Abstract
This study investigated whole‐brain dynamic lag pattern variations between neurotypical (NT) individuals and individuals with autism spectrum disorder (ASD) by applying a novel technique called dynamic lag analysis (DLA). The use of 3D magnetic resonance encephalography data with repetition time = 100 msec enables highly accurate analysis of the spread of activity between brain networks. Sixteen resting‐state networks (RSNs) with the highest spatial correlation between NT individuals (n = 20) and individuals with ASD (n = 20) were analyzed. The dynamic lag pattern variation between each RSN pair was investigated using DLA, which measures time lag variation between each RSN pair combination and statistically defines how these lag patterns are altered between ASD and NT groups. DLA analyses indicated that 10.8% of the 120 RSN pairs had statistically significant (P‐value <0.003) dynamic lag pattern differences that survived correction with surrogate data thresholding. Alterations in lag patterns were concentrated in salience, executive, visual, and default‐mode networks, supporting earlier findings of impaired brain connectivity in these regions in ASD. 92.3% and 84.6% of the significant RSN pairs revealed shorter mean and median temporal lags in ASD versus NT, respectively. Taken together, these results suggest that altered lag patterns indicating atypical spread of activity between large‐scale functional brain networks may contribute to the ASD phenotype. Autism Res 2020, 13: 244–258. © 2019 The Authors. Autism Research published by International Society for Autism Research published by Wiley Periodicals, Inc.
Lay Summary
Autism spectrum disorder (ASD) is characterized by atypical neurodevelopment. Using an ultra‐fast neuroimaging procedure, we investigated communication across brain regions in adults with ASD compared with neurotypical (NT) individuals. We found that ASD individuals had altered information flow patterns across brain regions. Atypical patterns were concentrated in salience, executive, visual, and default‐mode network areas of the brain that have previously been implicated in the pathophysiology of the disorder.
Keywords: dynamic lag analysis, MREG, resting state fMRI, lag pattern, ASD, human brain
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by repetitive behaviors, restricted interests, and impaired social interaction and communication skills [American Psychiatric Association, 2013]. ASD has long been associated with impaired communication among brain networks. Multiple studies have sought to characterize disordered brain function using resting‐state functional magnetic resonance imaging (rs‐fMRI) [Assaf et al., 2010; Ebisch et al., 2011; Just, Keller, Malave, Kana, & Varma, 2012; Khan et al., 2013; Rudie et al., 2012; Starck et al., 2013; Uddin, Supekar, & Menon, 2013; Uddin, 2015; von dem Hagen, Stoyanova, Baron‐Cohen, & Calder, 2013; Weng et al., 2010]. The recent proliferation of resting state fMRI investigations in ASD has provided mixed evidence for impaired communication among brain network in the disorder. Specifically, in the young adult age range of interest in the current study, there have been reports of null findings when comparing within‐ and between‐network functional connectivity in clinical and neurotypical (NT) groups [Nomi & Uddin, 2015; Tyszka, Kennedy, Paul, & Adolphs, 2013]. More recent work with larger samples exploring wider age ranges, however, provides evidence for both hypoconnectivity and hyperconnectivity in ASD [Abbott et al., 2016; Di Martino et al., 2013]. Another recent report using the Autism Brain Imaging Data Exchange (ABIDE) dataset provides evidence for globally reduced network cohesion and density and increased dispersion of networks in ASD compared with typically developing participants [Keown et al., 2017]. All of these previous studies used conventional measures of functional connectivity, including independent component analysis, seed‐based functional connectivity, and whole‐brain functional connectivity matrix computation. The authors take these findings as evidence of reduced network integration and differentiation across several brain networks in ASD. Although there is large consensus that altered brain circuitry underlies the atypical behavioral manifestations observed in ASD, the precise nature of these alterations continues to be debated.
Conventional analyses model resting‐state fMRI time series as a combination of network processes that evolve over time [Beckmann, DeLuca, Devlin, & Smith, 2005; Cordes & Nandy, 2006]. Most prior fMRI studies of intrinsic brain activity have used either spatial independent component analysis (sICA) [Beckmann et al., 2005; Kiviniemi, Kantola, Jauhiainen, Hyvärinen, & Tervonen, 2003] or seed‐based correlation approaches [Biswal et al., 2010] to define functional brain networks. These analyses address connectivity as a continuous steady state process rather than a dynamically altering propagation of signals between brain regions. Several rs‐fMRI studies in humans and rats suggest that intrinsic activity is a spatiotemporally dynamic, instead of continuous, phenomenon [Chang & Glover, 2010; Hutchison, Womelsdorf, Gati, Everling, & Menon, 2013; Kiviniemi et al., 2011; Liu & Duyn, 2013; Majeed, Magnuson, & Keilholz, 2009; Majeed et al., 2011]. However, more recent methods applying temporal independent component analysis (tICA) have shown that multiple “temporal functional nodes” exist in human resting‐state fMRI data [Raatikainen et al., 2017; Smith et al., 2012].
Recently, several studies investigated blood oxygen level dependent (BOLD) signal propagation by analyzing temporal lags across the brain [Mitra, Snyder, Blazey, & Raichle, 2015; Mitra et al., 2016; Mitra, Snyder, Hacker, & Raichle, 2014]. These studies defined the lag between two fMRI time series by computing the cross‐correlation function for the full‐time series and identifying the local extremum using parabolic interpolation. Temporal lags reflect that there exists a time delay in propagating brain activation between distinct brain areas. It has been demonstrated in healthy subjects that the BOLD signal exhibits highly reproducible temporal lag patterns; some regions are early, that is, “sources” of propagated BOLD activity, with respect to the rest of the brain, while other regions are systematically late, that is, “destinations” of propagated BOLD activity [Mitra, Snyder, Blazey, et al., 2015; Mitra et al., 2014]. It has also been shown that the lag pattern of rs‐fMRI is composed of multiple temporal sequences [Mitra, Snyder, Blazey, et al., 2015] and that experimental manipulations of behavioral state can focally alter the lag structure [Mitra, Snyder, Tagliazucchi, Laufs, & Raichle, 2015]. Mitra and co‐workers [Mitra, Snyder, Constantino, & Raichle, 2015] have also shown that the lag structure of intrinsic activity is focally altered in high functioning adults with autism, and the degree of abnormality in individuals was highly correlated with behavioral measures relevant to the diagnosis of ASD. However, there has been one technical limitation in the above‐mentioned studies complicating the accurate lag structure estimation of resting state BOLD signals: the relatively low temporal sampling resolution of fMRI. Furthermore, there have been theoretical questions regarding whether the ultra‐fast technology below 500 msec repetition time (TR) can benefit the lag and connectivity estimation due to regional lag variability and slow temporal response of the hemodynamic response to neuronal activity. Recently, it has been shown that cardiac power distributions start to overlap on top of respiratory frequency maps as TR is increased >0.5 sec, indicating aliasing [Huotari et al., 2019].
Over the years, ultra‐fast MRI technology has started to brake the technological barrier of estimating lag structure accurately; sequences such as inverse imaging (INI), generalized inverse imaging, and magnetic resonance encephalography (MREG) enable three dimensional (3D) whole brain scanning <150 msec, even down to 25 msec [Assländer et al., 2013; Boyacioglu, Beckmann, & Barth, 2013; Lin et al., 2010; Posse et al., 2013]. Task activation results of 100 msec INI have repeatedly shown temporally accurate hemodynamic responses that linearly correlate with magnetoencephalography responses [Lin et al., 2014]. Importantly, the ultrafast whole brain data indicate that the variability of the relative latency is only 0.03 sec between left and right visual cortices [Lin et al., 2018]. Resting state in vivo multiphoton microscopical imaging results also indicate a hemodynamic response peak variability of 0.03–0.05 sec after a lag of 0.86 sec from excitatory neuronal GCaMP6f auto‐fluorescence signal activation to [HbT] hemodynamic signal response in mice [Ma et al., 2016]. In our previous studies, we have also found evidence of precise mapping of individual resting state coactivation patterns using a whole brain MREG sequence with TR of 100 msec [Raatikainen et al., 2017; Rajna, Kananen, Keskinarkaus, Seppänen, & Kiviniemi, 2015]. Thus, the latest findings of neuroimaging data sampled at 10 Hz both at cellular and macroscopic levels indicate minimal variability of hemodynamic response, and strongly support the idea of lag analytics of individual resting state peak data.
In our previous study, we found a consistent temporal lag pattern across individuals by deriving the lag using lagged cross‐correlation function for full time series [Raatikainen et al., 2017]. However, there were lag values exceeding the threshold of significance when the lag values were computed for multiple time windows using a sliding‐window approach, suggesting that there might be biologically meaningful variation in the dynamic lag structure between functionally connected areas. In other words, the lag structure enables one to follow the BOLD signal propagation between networks based on the temporal order of succeeding activation peaks detected in the resting‐state networks (RSNs). With the novel ultrafast imaging techniques without cardiorespiratory aliasing, one can estimate resting‐state metrics and lag structure more precisely [Huotari et al., 2019; Raatikainen et al., 2017; Raitamaa et al., 2018; Rajna et al., 2015]. No previous investigations of autism have concentrated on dynamic lag pattern variations in individuals with the disorder compared with NT individuals.
In this study, we introduce a novel analysis technique called dynamic lag analysis (DLA) to detect whole‐brain dynamic lag pattern variations between NT individuals and individuals with ASD using an ultra‐fast MREG sequence. First, this method utilizes sICA to define RSNs. Second, all peaks of RSN time signals are detected. Finally, lag values are dynamically measured by calculating the time lag values between RSNs for each peak. Our aim was to measure the lag variations between each RSN and to statistically define how the lag pattern of the brain is altered between ASD and NT groups.
Materials and Methods
Subjects
Twenty young adults with childhood diagnosed ASD (age = 23.7 ± 3.2 years; five females) and 20 NT individuals (age = 25.3 ± 6.2 years; four females) participated in the study. Individuals with ASD originally participated in a longitudinal clinical ASD study conducted at Oulu University Hospital [Kuusikko et al., 2008, 2009; Weiss et al., 2009], or an epidemiological study in the Northern Ostrobothnia Hospital District [Mattila et al., 2007, 2011] between the years 2000 and 2003. NT individuals were randomly selected from the epidemiological study [Mattila et al., 2007, 2011], or randomly selected [Kuusikko et al., 2008, 2009] or recruited [Jansson‐Verkasalo et al., 2005] from mainstream schools in Oulu.
During the original recruiting processes between the years 2000 and 2003, ASD diagnoses were determined by a trained clinical psychologist, a pediatrician and a child psychiatrist using the Autism Diagnostic Interview Revised (ADI‐R) [Lord, Rutter, & LeCouteur, 1999] and the Autism Diagnostic Observation Schedule (ADOS) Module 3 or Module 4 [Lord, Rutter, Dilavore, & Risi, 2008] to obtain structured information from parents and for semistructured observation of individuals with ASD as well as clinical information [Kuusikko et al., 2008, 2009; Mattila et al., 2007, 2011]. The ADI‐R and ADOS were not used to make diagnostic classifications, that is, the diagnostic algorithms were not used. The diagnoses were clinical best estimates made according to the International Classification of Diseases‐10th Revision criteria [World Health Organization, 1993]. For the present study, conducted between the years 2014 and 2015, participants completed the Autism‐Spectrum Quotient (AQ) [Baron‐Cohen, Wheelwright, Skinner, Martin, & Clubley, 2001] as an online questionnaire and the Wechsler Adult Intelligence‐IV (WAIS‐IV, producing a General Ability Index [GAI]) [Wechsler, 1955] in a live situation. Three of the ASD participants and one NT participant were on current psychotropic medication. Those three ASD participants used mood stabilizers and one of them had also methylphenidate and antipsychotic drugs, and that one NT participant used mood stabilizers.
MRI data were not available (no flip angle [FA] 5 MREG data) in 3 of the 20 NT subjects. Therefore, three additional NT individuals were selected for this study. These subjects did not have AQ and Wechsler Adult Intelligence‐IV test results available. All the available participant characteristics and test results are shown in Table 1. In addition, P‐values between groups (age, AQ, GAI) were calculated using two‐sided unpaired t‐tests. In general, individuals with ASD had deficits in social behaviors and often with language expression and reception. All individuals with ASD had an IQ in the normal range and all were fluent speakers. None of the participants had any problems with vision or hearing, or any motor problems.
Table 1.
Participant Characteristics
| Characteristics | ASD group | NT group | ASD vs. NT | ||||
|---|---|---|---|---|---|---|---|
| n | μ | σ | n | μ | σ | P‐value | |
| Age in years | 20 | 23.7 | 3.2 | 20 | 25.3 | 6.2 | 0.317 |
| AQ | 18 | 20.3 | 9.1 | 12 | 10.5 | 5.1 | 0.002 |
| GAI | 20 | 110.7 | 13.1 | 16 | 107.7 | 10.5 | 0.462 |
Abbreviations: μ, mean; σ, standard deviation; ASD, autism spectrum disorder; AQ, autism‐spectrum quotient; GAI, General Ability Index; n, number of participants.
The study was approved by the Regional Ethics Committee of the Northern Ostrobothnia Hospital District and was conducted in accordance with the Declaration of Helsinki (53/2012). Participants gave their written informed consent to participating in the study.
Measurements
All subjects were scanned using a Siemens MAGNETOM Skyra 3T MRI scanner (Siemens Healthcare GmbH, Germany) with a 32‐channel head coil. We utilized an MREG sequence obtained from Freiburg University. MREG is a single‐shot three‐dimensional sequence that utilizes spherical stack of spirals and undersamples 3D k‐space trajectory [Assländer et al., 2013; Lee, Zahneisen, Hugger, LeVan, & Hennig, 2013; Zahneisen et al., 2012]. We used the following sequence parameters: repetition time (TR) = 100 msec, echo time (TE) = 36 msec, field of view (FOV) = (192 mm)3, voxel size = (3 mm)3, and flip angle (FA) = 5°. MREG data were reconstructed by L2‐Tikhonov regularization with lambda = 0.1, with the latter regularization parameter determined by the L‐curve method [Hugger et al., 2011]; the resulting effective spatial resolution was 4.5 mm. MREG includes a dynamic off‐resonance in k‐space (DORK) method which corrects the respiration induced dynamic field‐map changes in fMRI using 3D single shot techniques [Zahneisen et al., 2014]. High‐resolution T1‐weighted Magnetization Prepared Rapid Acquisition with Gradient Echo (MPRAGE) (TR = 1900 msec, TE = 2.49 msec, inversion time (TI) = 900 msec, FA 9°, FOV = 240, and slice thickness 0.9 mm) images were obtained for coregistration of the MREG data into subject's own anatomy during the preprocessing. During the 5 min (2961 volumes) MREG resting state study, subjects were instructed to lie still in the scanner with their eyes open fixating a cross on the screen. Soft pads were fitted over the study subjects' ears to protect hearing and to minimize motion.
Preprocessing
MREG data were preprocessed with a Oxford Centre for Functional MRI of the Brain (FMRIB) software library (FSL) pipeline [Jenkinson, Beckmann, Behrens, Woolrich, & Smith, 2012] prior to single‐session ICAs. The data were high‐pass filtered with cut‐off frequency of 0.0025 Hz (400 sec) and 80 time points (8 sec) were removed from the beginning of the data to minimize T1‐relaxation effects. Motion correction was performed using FSL MCFLIRT [Jenkinson, Bannister, Brady, & Smith, 2002]. Brain extraction for 3D MPRAGE volumes was carried out with FSL Brain Extraction Tool (BET) using the following parameters; fractional intensity = 0.25, threshold gradient = 0.22 with neck and bias‐field correction option [Smith, 2002]. Spatial smoothing was carried out using 5 mm full width at half maximum (FWHM) Gaussian Kernel. MREG images were aligned to three‐dimensional (MPRAGE) anatomical images (full‐search, 12 degree of freedom (DOF)) and to Montreal Neurologic Institute (MNI 152) 4 mm3 standard space (full‐search, 12 DOF) as a preprocessing step in FSL Multivariate Exploratory Linear Optimized Decomposition into Indepent Components (MELODIC) tool. Finally, the advanced ICA FIX (FMRIBs ICA‐based X‐noisifier) method [Griffanti et al., 2014; Salimi‐Khorshidi et al., 2014] was used for secondary artifact removal from the preprocessed MREG (single‐session ICA) data. FIX was trained with previously collected control MREG data. The applied FIX threshold was 10. The same FIX method was used for each subject.
DLA Workflow
A group level spatial ICA (multisession temporal concatenation in FSL) was performed for the FIX‐cleaned data with a model order of 40 (z‐threshold = 2.3), separately for NT and ASD data. Next, cross‐correlations (cc) were calculated between every spatials ICs of NT and ASD group using FSL's fslcc function. Those RSNs whose spatial correlation between two groups exceeded the cc threshold of 0.60 were chosen for DLA. The identification of the selected RSNs was based on FSL model order 40 ICA component templates from a previous research article on RSN model order [Abou Elseoud et al., 2011].
The DLA procedure is based on the following steps (Fig. 1). (1) Pairs of RSNs were selected from the NT and ASD data. (2) As cardiorespiratory signal peaks will introduce artificial peak timings, the data were low‐pass filtered (0.01–0.1 Hz frequency range) and detrended. The time locations of the local BOLD signal maximum of each peak of the time signals were determined individually from the time concatenated melodic_mix in FSL. The timing of BOLD signal activation was located with findpeaks in Matlab. No peak thresholding was applied. (3) The time lags of all peaks of the signals are calculated between each pair of the selected RSNs, that is, for each peak, the time lag between the nearest peak of the other signal is selected. Each lag value ≤±5.0 sec is assembled into a lag vector, similarly for both NT and ASD data. (4a) The Kolmogorov–Smirnov (kstest2 in Matlab) test is calculated between the NT and ASD lag vectors to determine which RSN pairs have statistically significant differences in the lag patterns between NT and ASD groups. The output value is assembled into a P‐value matrix. (4b) The mean (median respectively) value of the lag vectors is assembled into lag matrices. The source/destination relationship is marked with negative/positive values in the matrices, that is, the blue hues indicate networks (labeled in the y‐axis) that are on average early, that is, “sources” of propagated BOLD activity and red hues indicate networks that are late, that is, “destinations” of propagated BOLD activity. All these steps (1–4) are done for each selected RSN pair separately to construct the final P‐value and mean lag matrices. Sections 4a and 4b are independent analysis phases.
Figure 1.
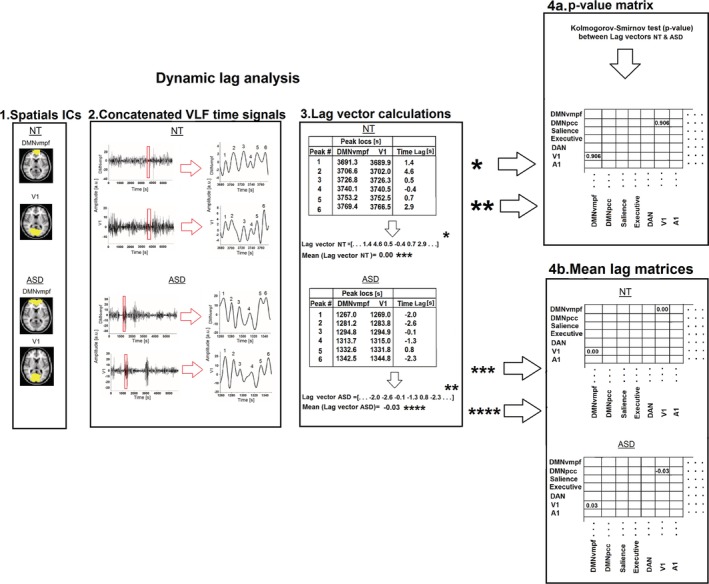
General workflow of dynamic lag analysis. (1) A given pair of resting‐state networks (RSNs) from the 16 RSNs is selected and (2) each peak of the time‐concatenated signals is determined with findpeaks function in MATLAB. (3) Next, the lag vector is formed by calculating the time lag values between each peak (between resting‐state networks) in the nearest neighbor principle (≤±5.0 sec lags). (4a) The Kolmogorov–Smirnov test is calculated between lag vectors of neurotypical and autism spectrum disorder to determine whether the lag variations are statistically different. (4b) The mean value of the lag vectors is calculated and assembled into a lag matrix. Steps 1–4 are applied to each selected resting‐state network pair separately to construct the final P‐value and mean lag matrices. Sections 4a and 4b are independent analysis phases.
Statistical Analysis
Surrogate data were created to evaluate the possibility of false positives in the P‐value matrix values. Data consisted of two groups, both including 16 surrogate time signals (59,220 samples, i.e., the length of concatenated 5 min signals of 20 subjects) created with “randi” function in Matlab. The same DLA workflow steps 1–4, as presented in Figure 1, were applied to surrogate data (two‐sided, two‐sample Kolmogorov Smirnov test). Benjamini–Hochberg false discovery rate (FDR) adjustment was used to correct for multiple comparisons. The smaller threshold, that is, either smallest P‐value in the P‐value matrix of surrogate data or Benjamin–Hochberg correction cut‐off, was selected as a P‐value threshold for significant network pairs.
Results
There were 16 RSNs whose spatial cross‐correlation exceeded the threshold of 0.60: precuneus, precuneus left (precuneus_left), posterior cingulate cortex default‐mode network (DMNpcc), ventromedial default‐mode network (DMNvmpf,) salience network, executive network, central executive network, dorsal attention network right (DAN_right), memory/attention, language network, language network right, primary visual cortex (V1), secondary visual cortex (V2), primary auditory cortex (A1), primary motor cortex left (M1_left), secondary somatosensory network (S2; Fig. 2).
Figure 2.
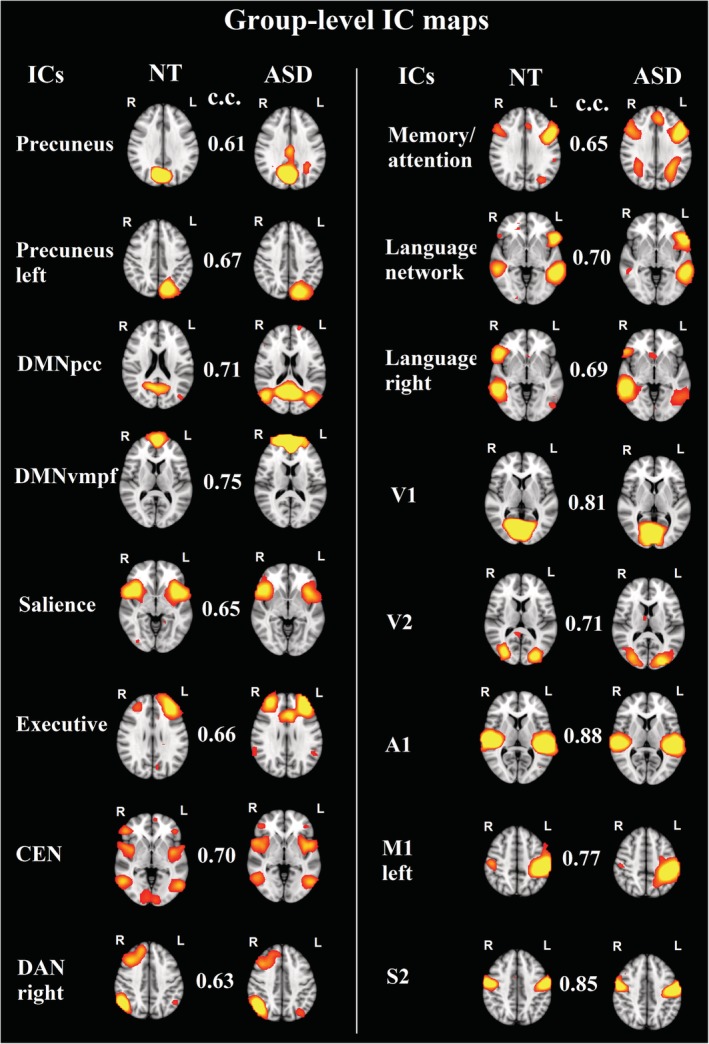
Group‐level IC maps for the 16 functional brain networks whose spatial cross‐correlation exceeded the threshold of 0.6 between neurotypical and autism spectrum disorder groups. Note: c.c., cross‐correlation; Z‐value in the IC‐maps is 2.3.
The smallest P‐value in the surrogate P‐value matrix was found to be 0.003, which was selected as a threshold to reject the possibility of false positives and to declare statistically highly significant network combinations (P‐value <0.003; Supplementary Fig. S1).This threshold was smaller than the P‐value calculated by the Benjamini–Hochberg procedure, that is, 0.007.
10.8% (13/120) of the RSN pairs had significant (P‐value <0.003) dynamic lag pattern that survived both Benjamini–Hochberg and surrogated network data corrections (Fig. 3). Altered lag pattern in ASD were concentrated especially in mutual connections of salience, executive, visual and default‐mode network pairs. Lag histograms of significant RSN pairs were formed for lag variability inspection (Fig. 4). In general, the lag pattern was more Gaussianic and skewed to the other side in NTs, referring to structured propagation patterns. In ASD, the time lag histograms were more symmetric or multimodal. Detailed lag characteristics of significant RSN pairs (%‐ratio, μ [mean lag value], ũ [median lag value], σ [standard deviation of lag values]) are shown in Table 2. Percentage of lags demonstrating preceding (−) versus lagging (+) values is reflected by a %‐ratio. In other words, the first value is the percentage of how many times the first mentioned network in the network pair has been the preceding network, that is, negative lag values, whereas the second value in the %‐ratio reflects the percentage of how many times it has been the lagging network, that is, positive lag values. In NT individuals, the ratio of preceding network (%‐ratio) in all 13/13 highly significant RSN pairs was observationally larger than 45/55, while in ASD the %‐ratio was in general close to a 50/50 ratio. Notably, the salience network was a source of activity to V1, V2, executive, precuneus, and DMNpcc networks in NT controls. In ASD, the source/relationship was nearly equivocal. Moreover, the results suggest that in ASD, the source/destination relationship is inversed in many RSN pairs compared with relationships observed in NTs. 92.3% (12/13 × 100%) and 84.6% (11/13 × 100%) of the significant RSN pairs revealed shorter mean and median temporal lags in ASD versus NT, respectively.
Figure 3.
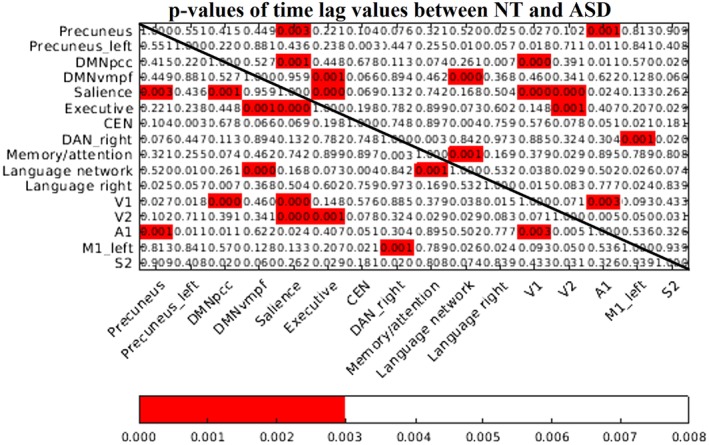
p‐value matrix between neurotypical (NT) and autism spectrum disorder groups. Red p‐values indicate that there are significant lag pattern variations (significance threshold of 0.003) between NT and autism spectrum disorder subjects that survived both Benjamini–Hochberg procedure and surrogate network data corrections.
Figure 4.
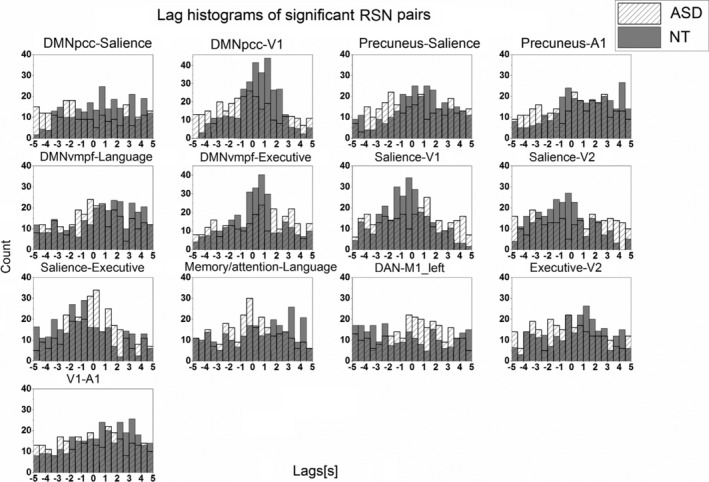
Lag histograms of 13 significant resting‐state network pairs. Neurotypical lag values are shown in dark gray bins, whereas autism spectrum disorder lag values are shown with bins filled with diagonal lines.
Table 2.
Detailed Lag Characteristics of Significant Resting‐State Network Pairs
| NT | ASD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % | μ |
|
σ | % | μ |
|
σ | |||
| DMNpcc‐Salience | 40/59 | 0.50 | 0.65 | 2.72 | 55/45 | −0.29 | −0.80 | 3.08 | ||
| DMNpcc‐V1 | 37/61 | 0.41 | 0.60 | 2.04 | 57/42 | −0.36 | −0.50 | 2.57 | ||
| Precuneus‐Salience | 38/60 | 0.56 | 0.60 | 2.47 | 48/51 | 0.18 | 0.15 | 2.70 | ||
| Precuneus‐A1 | 36/61 | 0.89 | 0.90 | 2.54 | 43/55 | 0.15 | 0.40 | 2.74 | ||
| DMNvmpf‐Language | 33/66 | 0.71 | 1.10 | 2.66 | 48/50 | 0.05 | 0.00 | 2.61 | ||
| DMNvmpf‐Executive | 41/57 | 0.23 | 0.30 | 2.12 | 41/57 | 0.33 | 0.70 | 2.71 | ||
| Salience‐V1 | 61/38 | −0.53 | −0.50 | 2.16 | 48/50 | −0.05 | 0.10 | 2.70 | ||
| Salience‐V2 | 61/38 | −0.47 | −0.60 | 2.36 | 48/50 | 0.02 | 0.15 | 2.90 | ||
| Salience‐Executive | 63/36 | −0.66 | −1.00 | 2.59 | 51/47 | −0.09 | −0.10 | 2.28 | ||
| Memory/attention‐Language | 43/56 | 0.34 | 0.80 | 2.81 | 53/45 | −0.15 | −0.20 | 2.55 | ||
| DAN_right‐M1_left | 58/41 | −0.50 | −0.70 | 3.05 | 45/53 | 0.10 | 0.25 | 2.59 | ||
| Executive‐V2 | 40/58 | 0.39 | 0.70 | 2.59 | 55/44 | −0.31 | −0.40 | 2.65 | ||
| V1‐A1 | 36/64 | 0.75 | 1.00 | 2.57 | 49/49 | −0.00 | 0.00 | 2.76 | ||
Note. %‐symbol reflects the percentage of lags demonstrating preceding (−) versus lagging (+) values, μ is the mean lag value, is the median lag value, and σ is the standard deviation of the lag values.
Supplementary Figure S2 shows mean lag value matrices between network pairs in NT (Supplementary Fig. S2A) and ASD groups (Supplementary Fig. S2B). In the lag matrices, the blue hues indicate networks (labeled in the y‐axis) that were on average early, that is, “sources” of propagated BOLD activity and red hues indicate networks that were late, that is, “destinations” of propagated BOLD activity. Although both groups had mean lag values ranging from −1 sec to 1 sec, NTs exhibited more structured lag pattern between RSN pairs, that is, RSN networks exhibited either preceding or lagging activity with respect to the other (cf. Table 2). Individuals with ASD had mean/median lag values close to zero, suggesting not so clear source/destination relationships between network pairs. Source/destination relationships seen in the real data were not present in the surrogate data (Supplementary Fig. S3).
Number of lag values (lags <5.0 sec) between each network were listed to explore whether the statistical significance could be explained by lag count variations between NT and ASD groups (Supplementary Fig. S4). The P‐value of the lag counts between NT and ASD groups was 0.94 (two‐sided Wilcoxon rank sum test), demonstrating that there were no significant variations in the lag counts, that is, in number of brain activation cycles between NT and ASD groups.
Finally, relative and absolute movement mean values were taken from FSL MCFLIRT to explore whether the lag differences between ASD and NT could be inferred by motion. However, no significant differences (absolute motion, P = 0.13; relative motion, P = 0.60) in motion values were seen between two groups (Fig. 5A). Furthermore, temporal signal‐to‐noise ratios (tSNR) were calculated before and after FIX denoising (Fig. 5B). No significant differences (tSNR before FIX, P = 0.11; tSNR after FIX, P = 0.10) in tSNR values were seen between two groups.
Figure 5.
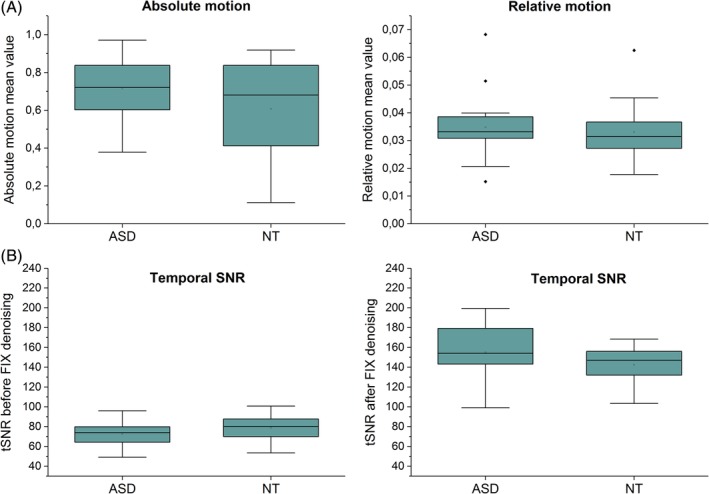
Absolute and relative movement mean values between autism spectrum disorder and controls in mm (A). Temporal signal‐to‐noise ratios before (left) and after (right) FIX denoising (B).
Discussion
Here we describe relationships between functional brain networks in ASD at a high level of temporal and spatial resolution. We used ultra‐fast MREG data (TR = 100 msec) and, a novel DLA method to reveal dynamic lag pattern variations between individuals with ASD and NT individuals. Sixteen out 40 RSNs examined showed adequate spatial similarity between ASD and NT groups. 10.8% of the possible 120 RSN pairs had significant dynamic lag pattern variations that survived both Benjamini–Hochberg FDR adjustment and surrogate network data corrections. 92.3% and 84.6% of the significant RSN pairs revealed shorter mean and median temporal lags in ASD versus NT, respectively. This is partly caused by the fact that NT individuals had more structured dynamic lag patterns, exhibiting a more distinct source/destination relationship of propagated BOLD activity between significantly connected networks. Spatially, the altered lag patterns were concentrated especially in mutual connections of salience, executive, visual, and default‐mode network nodes. Other highly significant lag pattern variations were seen in primary visual cortex (V1)‐primary auditory cortex (A1), primary auditory cortex‐precuneus, primary motor cortex (M1) left‐dorsal attention network (DAN) right and language network‐memory/attention RSN pairs.
Mitra and coworkers have recently introduced the concept of “one‐way streets” (or lag thread motifs), representing conserved regions of unidirectional propagation across distinct propagation sequences [Mitra, Snyder, Blazey, et al., 2015]. The lag thread motifs were shown to match the topographies of RSNs. Interestingly, they found that voxel‐wise propagation sequence correlation matrices also exhibit anticorrelations. These anticorrelations refer to “two‐way streets” demonstrating reciprocal bidirectional signaling between networks. Our results therefore suggest that this two‐way street activity could be more dominant in ASD. Moreover, in a conventional TR BOLD (2200 msec) study, Mitra and co‐workers [Mitra, Snyder, Constantino, et al., 2015] suggested that alterations in propagated intrinsic activity in ASD are more robust than alterations in static resting‐state functional connectivity. Group comparisons revealed focally altered lag differences in occipital cortex, frontopolar cortex and putamen, which strongly correlated with behavioral measures relevant to the diagnosis of ASD [Mitra, Snyder, Constantino, et al., 2015].
There are also other recent papers reporting variations in time‐varying patterns in autism. King et al. found increased durations of functional connections in both individual brain regions and distributed networks in autism, which were associated with metrics of disease severity [King et al., 2018]. They used a complementary method of analysis by introducing the width of cross‐correlation curves between resting‐state fMRI time series as a metric of relative duration of synchronous activity between brain regions (“sustained connectivity”) [King & Anderson, 2018]. The persistence of brain connectivity in autism may limit the ability to rapidly shift from one brain state to another [King et al., 2018]. Moreover, it has been also shown that there exists a negative correlation between processing speed and sustained connectivity [King & Anderson, 2018]. Processing speed is one aspect of cognition that is impaired in ASD compared with typical development [Haigh, Walsh, Mazefsky, Minshew, & Eack, 2018; Travers et al., 2014]. Interestingly, Watanabe and Rees studied brain network dynamics using an energy‐landscape analysis, and reported that high‐functioning adults with ASD show fewer neural transitions due to an unstable intermediate state [Watanabe & Rees, 2017]. Similar aberrant temporal dynamics were reported by the study by Rashid et al., who found longer dwell times related to a globally disconnected state in youth with higher autistic traits [Rashid et al., 2018]. The authors take these findings of impairment in processing speed and transitions between brain states in autism as an evidence that is in‐line with our results indicating diminished dynamic lag patterns between RSNs in individuals with ASD.
Instead of inspecting lag structure using lagged cross‐correlation (or covariance), between time series as in recent lag pattern analyses [Mitra, Snyder, Blazey, et al., 2015; Mitra, Snyder, Constantino, et al., 2015; Mitra et al., 2014; Raatikainen et al., 2017], we aimed to investigate dynamic lag pattern variations by taking into account each peak of the time signals. Lagged cross‐correlation using a sliding window approach can be used to explore lag dynamics [Raatikainen et al., 2017]. However, this approach requires that the minimum window length to avoid spurious fluctuations arising due to sliding window correlation itself should be at least equal to 1/f min, where f min is the minimum frequency in the simplified correlating signal [Leonardi & Van De Ville, 2015; Shakil, Keilholz, & Lee, 2015]. By using the frequency band of 0.01–0.1 Hz, the minimum window length should be 100 sec, which enables only few dynamic windows to be used in traditional 5–10 min BOLD studies. Therefore, as DLA approach determines lag separately for each peak, it offers more dynamic lag information compared with the lagged cross‐correlation approach that assumes the existence of a single temporal lag between brain regions over a wide time epoch.
The current results offer a new viewpoint based on dynamic lag pattern alterations in ASD. Our work suggests that some networks are early with respect to the rest of the brain while others are late, and this distinction is diminished between some functional brain networks in ASD. In other words, the dynamic lag structure between functional networks is less controlled in ASD, which can be observed in mutual connections between the salience network and other networks such as visual, executive and default‐mode networks. The salience network is thought to be involved in switching between the default mode network and central executive network [Goulden et al., 2014], and salience network dysfunction has been linked to autism [Uddin & Menon, 2009; Uddin et al., 2013; Uddin et al., 2014; Uddin, 2015]. The preliminary finding that atypical timing of salience network activity may be observed in autism is in line with these earlier empirical and theoretical accounts.
Strengths and Limitations
It has been shown that interregional lags are reproducibly present in resting‐state fMRI data and these time lags are not attributable to hemodynamic factors [Mitra et al., 2014]. However, a recent study by Yan et al., suggests that hemodynamic response function (HRF) parameters have been shown to vary in individuals with autism, specifically in the precuneus [Yan, Rangaprakash, & Deshpande, 2018]. Nevertheless, exact time‐to‐peak value differences in HRF still remain unclear [Yan et al., 2018]. Furthermore, like Yan et al. state, the number of methods capable of deconvolving resting state data is small. Therefore, a deeper understanding of potential HRF confounds to dynamic lag structure analysis would require complementary methods that take into account individual HRF response times, dynamically, for every peak of each RSN time signal in resting‐state time series. Although we strongly believe that potential HRF contribution to our between‐group findings is negligible, the potential confounding effects of HRF to lag structure analysis should be kept in mind in the future fMRI studies.
There has been a concern that head motion leads to systematic biases in functional connectivity analyses of fMRI data [Power, Barnes, Snyder, Schlaggar, & Petersen, 2012; Satterthwaite et al., 2012; Van Dijk, Sabuncu, & Buckner, 2012]. In this study, no significant differences in relative and absolute motion, and in tSNR values were seen between ASD and controls (Fig. 5.). Moreover, MREG sampled critically at 10 Hz enables accurate removal of cardiorespiratory signals by band‐pass filtering from data [Huotari et al., 2019]. This is an important step for the activation peak based lag pattern estimation, since it removes variance from the peak timings that are induced by cardiac and respiratory peaks from the data.
The modest sample size of the present study limits our power to detect differences between individuals with ASD and NT individuals. This could have been countered by collecting 10 min scans instead of 5 min scans, or by recruiting more subjects. However, it has been shown that drowsiness can affect dynamic BOLD‐correlations [Laumann et al., 2016] and that slow‐wave sleep alters intrinsic brain propagation [Mitra, Snyder, Tagliazucchi, et al., 2015]. Therefore, we wanted to ensure that the subjects stayed as vigilant as possible by keeping the resting‐state scanning time moderate and by checking the vigilance of the subjects verbally between each scan. Furthermore, MREG offers 3000 brain volumes for each 5 min scan, and this increases the power for statistical inferences in addition to permitting more accurate physiological signal analysis [Huotari et al., 2019]. We believe that the number of peaks (~230–330) in the analysis between each RSN pair in the low frequency band is enough for statistical lag variation analysis given the statistical power of MREG results that survived combined surrogate data and Benjamini–Hochberg procedure corrections. In addition, even though our study included individuals that were well matched on age and gender between NT and ASD groups, we cannot demonstrate the effects of gender and age on the lag structure with the current dataset. Furthermore, since only three of the ASD participants and one NT participant used psychotropic medication, we did not consider the effects of medication on our analyses, although it has been shown that psychostimulants can cause enhanced activation in bilateral inferior frontal cortex (IFC)/insula [Rubia et al., 2014].
It would be interesting to further investigate why there were only 16 RSNs with at least moderate (>0.6) spatial correlation between group ICA components of NT and ASD groups. One explanation could be altered structural brain network organization in autism, such as that suggested by Rudie and coworkers [Rudie et al., 2013]. Another potential explanation could be the utilization of temporally higher resolution data. In MREG (TR = 100 msec), our 5 min long data includes 2961 brain volumes compared with a traditional BOLD sequence with a TR that is typically between 2 sec and 3 sec (equivalent to 100–150 brain volumes). With ultra‐fast MREG, we have thus 20–30 times higher temporal resolution, which prevents aliasing of cardiac and respiratory signals. As the spatial ICA algorithm attempts to find spatially independent components with associated time courses, it is therefore likely that temporally more accurate data could further contribute to the minor variations of spatial topographies. It remains speculative if these minor spatial IC alterations could influence the lag results. Another approach would have been to use spatial regions of interests (ROIs) or ICA templates, or to do group ICA of the entire dataset rather than separately for each group. In these cases, however, factors like brain plasticity and individual differences in network organization could have interfered with the analysis. Also, the joined ICA analysis of both groups averages out the underlying group differences, reducing sensitivity to the deviant time lag structure.
These findings raise the question of whether brain dynamics should be studied in a more detailed manner, such as by utilizing every peak of the time signal in the analysis. One could argue whether each activation cycle of the network is of interest, or whether an approach such as thresholding of activity level might be preferable. An important outstanding question is how large activation lags should be interpreted; are they sharing mutual functional information with direct neuronal connectivity or are they influenced by joint information processing from several networks? How much are random intrinsic fluctuations of the neurovascular system also influencing the indirect BOLD signal? In this work, all positive signal peaks were used for between‐network comparison. In the future, we will explore lag variations between negative and positive signal peaks between temporally negatively correlated networks.
Conclusions
Significant dynamic lag pattern variations were found between individuals with ASD and NT individuals using novel fast fMRI analytics that can accurately be used for lag pattern analytics. We found that altered lag patterns were concentrated particularly in the mutual connections between salience, executive, visual, and default‐mode networks. NT individuals exhibited a more structured dynamic time lag pattern between network pairs compared with individuals with ASD. Understanding such propagation patterns will likely yield deeper insights into the role of spontaneous activity in brain function in health and disease.
Supporting information
Supplementary Fig. S1. P‐value matrix between NT and ASD groups. Small P‐values indicate that there are statistically significant lag pattern variations between NT and ASD subjects.
Supplementary Fig. S2. Lag value matrices of A) NT and B) ASD subjects. The blue hues indicate networks (labeled in the y‐axis) that are on average early, i.e. “sources” of propagated BOLD activity and red hues indicate networks that are late, i.e. “destinations” of propagated BOLD activity.
Supplementary Fig. S3. Lag value matrices of A) surrogate data 1 and B) surrogate data 2. The negative lag values indicate that the RSN labeled in the y‐axis is preceding.
Supplementary Fig. S4. Number of lag values between each RSN pair. The values above the bolded black line refer to the number of lag values in RSN pairs in ASD data, while the values below the bolded black line refer to values in NT data.
Acknowledgments
This study was financially supported by Academy of Finland Grant 275352 (MREG Analysis of Neuronal Avalanches), JAES grant, and MRC Oulu grant. Additional thanks to Instrumentarium Science Foundation, Finnish Epilepsy Association (FEA), Walter Ahlström Foundation, Finnish Foundation for Technology Promotion, and Tauno Tönning Foundation who supported this research by personal grant (V.R.). L.Q.U. is supported by the National Institute of Mental Health (R01MH107549).
References
- Abbott, A. E. , Nair, A. , Keown, C. L. , Datko, M. , Jahedi, A. , Fishman, I. , & Müller, R. (2016). Patterns of atypical functional connectivity and behavioral links in autism differ between default, salience, and executive networks. Cerebral Cortex, 26(10), 4034–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elseoud, A. , Littow, H. , Remes, J. , Starck, T. , Nikkinen, J. , Nissila, J. , … Kiviniemi, V. (2011). Group‐ICA model order highlights patterns of functional brain connectivity. Frontiers in Systems Neuroscience, 5, 37 10.3389/fnsys.2011.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . (2013). Diagnostic and statistical manual of mental disorders (DSM‐5). Philadelphia, PA: American Psychiatric Publishing. [Google Scholar]
- Assaf, M. , Jagannathan, K. , Calhoun, V. D. , Miller, L. , Stevens, M. C. , Sahl, R. , … Pearlson, G. D. (2010). Abnormal functional connectivity of default mode sub‐networks in autism spectrum disorder patients. NeuroImage, 53(1), 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assländer, J. , Zahneisen, B. , Hugger, T. , Reisert, M. , Lee, H. , LeVan, P. , & Hennig, J. (2013). Single shot whole brain imaging using spherical stack of spirals trajectories. NeuroImage, 73, 59–70. [DOI] [PubMed] [Google Scholar]
- Baron‐Cohen, S. , Wheelwright, S. , Skinner, R. , Martin, J. , & Clubley, E. (2001). The autism‐spectrum quotient (AQ): Evidence from asperger syndrome/high‐functioning autism, males and females, scientists and mathematicians. Journal of Autism and Developmental Disorders, 31(1), 5–17. [DOI] [PubMed] [Google Scholar]
- Beckmann, C. F. , DeLuca, M. , Devlin, J. T. , & Smith, S. M. (2005). Investigations into resting‐state connectivity using independent component analysis. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 360(1457), 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal, B. B. , Mennes, M. , Zuo, X. N. , Gohel, S. , Kelly, C. , Smith, S. M. , … Milham, M. P. (2010). Toward discovery science of human brain function. Proceedings of the National Academy of Sciences of the United States of America, 107(10), 4734–4739. 10.1073/pnas.0911855107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyacioglu, R. , Beckmann, C. , & Barth, M. (2013). An investigation of RSN frequency spectra using ultra‐fast generalized inverse imaging (GIN). Frontiers in Human Neuroscience, 7, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. , & Glover, G. H. (2010). Time–frequency dynamics of resting‐state brain connectivity measured with fMRI. NeuroImage, 50(1), 81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, D. , & Nandy, R. R. (2006). Estimation of the intrinsic dimensionality of fMRI data. NeuroImage, 29(1), 145–154. [DOI] [PubMed] [Google Scholar]
- Di Martino, A. , Zuo, X. , Kelly, C. , Grzadzinski, R. , Mennes, M. , Schvarcz, A. , … Milham, M. P. (2013). Shared and distinct intrinsic functional network centrality in autism and attention‐deficit/hyperactivity disorder. Biological Psychiatry, 74(8), 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisch, S. J. , Gallese, V. , Willems, R. M. , Mantini, D. , Groen, W. B. , Romani, G. L. , … Bekkering, H. (2011). Altered intrinsic functional connectivity of anterior and posterior insula regions in high‐functioning participants with autism spectrum disorder. Human Brain Mapping, 32(7), 1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulden, N. , Khusnulina, A. , Davis, N. J. , Bracewell, R. M. , Bokde, A. L. , McNulty, J. P. , & Mullins, P. G. (2014). The salience network is responsible for switching between the default mode network and the central executive network: Replication from DCM. NeuroImage, 99, 180–190. [DOI] [PubMed] [Google Scholar]
- Griffanti, L. , Salimi‐Khorshidi, G. , Beckmann, C. F. , Auerbach, E. J. , Douaud, G. , Sexton, C. E. , … Mackay, C. E. (2014). ICA‐based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. NeuroImage, 95, 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh, S. M. , Walsh, J. A. , Mazefsky, C. A. , Minshew, N. J. , & Eack, S. M. (2018). Processing speed is impaired in adults with autism spectrum disorder, and relates to social communication abilities. Journal of Autism and Developmental Disorders, 48(8), 2653–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugger, T. , Zahneisen, B. , LeVan, P. , Lee, K. J. , Lee, H. , Zaitsev, M. , & Hennig, J. (2011). Fast undersampled functional magnetic resonance imaging using nonlinear regularized parallel image reconstruction. PLoS One, 6(12), e28822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari, N. , Raitamaa, L. , Helakari, H. , Kananen, J. , Raatikainen, V. , Rasila, A. , … Kiviniemi, V. J. (2019). Sampling rate effects on resting state fMRI metrics. Frontiers in Neuroscience, 13, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, R. M. , Womelsdorf, T. , Gati, J. S. , Everling, S. , & Menon, R. S. (2013). Resting‐state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Human Brain Mapping, 34(9), 2154–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson‐Verkasalo, E. , Kujala, T. , Jussila, K. , Mattila, M. , Moilanen, I. , Näätänen, R. , … Korpilahti, P. (2005). Similarities in the phenotype of the auditory neural substrate in children with asperger syndrome and their parents. European Journal of Neuroscience, 22(4), 986–990. [DOI] [PubMed] [Google Scholar]
- Jenkinson, M. , Bannister, P. , Brady, M. , & Smith, S. (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage, 17(2), 825–841. [DOI] [PubMed] [Google Scholar]
- Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. , Woolrich, M. W. , & Smith, S. M. (2012). FSL. NeuroImage, 62(2), 782–790. [DOI] [PubMed] [Google Scholar]
- Just, M. A. , Keller, T. A. , Malave, V. L. , Kana, R. K. , & Varma, S. (2012). Autism as a neural systems disorder: A theory of frontal‐posterior underconnectivity. Neuroscience & Biobehavioral Reviews, 36(4), 1292–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown, C. L. , Datko, M. C. , Chen, C. P. , Maximo, J. O. , Jahedi, A. , & Müller, R. (2017). Network organization is globally atypical in autism: A graph theory study of intrinsic functional connectivity. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 2(1), 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. , Gramfort, A. , Shetty, N. R. , Kitzbichler, M. G. , Ganesan, S. , Moran, J. M. , … Kenet, T. (2013). Local and long‐range functional connectivity is reduced in concert in autism spectrum disorders. Proceedings of the National Academy of Sciences of the United States of America, 110(8), 3107–3112. 10.1073/pnas.1214533110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J. B. , & Anderson, J. S. (2018). Sustained versus instantaneous connectivity differentiates cognitive functions of processing speed and episodic memory. Human Brain Mapping, 39(12), 4949–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J. B. , Prigge, M. B. , King, C. K. , Morgan, J. , Dean, D. C. , Freeman, A. , … Alexander, A. L. (2018). Evaluation of differences in temporal synchrony between brain regions in individuals with autism and typical development. JAMA Network Open, 1(7), e184777–e184777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi, V. , Kantola, J. , Jauhiainen, J. , Hyvärinen, A. , & Tervonen, O. (2003). Independent component analysis of nondeterministic fMRI signal sources. NeuroImage, 19(2), 253–260. [DOI] [PubMed] [Google Scholar]
- Kiviniemi, V. , Vire, T. , Remes, J. , Elseoud, A. A. , Starck, T. , Tervonen, O. , & Nikkinen, J. (2011). A sliding time‐window ICA reveals spatial variability of the default mode network in time. Brain Connectivity, 1(4), 339–347. [DOI] [PubMed] [Google Scholar]
- Kuusikko, S. , Haapsamo, H. , Jansson‐Verkasalo, E. , Hurtig, T. , Mattila, M. , Ebeling, H. , … Moilanen, I. (2009). Emotion recognition in children and adolescents with autism spectrum disorders. Journal of Autism and Developmental Disorders, 39(6), 938–945. [DOI] [PubMed] [Google Scholar]
- Kuusikko, S. , Pollock‐Wurman, R. , Jussila, K. , Carter, A. S. , Mattila, M. , Ebeling, H. , … Moilanen, I. (2008). Social anxiety in high‐functioning children and adolescents with autism and asperger syndrome. Journal of Autism and Developmental Disorders, 38(9), 1697–1709. [DOI] [PubMed] [Google Scholar]
- Laumann, T. O. , Snyder, A. Z. , Mitra, A. , Gordon, E. M. , Gratton, C. , Adeyemo, B. , … Petersen, S. E. (2016). On the stability of BOLD fMRI correlations. Cerebral Cortex, 27(10), 4719–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Zahneisen, B. , Hugger, T. , LeVan, P. , & Hennig, J. (2013). Tracking dynamic resting‐state networks at higher frequencies using MR‐encephalography. NeuroImage, 65, 216–222. [DOI] [PubMed] [Google Scholar]
- Leonardi, N. , & Van De Ville, D. (2015). On spurious and real fluctuations of dynamic functional connectivity during rest. NeuroImage, 104, 430–436. [DOI] [PubMed] [Google Scholar]
- Lin, F. , Ahveninen, J. , Raij, T. , Witzel, T. , Chu, Y. , Jääskeläinen, I. P. , … Belliveau, J. W. (2014). Increasing fMRI sampling rate improves granger causality estimates. PLoS One, 9(6), e100319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. , Polimeni, J. R. , Lin, J. L. , Tsai, K. W. , Chu, Y. , Wu, P. , … Kuo, W. (2018). Relative latency and temporal variability of hemodynamic responses at the human primary visual cortex. NeuroImage, 164, 194–201. [DOI] [PubMed] [Google Scholar]
- Lin, F. , Witzel, T. , Chang, W. , Tsai, K. W. , Wang, Y. , Kuo, W. , & Belliveau, J. W. (2010). K‐space reconstruction of magnetic resonance inverse imaging (K‐InI) of human visuomotor systems. NeuroImage, 49(4), 3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , & Duyn, J. H. (2013). Time‐varying functional network information extracted from brief instances of spontaneous brain activity. Proceedings of the National Academy of Sciences, 110(11), 4392–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C. , Rutter, M. , Dilavore, P. C. , & Risi, S. (2008). ADOS: Autism diagnostic observation schedule. Boston, MA: Hogrefe. [Google Scholar]
- Lord, C. , Rutter, M. , & LeCouteur, A. (1999). Autism diagnostic interview‐revised: ADI‐R short edition. Developmental Disorders Clinic.
- Ma, Y. , Shaik, M. A. , Kozberg, M. G. , Kim, S. H. , Portes, J. P. , Timerman, D. , & Hillman, E. M. (2016). Resting‐state hemodynamics are spatiotemporally coupled to synchronized and symmetric neural activity in excitatory neurons. Proceedings of the National Academy of Sciences of the United States of America, 113(52), E8463–E8471. 10.1073/pnas.1525369113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed, W. , Magnuson, M. , Hasenkamp, W. , Schwarb, H. , Schumacher, E. H. , Barsalou, L. , & Keilholz, S. D. (2011). Spatiotemporal dynamics of low frequency BOLD fluctuations in rats and humans. NeuroImage, 54(2), 1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed, W. , Magnuson, M. , & Keilholz, S. D. (2009). Spatiotemporal dynamics of low frequency fluctuations in BOLD fMRI of the rat. Journal of Magnetic Resonance Imaging, 30(2), 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila, M. , Kielinen, M. , Jussila, K. , Linna, S. , Bloigu, R. , Ebeling, H. , & Moilanen, I. (2007). An epidemiological and diagnostic study of asperger syndrome according to four sets of diagnostic criteria. Journal of the American Academy of Child & Adolescent Psychiatry, 46(5), 636–646. [DOI] [PubMed] [Google Scholar]
- Mattila, M. , Kielinen, M. , Linna, S. , Jussila, K. , Ebeling, H. , Bloigu, R. , … Moilanen, I. (2011). Autism spectrum disorders according to DSM‐IV‐TR and comparison with DSM‐5 draft criteria: An epidemiological study. Journal of the American Academy of Child & Adolescent Psychiatry, 50(6), 583.e11–592.e11. [DOI] [PubMed] [Google Scholar]
- Mitra, A. , Snyder, A. Z. , Blazey, T. , & Raichle, M. E. (2015). Lag threads organize the brain's intrinsic activity. Proceedings of the National Academy of Sciences, 112(17), E2235–E2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. , Snyder, A. Z. , Constantino, J. N. , & Raichle, M. E. (2015). The lag structure of intrinsic activity is focally altered in high functioning adults with autism. Cerebral Cortex, 27(2), 1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. , Snyder, A. Z. , Hacker, C. D. , Pahwa, M. , Tagliazucchi, E. , Laufs, H. , … Raichle, M. E. (2016). Human cortical–hippocampal dialogue in wake and slow‐wave sleep. Proceedings of the National Academy of Sciences, 113(44), E6868–E6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. , Snyder, A. Z. , Hacker, C. D. , & Raichle, M. E. (2014). Lag structure in resting‐state fMRI. Journal of Neurophysiology, 111(11), 2374–2391. 10.1152/jn.00804.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. , Snyder, A. Z. , Tagliazucchi, E. , Laufs, H. , & Raichle, M. E. (2015). Propagated infra‐slow intrinsic brain activity reorganizes across wake and slow wave sleep. eLife, 4, 1–19. 10.7554/eLife.10781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi, J. S. , & Uddin, L. Q. (2015). Developmental changes in large‐scale network connectivity in autism. NeuroImage: Clinical, 7, 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posse, S. , Ackley, E. , Mutihac, R. , Zhang, T. , Hummatov, R. , Akhtari, M. , … Yonas, H. (2013). High‐speed real‐time resting‐state fMRI using multi‐slab echo‐volumar imaging. Frontiers in Human Neuroscience, 7, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Barnes, K. A. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage, 59(3), 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raatikainen, V. , Huotari, N. , Korhonen, V. , Rasila, A. , Kananen, J. , Raitamaa, L. , … Kiviniemi, V. (2017). Combined spatiotemporal ICA (stICA) for continuous and dynamic lag structure analysis of MREG data. NeuroImage, 1(148), 352–363. [DOI] [PubMed] [Google Scholar]
- Raitamaa, L. , Korhonen, V. , Huotari, N. , Raatikainen, V. , Hautaniemi, T. , Kananen, J. , … Myllylä, T. (2018). Breath hold effect on cardiovascular brain pulsations—A multimodal magnetic resonance encephalography study. Journal of Cerebral Blood Flow & Metabolism, 0271678X18798441, 1–15. 10.1177/0271678X18798441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajna, Z. , Kananen, J. , Keskinarkaus, A. , Seppänen, T. , & Kiviniemi, V. (2015). Detection of short‐term activity avalanches in human brain default mode network with ultrafast MR encephalography. Frontiers in Human Neuroscience, 9, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid, B. , Blanken, L. M. , Muetzel, R. L. , Miller, R. , Damaraju, E. , Arbabshirani, M. R. , … Jaddoe, V. W. (2018). Connectivity dynamics in typical development and its relationship to autistic traits and autism spectrum disorder. Human Brain Mapping, 39(8), 3127–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubia, K. , Alegria, A. A. , Cubillo, A. I. , Smith, A. B. , Brammer, M. J. , & Radua, J. (2014). Effects of stimulants on brain function in attention‐deficit/hyperactivity disorder: A systematic review and meta‐analysis. Biological Psychiatry, 76(8), 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudie, J. D. , Brown, J. , Beck‐Pancer, D. , Hernandez, L. , Dennis, E. , Thompson, P. , … Dapretto, M. (2013). Altered functional and structural brain network organization in autism. NeuroImage: Clinical, 2, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudie, J. D. , Shehzad, Z. , Hernandez, L. M. , Colich, N. L. , Bookheimer, S. Y. , Iacoboni, M. , & Dapretto, M. (2012). Reduced functional integration and segregation of distributed neural systems underlying social and emotional information processing in autism spectrum disorders. Cerebral Cortex, 22(5), 1025–1037. 10.1093/cercor/bhr171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi‐Khorshidi, G. , Douaud, G. , Beckmann, C. F. , Glasser, M. F. , Griffanti, L. , & Smith, S. M. (2014). Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. NeuroImage, 90, 449–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite, T. D. , Wolf, D. H. , Loughead, J. , Ruparel, K. , Elliott, M. A. , Hakonarson, H. , … Gur, R. E. (2012). Impact of in‐scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. NeuroImage, 60(1), 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil, S. , Keilholz, S. D. , & Lee, C. (2015). On frequency dependencies of sliding window correlation. Paper presented at the Bioinformatics and Biomedicine (BIBM), 2015 IEEE International Conference On, 363–368.
- Smith, S. M. (2002). Fast robust automated brain extraction. Human Brain Mapping, 17(3), 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Miller, K. L. , Moeller, S. , Xu, J. , Auerbach, E. J. , Woolrich, M. W. , … Ugurbil, K. (2012). Temporally‐independent functional modes of spontaneous brain activity. Proceedings of the National Academy of Sciences of the United States of America, 109(8), 3131–3136. 10.1073/pnas.1121329109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck, T. , Nikkinen, J. , Rahko, J. , Remes, J. , Hurtig, T. , Haapsamo, H. , … Jansson‐Verkasalo, E. (2013). Resting state fMRI reveals a default mode dissociation between retrosplenial and medial prefrontal subnetworks in ASD despite motion scrubbing. Frontiers in Human Neuroscience, 7, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, B. G. , Bigler, E. D. , Tromp, D. P. , Adluru, N. , Froehlich, A. L. , Ennis, C. , … Alexander, A. L. (2014). Longitudinal processing speed impairments in males with autism and the effects of white matter microstructure. Neuropsychologia, 53, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyszka, J. M. , Kennedy, D. P. , Paul, L. K. , & Adolphs, R. (2013). Largely typical patterns of resting‐state functional connectivity in high‐functioning adults with autism. Cerebral Cortex, 24(7), 1894–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin, L. Q. (2015). Salience processing and insular cortical function and dysfunction. Nature Reviews Neuroscience, 16(1), 55–61. [DOI] [PubMed] [Google Scholar]
- Uddin, L. Q. , & Menon, V. (2009). The anterior insula in autism: Under‐connected and under‐examined. Neuroscience & Biobehavioral Reviews, 33(8), 1198–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin, L. Q. , Supekar, K. , Lynch, C. J. , Cheng, K. M. , Odriozola, P. , Barth, M. E. , … Menon, V. (2014). Brain state differentiation and behavioral inflexibility in autism. Cerebral Cortex, 25(12), 4740–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin, L. Q. , Supekar, K. , Lynch, C. J. , Khouzam, A. , Phillips, J. , Feinstein, C. , … Menon, V. (2013). Salience network–based classification and prediction of symptom severity in children with autism. JAMA Psychiatry, 70(8), 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin, L. Q. , Supekar, K. , & Menon, V. (2013). Reconceptualizing functional brain connectivity in autism from a developmental perspective. Frontiers in Human Neuroscience, 7, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk, K. R. , Sabuncu, M. R. , & Buckner, R. L. (2012). The influence of head motion on intrinsic functional connectivity MRI. NeuroImage, 59(1), 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von dem Hagen, E. A. , Stoyanova, R. S. , Baron‐Cohen, S. , & Calder, A. J. (2013). Reduced functional connectivity within and between ‘social’ resting state networks in autism spectrum conditions. Social Cognitive and Affective Neuroscience, 8(6), 694–701. 10.1093/scan/nss053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T. , & Rees, G. (2017). Brain network dynamics in high‐functioning individuals with autism. Nature Communications, 8, 16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler, D. (1955). Manual for the wechsler adult intelligence scale. New York, NY: Psychological Corporation. [Google Scholar]
- Weiss, L. A. , Arking, D. E. , Daly, M. J. , Chakravarti, A. , Brune, C. W. , West, K. , … West, A. B. (2009). A genome‐wide linkage and association scan reveals novel loci for autism. Nature, 461(7265), 802–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, S. , Wiggins, J. L. , Peltier, S. J. , Carrasco, M. , Risi, S. , Lord, C. , & Monk, C. S. (2010). Alterations of resting state functional connectivity in the default network in adolescents with autism spectrum disorders. Brain Research, 1313, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (1993). The ICD‐10 classification of mental and behavioural disorders: Diagnostic criteria for research.
- Yan, W. , Rangaprakash, D. , & Deshpande, G. (2018). Aberrant hemodynamic responses in autism: Implications for resting state fMRI functional connectivity studies. NeuroImage: Clinical, 19, 320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahneisen, B. , Assländer, J. , LeVan, P. , Hugger, T. , Reisert, M. , Ernst, T. , & Hennig, J. (2014). Quantification and correction of respiration induced dynamic field map changes in fMRI using 3D single shot techniques. Magnetic Resonance in Medicine, 71(3), 1093–1102. [DOI] [PubMed] [Google Scholar]
- Zahneisen, B. , Hugger, T. , Lee, K. J. , LeVan, P. , Reisert, M. , Lee, H. , … Hennig, J. (2012). Single shot concentric shells trajectories for ultra fast fMRI. Magnetic Resonance in Medicine, 68(2), 484–494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. P‐value matrix between NT and ASD groups. Small P‐values indicate that there are statistically significant lag pattern variations between NT and ASD subjects.
Supplementary Fig. S2. Lag value matrices of A) NT and B) ASD subjects. The blue hues indicate networks (labeled in the y‐axis) that are on average early, i.e. “sources” of propagated BOLD activity and red hues indicate networks that are late, i.e. “destinations” of propagated BOLD activity.
Supplementary Fig. S3. Lag value matrices of A) surrogate data 1 and B) surrogate data 2. The negative lag values indicate that the RSN labeled in the y‐axis is preceding.
Supplementary Fig. S4. Number of lag values between each RSN pair. The values above the bolded black line refer to the number of lag values in RSN pairs in ASD data, while the values below the bolded black line refer to values in NT data.


