Abstract
This study investigated the co‐localization of the Middle East respiratory syndrome coronavirus (MERS‐CoV) and its receptor dipeptidyl peptidase‐4 (DPP4) by immunohistochemistry (IHC) across respiratory and lymphoid organs of experimentally MERS‐CoV infected pigs and llamas. Also, scanning electron microscopy was performed to assess the ciliary integrity of respiratory epithelial cells in both species. In pigs, on day 2 post‐inoculation (p.i.), DPP4‐MERS‐CoV co‐localization was detected in medial turbinate epithelium. On day 4 p.i., the virus/receptor co‐localized in frontal and medial turbinate epithelial cells in pigs, and epithelial cells distributed unevenly through the whole nasal cavity and in the cervical lymph node in llamas. MERS‐CoV viral nucleocapsid was mainly detected in upper respiratory tract sites on days 2 and 4 p.i. in pigs and day 4 p.i. in llamas. No MERS‐CoV was detected on day 24 p.i. in any tissue by IHC. While pigs showed severe ciliary loss in the nasal mucosa both on days 2 and 4 p.i. and moderate loss in the trachea on days 4 and 24 p.i., ciliation of respiratory organs in llamas was not significantly affected. Obtained data confirm the role of DPP4 for MERS‐CoV entry in respiratory epithelial cells of llamas. Notably, several nasal epithelial cells in pigs were found to express viral antigen but not DPP4, suggesting the possible existence of other molecule/s facilitating virus entry or down regulation of DPP4 upon infection.
Keywords: dipeptidyl peptidase‐4 (DPP4), immunohistochemistry, llama, Middle East respiratory syndrome coronavirus (MERS‐CoV), pig, scanning electron microscopy
1. INTRODUCTION
In June 2012, a novel human coronavirus named Middle East respiratory syndrome coronavirus (MERS‐CoV) emerged in the Kingdom of Saudi Arabia (Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, 2012). Since then, as per September 2018, 2,249 laboratory‐confirmed cases and at least 798 associated deaths have been reported to the World Health Organization (World Health Organization (WHO), 2018). Although the majority of cases were reported in the Middle East region, travel‐associated cases have been documented in other parts of the world (Cotten, Watson, et al., 2013; Hsieh, 2015; Tsiodras et al., 2014; WHO, 2015). On September 8th, 2018, South Korea reported the first MERS‐CoV case since the end of an outbreak in 2015, suggesting that MERS‐CoV is still a worldwide threat (WHO, 2018).
Dromedary camels have been considered as the main reservoir hosts for MERS‐CoV, as viral neutralizing antibodies have been reported in this species (Corman et al., 2014; Reusken et al., 2013). Moreover, animal‐to‐human transmissions have been described (Azhar et al., 2014). More recently, a surveillance study showed that a MERS‐CoV strain responsible for human outbreaks was isolated from the upper respiratory tract of dromedaries, demonstrating that the virus does not require mutations to jump between species (Sabir et al., 2016). Besides dromedaries, several animal species, including common marmosets, rhesus macaques, llamas, pigs and alpacas, are experimentally susceptible to MERS‐CoV infection (Adney, Bielefeldt‐Ohmann, Hartwig, & Bowen, 2016; Falzarano et al., 2014; Vergara‐Alert et al., 2017; de Wit et al., 2013).
Middle East respiratory syndrome coronavirus is a positive‐stranded RNA virus that belongs to the betacoronavirus genus (Zaki et al., 2012). It has a genome of approximately 30 Kb nucleotides that encodes four structural proteins: spike (S), nucleocapsid (N), membrane (M) and envelope (E) proteins, and a RNA polymerase (Cotten, Lam, et al., 2013; Cotten et al., 2014; Zhang, Shen, & Gu, 2016). The receptor binding domain (RBD) of the S‐protein mediates viral entrance through binding dipeptidyl peptidase‐4 (DPP4, also known as CD26), a serine protease expressed on the surface of many cell types (Raj et al., 2013). The tissue distribution of DPP4 has been described in some animal species, including dromedary camels, bats, pigs, llamas, sheep and horses (Vergara‐Alert et al., 2017; Widagdo et al., 2016, 2017). However, although DPP4 is decisive for MERS‐CoV entry in vitro (Raj et al., 2013; Chan et al., 2016), the role of the protein in determining tissue tropism with regards to MERS‐CoV pathogenesis in vivo has not been fully elucidated. MERS‐CoV antigen has been demonstrated in nasal epithelial cells expressing DPP4 (Haagmans et al., 2016; Vergara‐Alert et al., 2017). However, no double staining studies to detect potential co‐localization of both antigens were reported until very recently. Haverkamp et al. (2018) demonstrated that nasal epithelial cells infected with MERS‐CoV in dromedaries seemed to lose DPP4 expression, while adjacent non‐infected cells retained positivity for DPP4. Moreover, ciliary damage was also a feature of dromedary camels infected with MERS‐CoV (Haverkamp et al., 2018). These authors postulated that the mild and transient disease in dromedaries is, at least in part, potentially attributable to the down‐regulation of its own cell entry receptor, thus self‐limiting the viral infection.
Taking into account that a number of animal species may potentially act as reservoirs for MERS‐CoV (Vergara‐Alert et al., 2017), it is important to establish if DPP4 determines or not tissue tropism with regards to viral pathogenesis in vivo. Moreover, if the ciliary loss is a particular finding of dromedary camels infected with MERS‐CoV or may also affect other susceptible species is not known. Therefore, the objective of this study was to assess the co‐localization of MERS‐CoV and DPP4 by means of double immunostaining in two susceptible species: llama and pig. In addition, the degree of ciliation of the upper respiratory tract in both species was studied by scanning electron microscopy (SEM).
2. MATERIAL AND METHODS
2.1. Pig and llama tissue specimens
All paraffin blocks and fresh tissue samples used in the present work were obtained from a previous experimental study demonstrating that llamas and pigs were susceptible to MERS‐CoV infection (Vergara‐Alert et al., 2017). Briefly, 2‐month‐old pigs and 6–8‐month‐old llamas were intranasally inoculated with 107 50% tissue culture infective dose (TCID50) MERS‐CoV in 3 ml saline solution. Four pigs were euthanized on day 2 post‐inoculation (p.i.) with an intravenous overdose of pentobarbital followed by exsanguination, and four animals of each species were killed on day 4 p.i. The remaining animals (six pigs and four llamas) were euthanized on day 24 p.i. following the same protocol. Complete necropsies were performed and respiratory and lymphoid tissues (nasal turbinate, trachea, bronchus, lung, cervical lymph node, mediastinal lymph node, tonsil and thymus) were collected for IHC and RT‐qPCR examination. Formalin‐fixed samples of nasal turbinate, trachea and lung were used for SEM studies, including those from negative control pigs euthanized to prior to the start of the experiment.
2.2. Double immunohistochemistry for DPP4 and MERS‐CoV detection
Tissues collected from pigs on day 2, 4 and 24 p.i. and from llamas on day 4 and 24 p.i. were fixed by immersion in 10% neutral‐buffered formalin for 1 week and embedded in paraffin blocks. The tissues were sectioned (3 μm) onto coated glass slides (DAKO; Agilent Technologies Company, Santa Clara, CA, USA), deparaffinized in xylene and hydrated in decreasing grades of ethanol (100%, 96% and 70%). Endogenous peroxidase was blocked with 3% H2O2 solution in methanol for 30 min. Antigen retrieval was performed by incubating the slides in ethylenediaminetetraacetic acid (pH 9.0) at 98°C for 20 min. Slides were then incubated with blocking solution (DAKO; Agilent Technologies Company, Santa Clara, CA, USA) at room temperature (RT) for 1 hr.
Dipeptidyl peptidase‐4 protein was detected by IHC following a previously published protocol (Vergara‐Alert et al., 2017). Tissue sections were incubated with 5 μg/ml of polyclonal goat IgG anti‐human DPP4 primary antibody (R&D Systems, Abingdon, UK) in a humid chamber at 4°C overnight. On the following day, slides were washed with phosphate buffered saline with 0.1% tween 20 (pH 7.45), and incubated for 1 hr at RT with alkaline phosphatase‐labelled horse anti‐goat IgG (ImmPRESS™‐AP Polymer Anti‐Goat IgG Reagent, Burlingame, CA, USA). Each slide was then incubated with ImmPACT Vector Red Alkaline Phosphatase Substrate (Burlingame, CA, USA) for 10 min and presence of antigen produced a bright red reaction product. Subsequently, the same slides were subjected to MERS‐CoV IHC using a monoclonal primary antibody (final concentration of 0.8 μg/ml) that specifically reacts with the viral N protein (SinoBiological Inc., Beijing, China), according to a previously published method (Haagmans et al., 2016). After 4°C overnight incubation with the primary antibody, samples were then incubated with peroxidase labelled polymer goat anti‐mouse IgG (DAKO; Agilent Technologies Company, Santa Clara, CA, USA) for 1 hr at RT. The 3,3′‐Diaminobenzidine (DAB) substrate solution (0.1 g DAB in 200 ml PBS with 100 μl H2O2) was applied for 10 min to obtain positive signals with brown colour in the tissues. Finally, slides were counterstained with haematoxylin for 10 s and then coverslipped with mounting medium prior to microscopic examination (Nikon Eclipse 50i). Negative controls included the addition of blocking solution (DAKO; Agilent Technologies Company, Santa Clara, CA, USA) instead of primary antibodies for both immunohistochemical techniques. The studied tissues of pigs and llamas were considered as positive controls for the DPP4 IHC, since they constitutively express the studied viral receptor. Also, a nasal turbinate from a dromedary camel infected with MERS‐CoV (Haagmans et al., 2016) was used as positive control for MERS‐CoV IHC. A grading system for virus antigen expression was established based on the number of cells stained by IHC. Four scores were defined: −, no positive cells detected; +, less than 10 positive cells per tissue section; ++, 10 to 50 positive cells per tissue section; and +++, more than 50 positive cells per tissue section.
2.3. Scanning electron microscopy
Scanning electron microscopy was done following a previously published protocol (Haverkamp et al., 2018). For each necropsy day, formalin fixed samples of nasal mucosa, trachea and bronchus of two pigs and two llamas infected with MERS‐CoV were post‐fixed in 5% glutaraldehyde. Two additional negative control samples were obtained for non‐infected pigs. Afterwards the samples were dehydrated in a series of graded ethanol, dried and coated in a sputter‐coater (SCD 040; Oerlikon Balzers, Balzers, Liechtenstein) with gold. For visualization, a digital scanning microscope (DSM 940, Carl Zeiss Jena GmbH) was used. Per localization and time point post‐infection, eight fields at a magnification of 1,000 were evaluated; the percentage of ciliated area was analysed using GraphPad Prism 5.0 (GraphPad Software, Inc). Mann–Whitney Test was applied and results were considered statistically significant at p‐value < 0.05.
2.4. Viral RNA detection by real time quantitative PCR (RT‐qPCR)
A previously published RT‐qPCR protocol was used to detect MERS‐CoV genome (Vergara‐Alert et al., 2017). Briefly, lymphoid samples (cervical and mediastinal lymph nodes, tonsil and thymus) were placed in tubes containing 500 μl Dulbecco's modified Eagle medium (DMEM) supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin, 2 mmol/L glutamine and 2 mm glass beads (Fisher Scientific, USA), individually homogenized at 30 Hz for 2 min by using a TissueLyser II (QIAGEN, Hilden, Germany) and stored at −70°C until use. Viral RNA was extracted by using a NucleoSpin RNA virus kit (Macherey‐Nagel, Düren, Germany) following the manufacturer's recommendations. The extraction products were tested by RT‐qPCR, which was performed by using AgPath‐ID One‐Step RT‐PCR reagents (Applied Biosystems, Foster City, CA, USA). The amplification was conducted on a 7,500 Fast Real‐Time PCR System (Applied Biosystems, USA) programmed under the following conditions: 50°C for 5 min, 95°C for 20 s and 45 cycles at 95°C for 3 s and 60°C for 30 s. Samples with a cycle threshold <40 were considered positive for MERS‐CoV RNA (Vergara‐Alert et al., 2017).
3. RESULTS
3.1. Localization of DPP4 antigen
In pigs, DPP4 staining was observed in both nasal mucosa and submucosa including cytoplasm of epithelial (Figure 1a) and macrophage‐like cells (Figure 2a), Bowman's glands (Figures 1a, 2a and b) and endothelial cells (Figure 2a). In addition, DPP4 was also located in lining cilia and cytoplasm of pseudostratified columnar epithelial cells, and the cytoplasm of macrophages, and neutrophils and vascular endothelial cells in trachea (Figure 3a). The distribution of DPP4 in the bronchus was similar to that in the trachea, with some positive lymphocytes and lack of expression in the neutrophils (Figure 3b). It was also detected in the apical layers of the cuboidal epithelium in bronchioli (Figure 4a) and terminal bronchioli (Figure 4b). In the alveoli, cytoplasmic DPP4 was abundantly present in both type I and II pneumocytes (Figure 5a). Such expression was moderate in the mediastinal lymph node and bronchus‐associated lymphoid tissue (BALT, Figure 6a). DPP4 was extensively expressed in the cytoplasm of dendritic reticular cells of germinal centers as well as in the cytoplasm of macrophages and lymphocytes of the medulla in both cervical lymph nodes (Figure 7a) and tonsils. Also, DPP4 staining was seen in epithelial reticular cells in Hassall's corpuscles in the thymus. No differences in the distribution of DPP4 were observed when comparing tissues on different days p.i.
Figure 1.
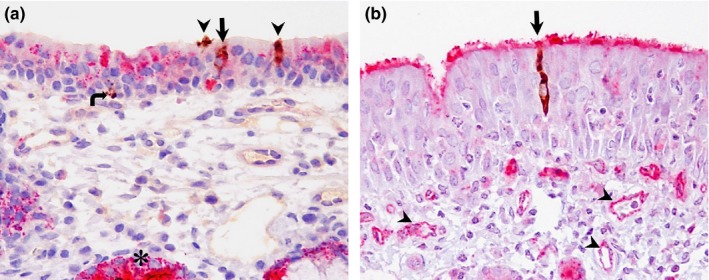
Double IHC staining of the frontal turbinate of pigs and llamas on day 4 p.i. (a) Frontal turbinate; pig. DPP4 (pink staining) is present in the cytoplasm of nasal epithelial cells and submucosal glands (asterisk). In this section, three individual epithelial cells (arrow and arrowheads) and a single basal cell (curved arrow) show MERS‐CoV antigen (brown cytoplasmic staining). One epithelial cell (arrow) shows MERS‐CoV and DPP4 co‐localization; the rest of cells containing viral antigen (arrowheads and curved arrow) do not show DPP4 expression. (b) Frontal turbinate; llama. DPP4 staining is present in the lining cilia as well as in the capillaries (arrowheads). In this section, a single epithelial cell harbours MERS‐CoV antigen (brown cytoplasmic staining) in the cytoplasm (arrow) in co‐localization with DPP4. Original magnification: ×400 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 2.

Double IHC staining in medial turbinate of pigs and medial/caudal turbinate of llamas at different time‐points post‐inoculation. (a) Medial turbinate; pig, day 2 p.i. DPP4 antigen (pink staining) is located mainly in the cytoplasm of epithelial cells, endothelium (thick arrow), macrophage‐like cells (curved arrows) and glandular cells in submucosa (asterisk). Several epithelial cells with cytoplasmic presence of MERS‐CoV antigen (brown) also contain DPP4 (arrows). Also, one MERS‐CoV positive cell does not display DPP4 staining (arrowhead). (b) Medial turbinate; pig, day 4 p.i. Both DPP4/MERS‐CoV double stained cell (arrow) and cells harbouring only MERS‐CoV antigen (arrowheads) are displayed. (c, d) Medial and caudal turbinates, respectively; llama, day 4 p.i. DPP4 expression (pink staining) is mostly restricted on lining cilia, submucosal glands (arrows) and capillaries (arrowheads). Co‐localization of DPP4 (pink) and MERS‐CoV (brown) antigens is observed in all infected cells. Original magnification: ×400 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
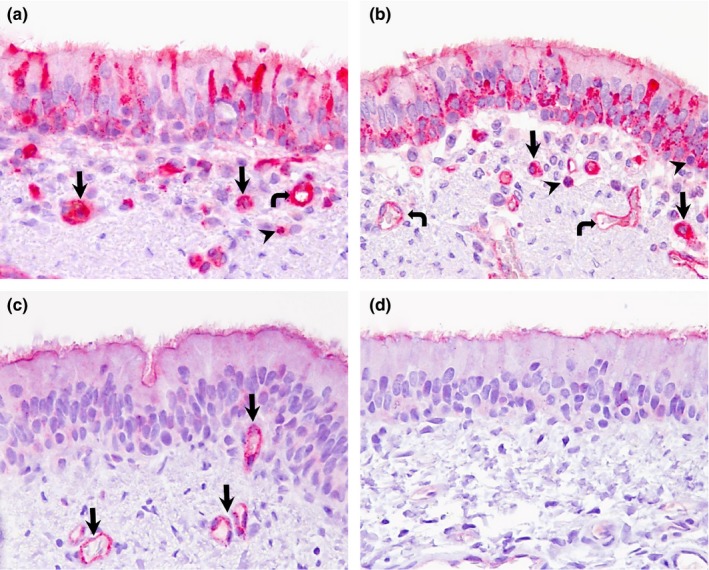
Double IHC staining in trachea and bronchus of pigs and llamas on day 4 p.i. (a) Trachea; pig. Extensive presence of DPP4 antigen (pink staining) in the cytoplasm and occasionally in cilia of pseudostratified epithelial cells, cytoplasm of macrophage like‐cells (arrows), neutrophils (arrowhead) and endothelial walls (curved arrow) in the trachea. (b) Bronchus; pig. Pseudostratified columnar epithelial cells, macrophage‐like cells (arrows), lymphocytes (arrowheads) and endothelial cells (curved arrows) show DPP4 antigen (pink staining) mainly in the cytoplasm and occasionally in cilia of respiratory epithelial cells. (c) Trachea; llama. DPP4 antigen (pink staining) is mainly located in the cilia and capillaries (arrows). (d) Bronchus; llama. DPP4 antigen (pink staining) was exclusively observed in the lining cilia. No MERS‐CoV antigen (brown) was detected in any of these tissues (a)‐(d). Original magnification: ×400 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 4.
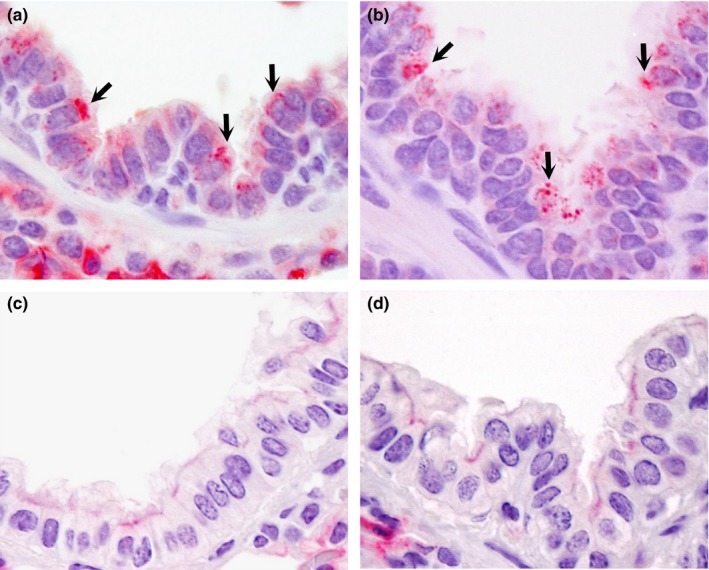
Double IHC staining in bronchioli and terminal bronchioli of pigs and llamas on day 4 p.i. In pigs, DPP4 antigen (pink staining) is present in the apical surface of epithelial cells in bronchioli (arrows in (a)) and terminal bronchioli (arrows in (b)). In llamas, DPP4 is absent in the epithelium of bronchioli (c) and terminal bronchioli (d). No MERS‐CoV antigen (brown) was detected in any of these tissues (a)‐(d). Original magnification: ×1,000 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 5.
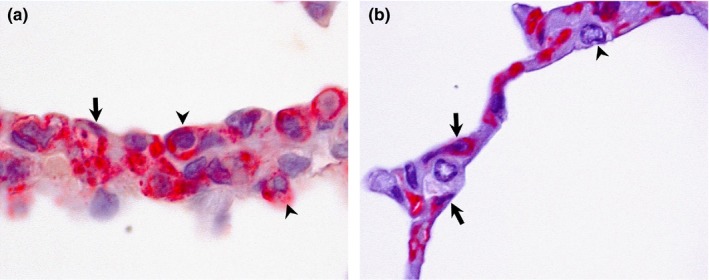
Double IHC staining in alveoli of pigs and llamas on day 4 p.i. (a) In pigs, both type I (arrow) and II (arrowheads) pneumocytes stained positive for DPP4 antigen (pink staining) in the cytoplasm. (b) In llamas, cytoplasmic DPP4 antigen (pink staining) is mainly located in type I pneumocytes (arrows) rather than type II pneumocytes (arrowhead). No MERS‐CoV antigen (brown) was detected in any of these tissues (a)‐(d). Original magnification: ×1,000 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 6.
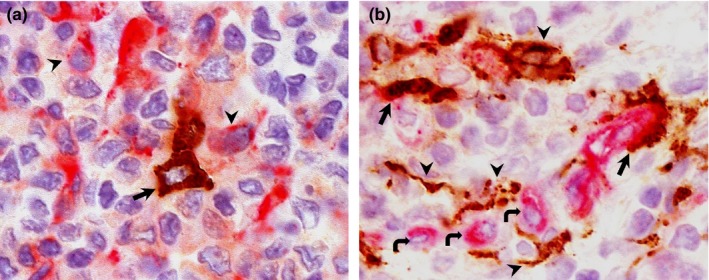
Double IHC staining in BALT of pigs and llamas at different time‐points post‐inoculation. (a) Pig, day 2 p.i. Positive staining for DPP4 antigen (pink staining) present in the cytoplasm of lymphocyte‐like cells (arrowheads). Some dendritic‐shaped cells (arrow) harbour MERS‐CoV antigen but lack of DPP4. (b) Llama, day 4 p.i. DPP4 antigen (pink staining) is present in the cytoplasm of lymphocyte‐like cells (curved arrows). Some dendritic‐shaped cells (arrowheads) show exclusively MERS‐CoV antigen (brown) while other had double positivity for MERS‐CoV/DPP4 (arrows). Original magnification: ×1,000 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 7.
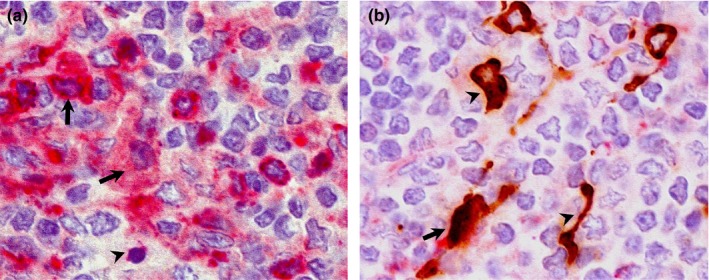
Double IHC staining in the cervical lymph node of pigs and llamas on day 4 p.i. (a) Paracortex‐medulla of cervical lymph node; pig. High amount of DPP4 antigen (pink staining) is located in the cytoplasm of macrophage‐like (arrows) and lymphocyte‐like cells (arrowhead). No MERS‐CoV antigen (brown) is present. (b) Paracortex‐medulla of cervical lymph node; llama. DPP4 antigen (pink staining) is slightly positive in one dendritic‐like cell co‐expressing MERS‐CoV antigen (arrow); rest of MERS‐CoV (brown) positive dendritic cells (arrowheads) do not show presence of DPP4 labelling (pink staining). Original magnification: ×1,000 for all tissues [Colour figure can be viewed at http://wileyonlinelibrary.com]
In llamas, in contrast, DPP4 was predominantly expressed in cilia of nasal epithelium, the cytoplasm of endothelial cells as well as on the apical surface of glandular cells of the submucosal glands in nasal turbinate; DPP4 antigen was not detected in the cytoplasm of nasal epithelial cells (Figures 1b, 2c and d). DPP4 expression was present in the cilia of tracheal and bronchial epithelial cells and in the cytoplasm of endothelial cells (Figures 3c and d). In the lung, although DPP4 was scarcely detected in the epithelium of bronchioli and terminal bronchioli (Figures 4c and d), it was observed in the cytoplasm of type I pneumocytes (Figure 5b). In the BALT, notable cytoplasmic DPP4 was located in the plasma cells (Figure 6b). In lymphoid tissues, DPP4 was rarely observable by IHC (Figure 7b). No differences on DPP4 distribution were observed when comparing tissues at different days p.i.
3.2. Detection of MERS‐CoV antigen and co‐localization with DPP4
In pigs, on day 2 p.i., MERS‐CoV antigen was detected in the cytoplasm of scattered epithelial cells in the medial part of the nasal turbinates (Figure 2a) and, occasionally, in the cytoplasm of dendritic shaped cells in the BALT (Figure 6a). On day 4 p.i., a few epithelial cells of frontal turbinates contained MERS‐CoV antigen in the cytoplasm (Figure 1a) and this number was slightly increased in the medial turbinates (Figure 2b). Although cytoplasmic co‐localization of DPP4 with MERS‐CoV antigen was observed in frontal and medial turbinates (Figures 1a, 2a and b), the majority of epithelial cells expressing viral N protein did not show apical DPP4 staining. No MERS‐CoV was detected in any other respiratory and lymphoid tissues on day 2 and 4 p.i. On day 24 p.i. none of the pig tissues examined in this study was positive for the MERS‐CoV N protein.
In llamas, on day 4 p.i., while only a few pseudostratified columnar epithelial cells in the frontal turbinate contained MERS‐CoV antigen in the cytoplasm (Figure 1b), the expression was remarkably intense in both medial and caudal nasal turbinate epithelia (Figure 2c and d). Notably, MERS‐CoV antigen was occasionally observed in stellate shaped cells resembling dendritic cells of BALT (Figure 6b) and cervical lymph nodes (Figure 7b). Along the nasal turbinate, MERS‐CoV antigen co‐localized in all cases with cells expressing DPP4 in the apical pole (Figures 1b, 2c and d). In contrast, only few dendritic‐like cells expressing MERS‐CoV were positive in the cytoplasm for DPP4 in BALT (Figure 6b) and cervical lymph nodes (Figure 7b). No MERS‐CoV was detected in any other respiratory and lymphoid tissues on day 4 p.i. On day 24 p.i none of the llama tissues used in this study was positive for the MERS‐CoV N protein.
Noteworthy, the number of MERS‐CoV positive cells as well as DPP4/MERS‐CoV double positive cells in llamas was much higher than that in pigs. Table 1 summarizes the semi‐quantitative scores of MERS‐CoV antigen found in the different studied tissues.
Table 1.
Summary of MERS‐CoV antigen distribution in respiratory and lymphoid tissues, RT‐qPCR Cq values and ciliary coverage (in percentage) of airways of pigs and llamas
| 2 dpi | ID of animal | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Pig No. 1 | Pig No. 2 | Pig No. 3 | Pig No. 4 | ||||||||
| IHC | Cq | SEM | IHC | Cq | SEM | IHC | Cq | SEM | IHC | Cq | SEM | |
| Turbinate | + | 39 | 35 | − | 28 | 35 | − | 27 | NP | − | 29 | NP |
| Trachea | − | ND | 100 | − | 36 | 100 | − | 36 | NP | − | ND | NP |
| Bronchus | − | ND | 100 | − | ND | 100 | − | ND | NP | − | ND | NP |
| Cervical LN | − | ND | NA | − | ND | NA | − | ND | NA | − | ND | NA |
| 4 dpi | ID of animal | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Pig No. 5 | Pig No. 6 | Pig No. 7 | Pig No. 8 | ||||||||
| IHC | Cq | SEM | IHC | Cq | SEM | IHC | Cq | SEM | IHC | Cq | SEM | |
| Turbinate | ++ | 25 | 51 | + | 24 | 50 | + | 32 | NP | + | 29 | NP |
| Trachea | − | 33 | 45 | − | 32 | 58 | − | ND | NP | − | ND | NP |
| Bronchus | − | ND | 100 | − | 36 | 100 | − | ND | NP | − | ND | NP |
| Cervical LN | − | 31 | NA | − | 35 | NA | − | 38 | NA | − | ND | NA |
| 4 dpi | ID of animal | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | llama No. 1 | llama No. 2 | llama No. 3 | llama No. 4 | ||||||||
| IHC | Cq | SEM | IHC | Cq | SEM | IHC | Cq | SEM | IHC | Cq | SEM | |
| Turbinate | +++ | 30 | 44 | +++ | 24 | 100 | +++ | 29 | NP | − | 30 | NP |
| Trachea | − | 34 | 86 | − | N | 100 | − | 37 | NP | − | 35 | NP |
| Bronchus | − | ND | 100 | − | ND | 100 | − | ND | NP | − | 34 | NP |
| Cervical LN | NS | ND | NA | + | 27 | NA | + | 31 | NA | − | ND | NA |
IHC, immunohistochemical score: −, negative; +, low (less than 10 cells/tissue); ++, moderate (10–50 cells/tissue); +++, high (more than 50 cells/tissues). Cq, quantification cycle obtained by RT‐qPCR. ND, non‐detectable. Cq value of turbinate, trachea and bronchus of both pigs and llamas is cited from a previously published work (Vergara‐Alert et al., 2017). SEM, percentage of ciliary coverage calculated through scanning electron microscopy. NS, no sample available. NP, not processed, NA, non‐applicable.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.3. Scanning electron microscopy
Pigs infected with MERS‐CoV showed severe ciliary loss in the nasal mucosa (Figure 8a, Table 1) on days 2 and 4 p.i. (p = 0.0082). In contrast, ciliation of the trachea was unaffected on day 2 p.i., but ciliary loss was significant on days 4 (Figure 8b, Table 1) and 24 p.i. (p = 0.008 and p = 0.0078, respectively). Bronchial ciliation was unaffected during the whole experiment (Figure 8, Table 1). On the other hand, llamas infected with MERS‐CoV showed almost no significant ciliary loss in nasal mucosa, trachea and bronchi at any time; only one animal exhibited moderate ciliary loss in the nasal mucosa (Figure 9a, Table 1) on day 4 p.i.
Figure 8.

SEM micrographs of ciliary damage in the respiratory tract of MERS‐CoV inoculated pigs and healthy control pigs. On day 4 p.i., severe ciliary loss was detected in nasal turbinate (a) and trachea (b), while ciliary density was unaffected in bronchus (c). Control pigs showed tightly packed ciliation in apical surface of epithelial cells of nasal turbinate, trachea and bronchus (scale bar, 10 μm)
Figure 9.
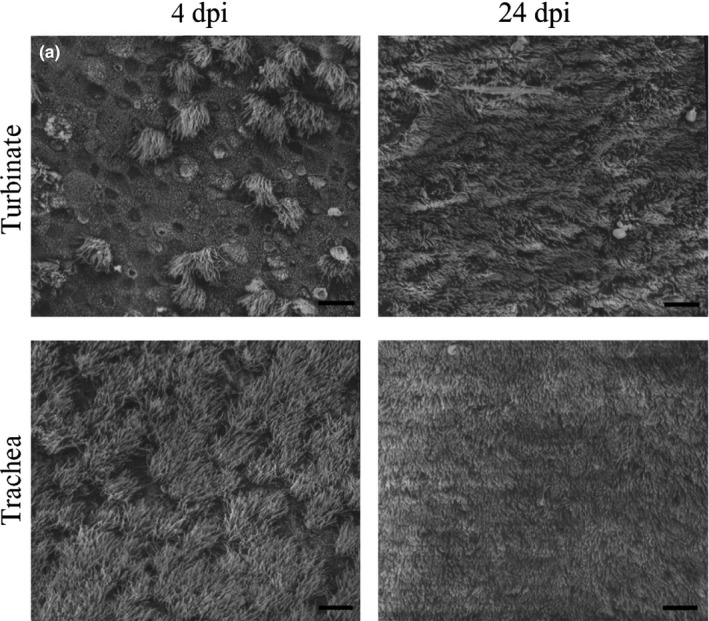
SEM micrographs of ciliary damage in the respiratory tract of MERS‐CoV inoculated llamas. On day 4 p.i., only one animal showed severe ciliary loss in nasal turbinate (a), while ciliary bundles were unaffected in trachea. On day 24 p.i., complete ciliation was seen in in apical surface of nasal turbinate and trachea (scale bar, 10 μm)
3.4. Viral RNA detection by RT‐qPCR in lymphoid tissues
In pigs, cervical and mediastinal lymph nodes and tonsil contained low viral RNA loads (Cq values ranged from 34.5 to 39) on day 4 p.i., but they were negative on days 2 and 24 p.i (Supporting information Figure S1a). In llamas, comparatively higher viral loads were observed in cervical lymph nodes on day 4 p.i., and to a lower extent, in mediastinal lymph nodes, tonsils and thymus (Supporting information Figure S1b). On day 24 p.i., a low amount of viral RNA (Cq = 32) was found in cervical lymph nodes, being even less (Cq = 36) in mediastinal lymph nodes and tonsils (Supporting information Figure S1b). MERS‐CoV RNA was not detected in the thymus of llamas on 24 day p.i (Supporting information Figure S1b).
4. DISCUSSION
Middle East respiratory syndrome coronavirus entry into target cells through interaction between the RBD and the DPP4 protein has been well documented (Raj et al., 2013). However, this is the first study in which co‐localization between the virus and DPP4 from respiratory and lymphoid organs of different MERS‐CoV susceptible species (pigs and llamas) has been studied.
In both species tested here, limited numbers of cells expressing DPP4 seemed to support MERS‐CoV replication. The number of MERS‐CoV positive cells and MERS‐CoV/DPP4 double positive cells was higher in llamas compared to that of pigs. Moreover, variation in labelling and localization of both DPP4 and MERS‐CoV were observed between pigs and llamas. In addition, the amount of MERS‐CoV antigen in these two species showed a consistent correlation with the MERS‐CoV genome levels detected by RT‐qPCR in respiratory and lymphoid tissues, indicating that host cell factors necessary for MERS‐CoV attachment, penetration and RNA and protein synthesis are present in both pigs and llamas.
Noteworthy, MERS‐CoV infection led to striking differences of ciliation in respiratory tracts between pigs and llamas. Based on the SEM study, experimental infection with MERS‐CoV in pigs was associated with a severe ciliary loss in the nasal turbinates in the early phase of the infection (days 2 and 4 p.i.). Tracheal deciliation, in contrast, was observed at later time points (days 4 and 24 p.i.), being more severe on day 4 p.i. Bronchial ciliation was unaffected during the whole experiment. On the other hand, experimental infection of llamas with MERS‐CoV led to almost no effect on nasal, tracheal and bronchial ciliation. These results contrast with recent data generated in dromedary camels, in which ciliary loss was very prominent (Haverkamp et al., 2018). In fact, these authors observed that MERS‐CoV infected epithelial cells lost cilia and showed lack of apical DPP4 antigen, while the adjacent non‐infected cells retained the DPP4 staining. Such evidence was not observed in pigs (DPP4 was detected in the cytoplasm mainly, with a significant loss of cilia) and llamas (infected cells retained apical and cytoplasmic DPP4 labelling and no significant cilia loss occurred) of this study. Ciliary loss of the upper respiratory tract has also been described in other viral infections such as Severe Acute Respiratory Syndrome‐CoV in humans (Nicholls et al., 2003), respiratory syncytial virus in humans (Jumat et al., 2015), influenza virus in humans and ferrets (Zeng et al., 2013) and canine respiratory coronavirus in dogs (Mitchell et al., 2013). A number of mechanisms have been proposed to explain such ciliary loss, including direct destruction of the ciliary apparatus, ionic variations during respiratory viral attachment and replication stages, damage through regulating the production of oxidizing metabolites, and apoptosis (Jumat et al., 2015; Mall, 2008; Vareille, Kieninger, Edwards, & Regamey, 2011). Therefore, the severe ciliary loss observed in nasal turbinates of pigs from this study was probably due to a bystander effect, since a very low amount of MERS‐CoV antigen and RNA was found in these affected turbinates.
Both pigs and llamas suffered from a subclinical, time‐limited infection by MERS‐CoV. Three out of 8 llamas showed moderate mucus secretion in 1 nostril on days 4–18 p.i while three pigs showed only mild mucus secretion on days 2–16 p.i (Vergara‐Alert et al., 2017). To date, decisive factors that restrict further replication of MERS‐CoV in the hosts are still poorly understood. It cannot be excluded that strong innate and/or adaptive immune responses may be responsible for virus clearance. In lymphoid tissues of both species (in the pigs only in the BALT), a limited number of cells were stained positive for MERS‐CoV antigen. Noteworthy, these cells had the morphology of dendritic cells (DCs). MERS‐CoV has been reported to productively infect human DCs in vitro, which can also serve as a vector for viral dissemination within the body (Chu et al., 2014). However, human DCs can secrete interferons (IFNs) and then trigger a potent innate/adaptive response upon MERS‐CoV infection (Scheuplein et al., 2015). Thus, those DC‐like cells could interfere with viral replication by releasing cytokines, like type I IFNs, or delivering antigens to other immune cells at the time MERS‐CoV reaches this area (Scheuplein et al., 2015). Future studies on this topic are therefore required to verify whether these cells are effectively DCs and if they are able to express DPP4 at the surface before infection.
Obtained results suggest the possibility that, besides DPP4, other surface molecules may facilitate virus entry; the potential absence or minimal expression of these latter putative molecules in pigs could be a cause for lower virus replication compared to llamas. Chan et al. (2016) estimated that carcinoembryonic antigen‐related cell adhesion molecule 5 (CEACAM5) is an important surface attachment factor that facilitates entry of MERS‐CoV in vitro. More recent evidence suggests that the speed and efficiency of viral antigen to attach the host cell is accelerated by an entry complex that includes DPP4, a CoV‐activating transmembrane protease serine 2 (TMPRSS2) and the tetraspanin Cd9 (Earnest et al., 2017). In their study, mice transfected with the human DPP4 gene became significantly less susceptible to MERS‐CoV infection after silencing Cd9 or TMPRSS2 with small RNAs. Furthermore, MERS‐CoV was reported to bind to sialic acid (Sia) and depletion of this molecule led to inhibition of MERS‐CoV entry in Calu‐3 human airway cells (Li et al., 2017). In consequence, it is possible that DPP4 is not the single host determinant for the virus entry, at least for some animal species. Another study has demonstrated that horses express DPP4 in the respiratory tract, but no viral RNA was detected (Vergara‐Alert et al., 2017) upon MERS‐CoV inoculation, suggesting the presence of host factors such as glycosyls (Peck et al., 2015) that block the entry of MERS‐CoV. Nevertheless, whether or not DPP4 is the only functional receptor for MERS‐CoV remains to be investigated in susceptible animals. Our observations and analyses will need to be further validated as some questions still remain. For instance, it is not known why ciliary loss did not occur in llamas despite the much higher amount of virus than in the pig. Moreover, Haverkamp et al. (2018) demonstrated down‐regulation of surface DPP4 upon MERs‐CoV infection, but this was not observed in llamas, since most infected cells still retained DPP4 on its cell surface.
In conclusion, the present work provides evidence that MERS‐CoV preferably infects respiratory epithelial cells expressing DPP4 in llamas, supporting that DPP4 is necessary for virus entry in these organs. However, based on our observations, DPP4 may not be the single host determinant in regulating virus entry in respiratory cells of pigs and lymphoid tissues of both species. Although pigs showed a significant expression of DPP4 (mostly in the cell cytoplasm), the number of cells permissive for MERS‐CoV in this species was lower than that of llamas.
AUTHOR CONTRIBUTIONS
Conceptualization: N. Te, J. Vergara‐Alert, A. Bensaid, J. Segalés, B.L. Haagmans. Formal analysis: N. Te, J. Segalés. Funding acquisition: A. Bensaid, J. Segalés. Investigation: N. Te, M. Pérez, A. Lehmbecker, W. Baumgärtner, J. Segalés. Supervision: J. Vergara‐Alert, A. Bensaid, J. Segalés. Writing ‐ original draft: N. Te. Writing ‐ review & editing: All co‐authors.
Supporting information
ACKNOWLEDGEMENTS
Xavier Abad, David Solanes and all animal caretakers from the IRTA‐CReSA biosecurity level 3 laboratories and animal facilities are greatly acknowledged for their support. The authors also thank Kerstin Rohn for excellent technical assistance with the scanning electron microscopic work. This research was performed as part of the Zoonoses Anticipation and Preparedness Initiative (ZAPI project; IMI Grant Agreement no. 115760), with the assistance and financial support of IMI and the European Commission, and in‐kind contributions from EFPIA partners. The funding from CERCA Programme/Generalitat de Catalunya to IRTA is also acknowledged. Nigeer Te is a recipient of a Chinese Scholarship Council grant (CSC NO. 201608150108).
Te N, Vergara‐Alert J, Lehmbecker A, et al. Co‐localization of Middle East respiratory syndrome coronavirus (MERS‐CoV) and dipeptidyl peptidase‐4 in the respiratory tract and lymphoid tissues of pigs and llamas. Transbound Emerg Dis. 2019;66:831–841. 10.1111/tbed.13092
REFERENCES
- Adney, D. R. , Bielefeldt‐Ohmann, H. , Hartwig, A. E. , & Bowen, R. A. (2016). Infection, replication, and transmission of Middle East Respiratory Syndrome Coronavirus in alpacas. Emerging Infectious Diseases, 22(6), 1031–1037. 10.3201/2206.160192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar, E. I. , El‐Kafrawy, S. A. , Farraj, S. A. , Hassan, A. M. , Al‐Saeed, M. S. , Hashem, A. M. , & Madani, T. A. (2014). Evidence for camel‐to‐human transmission of MERS coronavirus. New England Journal of Medicine, 370(26), 2499–2505. 10.1056/NEJMoa1401505 [DOI] [PubMed] [Google Scholar]
- Chan, C. M. , Chu, H. , Wang, Y. , Wong, B. H. , Zhao, X. , Zhou, J. , … Yuen, K. Y. (2016). Carcinoembryonic antigen‐related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of middle east respiratory syndrome coronavirus. Journal of Virology, 90(20), 9114–9127. 10.1128/JVI.01133-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Zhou, J. , Wong, B. H. , Li, C. , Cheng, Z. S. , Lin, X. , … Yuen, K. Y. (2014). Productive replication of Middle East respiratory syndrome coronavirus in monocyte‐derived dendritic cells modulates innate immune response. Virology, 454, 197–205. 10.1016/j.virol.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Jores, J. , Meyer, B. , Younan, M. , Liljander, A. , Said, M. Y. , … Müller, M. A. (2014). Antibodies against MERS coronavirus in dromedary camels, Kenya. Emerging Infectious Diseases, 20(8), 1319–1322. 10.3201/eid2008.140596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten, M. , Lam, T. T. , Watson, S. J. , Palser, A. L. , Petrova, V. , & Grant, P. , … Nastouli, E. (2013b). Full‐genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerging Infectious Diseases, 19(5), 736–742B 10.3201/eid1905.130057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten, M. , Watson, S. J. , Kellam, P. , Al‐Rabeeah, A. A. , Makhdoom, H. Q. , Assiri, A. , … Memish, Z. A. (2013a). Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. The Lancet, 382(9909), 1993–2002. 10.1016/S0140-6736(13)61887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten, M. , Watson, S. J. , Zumla, A. I. , Makhdoom, H. Q. , Palser, A. L. , & Ong, S. H. , … Memish, Z. A. (2014). Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio, 5(1), e01062‐13 10.1128/mbio.01062-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest, J. T. , Hantak, M. P. , Li, K. , McCray, P. B. Jr , Perlman, S. , & Gallagher, T. (2017). The tetraspanin CD9 facilitates MERS‐coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathogens, 13(7), e1006546 10.1371/journal.ppat.1006546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano, D. , de Wit, E. , Feldmann, F. , Rasmussen, A. L. , Okumura, A. , Peng, X. , … Munster, V. J. (2014). Infection with MERS‐CoV causes lethal pneumonia in the common marmoset. PLOS Pathogens, 10(8), e1004250 10.1371/journal.ppat.1004250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans, B. L. , van den Brand, J. M. , Raj, V. S. , Volz, A. , Wohlsein, P. , Smits, S. L. , … Osterhaus, A. D. (2016). An orthopoxvirus‐based vaccine reduces virus excretion after MERS‐CoV infection indromedary camels. Science, 351(6268), 77–81. 10.1126/science.aad1283 [DOI] [PubMed] [Google Scholar]
- Haverkamp, A. K. , Lehmbecker, A. , Spitzbarth, I. , Widagdo, W. , Haagmans, B. L. , Segalés, J. , … Baumgärtner, W. (2018). Experimental infection of dromedaries with Middle East respiratory syndrome‐Coronavirus is accompanied by massive ciliary loss and depletion of the cell surface receptor dipeptidyl peptidase 4. Scientific Reports, 8(1), 9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, Y. H. (2015). Middle East Respiratory Syndrome coronavirus (MERS‐CoV) nosocomial outbreak in South Korea: Insights from modeling. PeerJ, 3, e1505 10.7717/peerj.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumat, M. R. , Yan, Y. , Ravi, L. I. , Wong, P. , Huong, T. N. , Li, C. , … Sugrue, R. J. (2015). Morphogenesis of respiratory syncytial virus in human primary nasal ciliated epithelial cells occurs at surface membrane microdomains that are distinct from cilia. Virology, 484, 395–411. 10.1016/j.virol.2015.05.014 [DOI] [PubMed] [Google Scholar]
- Li, W. , Hulswit, R. J. G. , Widjaja, I. , Raj, V. S. , McBride, R. , Peng, W. , … Bosch, B. J. (2017). Identification of sialic acid‐binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proceedings of the National Academy of Sciences of the United States of America, 114(40), E8508–E8517. 10.1073/pnas.1712592114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall, M. A. (2008). Role of cilia, mucus, and airway surface liquid in mucociliary dysfunction: Lessons from mouse models. Journal of Aerosol Medicine and Pulmonary Drug Delivery, 21(1), 13–24. 10.1089/jamp.2007.0659 [DOI] [PubMed] [Google Scholar]
- Mitchell, J. A. , Brooks, H. W. , Szladovits, B. , Erles, K. , Gibbons, R. , Shields, S. , & Brownlie, J. (2013). Tropism and pathological findings associated with canine respiratory coronavirus (CRCoV). Veterinary Microbiology, 162(2–4), 582–594. 10.1016/j.vetmic.2012.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, J. M. , Poon, L. L. , Lee, K. C. , Ng, W. F. , Lai, S. T. , Leung, C. Y. , … Peiris, J. S. (2003). Lung pathology of fatal severe acute respiratory syndrome. The Lancet, 361(9371), 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, K. M. , Cockrell, A. S. , Yount, B. L. , Scobey, T. , Baric, R. S. , & Heise, M. T. (2015). Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndromecoronavirus infection. Journal of Virology, 89(8), 4696–4699. 10.1128/JVI.03445-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V. S. , Mou, H. , Smits, S. L. , Dekkers, D. H. , Müller, M. A. , Dijkman, R. , … Haagmans, B. L. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature, 495(7440), 251–254. 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken, C. B. , Haagmans, B. L. , Müller, M. A. , Gutierrez, C. , Godeke, G. J. , Meyer, B. , … Koopmans, M. P. (2013). Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. The Lancet Infectious Diseases, 13(10), 859–866. 10.1016/S1473-3099(13)70164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir, J. S. , Lam, T. T. , Ahmed, M. M. , Li, L. , Shen, Y. , Abo‐Aba, S. E. , … Guan, Y. (2016). Co‐circulation of three camel coronavirus species and recombination of MERS‐CoVs in Saudi Arabia. Science, 351(6268), 81–84. 10.1126/science.aac8608 [DOI] [PubMed] [Google Scholar]
- Scheuplein, V. A. , Seifried, J. , Malczyk, A. H. , Miller, L. , Höcker, L. , Vergara‐Alert, J. , … Mühlebach, M. D. (2015). High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. Journal of Virology, 89(7), 3859–3869. 10.1128/JVI.03607-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiodras, S. , Baka, A. , Mentis, A. , Iliopoulos, D. , Dedoukou, X. , Papamavrou, G. , … Saroglou, G. (2014). A case of imported Middle East Respiratory Syndrome coronavirus infection and public health response, Greece, April 2014. Euro Surveillance, 19(16), 20782. [DOI] [PubMed] [Google Scholar]
- Vareille, M. , Kieninger, E. , Edwards, M. R. , & Regamey, N. (2011). The airway epithelium: Soldier in the fight against respiratory viruses. Clinical Microbiology Reviews, 24(1), 210–229. 10.1128/CMR.00014-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara‐Alert, J. , vanden Brand, J. M. , Widagdo, W. , Muñoz, M. 5th. , Raj, S. , … Schipper, D. ,… Segalés, J. (2017). Livestock susceptibility to infection with Middle East Respiratory Syndrome coronavirus. Emerging Infectious Diseases, 23(2), 232–240. 10.3201/eid2302.161239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2015). Severe respiratory disease associated with Middle East respiratory syndrome coronavirus (MERS‐CoV). http://www.ecdc.europa.eu/en/publications/Publications/RRA-MERS-CoV-thirteenth-update.pdf.
- Widagdo, W. , Begeman, L. , Schipper, D. , Run, P. R. V. , Cunningham, A. A. , Kley, N. , … van den Brand, J. M. A. (2017). Tissue distribution of the MERS‐coronavirus receptor in bats. Scientific Reports, 7(1), 1193 10.1038/s41598-017-01290-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo, W. , Raj, V. S. , Schipper, D. , Kolijn, K. , van Leenders, G. J. , Bosch, B. J. , … Haagmans, B. L. (2016). Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. Journal of Virology, 90(9), 4838–4842. 10.1128/JVI.02994-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, E. , Rasmussen, A. L. , Falzarano, D. , Bushmaker, T. , Feldmann, F. , Brining, D. L. , … Munster, V. J. (2013). Middle East respiratory syndrome coronavirus (MERS‐CoV) causes transient lower respiratorytract infection in rhesus macaques. Proceedings of the National Academy of Sciences of the United States of America, 110(41), 16598–16603. 10.1073/pnas.1310744110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) . (2018). Middle East respiratory syndrome coronavirus (MERS‐CoV) ‐ update: August 2018. Retrieved from http://www.who.int/emergencies/mers-cov/en/. Accessed 14 September 2018.
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. , & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367(19), 1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- Zeng, H. , Goldsmith, C. S. , Maines, T. R. , Belser, J. A. , Gustin, K. M. , Pekosz, A. , … Tumpey, T. M. (2013). Tropism and infectivity of influenza virus, including highly pathogenic avian H5N1 virus, in ferret tracheal differentiated primary epithelial cell cultures. Journal of Virology, 87(5), 2597–2607. 10.1128/JVI.02885-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Shen, L. , & Gu, X. (2016). Evolutionary dynamics of MERS‐CoV: Potential recombination, positive selection and transmission. Scientific Reports, 6, 25049 10.1038/srep25049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


