Summary
Inhibition of hypocotyl growth is a well‐established UV‐B‐induced photomorphogenic response that is mediated by the UV‐B photoreceptor UV RESISTANCE LOCUS 8 (UVR8). However, the molecular mechanism by which UVR8 signaling triggers inhibition of hypocotyl growth is poorly understood. The bZIP protein ELONGATED HYPOCOTYL 5 (HY5) functions as the main positive regulatory transcription factor in the UVR8 signaling pathway, with HY5‐HOMOLOG (HYH) playing a minor role. However, here we demonstrate that hy5 hyh double mutants maintain significant UVR8‐dependent hypocotyl growth inhibition. We identify UVR8‐dependent inhibition of the activities of bHLH transcription factors PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5 as part of the UVR8 signaling pathway, which results in inhibition of hypocotyl growth. The UVR8‐mediated repression of several hypocotyl elongation‐related genes is independent of HY5 and HYH but largely associated with UVR8‐dependent degradation of PIF4 and PIF5, a process that consequently diminishes PIF4/5 target promoter occupancy. Taken together, our data indicate that UVR8‐mediated inhibition of hypocotyl growth involves degradation of PIF4 and PIF5. These findings contribute to our mechanistic understanding of UVR8‐induced photomorphogenesis and further support the function of PIFs as integrators of different photoreceptor signaling pathways.
Keywords: UVR8, COP1, PIF4, PIF5, HY5, photomorphogenesis, signal transduction, ultraviolet‐B, Arabidopsis thaliana
Significance Statement
This work describes a role for degradation of PIF4 and PIF5 in the core UVR8 signaling pathway associated with UV‐B‐induced gene repression and inhibition of hypocotyl growth.
Introduction
Plants are constantly exposed to a variety of environmental cues. A high degree of plasticity has evolved in plants, allowing them to integrate different signals to optimize their growth and development. Light is arguably one of the most fundamental environmental signals for plants. Light perception occurs through specific photoreceptors for red/far‐red, blue and UV‐B parts of the electromagnetic spectrum, with light commonly regulating physiological responses such as hypocotyl elongation (de‐etiolation) – a prime example of plant plasticity (Favory et al., 2009; Tilbrook et al., 2013; Fiorucci and Fankhauser, 2017; Jenkins, 2017; Gommers and Monte, 2018).
Hypocotyl elongation is a well‐established model system used to study how changes in the quality, quantity and direction of light can affect plant morphogenesis, otherwise referred to as plant photomorphogenesis (Fankhauser and Casal, 2004; Vandenbussche et al., 2005; Kretsch, 2010; Boron and Vissenberg, 2014). A wide range of mutants that exhibit defects in light‐regulated hypocotyl elongation, often compromised in downstream hormonal signaling (predominantly auxin) and cell expansion, are widely used to understand the molecular link between photoreceptor signaling and physiological adaptations of young seedlings (Kami et al., 2010). At the molecular level, this process is tightly regulated by the interplay of mainly three transcription factor families: ETHYLENE‐INSENSITIVE 3/EIN3‐LIKE 1 (EIN3/EIL1), PHYTOCHROME INTERACTING FACTORS (PIFs) and ELONGATED HYPOCOTYL 5 (HY5) (Shi et al., 2018). The latter protein is a key positive regulator of photomorphogenesis and functions antagonistically to the other two transcription factor families (Osterlund et al., 2000; Shi et al., 2018). The PIFs and EIN3/EIL1 are repressors of light responses and hence promote hypocotyl elongation (Lorrain et al., 2008; Zhong et al., 2009; Shi et al., 2018). Whereas HY5 accumulates in light to maintain photomorphogenesis, PIF proteins accumulate in the dark to maintain skotomorphogenesis (Osterlund et al., 2000; Leivar et al., 2008). In agreement with this, hy5 mutants maintain an elongated hypocotyl in the light (Koornneef et al., 1980; Oyama et al., 1997) whereas combinatorial pif mutants exhibit a short hypocotyl and open cotyledons in the dark (Leivar et al., 2008). In particular, PIF4 and PIF5 have a key role in elongation growth associated with shade avoidance responses (low red/far‐red ratio), directly regulating the expression of several elongation‐associated genes (Lorrain et al., 2008; Hornitschek et al., 2009; Lorrain et al., 2009; Hornitschek et al., 2012; Pfeiffer et al., 2014; Pacin et al., 2016; de Wit et al., 2016b).
Ultraviolet‐B is an important part of sunlight that influences plants and also serves as a regulatory signal for plant photomorphogenesis through the action of the UV RESISTANCE LOCUS 8 (UVR8) photoreceptor and its signaling pathway (Rizzini et al., 2011; Jenkins, 2017; Yin and Ulm, 2017; Demarsy et al., 2018; Liang et al., 2019). The UVR8‐mediated photomorphogenic responses include inhibition of hypocotyl elongation and accumulation of ‘sunscreen’ pigments (Kliebenstein et al., 2002; Favory et al., 2009; Stracke et al., 2010). The bZIP transcription factor HY5 plays a well‐established role in UVR8 signaling, contributing to both inhibition of hypocotyl growth and the regulation of changes in gene expression in response to UV‐B (Ulm et al., 2004; Brown et al., 2005; Oravecz et al., 2006; Brown and Jenkins, 2008; Stracke et al., 2010; Huang et al., 2012). The UV‐B‐mediated interaction of UVR8 and the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) induces stability of the HY5 protein and HY5 transcription, resulting in enrichment of HY5 at the promoters of UV‐B‐responsive genes (Favory et al., 2009; Huang et al., 2013; Binkert et al., 2014; Yin et al., 2015). The UVR8‐mediated inhibition of hypocotyl elongation is a UV‐B response commonly used to identify and characterize positive and negative regulators of UV‐B perception and signaling (Oravecz et al., 2006; Favory et al., 2009; Gruber et al., 2010; Heijde et al., 2013; Huang et al., 2013; Huang et al., 2014; Liang et al., 2018; Yang et al., 2018; Ren et al., 2019; Yadav et al., 2019). Despite this broad use as a UVR8 signaling readout, the mechanisms by which UVR8 triggers inhibition of hypocotyl growth have only recently begun to be elucidated. One mechanism relies on UVR8‐mediated induction of HY5 and HY5 stabilization (Brown et al., 2005; Oravecz et al., 2006), which results from inhibition of COP1 activity upon its interaction with UVR8, causing stabilization of HY5 and its binding to its own promoter (Binkert et al., 2014; Podolec and Ulm, 2018). Indeed, HY5, together with HYH, is required for UVR8‐induced activation of HY5 promoter activity (Binkert et al., 2014). Recently it was shown that WRKY DNA‐BINDING PROTEIN 36 (WRKY36) binds to the W‐box in the HY5 promoter and represses its transcription (Yang et al., 2018). In response to UV‐B, UVR8 interacts with WRKY36, thus inhibiting WRKY36 DNA‐binding and promoting expression of HY5 (Yang et al., 2018). Moreover, it was recently shown that UVR8 interferes with brassinosteroid (BR) signaling and inhibits BR‐related elongation by direct interaction with the transcription factors BRI1‐EMS‐SUPPRESSOR1 (BES1) and BES1‐INTERACTING MYC‐LIKE 1 (BIM1) and inhibiting their DNA‐binding (Liang et al., 2018).
It has recently been shown that UV‐B negatively regulates shade‐activated PIF4 and PIF5 to counteract shade avoidance responses and thermomorphogenesis, which are both associated with enhanced hypocotyl elongation (Hayes et al., 2014; Hayes et al., 2017). However, the role and regulation of PIF4 and PIF5 under conditions commonly used to study UVR8 signaling and UVR8‐mediated hypocotyl growth inhibition have not yet been reported. Here we show that UVR8‐mediated inhibition of PIF4 and PIF5 is indeed an important and intrinsic part of UVR8 signaling, which results in UV‐B‐mediated gene repression and inhibition of hypocotyl elongation.
Results
Mediation of hypocotyl growth inhibition by UVR8 is partially independent of HY5 and HYH
In order to investigate the role of HY5 and HYH in UV‐B‐mediated inhibition of hypocotyl elongation, we measured the hypocotyl length of 4‐day‐old wild‐type (WT), uvr8, hy5, hyh, hy5 hyh and uvr8 hy5 hyh seedlings grown under white light (WL) or WL supplemented with UV‐B. As described before (Favory et al., 2009), we observed UV‐B‐mediated growth inhibition in WT seedlings which was absent in uvr8 mutants (Figure 1a,b and Figure S1 in the online Supporting Information). hyh showed a similar response to the WT, whereas hy5 displayed longer hypocotyls even in the absence of UV‐B, as expected from its additional roles in cryptochrome and phytochrome signaling (Oyama et al., 1997; Osterlund et al., 2000), and showed a reduced growth inhibition response to UV‐B (Figure S1) (Oravecz et al., 2006; Huang et al., 2012). Interestingly, although affected, the hy5 hyh double mutant still showed obvious inhibition of hypocotyl growth under UV‐B (Figures 1a,b and S1), supporting the existence of HY5/HYH‐independent mechanism(s) that regulate hypocotyl elongation under UV‐B. Importantly, the UV‐B‐dependent hypocotyl growth inhibition apparent in hy5 hyh was absent in uvr8 hy5 hyh triple mutants (Figure 1a,b), demonstrating that the HY5/HYH‐independent signaling pathway is downstream of the UVR8 photoreceptor and the UV‐B phenotype is not due to an elevated UV‐B stress response associated with the combined impairment of phytochrome, cryptochrome and UVR8 signaling in hy5 hyh double mutants.
Figure 1.
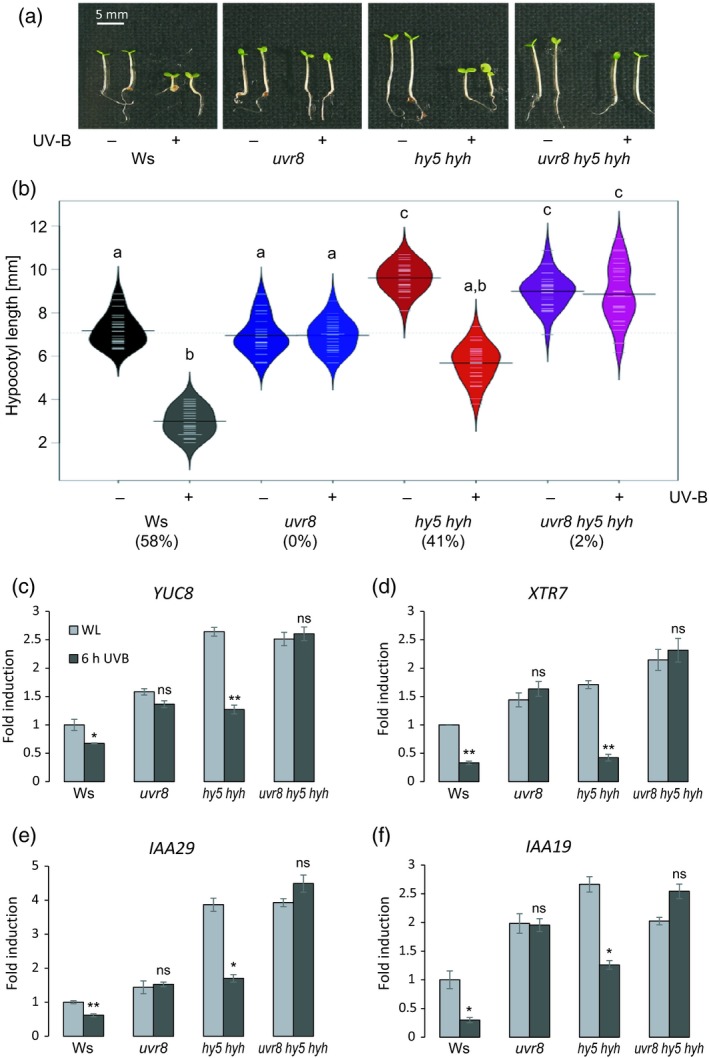
UVR8‐mediated inhibition of hypocotyl growth is partially independent on HY5 and HYH.
(a) Representative image showing the hypocotyl growth phenotype of 4‐day‐old wild‐type (Ws), uvr8‐7, hy5‐ks50 hyh‐1 and uvr8‐7 hy5‐ks50 hyh‐1 seedlings grown under white light (−UV‐B) or white light supplemented with UV‐B (+UV‐B). (b) Quantification of hypocotyl length. Beanplots represent data for n > 40 seedlings. Shared letters indicate no statistically significant difference in the means (P > 0.05). Percentages on the x‐axis indicate the relative hypocotyl growth inhibition induced by UV‐B. (c)–(f) Quantitative real‐time PCR analysis of (c) YUC8, (d) XTR7, (e) IAA29 and (f) IAA19 expression in 4‐day‐old wild‐type (Ws), uvr8‐7, hy5‐ks50 hyh‐1 and uvr8‐7 hy5‐ks50 hyh‐1 seedlings grown under white light and either exposed to narrowband UV‐B for 6 h (6 h UVB) or not (WL). Error bars represent the SE of three biological replicates. Asterisks indicate a significant decrease in transcript abundance when compared with that under white light (*P < 0.05; **P < 0.01; ns, no significant difference).
Genes linked to hypocotyl elongation include auxin biosynthesis and response genes such as YUC8, IAA19 and IAA29 (Sun et al., 2012; de Wit et al., 2016a; Gangappa and Kumar, 2017) as well as members of the xyloglucan endotransglycosylase‐related gene family such as XTR7 (Soy et al., 2014). To better understand UV‐B regulation of these genes under the light conditions generally used to study UV‐B inhibition of hypocotyl growth, we performed quantitative (q)PCR analysis to determine how expression levels of XTR7, YUC8, IAA19 and IAA29 in WT (Wassilewskija, Ws), uvr8, hy5 hyh and uvr8 hy5 hyh seedlings responded to 6 h of UV‐B. We observed that UV‐B repressed the expression of each of these four elongation‐related genes in the WT but not in uvr8 and uvr8 hy5 hyh (Figure 1c–f). Each of the analyzed genes exhibited a higher expression level in hy5 hyh compared with that in WT under WL conditions (Figure 1c–f), in agreement with the elongated hypocotyl phenotype of hy5 hyh (Figures 1a,b and S1). However, UV‐B repressed the expression of each analyzed gene to a similar level in both hy5 hyh and WT (Figure 1c–f). Together, our data support the presence of an HY5/HYH‐independent UVR8 signaling pathway that functions to repress genes associated with elongation and to inhibit hypocotyl growth in response to UV‐B.
UV‐B‐repressed genes partially overlap with PIF4 and PIF5 target genes
The PIF4 and PIF5 proteins regulate hypocotyl elongation through transcriptional regulation of elongation‐related genes (Leivar and Quail, 2011; Delker et al., 2014; Gangappa and Kumar, 2017). Previous work has shown that UV‐B negatively regulates PIF4 and PIF5 to antagonize thermomorphogenesis and shade‐avoidance responses (Hayes et al., 2014; Hayes et al., 2017). The selected elongation‐related genes – XTR7, YUC8, IAA19 and IAA29 – are direct targets of PIF4 and/or PIF5 (Hornitschek et al., 2012; Oh et al., 2012; Sun et al., 2012; Pfeiffer et al., 2014). To investigate the role of PIF4 and PIF5 in UVR8‐mediated inhibition of hypocotyl growth we first examined the expression of YUC8, XTR7, IAA19 and IAA29 in response to UV‐B in 4‐day‐old WT, uvr8 and pif4 pif5 mutant seedlings. We observed that UV‐B repressed the expression of all four genes in the WT (Columbia, Col) but not in uvr8 (Figure 2a–d), further supporting the data from the Ws background (Figure 1c–f). Interestingly, in pif4 pif5 grown under WL, these genes already had constitutively reduced expression levels when compared with those in the WT and uvr8 (Figure 2a–d). In order to more comprehensively understand the function of PIF4 and PIF5 in UV‐B signaling, and more specifically in the UV‐B‐mediated inhibition of hypocotyl elongation, we performed an RNA sequencing (RNA‐Seq) analysis to compare expression data in the WT and uvr8‐6 grown under WL with that in seedlings grown under WL with a final 6 h exposure to UV‐B (WL + UVB), as well as that in pif4 pif5 mutant seedlings grown under WL. We found 814 genes in the WT that were downregulated in response to UV‐B, of which the downregulation of 763 genes was UVR8 dependent (fold change > 2, adjusted P‐value <0.05) (Figures 2e and S2a, Table S1). We first compared the genes downregulated by UVR8 in the WT (763 genes, Col UV/WL ↓) with genes mis‐regulated in pif4 pif5 mutants when compared with those in the WT under WL (pif4pif5/Col WL) (Figure 2e). We found 153 genes significantly repressed by UV‐B in the WT that showed reduced expression in pif4 pif5 (20.1%), whereas only four genes showed enhanced expression in pif4 pif5 when compared with that in the WT (0.5%) (Figure 2e, Table S2). This indicates that UV‐B‐repressed genes partially overlap with genes whose expression depends on PIF4 and PIF5, among which were elongation‐related genes, including YUC8, XTR7, IAA19 and IAA29 (Table S2). In sharp contrast, of the 1070 genes that were upregulated in response to UV‐B in a UVR8‐dependent manner, only 14 (1.3%) and 24 (2.2%) genes were repressed or enhanced in the pif4 pif5 mutant, respectively (Figures 2f and S2b, Tables S1 and S3). Taken together, we conclude that part of the UVR8‐repressed transcriptome is positively regulated by PIF4 and PIF5, including several genes associated with hypocotyl elongation.
Figure 2.
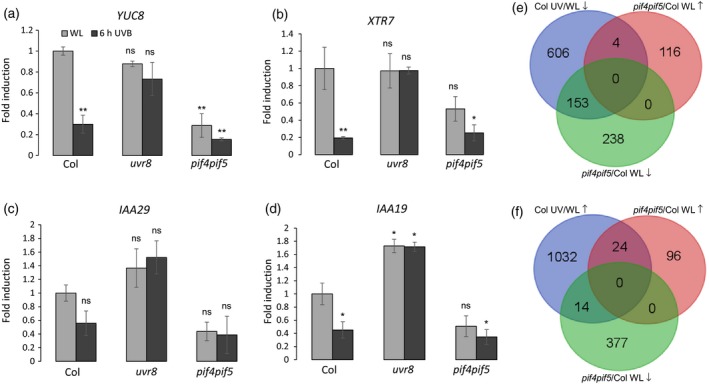
UV‐B‐repressed genes overlap with PIF4 and PIF5 target genes.
(a)–(d) Quantitative real‐time PCR analysis of (a) YUC8, (b) XTR7, (c) IAA29 and (d) IAA19 expression in 4‐day‐old wild‐type (Col), uvr8‐6 and pif4‐101 pif5‐3 seedlings grown in white light and either exposed to UV‐B for 6 h (6 h UVB) or not (WL). Error bars represent the SE of three biological replicates. Asterisks indicate a significant difference in transcript abundance when compared with that of WT under white light (*P < 0.05; **P < 0.01; ns, no significant difference). (e) Venn diagram representing the intersection between UV‐B‐repressed genes in the wild type (Col UV/WL ↓), genes with reduced expression in pif4‐101 pif5‐3 compared with that in the wild type under white light (pif4pif5/Col WL ↓) and genes with enhanced expression in pif4‐101 pif5‐3 compared with that in the wild type under white light (pif4pif5/Col WL ↑) (fold change > 2, adjusted P‐value <0.05). (f) Venn diagram representing the intersection between UV‐B‐induced genes in the wild type (Col UV/WL ↑), genes with reduced expression in pif4‐101 pif5‐3 compared with that in the wild type under white light (pif4pif5/Col WL ↓) and genes with enhanced expression in pif4‐101 pif5‐3 compared with that in the wild type under white light (pif4pif5/Col WL ↑) (fold change > 2, adjusted P‐value <0.05).
UVR8 negatively regulates PIF4 and PIF5 abundance
We tested whether UVR8 signaling affects the stability of PIF4 and PIF5 proteins under UV‐B. Immunoblot analysis showed that PIF4‐HA and PIF5‐HA were detectable under WL conditions in both WT and uvr8 mutant backgrounds and that UV‐B decreased their levels in the WT but not in uvr8 (Figure 3a,b). It is of note that UV‐B does not affect PIF4 and PIF5 mRNA levels (Figure S3, Table S1) (Favory et al., 2009), suggesting that the effect on levels of PIF4 and PIF5 protein is post‐transcriptional. Congruent with the role of COP1 in UVR8‐mediated UV‐B signaling, UV‐B‐mediated degradation of PIF4‐HA was strongly reduced in the cop1‐4 mutant (Figure 3c). Basal PIF4‐HA levels, however, were reduced in cop1‐4 (Figure 3c), in agreement with previous data (Gangappa and Kumar, 2017).
Figure 3.
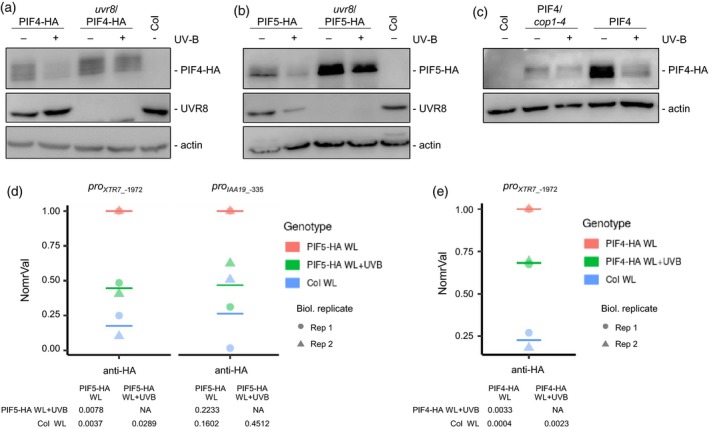
UVR8‐mediated degradation of PIF4 and PIF5 is associated with reduced target promoter occupancy in response to UV‐B.
(a)–(c) Anti‐hemagglutinin (HA) immunoblot analysis of HA‐tagged proteins in 4‐day‐old seedlings grown under white light (−UV‐B) or white light supplemented with a final 6 h of UV‐B (+UV‐B). The wild type (Col) is shown as a negative control. PIF4‐HA protein levels (a) were analyzed in Col/ProPIF4:PIF4‐3×HA (PIF4‐HA) and uvr8‐6/ProPIF4:PIF4‐3×HA (uvr8/PIF4‐HA). PIF5‐HA protein levels (b) were analyzed in Col/ProPIF5:PIF5‐3×HA (PIF5‐HA) and uvr8‐6/ProPIF5:PIF5‐3×HA (uvr8/PIF5‐HA). PIF4‐HA protein levels (c) were analyzed in Col/ProPIF4:PIF4‐3×HA (PIF4‐HA) and cop1‐4/ProPIF4:PIF4‐3×HA (cop1‐4/PIF4‐HA). Blots were reprobed with anti‐UVR8 (a,b), as well as anti‐actin as loading control (a–c).
(d) Chromatin immunopreciptation (ChIP) of DNA associated with PIF5‐HA. Chromatin immunopreciptation‐qPCR was performed for the XTR7 and IAA19 promoters and an intergenic region between the At4g26900 and At4g26910 genes using 5‐day‐old Col/ProPIF5:PIF5‐3×HA (PIF5‐HA) seedlings either exposed to narrowband UV‐B for 6 h (WL + UVB) or not (WL), and wild‐type (Col; negative control) seedlings grown only under white light (WL).
(e) Chromatin immunopreciptation of DNA associated with PIF4‐HA. Chromatin immunopreciptation‐qPCR was performed for the XTR7 promoter using 5‐day‐old Col/ProPIF4:PIF4‐3×HA (PIF4‐HA) seedlings either exposed to narrowband UV‐B for 6 h (WT + UVB) or not (WL) and wild‐type (Col; negative control) seedlings grown only under white light (WL). In (d) and (e) the ChIP experiments were performed with an anti‐HA antibody. The numbers of the analyzed DNA fragments (proXTR7_‐1972, proIAA19_‐335) indicate the positions of the base pair of the amplicon relative to the translation start site (referred to as position +1). The percentage of DNA associated with (d) PIF5‐HA and (e) PIF4‐HA relative to total input DNA of two independent biological replicates were normalized against the PIF5‐HA WL and PIF4‐HA WL sample, respectively (NormVal, PIF5‐HA WL = 1 and PIF4‐HA WL = 1). Means are represented as horizontal bars. P‐values from pairwise t‐tests using Holm's correction for multiple tests are shown.
Promoters of both XTR7 and IAA19 have been reported to contain G‐boxes approximately 1.9 kb and 0.33 kb, respectively, upstream of the start ATG, which are directly targeted by PIF4 and PIF5 (Hornitschek et al., 2012; Pfeiffer et al., 2014). Chromatin immunoprecipitation (ChIP) assays showed that the association of PIF5‐HA with XTR7 and IAA19 promoter fragments that include these elements was reduced in response to UV‐B (Figure 3d). Similarly, the association of PIF4‐HA with XTR7 promoter elements was reduced in response to UV‐B (Figure 3e). Together these data show that active UVR8 downregulates PIF4 and PIF5 at the post‐translational level, resulting in their reduced presence at target promoters and subsequently reduced target gene expression.
PIF4 and PIF5 are involved in UV‐B‐induced suppression of hypocotyl elongation
Our data strongly suggest that UVR8‐mediated degradation of PIF4 and PIF5 contributes to inhibition of hypocotyl growth in response to UV‐B. In support of such a mechanism, pif4 pif5 mutants exhibited reduced hypocotyl elongation compared with the WT in the absence of UV‐B (Figure 4a,b). Supplemental UV‐B resulted in inhibition of hypocotyl elongation in the WT but not in uvr8, whereas in pif4 pif5 seedlings the UV‐B inhibition effect was less pronounced, which is at least partially due to the underlying pif4 pif5 short hypocotyl phenotype under WL conditions (Figure 4a,b). We conclude that UVR8‐responsive reduction of PIF4 and PIF5 protein levels contributes to the UV‐B‐mediated inhibition of hypocotyl growth.
Figure 4.
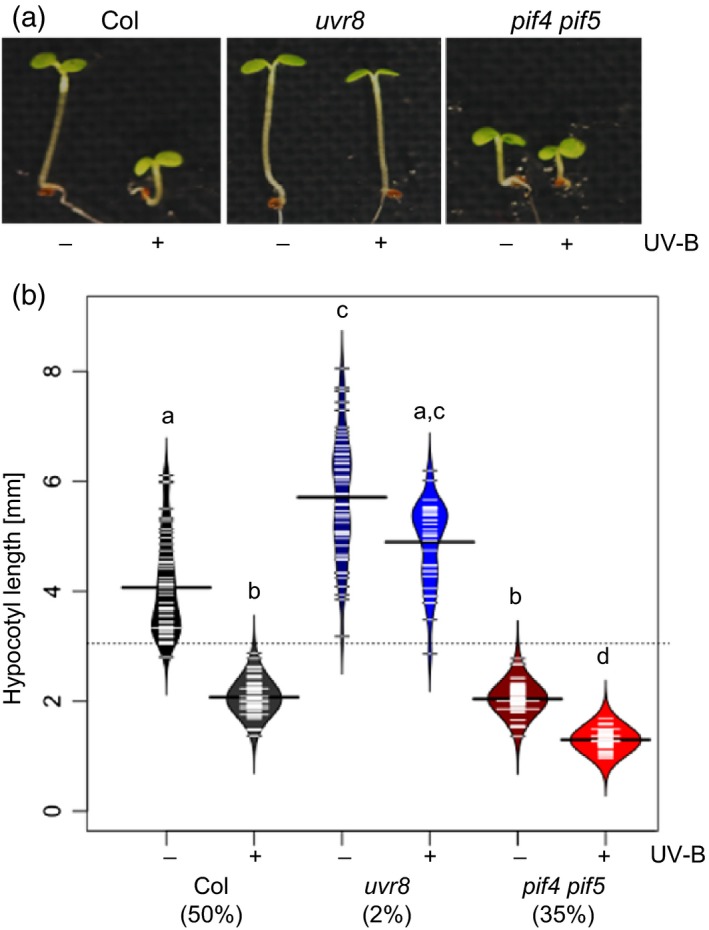
PIF4 and PIF5 contribute to UVB‐mediated hypocotyl elongation.
(a) Representative image showing the hypocotyl phenotype of 4‐day‐old wild‐type (Col), pif4‐101 pif5‐3 and uvr8‐6 seedlings grown under white light either supplemented with narrowband UV‐B (+) or not (−).
(b) Quantification of hypocotyl length. Data represent mean length ± SD for n > 40 samples. Shared letters indicate no statistically significant difference in the means (P > 0.05). Percentages on the x‐axis indicate the relative hypocotyl growth inhibition by UV‐B.
PIF4 and PIF5 are not involved in the UVR8‐mediated activation of UV‐B‐induced genes and anthocyanin accumulation
As the contribution of constitutively shorter hypocotyls to the reduced UV‐B‐responsive hypocotyl growth inhibition phenotype of pif4 pif5 resembles that previously described for cop1‐4 (Oravecz et al., 2006), we tested whether PIF4 and PIF5 are also involved in UVR8‐mediated activation of RUP2 and HY5. However, in contrast to uvr8 mutants, pif4 pif5 mutants showed UV‐B‐responsive activation of the two analyzed genes that was comparable to that in the WT (Figure 5a,b), as well as accumulation of HY5 protein (Figure 5c). Moreover, in pif4 pif5, UVR8‐induced enrichment of HY5 at chromatin of the CHS promoter in addition to anthocyanin accumulation was comparable to that in the WT (Figure 5d,e). Taken together, these data suggest that, in contrast to HY5 (Ulm et al., 2004; Brown et al., 2005; Oravecz et al., 2006; Huang et al., 2012; Binkert et al., 2014), PIF4 and PIF5 may not be required for UVR8‐mediated gene activation.
Figure 5.
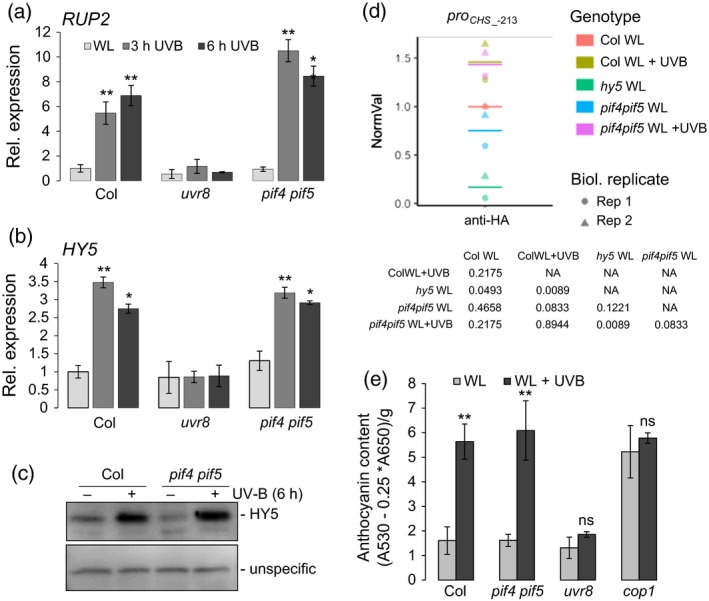
Induction of UV‐B‐inducible genes is independent of PIF4 and PIF5.
(a), (b) Quantitative real‐time PCR analysis of (a) RUP2 and (b) HY5 expression in 4‐day‐old wild‐type (Col), uvr8‐6 (uvr8) and pif4‐101 pif5‐3 (pif4pif5) seedlings exposed to narrowband UV‐B for 3 and 6 h (3 h/6 h UVB) or not (WL). Error bars represent the SE of three biological replicates. Asterisks indicate a significant increase in transcript abundance compared with that under WL (*P < 0.05; **P < 0.01). Error bars represent the SE of three biological replicates.
(c) Immunoblot analysis of HY5 levels in 4‐day‐old wild‐type (Col) and pif4‐101 pif5‐3 (pif4pif5) seedlings grown under white light (−) or white light supplemented with a final 6 h of UV‐B (+). An unspecific band is shown as a loading control.
(d) UV‐B‐responsive HY5 chromatin association in wild‐type plants (Col) was compared with that in the pif4 pif5 mutant, with that in hy5 plants being included as a negative control. Five‐day‐old seedlings were grown under white light and exposed to a final 6 h of narrowband UV‐B (+) or not (−). Chromatin immunoprecipitation‐qPCR was performed for the CHS promoter. The number of the analyzed DNA fragment (proCHS_‐213) indicates the position of the 5′ base pair of the amplicon relative to the translation start site (referred to as position +1). The percentage of DNA associated with HY5 relative to total input DNA of two independent biological replicates was normalized against the Col WL sample (NormVal, Col WL = 1). Means are represented as horizontal bars. P‐values from pairwise t‐tests using Holm's correction for multiple tests are shown.
(e) Anthocyanin accumulation in 4‐day‐old wild‐type (Col), pif4‐101 pif5‐3 (pif4pif5), uvr8‐6 (uvr8) and cop1‐4 (cop1) seedlings grown under white light either supplemented with narrowband UV‐B (WL + UVB) or not (WL). Error bars represent the SD of three biological replicates. Asterisks indicate a significant increase in anthocyanin levels under UV‐B compared with that under WL (*P < 0.05; **P < 0.01; ns, no significant difference).
Discussion
Inhibition of hypocotyl elongation is broadly used as a readout for UV‐B‐induced photomorphogenesis that is downstream of UVR8 photoreceptor signaling. In this work, we show that UVR8‐dependent degradation of PIF4 and PIF5 contributes to this UV‐B‐induced photomorphogenic response (see Figure 6 for our current working model).
Figure 6.
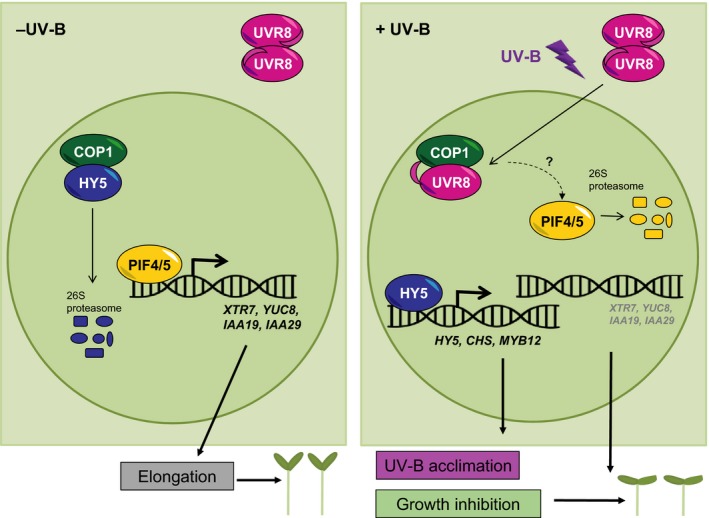
Working model for UVR8‐mediated regulation of hypocotyl elongation through degradation of PIF4 and PIF5.
In the absence of UV‐B, nuclear COP1 exerts it role as an E3 ubiquitin ligase by targeting HY5 for ubiquitination and proteasomal degradation, as well as facilitating accumulation of PIF4 and PIF5. PIF4 and PIF5 mediate the expression of genes associated with hypocotyl elongation (e.g. XTR7, YUC8, IAA19 and IAA29), thereby promoting elongation. In the presence of UV‐B, UVR8 monomers interact with and inhibit COP1, resulting in stabilization of HY5 and HY5‐dependent induction of genes associated with UV‐B acclimation, as well as degradation of PIF4 and PIF5 and hence reduced hypocotyl elongation. Note that UVR8‐mediated repression of brassinosteroid‐promoted plant growth is omitted for simplicity (see Liang et al., 2018).
PIF4 and PIF5 regulate hypocotyl elongation by binding and activating genes encoding proteins involved in auxin biosynthesis and auxin signaling (Hornitschek et al., 2012). PIF4 and PIF5 are key players that accumulate in response to environmental cues that favor elongation, such as shade (low red/far‐red ratio) and elevated temperature (Lorrain et al., 2008; Koini et al., 2009; Franklin et al., 2011; Quint et al., 2016; Iglesias et al., 2018). It has been shown that UV‐B antagonizes shade avoidance and thermomorphogenic responses through the degradation and inhibition of PIF4 and PIF5 (Hayes et al., 2014; Fraser et al., 2016; Hayes et al., 2017). Our data suggest that UVR8‐dependent degradation of PIF4 and PIF5 contributes to UV‐B‐responsive inhibition of hypocotyl growth. Levels of PIF4 and PIF5 are reduced under WL supplemented with UV‐B in a UVR8‐dependent manner, and pif4 pif5 double mutants already show short hypocotyls in the absence of UV‐B. Consistently, transcription of elongation‐related genes directly targeted by PIF4 and PIF5 is repressed by UV‐B. Although these observations clearly point to degradation of PIF4 and PIF5 underlying UVR8‐dependent growth inhibition, the mechanism by which UVR8 triggers their degradation remains unknown. A BLADE‐ON‐PETIOLE 1 (BOP1)‐ and BOP2‐based E3 ubiquitin ligase complex has been shown to ubiqitinate PIF4, thereby affecting photo‐ and thermomorphogenesis (Zhang et al., 2017); however, involvement of this complex in degradation of PIF4 under UV‐B remains to be demonstrated. Activity of COP1, on the other hand, is a direct target of UVR8 (Favory et al., 2009; Podolec and Ulm, 2018; Lau et al., 2019), and COP1 has been shown to facilitate the stability of PIF family proteins, including PIF4 and PIF5, in the dark (Bauer et al., 2004; Gangappa and Kumar, 2017; Pham et al., 2018). This UVR8‐mediated inhibition of COP1 may thus be linked to degradation of PIF4 and PIF5, although via a currently unknown mechanism.
How UVR8 regulates gene expression has remained rather enigmatic. Direct binding of UVR8 to chromatin at target genes to regulate gene transcription has previously been suggested, but remains debated (Brown et al., 2005; Binkert et al., 2016; Jenkins, 2017). However, direct interaction of UVR8 with transcription factors has recently been implicated in UVR8 signaling (Liang et al., 2018; Yang et al., 2018; Liang et al., 2019). UVR8 was shown to interact with BES1 and BIM1, two transcription factors that mediate BR‐regulated gene expression and plant growth. The UV‐B‐dependent nuclear accumulation of UVR8 (Kaiserli and Jenkins, 2007; Yin et al., 2016) leads to interference with BES1 and BIM1 activities through inhibition of their DNA‐binding capacities (Liang et al., 2018). This results in UV‐B‐mediated, UVR8‐dependent repression of BR‐responsive growth‐related genes (Liang et al., 2018). The prevention of DNA binding by BES1 and BIM1 thus constitutes another mechanism in addition to PIF4 and PIF5 degradation through which UVR8 can mediate gene repression. Additional mechanisms contributing to UV‐B‐responsive gene repression remain to be identified. Moreover, future studies of great interest are to further elucidate the complex interplay between PIF4, PIF5, HY5, HYH, BES1 and BIM1 transcription factors underlying UV‐B‐induced changes in gene expression and photomorphogenesis in order to understand how plants dynamically regulate their growth and development in response to their light environment.
Experimental procedures
Plant material and generation of transgenic lines
The uvr8‐6, cop1‐4, hy5‐215 and pif4‐101 pif5‐3 mutants are in the Col accession (McNellis et al., 1994; Oyama et al., 1997; Lorrain et al., 2008; Favory et al., 2009). Mutants hy5‐ks50, hyh‐1, hy5‐ks50 hyh‐1 and uvr8‐7 are in the Ws background (Oyama et al., 1997; Holm et al., 2002; Favory et al., 2009). The triple mutant hy5 hyh uvr8 was generated by genetic crossing of hy5‐ks50 hyh‐1 with uvr8‐7. The uvr8‐7 genotype was selected by anti‐UVR8 protein gel blots to identify the absence of UVR8, and the UVR8 Q124‐to‐Stop mutation was verified by sequencing. hy5‐ks50 and hyh‐1 were genotyped as follows:
hy5‐ks50: hy5ks50_LP (5′‐TCC ACC CAC GTT CCA ATC TC‐3′) + hy5ks50_RP (5′‐GAC ACC TCT TCA GCC GCT TG‐3′) + T‐DNA pGV3850 LB1 primer (5′‐GCG TGG ACC GCT TGC TGC AAC T‐3′); 0.8 kb for WT, 0.5 kb for hy5‐ks50.
hyh‐1: hyh1_LP (5′‐GGA CCC ACC ACG GCA TTT TA‐3′) + hyh1_RP (5′‐CGC GTC CAT TCC ATA CGA CT‐3′) + T‐DNA pD991 LB primer (5′‐TAA TAA CGC TGC GGA CAT CTA C‐3′); 1.0 kb for WT, 0.8 kb for hyh‐1.
The pif5‐1/ProPIF5:PIF5‐3×HA and pif4‐101/ProPIF4:PIF4‐3×HA lines have been described previously (de Wit et al., 2016b; Zhang et al., 2017). uvr8‐6/ProPIF4:PIF4‐3×HA, uvr8‐6/ProPIF5:PIF5‐3xHA were generated by genetic crossing and genotyping F2 plants for the absence of UVR8; Western blot analysis confirmed the presence of the transgene. cop1‐4/ProPIF4:PIF4‐3×HA was generated by genetic crossing followed by phenotypic selection for cop1‐4 phenotypes and confirmation by sequencing; Western blot analysis was used to check for the presence of the transgene.
Growth conditions and light treatments
Arabidopsis seeds were surface sterilized with sodium hypochlorite and sown on half‐strength Murashige and Skoog basal salt medium (Duchefa, https://www.duchefa-biochemie.com/) containing 1% (w/v) agar (Applichem, https://www.itwreagents.com/) and 1% (w/v) sucrose. Seeds were germinated aseptically at 22°C in a WL field under continuous irradiation provided by Osram L18W/30 tubes (3.6 μmol m−2 sec−1; measured with an LI‐250 light meter). The UV‐B was supplemented for the indicated times with Philips TL20W/01RS narrowband UV‐B tubes (1.5 μmol m−2 sec−1; measured with a VLX‐3W ultraviolet light meter equipped with a CX‐312 sensor; Vilber Lourmat, https://www.vilber.com/). The UV‐B range was modulated by the use of 3‐mm transmission cut‐off filters from the WG series (Schott Glaswerke, https://www.schott.com/) with half‐maximal transmission at the indicated wavelength (WG304 for UV‐B‐treated samples and WG368 for non‐UV‐B‐treated controls).
Hypocotyl length measurements
Measurement of hypocotyl length was performed with ImageJ software (http://www.rsb.info.gov/ij). A minimum of 40 seedlings were measured per treatment and genotype, with at least two independent experimental repetitions. For each repeat, anova type II was performed (R ‘CAR’ package v.2.1‐4) followed by the Tukey honestly significant difference post‐hoc test.
Anthocyanin measurement
Anthocyanin measurement was performed as described previously (Yin et al., 2012). Two hundred and fifty microliters of acidic methanol (HCl 1% v/v) was added to about 50 mg of 4‐day‐old seedlings grown with or without supplemental UV‐B. Plant material was homogenized and incubated with rotation for 1 h in the dark. Absorbance of the extracts was measured at 535 and 655 nm and anthocyanin content was determined according to the equation A535 – (0.25 × A655)/g (A535 and A655 = absorbance at the indicated wavelength, g = gram fresh weight).
Protein extraction and gel blot analysis
For PIF4 and PIF5 immunoblot analyses, total proteins were extracted in extraction buffer [50 mm 2‐amino‐2‐(hydroxymethyl)‐1,3‐propanediol Tris‐HCl pH 7.6, 150 mm NaCl, 5 mm MgCl2, 30% [v/v] glycerol, 10 μm MG132, 10μm 3,4‐dichloroisocoumarin, 1% [v/v] Protease Inhibitor Cocktail for plant extracts [Sigma‐Aldrich], 0.1% [v/v] Igepal. [Correction added on 29 January 2020, after first online publication: concentrations are added.] Total protein was quantified by a Bio‐Rad Protein Assay (Bio‐Rad, https://www.bio-rad.com/). For HY5 immunoblot analysis, total proteins were extracted in extraction buffer (50 μm EDTA, 0.1 m TRIS‐HCl pH 8, 0.7% w/v SDS, 10 mm NaF, protease inhibitor tablet, 1 mm DTT, 0.25 m NaCl, 15 mm β‐glycerolphosphate, 15 mm p‐nitrophenyl phosphate). Proteins were separated by SDS/PAGE and transferred to polyvinylidene difluoride membranes according to the manufacturer’s instructions (Bio‐Rad). Anti‐HA (HA.11, Covance, https://www.covance.com/), anti‐actin (Sigma‐Aldrich, https://www.sigmaaldrich.com/), anti‐UVR8(426–440) (Favory et al., 2009) and anti‐HY5 (Oravecz et al., 2006) were used as primary antibodies, with horseradish peroxidase‐conjugated anti‐mouse and anti‐rabbit immunoglobulins as secondary antibodies. Chemiluminescent signals were generated by using the Amersham ECL Select Western Blotting Detection Reagent kit (GE Healthcare, https://www.gehealthcare.com/) and detected with an ImageQuant LAS 4000 mini CCD camera system (GE Healthcare).
Quantitative real‐time PCR
Arabidopsis total RNA was isolated with Plant RNeasy kit (Qiagen, https://www.qiagen.com/) and treated with DNaseI according to the manufacturer’s instructions. Complementary DNA synthesis and quantitative real‐time PCR using PowerUp SYBR Green Master Mix reagents and a QuantStudio 5 real‐time PCR system (Thermo Fisher Scientific, https://www.thermofisher.com/) were performed as previously described (Arongaus et al., 2018). Gene‐specific primers used for XTR7, YUC8, IAA19, IAA29, RUP2, HY5 and PP2A (reference gene) are listed in Table S4. Expression values were calculated using the ΔΔCt method (Livak and Schmittgen, 2001) and normalized to that in the WT. Each reaction was performed in technical triplicate; data shown are from three biological repetitions.
RNA‐Seq and transcriptome analysis
Total RNA was extracted from three biological replicates of WT (Col), uvr8 and pif4 pif5 using the Plant RNeasy Kit (Qiagen). The RNA quality control, library preparation using TruSeqUD Stranded mRNA (Illumina, https://www.illumina.com/) and sequencing on an Illumina HiSeq 4000 System using 100‐bp single‐end reads protocol were performed at the iGE3 genomics platform of the University of Geneva (https://ige3.genomics.unige.ch/).
Quality control was performed with FastQC v.0.11.5. Reads were mapped to the Arabidopsis TAIR‐10 genome using STAR v.2.5.3a software with average alignment around 92%. Biological quality control was done with PicardTools v.2.9.0. Raw counts obtained using HTSeq v.0.9.1 were filtered for genes with low expression (to 21 655 genes) and normalized according to library size. Normalization and differential expression analysis was performed with the R/Bioconductor package edgeR v.3.22.3. Annotations were obtained from Araport11 v.1.10.4 (Krishnakumar et al., 2015).
The differentially expressed genes were estimated using a general linear model (GLM) approach, negative binomial distribution and a quasi‐likelihood F‐test. Pairwise comparisons (GLM, quasi‐likelihood F‐test) were performed on the filtered dataset with two factors defined (genotype and condition). Genes with a fold change > 2 and P < 0.05 (with a false discovery rate of 5%) were considered differentially expressed. For further analysis, data sets were filtered for genes differentially expressed in the WT but not in uvr8. Venn diagrams were generated using a webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Chromatin Immunoprecipitation assays
Chromatin was immunoprecipitated with polyclonal ChIP‐grade anti‐HA antibodies (Abcam, https://www.abcam.com/), as previously described (Stracke et al., 2010). The ChIP‐qRT data were obtained using PowerUP SYBR Green Master Mix Kit and a QuantStudio 5 Real‐Time PCR system (Applied Biosystems), with primers as listed in Table S4. The qPCR data were analyzed according to the percentage of input method (Haring et al., 2007).
Accession numbers
The RNA‐Seq data reported in this article have been deposited in the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132169. Sequence data from this article can be found in The Arabidopsis Information Resource (https://www.arabidopsis.org/) database under the following accession numbers: AT5G13930 (CHS), AT2G32950 (COP1), AT5G11260 (HY5), AT3G17609 (HYH), AT3G15540 (IAA19), AT4G32280 (IAA29), AT2G43010 (PIF4), AT3G59060 (PIF5), AT5G63860 (UVR8), AT4G14130 (XTR7), and AT4G28720 (YUC8).
Author contributions
ET, MP and RU designed the research. ET performed hypocotyl measurements, qPCR, Western blots and ChIP assays. MP performed hypocotyl measurements, qPCR and bioinformatic analysis. ET, MP and RU wrote the paper. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflict of interests.
Supporting information
Figure S1. UVR8‐mediated inhibition of hypocotyl growth is partially independent of HY5 and HYH.
Figure S2. UVR8‐dependent, UV‐B‐regulated genes.
Figure S3. UV‐B does not affect expression of PIF4 and PIF5.
Table S1. Genes regulated by narrowband UV‐B in Col wild type and/or uvr8‐6 (lists correspond to Figure S2).
Table S2. UV‐B‐repressed genes in Col wild type and/or genes misexpressed in pif4‐101 pif5‐3 in white light‐grown plants (lists correspond to Figure 2e).
Table S3. UV‐B‐induced genes in Col wild type and/or genes misexpressed in pif4‐101 pif5‐3 in white light‐grown plants (lists correspond to Figure 2f).
Table S4. Oligonucleotide sequences used in this study.
Acknowledgements
We are grateful to Christian Fankhauser for helpful comments, Jose Manuel Nunes of the platform for biomathematical and biostatistical analyses (BioSC) for help with ChIP data analyses, and Séverine Lorrain and Christian Fankhauser for providing the pif4‐101/ProPIF4:PIF4‐3×HA and pif5‐1/ProPIF5:PIF5‐3×HA lines. The RNA‐Seq experiments were performed at the iGE3 genomics platform of the University of Geneva (https://ige3.genomics.unige.ch/). We thank Natacha Civic and Céline Delucinge‐Vivier of the iGE3 genomics platform for bioinformatic analysis of the RNA‐Seq data. This work was supported by the University of Geneva and the Swiss National Science Foundation (grant nos 31003A_175774 and CRSII3_154438).
Linked article: This paper is the subject of a Research Highlight article. To view this Research Highlight article visit https://doi.org/10.1111/tpj.14644.
References
- Arongaus, A.B. , Chen, S. , Pireyre, M. , Glockner, N. , Galvao, V.C. , Albert, A. , Winkler, J.B. , Fankhauser, C. , Harter, K. and Ulm, R. (2018) Arabidopsis RUP2 represses UVR8‐mediated flowering in noninductive photoperiods. Genes Dev. 32, 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, D. , Viczian, A. , Kircher, S. et al . (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell, 16, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert, M. , Kozma‐Bognar, L. , Terecskei, K. , De Veylder, L. , Nagy, F. and Ulm, R. (2014) UV‐B‐responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell, 26, 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert, M. , Crocco, C.D. , Ekundayo, B. , Lau, K. , Raffelberg, S. , Tilbrook, K. , Yin, R. , Chappuis, R. , Schalch, T. and Ulm, R. (2016) Revisiting chromatin binding of the Arabidopsis UV‐B photoreceptor UVR8. BMC Plant Biol. 16, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron, A.K. and Vissenberg, K. (2014) The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep. 33, 697–706. [DOI] [PubMed] [Google Scholar]
- Brown, B.A. and Jenkins, G.I. (2008) UV‐B signaling pathways with different fluence‐rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146, 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, B.A. , Cloix, C. , Jiang, G.H. , Kaiserli, E. , Herzyk, P. , Kliebenstein, D.J. and Jenkins, G.I. (2005) A UV‐B‐specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA, 102, 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker, C. , Sonntag, L. , James, G.V. et al . (2014) The DET1‐COP1‐HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9, 1983–1989. [DOI] [PubMed] [Google Scholar]
- Demarsy, E. , Goldschmidt‐Clermont, M. and Ulm, R. (2018) Coping with 'dark sides of the sun' through photoreceptor signaling. Trends Plant Sci. 23, 260–271. [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. and Lash, A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C. and Casal, J.J. (2004) Phenotypic characterization of a photomorphogenic mutant. Plant J. 39, 747–760. [DOI] [PubMed] [Google Scholar]
- Favory, J.J. , Stec, A. , Gruber, H. et al . (2009) Interaction of COP1 and UVR8 regulates UV‐B‐induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci, A.S. and Fankhauser, C. (2017) Plant strategies for enhancing access to sunlight. Curr. Biol. 27, R931–R940. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A. , Lee, S.H. , Patel, D. et al . (2011) Phytochrome‐interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA, 108, 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, D.P. , Hayes, S. and Franklin, K.A. (2016) Photoreceptor crosstalk in shade avoidance. Curr. Opin. Plant Biol. 33, 1–7. [DOI] [PubMed] [Google Scholar]
- Gangappa, S.N. and Kumar, S.V. (2017) DET1 and HY5 control PIF4‐mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 18, 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers, C.M.M. and Monte, E. (2018) Seedling establishment: a dimmer switch‐regulated process between dark and light signaling. Plant Physiol. 176, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, H. , Heijde, M. , Heller, W. , Albert, A. , Seidlitz, H.K. and Ulm, R. (2010) Negative feedback regulation of UV‐B‐induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA, 107, 20132–20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring, M. , Offermann, S. , Danker, T. , Horst, I. , Peterhansel, C. and Stam, M. (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods, 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, S. , Velanis, C.N. , Jenkins, G.I. and Franklin, K.A. (2014) UV‐B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. USA, 111, 11894–11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, S. , Sharma, A. , Fraser, D.P. , Trevisan, M. , Cragg‐Barber, C.K. , Tavridou, E. , Fankhauser, C. , Jenkins, G.I. and Franklin, K.A. (2017) UV‐B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr. Biol. 27, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde, M. , Binkert, M. , Yin, R. et al . (2013) Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl. Acad. Sci. USA, 110, 20326–20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M. , Ma, L.G. , Qu, L.J. and Deng, X.W. (2002) Two interacting bZIP proteins are direct targets of COP1‐mediated control of light‐dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek, P. , Lorrain, S. , Zoete, V. , Michielin, O. and Fankhauser, C. (2009) Inhibition of the shade avoidance response by formation of non‐DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek, P. , Kohnen, M.V. , Lorrain, S. et al . (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Ouyang, X. , Yang, P. , Lau, O.S. , Li, G. , Li, J. , Chen, H. and Deng, X.W. (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV‐B light. Plant Cell, 24, 4590–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Ouyang, X. , Yang, P. , Lau, O.S. , Chen, L. , Wei, N. and Deng, X.W. (2013) Conversion from CUL4‐based COP1‐SPA E3 apparatus to UVR8‐COP1‐SPA complexes underlies a distinct biochemical function of COP1 under UV‐B. Proc. Natl. Acad. Sci. USA, 110, 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yang, P. , Ouyang, X. , Chen, L. and Deng, X.W. (2014) Photoactivated UVR8‐COP1 module determines photomorphogenic UV‐B signaling output in Arabidopsis. PLoS Genet. 10, e1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, M.J. , Sellaro, R. , Zurbriggen, M.D. and Casal, J.J. (2018) Multiple links between shade avoidance and auxin networks. J. Exp. Bot. 69, 213–228. [DOI] [PubMed] [Google Scholar]
- Jenkins, G.I. (2017) Photomorphogenic responses to ultraviolet‐B light. Plant Cell Environ. 40, 2544–2557. [DOI] [PubMed] [Google Scholar]
- Kaiserli, E. and Jenkins, G.I. (2007) UV‐B promotes rapid nuclear translocation of the Arabidopsis UV‐B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell, 19, 2662–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami, C. , Lorrain, S. , Hornitschek, P. and Fankhauser, C. (2010) Light‐regulated plant growth and development. Curr. Top Dev. Biol. 91, 29–66. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J. , Lim, J.E. , Landry, L.G. and Last, R.L. (2002) Arabidopsis UVR8 regulates ultraviolet‐B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini, M.A. , Alvey, L. , Allen, T. , Tilley, C.A. , Harberd, N.P. , Whitelam, G.C. and Franklin, K.A. (2009) High temperature‐mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. , Rolff, E. and Spruit, C.J.P. (1980) Genetic control of light‐inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Kretsch, T. (2010) Phenotypic characterization of photomorphogenic responses during plant development. Methods Mol. Biol. 655, 189–202. [DOI] [PubMed] [Google Scholar]
- Krishnakumar, V. , Hanlon, M.R. , Contrino, S. et al . (2015) Araport: the Arabidopsis information portal. Nucleic Acids Res. 43, D1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, K. , Podolec, R. , Chappuis, R. , Ulm, R. and Hothorn, M. (2019) Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 38, e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P. and Quail, P.H. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P. , Monte, E. , Oka, Y. , Liu, T. , Carle, C. , Castillon, A. , Huq, E. and Quail, P.H. (2008) Multiple phytochrome‐interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, T. , Mei, S. , Shi, C. et al. (2018) UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell, 44, 512–523. [DOI] [PubMed] [Google Scholar]
- Liang, T. , Yang, Y. and Liu, H. (2019) Signal transduction mediated by the plant UV‐B photoreceptor UVR8. New Phytol. 221, 1247–1252. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2− ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorrain, S. , Allen, T. , Duek, P.D. , Whitelam, G.C. and Fankhauser, C. (2008) Phytochrome‐mediated inhibition of shade avoidance involves degradation of growth‐promoting bHLH transcription factors. Plant J. 53, 312–323. [DOI] [PubMed] [Google Scholar]
- Lorrain, S. , Trevisan, M. , Pradervand, S. and Fankhauser, C. (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de‐etiolation in continuous far‐red light. Plant J. 60, 449–461. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W. , von Arnim, A.G. , Araki, T. , Komeda, Y. , Miséra, S. and Deng, X.W. (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell, 6, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E. , Zhu, J.Y. and Wang, Z.Y. (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz, A. , Baumann, A. , Mate, Z. , Brzezinska, A. , Molinier, J. , Oakeley, E.J. , Adam, E. , Schafer, E. , Nagy, F. and Ulm, R. (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV‐B response in Arabidopsis. Plant Cell, 18, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T. , Hardtke, C.S. , Wei, N. and Deng, X.W. (2000) Targeted destabilization of HY5 during light‐regulated development of Arabidopsis. Nature, 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T. , Shimura, Y. and Okada, K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus‐induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacin, M. , Semmoloni, M. , Legris, M. , Finlayson, S.A. and Casal, J.J. (2016) Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 211, 967–979. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, A. , Shi, H. , Tepperman, J.M. , Zhang, Y. and Quail, P.H. (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant, 7, 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, V.N. , Kathare, P.K. and Huq, E. (2018) Dynamic regulation of PIF5 by COP1‐SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 96, 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec, R. and Ulm, R. (2018) Photoreceptor‐mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45, 18–25. [DOI] [PubMed] [Google Scholar]
- Quint, M. , Delker, C. , Franklin, K.A. , Wigge, P.A. , Halliday, K.J. and van Zanten, M. (2016) Molecular and genetic control of plant thermomorphogenesis. Nat. Plants, 2, 15190. [DOI] [PubMed] [Google Scholar]
- Ren, H. , Han, J. , Yang, P. et al . (2019) Two E3 ligases antagonistically regulate the UV‐B response in Arabidopsis. Proc. Natl. Acad. Sci. USA, 116, 4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini, L. , Favory, J.J. , Cloix, C. et al . (2011) Perception of UV‐B by the Arabidopsis UVR8 protein. Science, 332, 103–106. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Lyu, M. , Luo, Y. , Liu, S. , Li, Y. , He, H. , Wei, N. , Deng, X.W. and Zhong, S. (2018) Genome‐wide regulation of light‐controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. USA, 115, 6482–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy, J. , Leivar, P. and Monte, E. (2014) PIF1 promotes phytochrome‐regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J. Exp. Bot. 65, 2925–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R. , Favory, J.J. , Gruber, H. , Bartelniewoehner, L. , Bartels, S. , Binkert, M. , Funk, M. , Weisshaar, B. and Ulm, R. (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet‐B radiation. Plant Cell Environ. 33, 88–103. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Qi, L. , Li, Y. , Chu, J. and Li, C. (2012) PIF4‐mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook, K. , Arongaus, A.B. , Binkert, M. , Heijde, M. , Yin, R. and Ulm, R. (2013) The UVR8 UV‐B photoreceptor: perception, signaling and response. Arabidopsis Book, 11, e0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R. , Baumann, A. , Oravecz, A. , Mate, Z. , Adam, E. , Oakeley, E.J. , Schafer, E. and Nagy, F. (2004) Genome‐wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV‐B response of Arabidopsis. Proc. Natl. Acad. Sci. USA, 101, 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, F. , Verbelen, J.P. and Van Der Straeten, D. (2005) Of light and length: regulation of hypocotyl growth in Arabidopsis. BioEssays, 27, 275–284. [DOI] [PubMed] [Google Scholar]
- de Wit, M. , Galvao, V.C. and Fankhauser, C. (2016a) Light‐mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67, 513–537. [DOI] [PubMed] [Google Scholar]
- de Wit, M. , Keuskamp, D.H. , Bongers, F.J. , Hornitschek, P. , Gommers, C.M.M. , Reinen, E. , Martinez‐Ceron, C. , Fankhauser, C. and Pierik, R. (2016b) Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 26, 3320–3326. [DOI] [PubMed] [Google Scholar]
- Yadav, A. , Bakshi, S. , Yadukrishnan, P. , Lingwan, M. , Dolde, U. , Wenkel, S. , Masakapalli, S.K. and Datta, S. (2019) The B‐box‐containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV‐B protection. Plant Physiol. 179, 1876–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Liang, T. , Zhang, L. , Shao, K. , Gu, X. , Shang, R. , Shi, N. , Li, X. , Zhang, P. and Liu, H. (2018) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants, 4, 98–107. [DOI] [PubMed] [Google Scholar]
- Yin, R. and Ulm, R. (2017) How plants cope with UV‐B: from perception to response. Curr. Opin. Plant Biol. 37, 42–48. [DOI] [PubMed] [Google Scholar]
- Yin, R. , Messner, B. , Faus‐Kessler, T. , Hoffmann, T. , Schwab, W. , Hajirezaei, M.R. , von Saint Paul, V. , Heller, W. and Schaffner, A.R. (2012) Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol‐3‐O‐glycosylation. J. Exp. Bot. 63, 2465–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, R. , Arongaus, A.B. , Binkert, M. and Ulm, R. (2015) Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV‐B signaling in Arabidopsis. Plant Cell, 27, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, R. , Skvortsova, M.Y. , Loubery, S. and Ulm, R. (2016) COP1 is required for UV‐B‐induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. USA, 113, E4415–E4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Holmlund, M. , Lorrain, S. , Norberg, M. , Bako, L. , Fankhauser, C. and Nilsson, O. (2017) BLADE‐ON‐PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. Elife, 6, e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S. , Zhao, M. , Shi, T. , Shi, H. , An, F. , Zhao, Q. and Guo, H. (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo‐oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA, 106, 21431–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. UVR8‐mediated inhibition of hypocotyl growth is partially independent of HY5 and HYH.
Figure S2. UVR8‐dependent, UV‐B‐regulated genes.
Figure S3. UV‐B does not affect expression of PIF4 and PIF5.
Table S1. Genes regulated by narrowband UV‐B in Col wild type and/or uvr8‐6 (lists correspond to Figure S2).
Table S2. UV‐B‐repressed genes in Col wild type and/or genes misexpressed in pif4‐101 pif5‐3 in white light‐grown plants (lists correspond to Figure 2e).
Table S3. UV‐B‐induced genes in Col wild type and/or genes misexpressed in pif4‐101 pif5‐3 in white light‐grown plants (lists correspond to Figure 2f).
Table S4. Oligonucleotide sequences used in this study.


