiversity and neo‐ and paleo‐endemism in Australian Acacia. Nature Communications 5: 4473. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. 2007. Using plastid genome‐scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences, USA 104: 19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Funk V, Sakai AK. 2002. Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology 51: 238–254. [DOI] [PubMed] [Google Scholar]
- Muasya AM, Simpson DA, Verboom GA, Goetghebeur P, Naczi RFC, Chase MW, Smets E. 2009. Phylogeny of cyperaceae based on DNA sequence data: current progress and future prospects. Botanical Review 75: 2–21. [Google Scholar]
- Muellner‐Riehl AN. 2019. Mountains as evolutionary arenas: patterns, emerging approaches, paradigm shifts, and their implications for plant phylogeographic research in the Tibeto‐Himalayan region. Frontiers in Plant Science 10: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murienne J, Benavides LR, Prendini L, Hormiga G, Giribet G. 2013. Forest refugia in Western and Central Africa as ‘museums' of Mesozoic biodiversity. Biology Letters 9: 20120932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Nekola JC. 1999. Paleorefugia and neorefugia: the influence of colonization history on community pattern and process. Ecology 80: 2459–2473. [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D'amico JA, Itoua I, Strand HE, Morrison JC et al 2001. Terrestrial ecoregions of the world: a new map of life on earth. BioScience 51: 933. [Google Scholar]
- Peterson DL, Schreiner EG, Buckingham NM. 1997. Gradients, vegetation and climate: spatial and temporal dynamics in the Olympic Mountains, U.S.A. Global Ecology and Biogeography Letters 6: 7. [Google Scholar]
- Plana V. 2004. Mechanisms and tempo of evolution in the African Guineo‐Congolian rainforest. Philosophical Transactions of the Royal Society B: Biological Sciences 359: 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plana V, Gascoigne A, Forrest LL, Harris D, Pennington RT. 2004. Pleistocene and pre‐Pleistocene Begonia speciation in Africa. Molecular Phylogenetics and Evolution 31: 449–461. [DOI] [PubMed] [Google Scholar]
- Plumptre AJ, Davenport TRB, Behangana M, Kityo R, Eilu G, Ssegawa P, Ewango C, Meirte D, Kahindo C, Herremans M et al 2007. The biodiversity of the Albertine Rift. Biological Conservation 134: 178–194. [Google Scholar]
- Pollock LJ, Rosauer DF, Thornhill AH, Kujala H, Crisp MD, Miller JT, McCarthy MA. 2015. Phylogenetic diversity meets conservation policy: small areas are key to preserving eucalypt lineages. Philosophical Transactions of the Royal Society B: Biological Sciences 370: 20140007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock LJ, Thuiller W, Jetz W. 2017. Large conservation gains possible for global biodiversity facets. Nature 546: 141–144. [DOI] [PubMed] [Google Scholar]
- Qian H, Jin Y. 2016. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. Journal of Plant Ecology 9: 233–239. [Google Scholar]
- Rahbek C, Borregaard MK, Antonelli A, Colwell RK, Holt BG, Nogues‐Bravo D, Rasmussen CMØ, Richardson K, Rosing MT, Whittaker RJ et al 2019. Building mountain biodiversity: Geological and evolutionary processes. Science 365: 1114–1119. [DOI] [PubMed] [Google Scholar]
- Robbrecht E. 1996. Geography of African Rubiaceae with reference to glacial rain forest refuges In: van der Maesen LJG, van der Burgt XM, van Medenbach de Rooy JM, eds. The biodiversity of African plants: Proceedings XIVth AETFAT Congress 22–27 August 1994, Wageningen, the Netherlands. Dordrecht, the Netherlands: Springer, 564–581. [Google Scholar]
- Rosauer D, Laffan SW, Crisp MD, Donnellan SC, Cook LG. 2009. Phylogenetic endemism: a new approach for identifying geographical concentrations of evolutionary history. Molecular Ecology 18: 4061–4072. [DOI] [PubMed] [Google Scholar]
- Roy MS. 1997. Recent diversification in African greenbuls (Pycnonotidae: Andropadus) supports a montane speciation model. Proceedings of the Royal Society of London B: Biological Sciences 264: 1337–1344. [Google Scholar]
- Scherson RA, Thornhill AH, Urbina‐Casanova R, Freyman WA, Pliscoff PA, Mishler BD. 2017. Spatial phylogenetics of the vascular flora of Chile. Molecular Phylogenetics and Evolution 112: 88–95. [DOI] [PubMed] [Google Scholar]
- Schwery O, Onstein RE, Bouchenak‐Khelladi Y, Xing Y, Carter RJ, Linder HP. 2014. As old as the mountains: the radiations of the Ericaceae. New Phytologist 207: 355–367. [DOI] [PubMed] [Google Scholar]
- Sechrest W, Brooks TM, da Fonseca GA, Konstant WR, Mittermeier RA, Purvis A, Rylands AB, Gittleman JL. 2002. Hotspots and the conservation of evolutionary history. Proceedings of the National Academy of Sciences, USA 99: 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes AR, Staples G. 2017. Dissolution of Convolvulaceae tribe Merremieae and a new classification of the constituent genera. Botanical Journal of the Linnean Society 183: 561–586. [Google Scholar]
- Smith SA, Brown JW. 2018. Constructing a broadly inclusive seed plant phylogeny. American Journal of Botany 105: 1–13. [DOI] [PubMed] [Google Scholar]
- Smith SA, O'Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Savolainen V, Crane PR, Barraclough TG. 2002. Rate heterogeneity among lineages of tracheophytes: Integration of molecular and fossil data and evidence for molecular living fossils. Proceedings of the National Academy of Sciences, USA 99: 4430–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosef MSM. 1996. Begonias and African rain forest refuges: general aspects and recent progress In: van der Maesen LJG, van der Burgt XM, van Medenbach de Rooy JM, eds. The biodiversity of African plants: Proceedings XIVth AETFAT Congress 22–27 August 1994, Wageningen, the Netherlands. Dordrecht, the Netherlands: Springer, 602–611. [Google Scholar]
- Sosef MSM, Dauby G, Blach‐Overgaard A, van der Burgt X, Catarino L, Damen T, Deblauwe V, Dessein S, Dransfield J, Droissart V et al 2017. Exploring the floristic diversity of tropical Africa. BMC Biology 15: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML‐VI‐HPC: maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stropp J, Ladle RJ, Malhado ACM, Hortal J, Gaffuri J, Temperley WH, Skøien JO, Mayaux P. 2016. Mapping ignorance: 300 years of collecting flowering plants in Africa. Global Ecology and Biogeography 25: 1085–1096. [Google Scholar]
- Swenson NG, Enquist BJ, Thompson J, Zimmerman JK. 2007. The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology 88: 1770–1780. [DOI] [PubMed] [Google Scholar]
- Thornhill AH, Baldwin BG, Freyman WA, Nosratinia S, Kling MM, Morueta‐Holme N, Madsen TP, Ackerly DD, Mishler BD. 2017. Spatial phylogenetics of the native California flora. BMC Biology 15: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill AH, Mishler BD, Knerr NJ, González‐Orozco CE, Costion CM, Crayn DM, Laffan SW, Miller JT. 2016. Continental‐scale spatial phylogenetics of Australian angiosperms provides insights into ecology, evolution and conservation. Journal of Biogeography 43: 2085–2098. [Google Scholar]
- Tolley KA, Tilbury CR, Measey GJ, Menegon M, Branch WR, Matthee CA. 2011. Ancient forest fragmentation or recent radiation? Testing refugial speciation models in chameleons within an African biodiversity hotspot: Palaeoendemic chameleon lineages in East Africa. Journal of Biogeography 38: 1748–1760. [Google Scholar]
- Tzedakis PC, Lawson IT, Frogley MR, Hewitt GM, Preece RC. 2002. Buffered tree population changes in a quaternary refugium: evolutionary implications. Science 297: 2044–2047. [DOI] [PubMed] [Google Scholar]
- Voelker G, Outlaw RK, Bowie RCK. 2010. Pliocene forest dynamics as a primary driver of African bird speciation. Global Ecology and Biogeography 19: 111–121. [Google Scholar]
- Wasser SK, Lovett JC. 1993. Introduction to the biogeography and Ecology of the rain forests of eastern Africa In: Lovett JC, Wasser SK, eds. Biogeography and ecology of the rain forests of Eastern Africa. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Webb CO. 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. The American Naturalist 156: 145–155. [DOI] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annual Review of Ecology and Systematics 33: 475–505. [Google Scholar]
- Wen J, Zhang J, Nie Z‐L, Zhong Y, Sun H. 2014. Evolutionary diversifications of plants on the Qinghai‐Tibetan Plateau. Frontiers in Genetics 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F. 1983. The vegetation of Africa: a descriptive memoir to accompany the Unesco/AETFAT/UNSO vegetation map of Africa. Paris, France: UNESCO. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Relationship between weighted endemism (WE) and genus richness (GR).
Fig. S2 Relationship between phylogenetic diversity (PD) and genus richness (GR).
Fig. S3 Map of observed phylogenetic diversity.
Fig. S4 Boxplot of the distributions of the mean ruggedness (in metres) of the SUs depending on their phylogenetic diversity (PD) significance class.
Fig. S5 Relationship between phylogenetic endemism (PE) and weighted endemism (WE).
Fig. S6 Maps of phylogenetic endemism.
Fig. S7 Maps of CANAPE results with different SU sizes.
Fig. S8 Boxplot of the distribution of the percentage of montane plots of the sampling units depending on their CANAPE category.
Fig. S9 Boxplot of the distribution of the percentage of overlap of the SUs with ‘montane areas' depending on their CANAPE category.
Table S1 P‐values resulting from the pairwise comparisons of mean ruggedness per sampling unit in each PD significance class.
Table S2 P‐values resulting from the pairwise comparisons of mean elevation per sampling unit in each RPD significance class.
Table S3 P‐values resulting from the pairwise comparisons of mean ruggedness per sampling unit in each CANAPE significance class.
Table S4 P‐values resulting from the pairwise comparison of the percentage of montane plots of the sampling units depending on their CANAPE category.
Table S5 P‐values resulting from the pairwise comparison of the distribution of the percentage of overlap of the SUs with ‘montane areas' depending on their CANAPE category.
Table S6 Number of significant CANAPE SUs overlaid by at least one protected area.
Table S7 Surface intersection between significant CANAPE SUs and protected areas.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


