Abstract
Life on earth is assumed to have developed in tropical regions that are characterized by regular 24 hr cycles in irradiance and temperature that remain the same throughout the seasons. All organisms developed circadian clocks that predict these environmental cycles and prepare the organisms in advance for them. A central question in chronobiology is how endogenous clocks changed in order to anticipate very different cyclical environmental conditions such as extremely short and long photoperiods existing close to the poles. Flies of the family Drosophilidae can be found all over the world—from the tropics to subarctic regions—making them unprecedented models for studying the evolutionary processes that underlie the adaptation of circadian clocks to different latitudes. This review summarizes our current understanding of these processes. We discuss evolutionary changes in the clock genes and in the clock network in the brain of different Drosophilids that may have caused behavioural adaptations to high latitudes.
Keywords: circadian rhythm, cryptochrome, latitudinal adaptations, locomotor activity, pigment‐dispersing factor
This review summarizes our current understanding of the evolutionary processes that underlie the molecular, neuronal, and behavioural adaptation of fly circadian clocks to different latitudes.
Abbreviations
- 5th LNv
fifth ventrolateral neuron
- Ca
calcium
- Clk
Clock gene
- CLK
Clock protein
- CRY
Cryptochrome
- CYC
CYC protein
- cyc
cycle gene
- DN
dorsal neurons
- l‐LNv
large ventrolateral neurons
- LN
lateral neurons
- LNd
dorsolateral neurons
- ls‐tim
long‐short splice form of timeless
Pigment‐Dispersing Factor
- Pdp1ε
gene coding for the PAR Domain Protein 1ε
- PDP1ε
PAR Domain Protein 1ε
- per
period gene
- PER
Period protein
- POC
posterior optic commissure
- SCN
suprachiasmatic nucleus
- s‐LNv
small ventrolateral neurons
- s‐tim
short splice form of timeless
- Thr‐Gly
threonine‐glycine
- tim
timeless gene
- TIM
Timeless protein
- vri
vrille gene
1. INTRODUCTION
Circadian clocks enable animals to anticipate the regular daily changes in the environment and to time specific activities to specific times of the day (Daan, 2010). On a different timescale, circannual clocks evolved to anticipate yearly environmental changes that are most evident in the north and south of our planet (Helm & Lincoln, 2017). The circannual and circadian clocks may be linked/interlocked, as circadian clocks can provide the necessary time reference for measuring day length and prepare for the forthcoming winter or summer (Bünning, 1960; Denlinger, Hahn, Merlin, Holzapfel, & Bradshaw, 2017; Goldman, 2001; Pittendrigh & Minis, 1964; Saunders, 2013). Seasonal changes do not only induce photoperiodic responses such as overwintering or reproduction, they have also a strong impact on animal's daily activity pattern. This is most evident in small‐sized animals, which are more subjected to rapid heat and energy loss than larger animals (Daan & Aschoff, 1975; Halle & Stemseth, 2000; Hoogenboom, Daan, Dallinga, & Schoenmakers, 1984; Ikeda et al., 2016). Insects highly depend on ambient temperature. On cold days, it is favourable for them to be active during the warmer parts of the day that occur during the photophase. In contrast, on hot days it is favourable for them to shift activity to the morning, the late evening, or even the night to avoid overheating (e.g., Fowler & Robinson, 1979). At least two mechanisms might help animals to be active at the best time of the day: (a) the quick response to acute ambient environment and (b) their circadian clocks. Here, we will refer to the latter.
For several small vertebrates it was observed that the clock has different waveforms under different day lengths (Aschoff, 1966; Daan & Aschoff, 1975). Birds, fishes, and several mammals show two activity bouts, one in the morning (M) and one in the evening (E). M and E activity bouts are close together under short days and separated by a pronounced siesta under long days. This pattern persists under subsequent constant conditions highlighting its endogenous nature. In hamsters, the bimodal activity is reflected by a bimodal rhythm of electrical firing in cultured slices of the brain master clock—the suprachiasmatic nuclei (SCN) (Jagota, de la Iglesia, & Schwartz, 2000). The two maxima in electrical firing are close together when the animals have previously been kept under a short photoperiod and far apart, when they have been kept under a long photoperiod (Jagota et al., 2000). Among insects, M and E activity bouts have been reported in fruit and house flies (Drosophila melanogaster and Musca domestica; Bazalova & Dolezel, 2017; Hamblen‐Coyle, Wheeler, Rutila, Rosbash, & Hall, 1992; Helfrich‐Förster, 2000; Prabhakaran & Sheeba, 2012, 2013; Wheeler, Hamblen‐Coyle, Dushay, & Hall, 1993) as well as in some mosquitoes (Chiba, 1971; Chiba, Kubota, & Nakamura, 1982). Although, in fruit flies, M and E activity bouts usually merge to one major peak under constant conditions, the M bout remains visible as a shoulder on the E bout in most individuals and in some even as separate activity bout (Helfrich‐Förster, 2000). Therefore, similar to vertebrates both activity bouts appear of endogenous nature. Comparable to vertebrates, the two fly activity bouts are closer together during cold and/or short days and further apart under hot and/or long days (Bazalova & Dolezel, 2017; Chiba, 1971; Majercak, Sidote, Hardin, & Edery, 1999; Rieger, Stanewsky, & Helfrich‐Förster, 2003; Rieger, Peschel, Dusik, Glotz, & Helfrich‐Förster, 2012). Thus, the adaptation of the daily activity patterns to different environmental conditions can be easily observed in flies, although individual animals are relatively short‐lived and do usually not experience several seasons (only the individuals that overwinter experience autumn, winter, and spring). The great advantage of studying flies is that the molecular and neuronal mechanisms of the circadian clock are well‐understood in D. melanogaster (see King & Sehgal, this issue) and start to emerge also in other Diptera (Bazalova & Dolezel, 2017; Bertolini et al., 2018; Codd et al., 2007; Gentile, Rivas, Meireles‐Filho, Lima, & Peixoto, 2009; Gesto et al., 2015; Kaiser et al., 2016; Kyriacou, 2014; Meireles‐Filho & Kyriacou, 2013; Meuti, Stone, Ikeno, & Denlinger, 2015; Noreen, Pegoraro, Nouroz, Tauber, & Kyriacou, 2018; Rivas et al., 2018; Rund, Hou, Ward, Collins, & Duffield, 2011). In D. melanogaster, recent studies have shown that M and E activity bouts are reflected by Ca2+ rhythms in the relevant clock neurons controlling M and E activity, respectively (Liang, Holy, & Taghert, 2016, 2017). As shown for the electrical activity in the SCN of hamsters, the Ca2+ peaks in the M and E neurons are close together in flies that have been kept under short‐day conditions and far apart in flies that have been kept under long‐day conditions (Liang et al., 2016). Thus, the different activity patterns of flies in short and long photoperiods have a neuronal basis (reviewed in Helfrich‐Förster, 2017; Yoshii, Rieger, & Helfrich‐Förster, 2012).
In this review, we focus on the circadian clocks of insects within the family Drosophilidae, which is part of the order Diptera (flies and mosquitoes; Figure 1a); they alone comprise many species that colonized almost all parts of the world. Currently, about 4,000 species are described (Brake & Bächli, 2008; O'Grady & DeSalle, 2018) but a number of surveys have projected the eventual number of Drosophilidae species to 5,200 (summarized in O'Grady & DeSalle, 2018). This is a huge number of species in comparison to the overall 6,399 known mammals (Burgin, Colella, Kahn, & Upham, 2018). Drosophilidae are thought to have originated in the tropics (as assumed for mammals) about 50 million years ago and have subsequently colonized virtually all environments including subarctic regions (Markow & O'Grady, 2007; Obbard et al., 2012; O'Grady & DeSalle, 2018; Russo, Mello, Frazão, & Voloch, 2013; Throckmorton, 1975). These elements make the Drosophilidae family an unprecedented model for studying the evolutionary processes that underlie the adaptation of circadian clocks to different environments. Here, we will review our current understanding of these processes.
Figure 1.
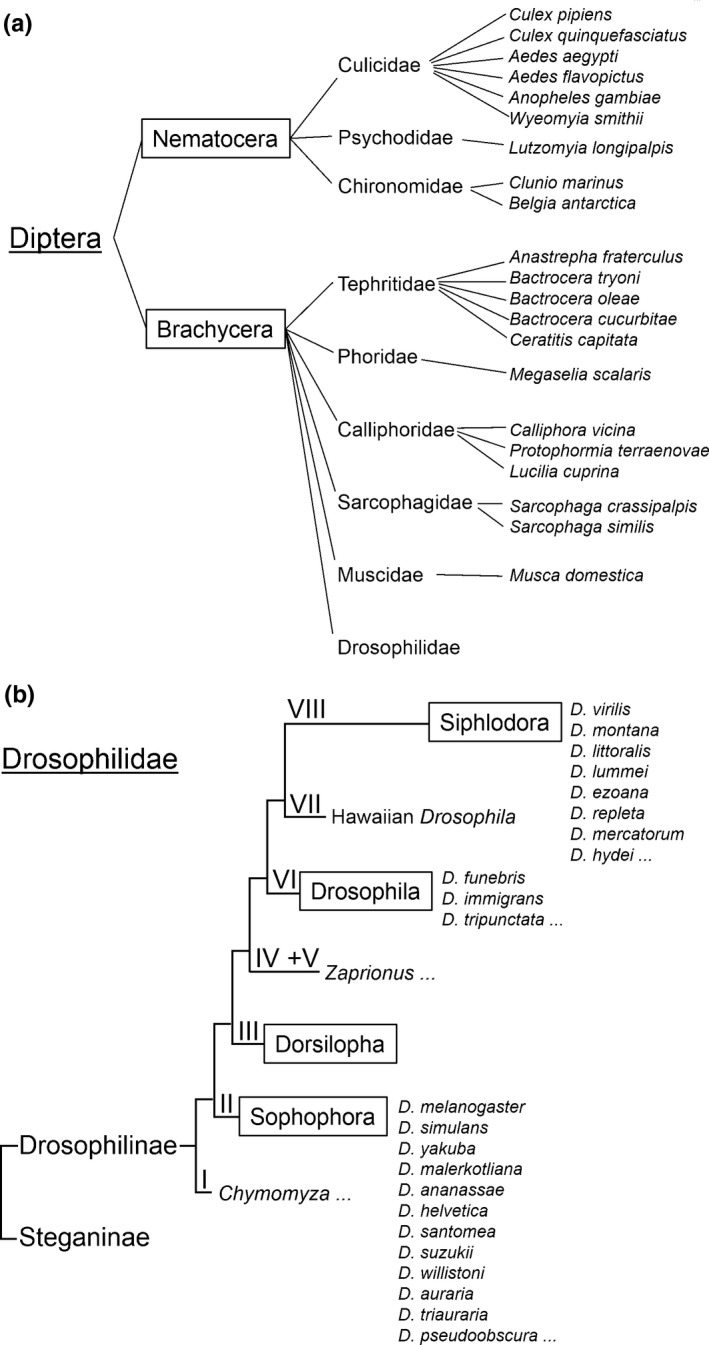
Rough classification of Diptera with a focus on the phylogeny of the family Drosophilidae. (a) The Diptera are historically divided in the Nematocera and Brachycera. The Nematocera contain ~34 and the Brachycera ~110 families. Here, only the families are shown, in which some species have been investigated in respect of their rhythms and/or clock genes. (b) Phylogeny of the family Drosophilidae (after Yassin, 2013 and O'Grady & DeSalle, 2018). They are divided into two sister groups, the Drosophilininae and Steganinae. The subfamily Drosophilinae is divided in eight clades (I–VIII) of which always the groups that are on the same “vertical” level are sister groups (e.g., Chymomyza is the sister group of the remaining Drosophilinae (clades II–VIII) and the group of Sophophora can be regarded as the sister group of clade III–VIII, etc.). As in a, emphasis is laid on those species that have been investigated from the chronobiological point of view. These stem predominantly from the Subgenera Sophophora and Siphlodora and are listed on the right. Furthermore, rhythms of the species from the genus Chymomyza (clade I) and Zaprionus (clade IV + V) have been studied. So far none of the species belonging to the subgenus Drosophila has been investigated for rhythms. The three species listed here are the key species of this group. Details see text
2. PHYLOGENY OF FLIES (DIPTERA) WITH A SPECIAL FOCUS ON THE DROSOPHILIDAE
The flies (Diptera) are historically divided into the Nematocera (lower Diptera, mosquitoes + others) and Brachycera (higher Diptera, “real” flies), but the Nematocera classification is controversial and this suborder was recently identified as paraphyletic to the Brachycera (Amorim & Yeates, 2006; Beckenbach, 2012). Here, we mention only few families of which some members have been studied in respect of rhythmic behaviour and/or clock genes (Figure 1a). Among these are mosquitoes (Culicidae) (Chiba, 1971; Chiba et al., 1982; Gentile et al., 2009; Meuti et al., 2015) and sand flies (Psychodidae) (Meireles‐Filho, Amoretty, De Souza, Kyriacou, & Peixoto, 2006; Meireles‐Filho, da S. Rivas, et al., 2006). Mosquitoes and sand flies are very interesting because most of them are blood‐sucking and potential disease vectors (Meireles‐Filho & Kyriacou, 2013). Furthermore, they have undergone rapid speciation colonizing very different habitats all over the world. Their success may be partly due to rapidly evolving clock genes that allowed them to colonize different temporal niches (Kyriacou, 2014). Particularly interesting is also the pitcher plant mosquito, Wyeomyia smithii, which has undergone an evolutionary transition from blood feeding to obligate nonbiting and at the same time an expansion from the south to the north of North America (Bradshaw et al., 2018; Merz et al., 2013). This species has become a model species to investigate the genetic mechanisms underlying photoperiodic time measurement (Bradshaw, Emerson, Catchen, Cresko, & Holzapfel, 2012). In the family of Chironomidae, Clunio marinus and Belgica antarctica, both living in intriguing environments, are studied for their endogenous clocks. Clunio marinus, is the only fly that colonized sea water and it possesses circadian and circatidal clocks (Neumann, 1989; Kaiser et al., 2016). Belgica Antarctica the only insect endemic to Antarctica (Kobelková et al., 2015).
Thus far, the neuronal basis of the circadian clock has not been revealed in any of the “lower Diptera” species.
The Brachycera comprise about 120 families. Species of the following families have been investigated with respect to their rhythms: Tephritidae (An et al., 2002; An, Tebo, Song, Frommer, & Raphael, 2004; Bertolini et al., 2018; Chahad‐Ehlers et al., 2017; Fuchikawa et al., 2010; Matsumoto et al., 2008; Mazzotta et al., 2005; Miyatake et al., 2002), Phoridae (Bostock, Green, Kyriacou, & Vanin, 2017), Calliphoridae (Muguruma, Goto, Numata, & Shiga, 2010; Saunders, 1997; Smith, 1987; Shiga & Numata, 2009; Warman, Newcomb, Lewis, & Evans, 2000; Yasuyama, Hase, & Shiga, 2015), Sarcophagidae (Goto & Denlinger, 2002; Koštál, Závodská, & Denlinger, 2009; Yamamoto, Nishimura, & Shiga, 2017; Yamamoto, Shiga, & Goto, 2017), Muscidae (Codd et al., 2007; Bazalova & Dolezel, 2017; Pyza & Meinertzhagen, 2003; Pyza, Siuta, & Tanimura, 2003), and Drosophilidae (see below) (Figure 1a). Among these, the genetic and neuronal basis of the circadian clock was revealed for the house fly M. domestica (Codd et al., 2007), the blow fly Protophormia terraenovae (Muguruma et al., 2010) and recently also for the olive fly, Bactrocera oleae (Bertolini et al., 2018). In all three fly species, it turned out to be rather similar to that of the fruit fly D. melanogaster, although the families of Tephritidae, Muscidae, Calliphoridae, and Drosophilidae have separated millions of years ago.
The phylogeny of the Drosophilidae is complex and still not completely resolved. A recent elaborate review can be found in O'Grady and DeSalle (2018). Most researchers agree to the idea that the Drosophilidae can be divided into two sister subfamilies, the Steganinae and the Drosophilinae (discussed in O'Grady & DeSalle, 2018) (for the definition of sister groups, see Figure 1b). Since all of the species investigated for their endogenous clocks are within the Drosophilinae, we will only discuss this subfamily (Figure 1b). Yassin (2013) divided the Drosophilinae into eight major clades (see Figure 1b). Clade I can be regarded as sister clade of the other seven clades (II–VII; which cover the main Drosophila groups) and contains the genus Chymomyza (which is investigated from the chronobiological point of view (e.g., Koštál, 2011a,b; Stehlík, Závodská, Shimada, Sauman, & Kostál, 2008)) and several other genera that are not important for this review. Within the main Drosophila groups, clade II is again the sister group of the remaining six clades. Clade II coincides with the subgenus Sophophora and contains the “model” species D. melanogaster and other species that have been investigated from the chronobiological point of view such as D. suzukii, D. yakuba, D. simulans, D. pseudoobscura, D. helvetica, D.ananassae, D. willistoni, D. auraria, and D. triauraria (Gleason & Powell, 1997; Hamby, Kwok, Zalom, & Chiu, 2013; Hermann et al., 2013; Joshi, 1999; Low, Lim, Ko, & Edery, 2008;Nishinokubi et al., 2003; Nishinokubi, Shimoda, & Ishida, 2006; Ousley et al., 1998; Pittendrigh, 1967; Pittendrigh & Takamura, 1989; Prabhakaran, De, & Sheeba, 2013; Prabhakaran & Sheeba, 2012, 2013, 2014; Vanlalhriatpuia et al., 2007; Wheeler et al., 1991; Yamada & Yamamoto, 2011). Important for the present review is that the subgenus Sophophora has separated from the remaining groups (clades III–VIII), among which is the Drosophila subgenus (clade VI), about 30 million years ago. Clade III that contains the subgenus Dorsilopha is again the sister clade of the remaining clades (clades IV–VIII). Clades IV and V are heterogenous and the lineages within this groups are still under debate (O'Grady & DeSalle, 2018; Russo et al., 2013; Yassin, 2013). Therefore, we will treat them as one group (Figure 1b). This group includes the tropical genus Zaprionus that has been investigated from the chronobiological point of view (Beauchamp et al., 2018; Prabhakaran & Sheeba, 2013, 2014). The subgenus Drosophila (clade VI) has been redefined recently by Yassin (2013) and corresponds roughly to the immigrans‐tripunctata radiation of Throckmorton (1975). Whereas the immigrans radiation consists of New World species, the tripunctata radiation contains Old World species. None of these species has been studied so far for their rhythmic behaviour. The last two clades are the Hawaiian Drosophila species and the subgenus Siphlodora. The subgenus Siphlodora was also recently redefined (Yassin, 2013) and corresponds roughly to the repleta‐virilis radiation of Throckmorton (1975). Several species within the virilis radiation have colonized high‐latitudes and were investigated for their rhythmic behaviour and neuronal clock network (Bahn, Lee, & Park, 2009; Beauchamp et al., 2018; Hermann et al., 2013; Kauranen et al., 2012; Lankinen, 1986; Lankinen & Forsman, 2006; Menegazzi et al., 2017).
3. THE MOLECULAR BASIS OF THE CIRCADIAN CLOCK IN D. melanogaster AND ITS ADAPTATION TO DIFFERENT ENVIRONMENTAL CONDITIONS
In 2017, the Nobel Prize in Physiology/Medicine was awarded to Jeffrey Hall, Michael Young, and Michael Rosbash for their work that led to the understanding of the molecular basis of circadian rhythms in D. melanogaster (Callaway & Ledford, 2017). This work was initiated by Konopka and Benzer in the 1970s of the last century by the isolation of the period mutants (Konopka & Benzer, 1971). The period gene participates in interlocked molecular transcriptional/translational feedback loops (Hardin, 2011; Glossop, Lyons, & Hardin, 1999).
A first feedback loop involves the clock genes period (per), timeless (tim), cycle (cyc), and Clock (Clk), and their respective products. CLK and CYC form heterodimers and bind to E‐box regulatory elements in the promoters of per and tim, activating their transcription. Consequently, per and tim mRNA levels rise and are translated in the cytoplasm, where their products PER and TIM are subjected to posttranslational modification, dimerize, and after a while enter the nucleus as a complex. In the nucleus, PER/TIM complexes bind to CLK/CYC and repress their transcriptional activity. Doing so, they negatively regulate their own expression. Subsequent PER and TIM destabilization and degradation stops the repression on CLK/CYC activity, and a new transcriptional‐translational cycle restarts.
A second feedback loop involves the clock genes, cycle (cyc), Clock (Clk), Vrille (Vri), and PAR Domain Protein 1ε (Pdp1ε), and their respective products. Vri and Pdp1ε carry E‐box regulatory elements in their promoters, therefore their expression is also activated by the active CLK/CYC complex. VRI accumulates earlier than PDP1ε and it represses the expression of Clk, acting at the level of VP‐boxes (VRI/PDP regulatory elements (Emery & Reppert, 2004) present in its promoter region. PDP1ε accumulates later than VRI and finally promotes Clk expression. The synergistic activity of VRI and PDP1ε generates circadian transcription of Clk. Several additional clock factors, that are not focus of this review, fine‐tune the transcriptional/translational feedback loops (for deeper insights see Glossop et al., 1999; Hardin, 2011; Hardin & Panda, 2013; Helfrich‐Förster, 2017; Ozkaya & Rosato, 2012).
Environmental light‐dark cycles synchronize the molecular oscillations via Rhodopsins and Cryptochrome (CRY) (reviewed in Helfrich‐Förster, 2017; Yoshii, Hermann‐Luibl, & Helfrich‐Förster, 2016). CRY makes the Drosophila extraordinary light sensitive (Vinayak et al., 2013), because it interacts directly with the core clock proteins: light‐activated CRY binds to TIM and leads to its degradation in the proteasome (Ceriani et al., 1999; Naidoo, Song, Hunter‐Ensor, & Sehgal, 1999). Without TIM, PER is also destabilized and degraded, which immediately resets the clock. When flies are exposed to constant light instead of light‐dark cycles, TIM is permanently degraded, which makes the flies arrhythmic, whereas they remain rhythmic without functional CRY (Emery, So, Kaneko, Hall, & Rosbash, 1998; Emery, Stanewsky, Hall, & Rosbash, 2000a,b; Stanewsky et al., 1998). Due to the high light sensitivity of the flies, arrhythmicity of wild‐type flies occurs already at rather low irradiances (Konopka, Pittendrigh, & Orr, 1989) and flies with CRY overexpressed show even a higher sensitivity towards light (Emery et al., 1998).
3.1. Polymorphisms in TIM and PER affect clock sensitivity to light and temperature
The original tim gene gives rise to a short form of TIM (= S‐TIM) that strongly interacts with CRY and is therefore sensitive to degradation by light. However, ~10,000 years ago, after D. melanogaster colonized Europe, a mutation occurred in the south eastern Italian populations, in which a single guanosine was inserted in the 5′ coding region of tim that resulted in the production of a long TIM isoform in addition to a short one (= LS‐TIM). The ls‐tim mutants are less light sensitive due to a reduced ability of LS‐TIM to interact with CRY, and the ls‐tim mutants gradually invaded Northern Europe (Sandrelli et al., 2007; Tauber et al., 2007). The lower light sensitivity of the flies prevents the flies from enhanced TIM degradation and from getting arrhythmic during the long exposure to light in northern summers. On the other hand, the ls‐tim mutation causes females to enter diapause (reproductive arrest) earlier in autumn than the s‐tim allele. Together with the reduced light‐sensitivity of ls‐tim flies, the earlier diapause induction is advantageous for a life in the north. Thus, the observed latitudinal cline in TIM polymorphism appears to be an evolutionary adaptation to the conditions at higher latitudes (Kyriacou, Peixoto, Sandrelli, Costa, & Tauber, 2008). Polymorphisms appear also present in CRY, but a clear latitudinal cline was so far not observed (Pegoraro et al., 2015).
Nevertheless, a latitudinal cline in PER polymorphism exists, but in contrast with the TIM polymorphism, the PER polymorphism does not affect light‐sensitivity but has a role in temperature adaptation (reviewed in Costa & Kyriacou, 1998; Kyriacou et al., 2008). PER possesses an uninterrupted stretch of alternating threonine‐glycine (Thr‐Gly) repeats at its C‐terminus (Yu, Colot, Kyriacou, Hall, & Rosbash, 1987). This unusual sequence is shorter in flies stemming from southern D. melanogaster populations and longer in flies stemming from northern populations in Europe. This latitudinal cline in the Thr‐Gly length turned out to be important for temperature compensation of the clock. The northern populations with long Thr‐Gly stretch had the most thermally stable periods that were close to 24 hr at all temperatures tested. In contrast, southern populations, with short Thr‐Gly stretch, had free‐running periods of 24 hr at 29°C, but the periods shortened significantly at cooler temperatures. Thus, the “temperature compensated” PER protein with long Thr‐Gly stretch appears better adapted to the colder and thermally variable higher latitudes, whereas the PER protein with short Thr‐Gly stretch is more suitable for the warmer Mediterranean region. However, this latitudinal cline of the PER Thr‐Gly stretch was not observed in D. melanogaster populations collected along the coast of Australia, suggesting that there exist other adaptations to latitudinal clines in temperature (Weeks, McKechnie, & Hoffmann, 2006).
Nevertheless, the polymorphisms in PER and TIM offer a fine‐tuning of daily rhythms to the environment and can be regarded as evolutionary adaptations of the clock to a life in warmer or colder regions.
3.2. Differential splicing of the clock genes contributes to the adaptation to different temperatures
In addition to polymorphisms in the clock genes, D. melanogaster flies can adapt their activity pattern to changes in the environment by splicing per and tim differently. The mRNAs of both genes are differentially spliced in their 3′ untranslated regions, and in both cases the degree of splicing is dependent on the environmental temperature. Per shows an enhanced splicing at low and an attenuated splicing at higher environmental temperatures and long days (Majercak et al., 1999; Majercak, Chen, & Edery, 2004). For tim it is the opposite: it is spliced to a larger degree at high temperatures than at low ones (Montelli et al., 2015). Enhanced per splicing at low temperatures leads to a quicker accumulation of per mRNA and PER protein, which accelerates the molecular cycle and advances the evening activity of the flies, whereas reduced per‐splicing at high temperatures slows down the molecular cycle and delays the evening activity. This explains activity timing of the flies under cold and short autumn days and long and hot summer days (Majercak et al., 2004; Montelli et al., 2015): under autumn condition the flies start evening activity early in the day and lack the midday siesta almost completely, whereas under summer conditions they have a pronounced siesta and start evening activity late. Most interestingly, differential tim splicing enhances these seasonal adaptations. The unspliced TIM isoforms at low temperatures have a higher affinity for CRY what leads to an earlier degradation of TIM and an advance of evening activity under cold days (Montelli et al., 2015). Under warm summer days the spliced TIM isoforms have less affinity to CRY, TIM is degraded later and evening activity is also later. Thus, per‐ and tim‐splicing work in the same direction—advancing evening activity under short winter days and delaying it under long summer days.
The thermal regulation of per‐splicing can be regarded as a mechanism that facilitated the radiation of D. melanogaster from the tropics to temperate climates (Figure 2a). Consistent with this hypothesis, Low et al. (2008), Low, Chen, Yildirim, and Edery (2012) discovered several single nucleotide polymorphisms (SNPs) in per's 3′ untranslated region of D. melanogaster that modulate its splicing efficiency. Most significantly, there was a latitudinal cline in these SNPs in wild‐caught populations of flies originating along the east coast of the United States with the least efficiently spliced versions associated with a longer midday siesta in regions where temperatures can reach high levels. Similarly, an altitudinal cline in per splicing was associated in D. melanogaster that influences midday siesta (Cao & Edery, 2017). This demonstrates that natural selection can work at the level of splicing signals and that differential splicing plays an important role in the thermal adaptation of life forms. Without doubt there are selection mechanisms that are independent of the per gene that adapted the rhythms of D. melanogaster flies to different temperatures as was shown by Maguire, Schmidt, and Sehgal (2014) for natural populations living at different altitudes in Africa. Nonetheless, we will continue to focus our review on the clock genes and latitudinal adaptations.
Figure 2.
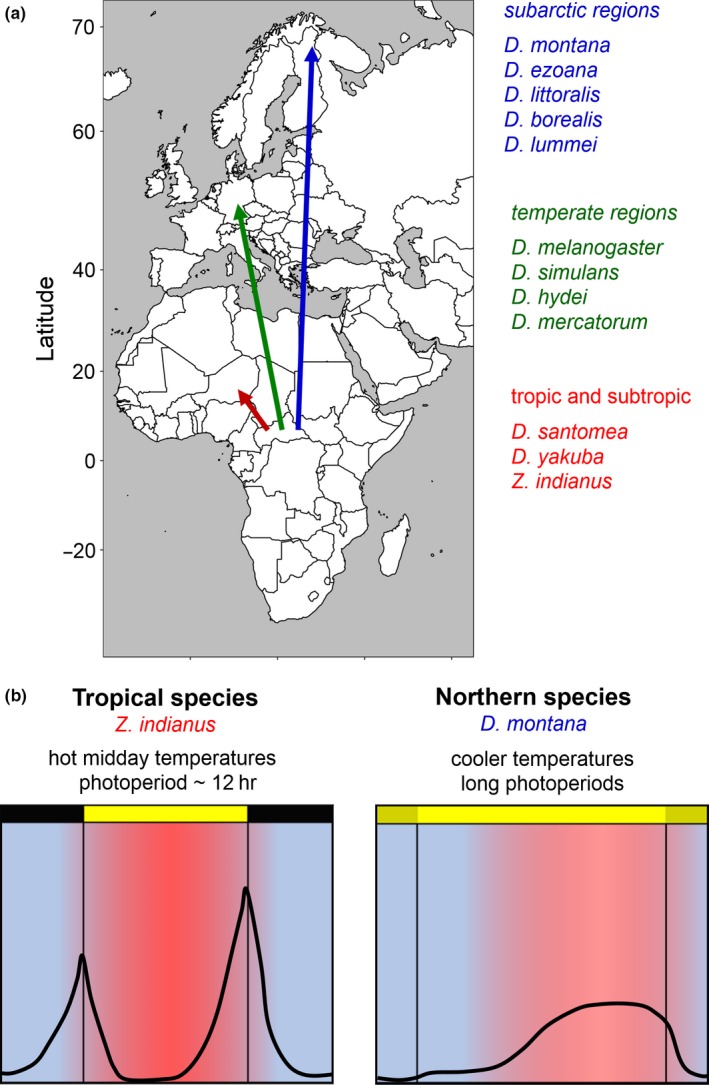
Colonization of northern latitudes by Drosophilidae and simplified activity patterns of tropical and subarctic flies. (a) Drosophilids are thought to have their origin in the tropics. Certain species still have a tropical or subtropical habitat (red). Others have colonized temperate regions (green) and still others colonized subarctic regions (blue). (b) The tropics are characterized by photoperiods that remain close to 12 hr throughout the year and hot midday temperatures. Flies living in this region (e.g., Zaprionus indianus) avoid the midday heat by taking a siesta and being active during the morning and evening. The summers in subarctic regions are characterized by very long photoperiods with moderate temperatures that are most pleasant during the afternoon. Flies living in this region (e.g., Drosophila montana) show a broad activity band in the afternoon that extends until dusk
4. EVOLUTIONARY ADAPTATIONS OF THE MOLECULAR CLOCK IN SPECIES OTHER THAN D. melanogaster
Like in D. melanogaster, there is thermal regulation of per‐splicing in D. simulans that also invaded temperate regions, but not in D. yakuba and D. santomea, which have a more ancestral distribution in equatorial regions of Africa (Low et al., 2008), where day length and temperature exhibit little fluctuation throughout the year (Figure 2). D. simulans flies also show similar PER polymorphisms in the Thr‐Gly repeats (Rosato, Peixoto, Barbujani, Costa, & Kyriacou, 1994). Thus, the thermal adaptations in form of per‐splicing and PER polymorphisms have most likely facilitated colonization of temperate northern zones on earth. The repetitive threonine‐glycine (Thr‐Gly) region at the PER C‐terminus has been investigated in several flies and found to be the longest in D. pseudoobscura (Colot, Hall, & Rosbash, 1988; Costa, Peixoto, Thackeray, Dalgleish, & Kyriacou, 1991; Nielsen et al., 1994; Peixoto, Costa, Wheeler, Hall, & Kyriacou, 1992; Peixoto et al., 1998). D. pseudoobscura flies have a wide geographic distribution and extend from Mexico along the western third of the North American continent to British Columbia (Wang & Hey, 1996). Thus, this species is exposed to very different climatic zones and needs to have a well temperature compensated clock, which was demonstrated by generating hybrid per transgenes between Drosophila pseudoobscura and D. melanogaster (Noreen et al., 2018; Peixoto et al., 1998). Nevertheless, not all species with wide geographic distribution have a long Thr‐Gly region in PER. For example, the Thr‐Gly region is short in the D. virilis group (subgenus Siphlodora; Figure 1b) and seems to encompass just two pairs of Thr‐Gly (Hilton & Hey, 1996; Lankinen & Forsman, 2006). Drosophila littoralis flies, which are latitudinally exceptionally widespread from Mediterranean regions to the northern side of the Arctic Circle, do not even show a latitudinal cline in the length of the Thr‐Gly stretch (Lankinen & Forsman, 2006). This shows that this region is not always included in the adaptive clock variability; obviously, these flies found other solutions to adapt their clocks to northern zones on earth.
Evolutionary modifications in TIM or CRY have so far not been demonstrated for fly species other than D. melanogaster, but they are most likely. At least a reduction in circadian photosensitivity of northern fly populations has been documented in Japanese D. auraria flies and was interpreted as being an adaptation of the circadian system to the long summer day lengths (Pittendrigh & Takamura, 1989). It is important to mention that all so far reported results are gained in species that belong to the Sophophora subgenus and are consequently relatively close to D. melanogaster (Figure 1b).
5. EVOLUTIONARY ADAPTATIONS OF THE NEURONAL CLOCK NETWORK ACROSS SPECIES
Only a few fruit fly species have colonized subarctic regions (Figure 2a). Among these are D. littoralis, D. montana, D. ezoana, and D. lummei, all belonging to the virilis‐repleta radiation that corresponds to the newly introduced subgenus Siphlodora (Figure 1b; O'Grady & DeSalle, 2018). In subarctic regions, the flies are not only exposed to cold winters that require effective overwintering strategies (e.g., a strictly photoperiodically controlled diapause), but also to very long photoperiods up to constant light in spring/summer (Figure 2b). The activity pattern of the high latitude species is quite different from the bimodal activity pattern of flies that inhabit lower latitudes (Beauchamp et al., 2018; Kauranen et al., 2012; Menegazzi et al., 2017): they show reduced activity in the morning, no siesta, and broad activity in the afternoon that extends until dusk (Figure 2b). Most interestingly, this activity pattern even persists to a certain extent under laboratory conditions with constant pleasant temperatures, demonstrating that the activity pattern is genetically determined; most probably, an altered circadian clock in the brain causes it. We do not know yet, to what degree alterations in the clock proteins contribute to the altered activity patterns, but most likely, they significantly do so. A recent study has conducted a phylogenetic analysis of the clock genes period, timeless, Clock, cycle, and the cryptochrome gene in 12 Drosophila species, among which was D. virilis that belongs to the subgenus Siphlodora (Noreen et al., 2018). As may be expected from the phylogenetic tree (Figure 1b), period, Clock, and Cycle of D. virilis showed the largest genetic distance to the respective genes in D. melanogaster. Nevertheless, this was slightly different for timeless, where the largest distance was present in D. pseudoobscura that belongs to the Sophophora subgenus (Figure 1b). For cryptochrome the largest genetic difference was revealed for D. willistoni, another species of the Sophophora subgenus that is assumed to be closely related to D. melanogaster (Figure 1b). Altogether, this indicates that the clock genes have differentially evolved, most probably due to different environmental challenges as already nicely revealed in mosquitoes (see above).
In addition to a putatively altered molecular clock, high latitude flies possess an altered neuronal clock network in comparison to Drosophila species that remained in tropic regions or colonized temperate regions on earth (Bahn et al., 2009; Hermann et al., 2013; Kauranen et al., 2012; Kauranen, Ala‐Honkola, Kankare, & Hoikkala, 2016; Menegazzi et al., 2017; Beauchamp et al., 2018). In the following, we will summarize the most important aspects of the clock network in the brain of D. melanogaster and then describe the observed alterations in the brains of high latitude species.
In D. melanogaster, the central clock consists of dorsal and lateral neurons that express the core clock genes and form an extensive neuropeptidergic network in the brain (Figure 3a; reviewed in Helfrich‐Förster et al., 2007; Hermann‐Luibl & Helfrich‐Förster, 2015; Helfrich‐Förster, 2017; Schubert, Hagedorn, Yoshii, Helfrich‐Förster, & Rieger, 2018). One of the best conserved and most important neuropeptide in the insect circadian clock is the pigment‐dispersing factor (PDF) (e.g., Beer et al., 2018; Helfrich‐Förster et al., 2000; Helfrich‐Förster, 2014; Ikeno, Numata, Goto, & Shiga, 2014; Renn, Park, Rosbash, Hall, & Taghert, 1999; Shafer & Yao, 2014; Wei et al., 2014). In D. melanogaster, PDF is expressed in four small ventro‐lateral neurons (s‐LNv) and in four large ventro‐lateral neurons (l‐LNv) (Figure 3a), which have different roles in the clock network. The s‐LNv are major pacemaker neurons that are essential for robust rhythmic activity under constant darkness and for pronounced morning activity under cycling environmental conditions. Furthermore, they are connected in a light‐dependent manner to the dorsal lateral neurons (LNd) and to one group of the dorsal neurons, the DN1p, and by this way shape the activity pattern under different light conditions (Chatterjee et al., 2018). The l‐LNv are dispensable for rhythmic activity, but they are part of the light‐input pathway to the clock and they control the phase of evening activity, especially under long photoperiods (Schlichting et al., 2016; Menegazzi et al., 2017). PDF is released in a rhythmic manner from the s‐LNv to the dorsal brain, whereas such a rhythm has not been demonstrated in the l‐LNv (Park et al., 2000; Fernández, Berni, & Ceriani, 2008). The l‐LNv produce amidated PDF that is more stable than the PDF from the s‐LNv (Park et al., 2008) and that may have long‐lasting effects on other clock neurons expressing the PDF‐receptor (Im, Li, & Taghert, 2011; reviewed in Helfrich‐Förster, 2014). Most importantly, PDF from the l‐LNv (or amidated synthetic PDF) phase‐delays the clock neurons that control evening activity (LNd and 5th LNv) (Yoshii et al., 2009; Liang et al., 2017; Menegazzi et al., 2017). Flies without functional PDF signalling cannot phase‐delay the Ca2+ oscillations in the evening neurons (Liang et al., 2016) and consequently cannot phase‐delay evening activity (Renn et al., 1999; Yoshii et al., 2009). Thus, their evening activity occurs in the middle of the day instead of in the late afternoon, and this is most evident under long photoperiods (Menegazzi et al., 2017; Schlichting et al., 2016; Yoshii et al., 2009). In contrast, flies in which the l‐LNv run and secrete PDF into the central brain instead of into the optic lobes exhibit a long siesta and late evening activity already under 12:12 hr light‐dark cycles (Wülbeck, Grieshaber, & Helfrich‐Förster, 2008). All this fits into the picture that PDF from the l‐LNv phase‐delays evening activity, although we cannot exclude that also PDF from the s‐LNv contribute to this effect.
Figure 3.
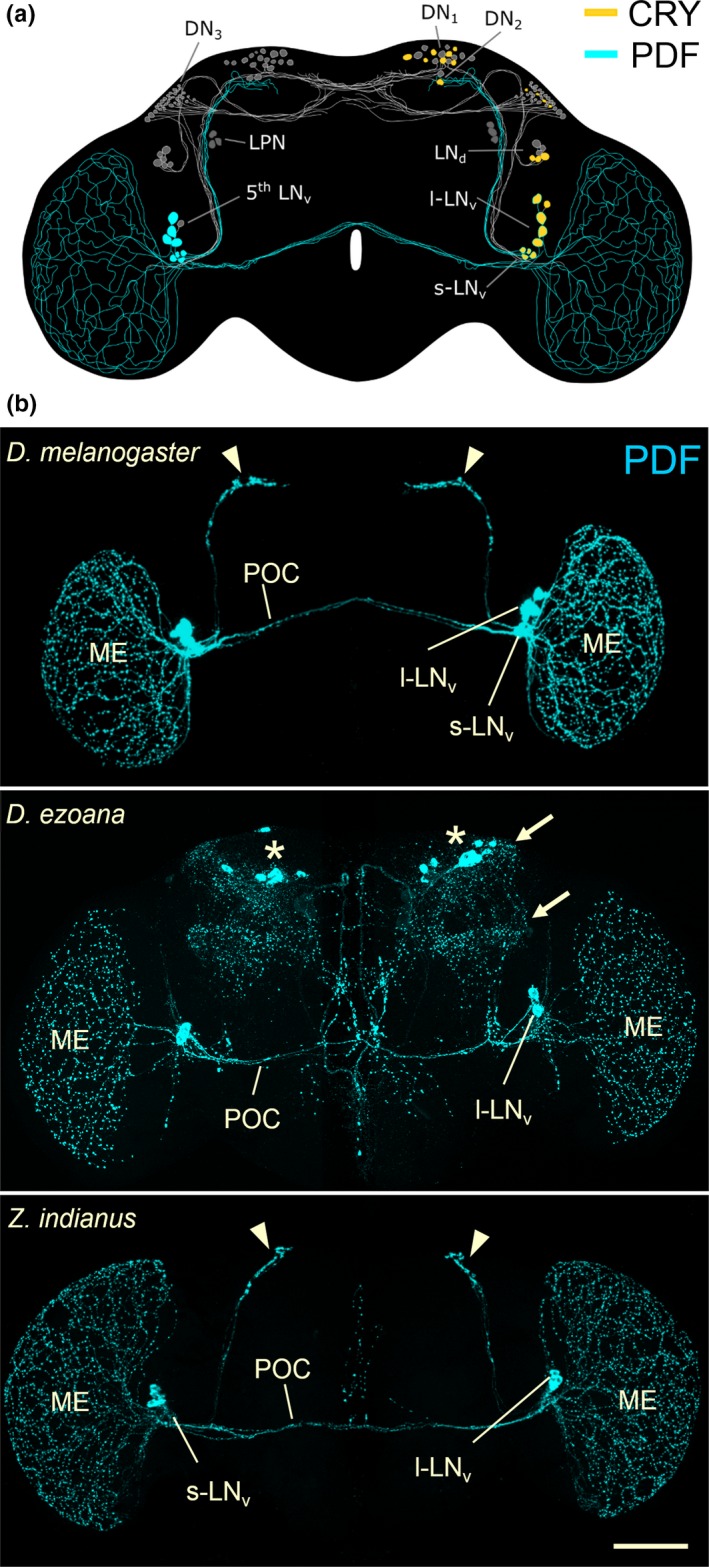
Clock network in the fruit fly brain. (a) Clock neurons and their neurites in Drosophila melanogaster. The neurons consist of lateral neurons (s‐LN v, l‐LN v, LN d, 5th LN v, LPN) and dorsal neurons (DN 1, DN 2, DN 3) that are heterogeneous in respect of neuropeptide and Cryptochrome (CRY) expression. Here, we focus on the s‐LN v and l‐LN v that express the Pigment‐Dispersing Factor (PDF; cyan, left hemisphere) and CRY (yellow, right hemisphere). (b) Confocal pictures showing the PDF neurons and their arborizations in the brain and the medulla of the optic lobe for D. melanogaster, the high‐latitude species D. ezoana and the tropic species Zaprionus indianus. D. melanogaster expresses PDF in the s‐LN v and in the l‐LN v. The l‐LN v send arborizations to the medulla (ME) and via the posterior optic commissure (POC) to the other brain hemisphere. The s‐LN v project into the central brain and terminate close to the dorsal neurons (arrowheads in b, compare with a). D. ezoana expresses PDF only in the l‐LN v. These also project via the POC to the other brain hemisphere, but many fibres leave the POC and invade the central brain. Here, the latter come close to the LN d and dorsal neurons (arrows in b, compare with a). In the dorsal central brain PDF is additionally expressed in non‐clock neurons (asterisks). Z. indianus expresses PDF in the s‐LN v and the l‐LN v as does D. melanogaster and the overall arborization pattern of these two groups of neurons also largely resembles D. melanogaster. The cell bodies of the s‐LN v are only weakly stained in this individual, but the terminals of the s‐LN v in the dorsal brain are nicely visible (arrow heads). Scale bar: 100 μm
As mentioned above, CRY is a similarly important molecule in the circadian clock of D. melanogaster, because it interacts directly with TIM and synchronizes the molecular clock to environmental light‐dark cycles. CRY is not the only fly circadian photoreceptor. The compound eyes and the extraretinal eyelets contribute essential aspects of entrainment and suffice to synchronize flies that lack functional CRY (reviewed in Yoshii et al., 2016). Nevertheless, CRY is responsible for the extraordinary light sensitivity of the circadian clock. Flies without functional CRY are not able to phase‐shift their activity rhythms in response to short light‐pulses and they do not become arrhythmic under constant light (Emery et al., 1998, 2000a,b; Kistenpfennig, Hirsh, Yoshii, & Helfrich‐Förster, 2012). CRY is expressed in about half of the clock neurons, among which are the PDF‐positive neurons (Figure 3a).
High latitude Drosophila species show evident differences in the expression of PDF and CRY in the clock neurons in comparison to D. melanogaster: They lack PDF in the s‐LNv and CRY in the l‐LNv (Bahn et al., 2009; Beauchamp et al., 2018; Hermann et al., 2013; Kauranen et al., 2012; Menegazzi et al., 2017). Furthermore, their l‐LNv invade the central brain in addition to the optic lobes (Figure 3b). The lack of PDF in the s‐LNv explains the reduced morning activity of the flies, and the PDF arborizations of the l‐LNv in the central brain may explain their broad late evening activity. In addition, they do not become completely arrhythmic under constant light that might be caused by the absence of CRY in the l‐LNv. Most interestingly, we could partly mimic the activity patterns of the high‐latitude Drosophila species in D. melanogaster by expressing PDF only in the l‐LNv and the central brain and by additionally downregulating CRY in the l‐LNv (Menegazzi et al., 2017). This indicates that there is indeed a causal relation between PDF/CRY expression and activity patterns.
Further evidence comes from the comparison of the PDF network and CRY expression in fly species that are also from the Siphlodora subgenus, but did not invade subarctic regions, such as D. hydei and D. mercatorum (Figure 1b). These showed a very similar clock network as D. melanogaster in which CRY and PDF was present in the s‐LNv and l‐LNv and no or very little PDF in the central brain (Beauchamp et al., 2018). Very similarly, Zaprionus indianus and Z. camerounensis that are also distantly related to the Sophophora subgenus, but restricted to tropic regions, exhibit a CRY/PDF expression pattern that is undistinguishable from D. melanogaster (Figure 3b) (Beauchamp et al., 2018). Even subtropic species outside the Drosophilidae such as olive flies, B. oleae (Diptera: Tephritidae), have a clock network that strongly resembled that of D. melanogaster (Bertolini et al., 2018). Together, these results strongly suggest that the D. melanogaster‐like clock network is the ancestral one that is preserved in all fly species that did not invade high latitudes. Only flies that colonized the very north, may have lost PDF in the s‐LNv, gained PDF fibres from the l‐LNv running into the central brain, and have lost CRY in the l‐LNv. If our hypothesis is true, there might even exist fly species in the Sophophora group that colonized the north and developed the clock network typical for high latitude species. Indeed, the latter appears to apply for D. pseudoobscura that have the typical high latitude clock network (Hermann et al., 2013). D. pseudoobscura did not invade subarctic regions, but they colonized regions in Canada up to 60°N.
Nevertheless, as already discussed for the Thr‐Gly repeat in PER of the Siphlodora subgenus, there are different ways how the circadian clock can adapt to high latitude habitats. The species Chymomyza costata, which is at the base of the Drosophilinae (Figure 1b) is well‐known for its extreme cold resistance (Koštál, 2011a,b). C. costata flies are distributed in Eastern Siberia, Northern Lapland, Iceland, and from northern Japan to the Artic Cycle (Hackman, Lakovaara, Saura, Sorsa, & Vepsalainen, 1970). Nevertheless, they possess a D. melanogaster‐like clock network and their rhythmic behaviour under artificial long photoperiods in the lab is also D. melanogaster‐like (Bertolini and Menegazzi, unpublished observations). This confirms that the clock network determines rhythmic behaviour, but it also shows that the circadian clock of C. costata flies has found other ways to adapt to high‐latitudes. Future studies have to reveal these mechanisms.
6. CONCLUSION
The dissection of the circadian clock into its molecular components propelled the understanding of circadian timing mechanisms. Clock genes evolve rapidly (Colot et al., 1988) permitting latitude‐specific modifications that enable organisms to maintain an optimal timing while colonizing higher latitudes (Hut, Paolucci, Dor, Kyriacou, & Daan, 2013; Kyriacou et al., 2008). Here, we discussed additional modifications of the brain clock network evolved in Drosophila species that colonized very high latitudes. These modifications concern the clock factors CRY and PDF that are involved in circadian photoreception and the timing of activity. It will be most promising to investigate, whether similar modifications are also present in other insect species that colonized very high latitudes and if not, which solutions these found to cope with the environmental challenges at these regions from a clock watcher's perspective.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
C.H.F., E.B., and P.M. planned and discussed the outline of the review, E.B. designed Figure 3a and the graphical abstract, C.H.F. wrote and E.B. and P.M. corrected the manuscript.
Supporting information
ACKNOWLEDGEMENTS
We thank the German Research Foundation (DFG) for funding our research (SFB 1047 “Insect Timing”, Project A1, as well as FO 207‐15 to CHF and ME 4866‐1 to PM).
Helfrich‐Förster C, Bertolini E, Menegazzi P. Flies as models for circadian clock adaptation to environmental challenges. Eur J Neurosci. 2020;51:166–181. 10.1111/ejn.14180
Edited by Rae Silver. Reviewed by Amita Sehgal.
All peer review communications can be found with the online version of the article.
DATA ACCESSIBILITY
This review does not report unpublished primary data.
REFERENCES
- Amorim, D. D. S. , & Yeates, D. K. (2006). Pesky gnats: Ridding dipteran classification of the Nematocera. Studia Dipterologica, 13, 3–9. [Google Scholar]
- An, X. , Tebo, M. , Song, S. , Frommer, M. , & Raphael, K. A. (2004). The cryptochrome (cry) gene and a mating isolation mechanism in tephritid fruit flies. Genetics, 168, 2025–2036. 10.1534/genetics.104.028399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, X. , Wilkes, K. , Bastian, Y. , Morrow, J. L. , Frommer, M. , & Raphael, K. A. (2002). The period gene in two species of tephritid fruit fly differentiated by mating behaviour. Insect Molecular Biology, 11, 419–430. 10.1046/j.1365-2583.2002.00351.x [DOI] [PubMed] [Google Scholar]
- Aschoff, J. (1966). Circadian activity pattern with two peaks. Ecology, 47, 657–662. 10.2307/1933949 [DOI] [Google Scholar]
- Bahn, J. H. , Lee, G. , & Park, J. H. (2009). Comparative analysis of Pdf‐mediated circadian behaviors between Drosophila melanogaster and D. virilis . Genetics, 181, 965–975. 10.1534/genetics.108.099069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazalova, O. , & Dolezel, D. (2017). Daily activity of the house fly, Musca domestica, is influenced by temperature independent of 3′ UTR period gene splicing. G3 (Bethesda, MD), 7, 2637–2649. 10.1534/g3.117.042374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp, M. , Bertolini, E. , Deppisch, P. , Steubing, J. , Menegazzi, P. , & Helfrich‐Förster, C. (2018). Closely related fruit fly species living at different latitudes diverge in their circadian clock anatomy and rhythmic behaviour. Journal of Biological Rhythms. 10.1177/0748730418798096. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Beckenbach, A. T. (2012). Mitochondrial genome sequences of nematocera (lower diptera): Evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biology and Evolution, 4, 89–101. 10.1093/gbe/evr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer, K. , Kolbe, E. , Kahana, N. B. , Yayon, N. , Weiss, R. , Menegazzi, P. , … Helfrich‐Förster, C. (2018). Pigment‐dispersing factor‐expressing neurons convey circadian information in the honey bee brain. Open Biology, 8 10.1098/rsob.170224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini, E. , Kistenpfennig, C. , Menegazzi, P. , Keller, A. , Koukidou, M. , & Helfrich‐Förster, C. (2018). The characterization of the circadian clock in the olive fly Bactrocera oleae (Diptera: Tephritidae) reveals a Drosophila‐like organization. Scientific Reports, 8, 816 10.1038/s41598-018-19255-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock, E. , Green, E. W. , Kyriacou, C. P. , & Vanin, S. (2017). Chronobiological studies on body search, oviposition and emergence of Megaselia scalaris (Diptera, Phoridae) in controlled conditions. Forensic Science International, 275, 155–159. 10.1016/j.forsciint.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Bradshaw, W. E. , Burkhart, J. , Colbourne, J. K. , Borowczak, R. , Lopez, J. , Denlinger, D. L. , … Holzapfel, C. M. (2018). Evolutionary transition from blood feeding to obligate nonbiting in a mosquito. Proceedings of the National Academy of Sciences of the United States of America, 115, 1009–1014. 10.1073/pnas.1717502115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, W. E. , Emerson, K. J. , Catchen, J. M. , Cresko, W. A. , & Holzapfel, C. M. (2012). Footprints in time: Comparative quantitative trait loci mapping of the pitcher‐plant mosquito, Wyeomyia smithii . Proceedings of the Royal Society of London. Series B, Biological sciences, 279, 4551–4558. 10.1098/rspb.2012.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake, I. , & Bächli, G. (2008). World catalogue of insects in Drosophilidae (Diptera). Stenstrupt, Denmark: Apollo Books. [Google Scholar]
- Bünning, E. (1960). Circadian rhythms and time measurement in photoperiodism. Cold Spring Harbor Symposia on Quantitative Biology, 25, 249–256. 10.1101/SQB.1960.025.01.026 [DOI] [PubMed] [Google Scholar]
- Burgin, C. J. , Colella, J. P. , Kahn, P. L. , & Upham, N. S. (2018). How many species of mammals are there? Journal of Mammalogy, 99, 1–14. 10.1093/jmammal/gyx147 [DOI] [Google Scholar]
- Callaway, E. , & Ledford, H. (2017). Medicine Nobel awarded for work on circadian clocks. Nature, 550, 18. [DOI] [PubMed] [Google Scholar]
- Cao, W. , & Edery, I. (2017). Mid‐day siesta in natural populations of D. melanogaster from Africa exhibits an altitudinal cline and is regulated by splicing of a thermosensitive intron in the period clock gene. BMC Evolutionary Biology, 17, 32 10.1186/s12862-017-0880-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani, M. F. , Darlington, T. K. , Staknis, D. , Más, P. , Petti, A. A. , Weitz, C. J. , & Kay, S. A. (1999). Light‐dependent sequestration of timeless by cryptochrome. Science, 285, 553–556. 10.1126/science.285.5427.553 [DOI] [PubMed] [Google Scholar]
- Chahad‐Ehlers, S. , Arthur, L. P. , Lima, A. L. A. , Gesto, J. S. M. , Torres, F. R. , Peixoto, A. A. , & de Brito, R. A. (2017). Expanding the view of Clock and cycle gene evolution in Diptera. Insect Molecular Biology, 26, 317–331. 10.1111/imb.12296 [DOI] [PubMed] [Google Scholar]
- Chatterjee, A. , Lamaze, A. , De, J. , Mena, W. , Chélot, E. , Martin, B. , … Rouyer, F. (2018). Reconfiguration of a multi‐oscillator network by light in the Drosophila circadian clock. Current Biology, 28, 2007–2017.e4. 10.1016/j.cub.2018.04.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, Y. (1971). Phase angle relationship between the circadian activity of the mosquito, Aedes flavopictus, and 24 hr LD cycles. Japanese Journal of Ecology, 21, 221–226. [Google Scholar]
- Chiba, Y. , Kubota, M. , & Nakamura, Y. (1982). Differential effects of temperature upon evening and morning peaks in the circadian activity of mosquitoes. Journal of Interdisciplinary Cycle Research, 13, 55–60. 10.1080/09291018209359763 [DOI] [Google Scholar]
- Codd, V. , Dolezel, D. , Stehlik, J. , Piccin, A. , Garner, K. J. , Racey, S. N. , … Rosato, E. (2007). Circadian rhythm gene regulation in the housefly Musca domestica . Genetics, 177, 1539–1551. 10.1534/genetics.107.079160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, H. V. , Hall, J. C. , & Rosbash, M. (1988). Interspecific comparison of the period gene of Drosophila reveals large blocks of non‐conserved coding DNA. EMBO Journal, 7, 3929–3937. 10.1002/j.1460-2075.1988.tb03279.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, R. , & Kyriacou, C. P. (1998). Functional and evolutionary implications of natural variation in clock genes. Current Opinion in Neurobiology, 8, 659–664. 10.1016/S0959-4388(98)80096-2 [DOI] [PubMed] [Google Scholar]
- Costa, R. , Peixoto, A. A. , Thackeray, J. R. , Dalgleish, R. , & Kyriacou, C. P. (1991). Length polymorphism in the threonine‐glycine‐encoding repeat region of the period gene in Drosophila . Journal of Molecular Evolution, 32, 238–246. 10.1007/BF02342746 [DOI] [PubMed] [Google Scholar]
- Daan, S. (2010). A history of chronobiological concepts In Albrecht U. (Ed.), The circadian clock (pp. 1–35). New York, NY: Springer. [Google Scholar]
- Daan, S. , & Aschoff, J. (1975). Circadian rhythms of locomotor activity in captive birds and mammals: Their variations with season and latitude. Oecologia, 18, 269–316. 10.1007/BF00345851 [DOI] [PubMed] [Google Scholar]
- Denlinger, D. L. , Hahn, D. A. , Merlin, C. , Holzapfel, C. M. , & Bradshaw, W. E. (2017). Keeping time without a spine: What can the insect clock teach us about seasonal adaptation? Philosophical Transactions of the Royal Society of London. Series B, Biological sciences, 37, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, P. , & Reppert, S. M. (2004). A rhythmic Ror. Neuron, 43, 443–446. 10.1016/j.neuron.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Emery, P. , So, W. V. , Kaneko, M. , Hall, J. C. , & Rosbash, M. (1998). CRY, a Drosophila clock and light‐regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell, 95, 669–679. 10.1016/S0092-8674(00)81637-2 [DOI] [PubMed] [Google Scholar]
- Emery, P. , Stanewsky, R. , Hall, J. C. , & Rosbash, M. (2000a). Drosophila cryptochromes: A unique circadian‐rhythm photoreceptor. Nature, 404, 456–457. 10.1038/35006558 [DOI] [PubMed] [Google Scholar]
- Emery, P. , Stanewsky, R. , Hall, J. C. , & Rosbash, M. (2000b). A unique circadian‐rhythm photoreceptor. Nature, 404, 456–457. 10.1038/35006558 [DOI] [PubMed] [Google Scholar]
- Fernández, M. P. , Berni, J. , & Ceriani, M. F. (2008). Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biology, 6, e69 10.1371/journal.pbio.0060069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, H. G. , & Robinson, S. W. (1979). Foraging by Atta sexdens (Formicidae: Attini): Seasonal patterns, caste and efficiency. Ecological Entomology, 4, 239–247. 10.1111/j.1365-2311.1979.tb00581.x [DOI] [Google Scholar]
- Fuchikawa, T. , Sanada, S. , Nishio, R. , Matsumoto, A. , Matsuyama, T. , Yamagishi, M. , … Miyatake, T. (2010). The clock gene cryptochrome of Bactrocera cucurbitae (Diptera: Tephritidae) in strains with different mating times. Heredity, 104, 387–392. 10.1038/hdy.2009.167 [DOI] [PubMed] [Google Scholar]
- Gentile, C. , Rivas, G. B. S. , Meireles‐Filho, A. C. A. , Lima, J. B. P. , & Peixoto, A. A. (2009). Circadian expression of clock genes in two mosquito disease vectors: cry2 is different. Journal of Biological Rhythms, 24, 444–451. 10.1177/0748730409349169 [DOI] [PubMed] [Google Scholar]
- Gesto, J. S. M. , da S Rivas, G. B. , Pavan, M. G. , Meireles‐Filho, A. C. A. , de Amoretty, P. R. , de Souza, N. A. , … Peixoto, A. A. (2015). Clocks do not tick in unison: Isolation of Clock and vrille shed new light on the clockwork model of the sand fly Lutzomyia longipalpis . Parasites & Vectors, 8, 505 10.1186/s13071-015-1117-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, J. M. , & Powell, J. R. (1997). Interspecific and intraspecific comparisons of the period locus in the Drosophila willistoni sibling species. Molecular Biology and Evolution, 14, 741–753. 10.1093/oxfordjournals.molbev.a025814 [DOI] [PubMed] [Google Scholar]
- Glossop, N. R. , Lyons, L. C. , & Hardin, P. E. (1999). Interlocked feedback loops within the Drosophila circadian oscillator. Science, 286, 766–768. 10.1126/science.286.5440.766 [DOI] [PubMed] [Google Scholar]
- Goldman, B. D. (2001). Mammalian photoperiodic system: Formal properties and neuroendocrine mechanisms of photoperiodic time measurement. Journal of Biological Rhythms, 16, 283–301. 10.1177/074873001129001980 [DOI] [PubMed] [Google Scholar]
- Goto, S. G. , & Denlinger, D. L. (2002). Short‐day and long‐day expression patterns of genes involved in the flesh fly clock mechanism: Period, timeless, cycle and cryptochrome. Journal of Insect Physiology, 48, 803–816. 10.1016/S0022-1910(02)00108-7 [DOI] [PubMed] [Google Scholar]
- Hackman, W. , Lakovaara, S. , Saura, A. , Sorsa, M. , & Vepsalainen, K. (1970). On the biology and karylogy of Chymomyza costata Zetterstedt, with reference to the taxonomy and distribution of various species of Chymomyza (Dipt., Drosophilidae). Ann Ent Fenn, 36, 1–9. [Google Scholar]
- Halle, S. , & Stemseth, N. C. (2000). Activity patterns in small mammals. An ecological approach. Ecological Studies Berlin, Heidelberg: Springer; 10.1007/978-3-642-18264-8 [DOI] [Google Scholar]
- Hamblen‐Coyle, M. J. , Wheeler, D. A. , Rutila, J. E. , Rosbash, M. , & Hall, J. C. (1992). Behavior of period‐altered circadian rhythm mutants of Drosophila in light: Dark cycles (Diptera: Drosophilidae). Journal of Insect Behavior, 5, 417–446. 10.1007/BF01058189 [DOI] [Google Scholar]
- Hamby, K. A. , Kwok, R. S. , Zalom, F. G. , & Chiu, J. C. (2013). Integrating circadian activity and gene expression profiles to predict chronotoxicity of Drosophila suzukii response to Insecticides. PLoS ONE, 8, e68472 10.1371/journal.pone.0068472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin, P. E. (2011). Molecular genetic analysis of circadian timekeeping in Drosophila . Advances in Genetics, 74, 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin, P. E. , & Panda, S. (2013). Circadian timekeeping and output mechanisms in animals. Current Opinion in Neurobiology, 23, 724–731. 10.1016/j.conb.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich‐Förster, C. (2000). Differential control of morning and evening components in the activity rhythm of Drosophila melanogaster–sex‐specific differences suggest a different quality of activity. Journal of Biological Rhythms, 15, 135–154. 10.1177/074873040001500208 [DOI] [PubMed] [Google Scholar]
- Helfrich‐Förster, C. (2014). From neurogenetic studies in the fly brain to a concept in circadian biology. Journal of Neurogenetics, 28, 329–347. 10.3109/01677063.2014.905556 [DOI] [PubMed] [Google Scholar]
- Helfrich‐Förster, C. (2017). The Drosophila clock system In Biological timekeeping: Clocks, rhythms and behaviour (pp. 133–176). New Delhi: Springer; 10.1007/978-81-322-3688-7 [DOI] [Google Scholar]
- Helfrich‐Förster, C. , Täuber, M. , Park, J. H. , Mühlig‐Versen, M. , Schneuwly, S. , & Hofbauer, A. (2000). Ectopic expression of the neuropeptide pigment‐dispersing factor alters behavioral rhythms in Drosophila melanogaster . Journal of Neuroscience, 20, 3339–3353. 10.1523/JNEUROSCI.20-09-03339.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich‐Förster, C. , Yoshii, T. , Wülbeck, C. , Grieshaber, E. , Rieger, D. , Bachleitner, W. , … Rouyer, F. (2007). The lateral and dorsal neurons of Drosophila melanogaster: New insights about their morphology and function. Cold Spring Harbor Symposia on Quantitative Biology, 72, 517–525. 10.1101/sqb.2007.72.063 [DOI] [PubMed] [Google Scholar]
- Helm, B. , & Lincoln, G. A. (2017). Circannual rhythms anticipate the earth's annual periodicity In Vinod Kumar (Ed.), Biological timekeeping: Clocks, rhythms and behaviour (pp. 545–569). 10.1007/978-81-322-3688-7 [DOI] [Google Scholar]
- Hermann, C. , Saccon, R. , Senthilan, P. R. , Domnik, L. , Dircksen, H. , Yoshii, T. , & Helfrich‐Förster, C. (2013). The circadian clock network in the brain of different Drosophila species. Journal of Comparative Neurology, 521, 367–388. 10.1002/cne.23178 [DOI] [PubMed] [Google Scholar]
- Hermann‐Luibl, C. , & Helfrich‐Förster, C. (2015). Clock network in Drosophila . Current Opinion in Insect Science, 7, 65–70. 10.1016/j.cois.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Hilton, H. , & Hey, J. (1996). DNA sequence variation at the period locus reveals the history of species and speciation events in the Drosophila virilis group. Genetics, 144, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom, I. , Daan, S. , Dallinga, J. H. , & Schoenmakers, M. (1984). Seasonal change in the daily timing of behaviour of the common vole, Microtus arvalis . Oecologia, 61, 18–31. 10.1007/BF00379084 [DOI] [PubMed] [Google Scholar]
- Hut, R. A. , Paolucci, S. , Dor, R. , Kyriacou, C. P. , & Daan, S. (2013). Latitudinal clines: An evolutionary view on biological rhythms. Proceedings of the Royal Society of London. Series B, Biological sciences, 280, 20130433 10.1098/rspb.2013.0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, T. , Uchida, K. , Matsuura, Y. , Takahashi, H. , Yoshida, T. , Kaji, K. , & Koizumi, I. (2016). Seasonal and diel activity patterns of eight sympatric mammals in northern Japan revealed by an intensive camera‐trap survey. PLoS ONE, 11, e0163602 10.1371/journal.pone.0163602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno, T. , Numata, H. , Goto, S. G. , & Shiga, S. (2014). Involvement of the brain region containing pigment‐dispersing factor‐immunoreactive neurons in the photoperiodic response of the bean bug, Riptortus pedestris . Journal of Experimental Biology, 217, 453–462. 10.1242/jeb.091801 [DOI] [PubMed] [Google Scholar]
- Im, S. H. , Li, W. , & Taghert, P. H. (2011). PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila . PLoS ONE, 6, e18974 10.1371/journal.pone.0018974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagota, A. , de la Iglesia, H. O. , & Schwartz, W. J. (2000). Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nature Neuroscience, 3, 372–376. 10.1038/73943 [DOI] [PubMed] [Google Scholar]
- Joshi, D. S. (1999). Latitudinal variation in locomotor activity rhythm in adult Drosophila ananassae . Canadian Journal of Zoology, 77, 865–870. 10.1139/z99-051 [DOI] [Google Scholar]
- Kaiser, T. S. , Poehn, B. , Szkiba, D. , Preussner, M. , Sedlazeck, F. J. , Zrim, A. , … Tessmar‐Raible, K. (2016). The genomic basis of circadian and circalunar timing adaptations in a midge. Nature, 540, 69–73. 10.1038/nature20151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauranen, H. , Ala‐Honkola, O. , Kankare, M. , & Hoikkala, A. (2016). Circadian clock of Drosophila montana is adapted to high variation in summer day lengths and temperatures prevailing at high latitudes. Journal of Insect Physiology, 89, 9–18. 10.1016/j.jinsphys.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Kauranen, H. , Menegazzi, P. , Costa, R. , Helfrich‐Förster, C. , Kankainen, A. , & Hoikkala, A. (2012). Flies in the north: Locomotor behavior and clock neuron organization of Drosophila montana . Journal of Biological Rhythms, 27, 377–387. 10.1177/0748730412455916 [DOI] [PubMed] [Google Scholar]
- Kistenpfennig, C. , Hirsh, J. , Yoshii, T. , & Helfrich‐Förster, C. (2012). Phase‐shifting the fruit fly clock without cryptochrome. Journal of Biological Rhythms, 27, 117–125. 10.1177/0748730411434390 [DOI] [PubMed] [Google Scholar]
- Kobelková, A. , Goto, S. G. , Peyton, J. T. , Ikeno, T. , Lee, R. E. , & Denlinger, D. L. (2015). Continuous activity and no cycling of clock genes in the Antarctic midge during the polar summer. Journal of Insect Physiology, 81, 90–96. 10.1016/j.jinsphys.2015.07.008 [DOI] [PubMed] [Google Scholar]
- Konopka, R. J. , & Benzer, S. (1971). Clock mutants of Drosophila melanogaster . Proceedings of the National Academy of Sciences of the United States of America, 68, 2112–2116. 10.1073/pnas.68.9.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka, R. J. , Pittendrigh, C. , & Orr, D. (1989). Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. Journal of Neurogenetics, 6, 1–10. 10.3109/01677068909107096 [DOI] [PubMed] [Google Scholar]
- Koštál, V. (2011a). Insect photoperiodic calendar and circadian clock: Independence, cooperation, or unity? Journal of Insect Physiology, 57, 538–556. [DOI] [PubMed] [Google Scholar]
- Koštál, V. (2011b). Hyperprolinemic larvae of the drosophilid fly, Chymomyza costata, survive cryopreservation in liquid nitrogen. Proceedings of the National Academy of Sciences of the United States of America, 108, 13041–13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koštál, V. , Závodská, R. , & Denlinger, D. (2009). Clock genes period and timeless are rhythmically expressed in brains of newly hatched, photosensitive larvae of the fly, Sarcophaga crassipalpis . Journal of Insect Physiology, 55, 408–414. [DOI] [PubMed] [Google Scholar]
- Kyriacou, C. P. (2014). Sex and rhythms in sandflies and mosquitoes: An appreciation of the work of Alexandre Afranio Peixoto (1963–2013). Infection, Genetics and Evolution, 28, 662–665. 10.1016/j.meegid.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou, C. P. , Peixoto, A. A. , Sandrelli, F. , Costa, R. , & Tauber, E. (2008). Clines in clock genes: Fine‐tuning circadian rhythms to the environment. Trends in Genetics, 24, 124–132. 10.1016/j.tig.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Lankinen, P. (1986). Genetic correlation between circadian eclosion rhythm and photoperiodic diapause in Drosophila littoralis . Journal of Biological Rhythms, 1, 101–118. 10.1177/074873048600100202 [DOI] [PubMed] [Google Scholar]
- Lankinen, P. , & Forsman, P. (2006). Independence of genetic geographical variation between photoperiodic diapause, circadian eclosion rhythm, and Thr‐Gly repeat region of the period gene in Drosophila littoralis . Journal of Biological Rhythms, 21, 3–12. 10.1177/0748730405283418 [DOI] [PubMed] [Google Scholar]
- Liang, X. , Holy, T. E. , & Taghert, P. H. (2016). Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science, 351, 976–981. 10.1126/science.aad3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Holy, T. E. , & Taghert, P. H. (2017). A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron, 94(6), 1173–1189.e4. 10.1016/j.neuron.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, K. H. , Chen, W.‐F. , Yildirim, E. , & Edery, I. (2012). Natural variation in the Drosophila melanogaster clock gene period modulates splicing of its 3′‐terminal intron and mid‐day siesta. PLoS ONE, 7, e49536 10.1371/journal.pone.0049536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, K. H. , Lim, C. , Ko, H. W. , & Edery, I. (2008). Natural variation in the splice site strength of a clock gene and species‐specific thermal adaptation. Neuron, 60, 1054–1067. 10.1016/j.neuron.2008.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, S. E. , Schmidt, P. S. , & Sehgal, A. (2014). Natural populations of Drosophila melanogaster reveal features of an uncharacterized circadian property: The lower temperature limit of rhythmicity. Journal of Biological Rhythms, 29, 167–180. 10.1177/0748730414537801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak, J. , Chen, W.‐F. , & Edery, I. (2004). Splicing of the period gene 3′‐terminal intron is regulated by light, circadian clock factors, and phospholipase C. Molecular and Cellular Biology, 24, 3359–3372. 10.1128/MCB.24.8.3359-3372.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak, J. , Sidote, D. , Hardin, P. E. , & Edery, I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron, 24, 219–230. 10.1016/S0896-6273(00)80834-X [DOI] [PubMed] [Google Scholar]
- Markow, T. A. , & O'Grady, P. M. (2007). Drosophila biology in the genomic age. Genetics, 177, 1269–1276. 10.1534/genetics.107.074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, A. , Ohta, Y. , Itoh, T. , Sanada‐Morimura, S. , Matsuyama, T. , Fuchikawa, T. , & Miyatake, T. (2008). Period gene of Bactrocera cucurbitae (Diptera: Tephritidae) among strains with different mating times and sterile insect technique. Annals of the Entomological Society of America, 101, 1121–1130. 10.1603/0013-8746-101.6.1121 [DOI] [Google Scholar]
- Mazzotta, G. M. , Sandrelli, F. , Zordan, M. A. , Mason, M. , Benna, C. , Cisotto, P. , … Costa, R. (2005). The clock gene period in the medfly Ceratitis capitata . Genetical Research, 86, 13–30. 10.1017/S0016672305007664 [DOI] [PubMed] [Google Scholar]
- Meireles‐Filho, A. C. A. , Amoretty, P. R. , De Souza, N. A. , Kyriacou, C. P. , & Peixoto, A. A. (2006). Rhythmic expression of the cycle gene in a hematophagous insect vector. BMC Molecular Biology, 7, 38 10.1186/1471-2199-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles‐Filho, A. C. A. , da S. Rivas, G. B. , Gesto, J. S. M. , Machado, R. C. , Britto, C. , De Souza, N. A. , & Peixoto, A. A. (2006). The biological clock of an hematophagous insect: Locomotor activity rhythms, circadian expression and downregulation after a blood meal. FEBS Letters, 580, 2–8. 10.1016/j.febslet.2005.11.031 [DOI] [PubMed] [Google Scholar]
- Meireles‐Filho, A. C. A. , & Kyriacou, C. P. (2013). Circadian rhythms in insect disease vectors. Memorias do Instituto Oswaldo Cruz, 108, 48–58. 10.1590/0074-0276130438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegazzi, P. , Dalla Benetta, E. , Beauchamp, M. , Schlichting, M. , Steffan‐Dewenter, I. , & Helfrich‐Förster, C. (2017). Adaptation of circadian neuronal network to photoperiod in high‐latitude European Drosophilids. Current Biology, 27, 833–839. 10.1016/j.cub.2017.01.036 [DOI] [PubMed] [Google Scholar]
- Merz, C. , Catchen, J. M. , Hanson‐Smith, V. , Emerson, K. J. , Bradshaw, W. E. , & Holzapfel, C. M. (2013). Replicate phylogenies and post‐glacial range expansion of the pitcher‐plant mosquito, Wyeomyia smithii, in North America. PLoS ONE, 8, e72262 10.1371/journal.pone.0072262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuti, M. E. , Stone, M. , Ikeno, T. , & Denlinger, D. L. (2015). Functional circadian clock genes are essential for the overwintering diapause of the Northern house mosquito, Culex pipiens . Journal of Experimental Biology, 218, 412–422. 10.1242/jeb.113233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake, T. , Matsumoto, A. , Matsuyama, T. , Ueda, H. R. , Toyosato, T. , & Tanimura, T. (2002). The period gene and allochronic reproductive isolation in Bactrocera cucurbitae . Proceedings of the Royal Society of London. Series B, Biological sciences, 269, 2467–2472. 10.1098/rspb.2002.2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelli, S. , Mazzotta, G. , Vanin, S. , Caccin, L. , Corrà, S. , De Pittà, C. , … Costa, R. (2015). Period and timeless mRNA splicing profiles under natural conditions in Drosophila melanogaster . Journal of Biological Rhythms, 30, 217–227. 10.1177/0748730415583575 [DOI] [PubMed] [Google Scholar]
- Muguruma, F. , Goto, S. G. , Numata, H. , & Shiga, S. (2010). Effect of photoperiod on clock gene expression and subcellular distribution of PERIOD in the circadian clock neurons of the blow fly Protophormia terraenovae . Cell and Tissue Research, 340, 497–507. 10.1007/s00441-010-0966-8 [DOI] [PubMed] [Google Scholar]
- Naidoo, N. , Song, W. , Hunter‐Ensor, M. , & Sehgal, A. (1999). A role for the proteasome in the light response of the timeless clock protein. Science, 285, 1737–1741. 10.1126/science.285.5434.1737 [DOI] [PubMed] [Google Scholar]
- Neumann, D. (1989). Circadian components of semilunar and lunar timing mechanisms. Journal of Biological Rhythms, 4, 285–294. [PubMed] [Google Scholar]
- Nielsen, J. , Peixoto, A. A. , Piccin, A. , Costa, R. , Kyriacou, C. P. , & Chalmers, D. (1994). Big flies, small repeats: The “Thr‐Gly” region of the period gene in Diptera. Molecular Biology and Evolution, 11, 839–853. [DOI] [PubMed] [Google Scholar]
- Nishinokubi, I. , Shimoda, M. , & Ishida, N. (2006). Mating rhythms of Drosophila: Rescue of tim01 mutants by D. ananassae timeless. Journal of Circadian Rhythms, 4, 4 10.1186/1740-3391-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinokubi, I. , Shimoda, M. , Kako, K. , Sakai, T. , Fukamizu, A. , & Ishida, N. (2003). Highly conserved Drosophila ananassae timeless gene functions as a clock component in Drosophila melanogaster . Gene, 307, 183–190. 10.1016/S0378-1119(03)00468-2 [DOI] [PubMed] [Google Scholar]
- Noreen, S. , Pegoraro, M. , Nouroz, F. , Tauber, E. , & Kyriacou, C. P. (2018). Interspecific studies of circadian genes period and timeless in Drosophila . Gene, 648, 106–114. 10.1016/j.gene.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard, D. J. , Maclennan, J. , Kim, K.‐W. , Rambaut, A. , O'Grady, P. M. , & Jiggins, F. M. (2012). Estimating divergence dates and substitution rates in the Drosophila phylogeny. Molecular Biology and Evolution, 29, 3459–3473. 10.1093/molbev/mss150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Grady, P. M. , & DeSalle, R. (2018). Phylogeny of the Genus Drosophila . Genetics, 209, 1–25. 10.1534/genetics.117.300583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousley, A. , Zafarullah, K. , Chen, Y. , Emerson, M. , Hickman, L. , & Sehgal, A. (1998). Conserved regions of the timeless (tim) clock gene in Drosophila analyzed through phylogenetic and functional studies. Genetics, 148, 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaya, O. , & Rosato, E. (2012). The circadian clock of the fly: A neurogenetics journey through time. Advances in Genetics, 77, 79–123. [DOI] [PubMed] [Google Scholar]
- Park, J. H. , Helfrich‐Förster, C. , Lee, G. , Liu, L. , Rosbash, M. , & Hall, J. C. (2000). Differential regulation of circadian pacemaker output by separate clock genes in Drosophila . Proceedings of the National Academy of Sciences of the United States of America, 97, 3608–3613. 10.1073/pnas.97.7.3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D. , Shafer, O. T. , Shepherd, S. P. , Suh, H. , Trigg, J. S. , & Taghert, P. H. (2008). The Drosophila basic helix‐loop‐helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide‐amidating enzyme. Molecular and Cellular Biology, 28, 410–421. 10.1128/MCB.01104-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro, M. , Picot, E. , Hansen, C. N. , Kyriacou, C. P. , Rosato, E. , & Tauber, E. (2015). Gene expression associated with early and late chronotypes in Drosophila melanogaster . Frontiers in Neurology, 6, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto, A. A. , Costa, R. , Wheeler, D. A. , Hall, J. C. , & Kyriacou, C. P. (1992). Evolution of the threonine‐glycine repeat region of the period gene in the melanogaster species subgroup of Drosophila . Journal of Molecular Evolution, 35, 411–419. [DOI] [PubMed] [Google Scholar]
- Peixoto, A. A. , Hennessy, J. M. , Townson, I. , Hasan, G. , Rosbash, M. , Costa, R. , & Kyriacou, C. P. (1998). Molecular coevolution within a Drosophila clock gene. Proceedings of the National Academy of Sciences of the United States of America, 95, 4475–4480. 10.1073/pnas.95.8.4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh, C. S. (1967). Circadian systems. I. The driving oscillation and its assay in Drosophila pseudoobscura . Proceedings of the National Academy of Sciences of the United States of America, 58, 1762–1767. 10.1073/pnas.58.4.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh, C. S. , & Minis, D. H. (1964). The entrainment of circadian oscillations by light and their role as photoperiodic clocks. American Naturalist, 98, 261–294. 10.1086/282327 [DOI] [Google Scholar]
- Pittendrigh, C. S. , & Takamura, T. (1989). Latitudinal clines in the properties of a circadian pacemaker. Journal of Biological Rhythms, 4, 217–235. [PubMed] [Google Scholar]
- Prabhakaran, P. M. , De, J. , & Sheeba, V. (2013). Natural conditions override differences in emergence rhythm among closely related drosophilids. PLoS ONE, 8, e83048 10.1371/journal.pone.0083048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakaran, P. M. , & Sheeba, V. (2012). Sympatric Drosophilid species melanogaster and ananassae differ in temporal patterns of activity. Journal of Biological Rhythms, 27, 365–376. 10.1177/0748730412458661 [DOI] [PubMed] [Google Scholar]
- Prabhakaran, P. M. , & Sheeba, V. (2013). Insights into differential activity patterns of drosophilids under semi‐natural conditions. Journal of Experimental Biology, 216, 4691–4702. 10.1242/jeb.092270 [DOI] [PubMed] [Google Scholar]
- Prabhakaran, P. M. , & Sheeba, V. (2014). Temperature sensitivity of circadian clocks is conserved across Drosophila species melanogaster, malerkotliana and ananassae . Chronobiology International, 31, 1008–1016. 10.3109/07420528.2014.941471 [DOI] [PubMed] [Google Scholar]
- Pyza, E. , & Meinertzhagen, I. A. (2003). The regulation of circadian rhythms in the fly's visual system: Involvement of FMRFamide‐like neuropeptides and their relationship to pigment dispersing factor in Musca domestica and Drosophila melanogaster . Neuropeptides, 37, 277–289. 10.1016/j.npep.2003.06.001 [DOI] [PubMed] [Google Scholar]
- Pyza, E. , Siuta, T. , & Tanimura, T. (2003). Development of PDF‐immunoreactive cells, possible clock neurons, in the housefly Musca domestica . Microscopy Research and Technique, 62, 103–113. 10.1002/(ISSN)1097-0029 [DOI] [PubMed] [Google Scholar]
- Renn, S. C. , Park, J. H. , Rosbash, M. , Hall, J. C. , & Taghert, P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila . Cell, 99, 791–802. 10.1016/S0092-8674(00)81676-1 [DOI] [PubMed] [Google Scholar]
- Rieger, D. , Peschel, N. , Dusik, V. , Glotz, S. , & Helfrich‐Förster, C. (2012). The ability to entrain to long photoperiods differs between 3 Drosophila melanogaster wild‐type strains and is modified by twilight simulation. Journal of Biological Rhythms, 27, 37–47. 10.1177/0748730411420246 [DOI] [PubMed] [Google Scholar]
- Rieger, D. , Stanewsky, R. , & Helfrich‐Förster, C. (2003). Cryptochrome, compound eyes, Hofbauer‐Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster . Journal of Biological Rhythms, 18, 377–391. 10.1177/0748730403256997 [DOI] [PubMed] [Google Scholar]
- Rivas, G. B. S. , Teles‐de‐Freitas, R. , Pavan, M. G. , Lima, J. B. P. , Peixoto, A. A. , & Bruno, R. V. (2018). Effects of light and temperature on daily activity and clock gene expression in two mosquito disease vectors. Journal of Biological Rhythms, 33, 272–288. 10.1177/0748730418772175 [DOI] [PubMed] [Google Scholar]
- Rosato, E. , Peixoto, A. A. , Barbujani, G. , Costa, R. , & Kyriacou, C. P. (1994). Molecular polymorphism in the period gene of Drosophila simulans . Genetics, 138, 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund, S. S. C. , Hou, T. Y. , Ward, S. M. , Collins, F. H. , & Duffield, G. E. (2011). Genome‐wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae . Proceedings of the National Academy of Sciences, 108, E421–E430. 10.1073/pnas.1100584108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, C. A. M. , Mello, B. , Frazão, A. , & Voloch, C. M. (2013). Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zoological Journal of the Linnean Society, 169, 765–775. 10.1111/zoj.12062 [DOI] [Google Scholar]
- Sandrelli, F. , Tauber, E. , Pegoraro, M. , Mazzotta, G. , Cisotto, P. , Landskron, J. , … Kyriacou, C. P. (2007). A molecular basis for natural selection at the timeless locus in Drosophila melanogaster . Science, 316, 1898–1900. 10.1126/science.1138426 [DOI] [PubMed] [Google Scholar]
- Saunders, D. S. (1997). Insect circadian rhythms and photoperiodism. Invertebrate Neuroscience, 3, 155–164. 10.1007/BF02480370 [DOI] [PubMed] [Google Scholar]
- Saunders, D. S. (2013). Insect photoperiodism: Measuring the night. Journal of Insect Physiology, 59, 1–10. 10.1016/j.jinsphys.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Schlichting, M. , Menegazzi, P. , Lelito, K. R. , Yao, Z. , Buhl, E. , Dalla Benetta, E. , … Shafer, O. T. (2016). A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila . Journal of Neuroscience, 36, 9084–9096. 10.1523/JNEUROSCI.0992-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, F. K. , Hagedorn, N. , Yoshii, T. , Helfrich‐Förster, C. , & Rieger, D. (2018). Neuroanatomical details of the lateral neurons of Drosophila melanogaster support their functional role in the circadian system. Journal of Comparative Neurology, 526, 1209–1231. 10.1002/cne.24406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer, O. T. , & Yao, Z. (2014). Pigment‐dispersing factor signaling and circadian rhythms in insect locomotor activity. Current Opinion in Insect Science, 1, 73–80. 10.1016/j.cois.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga, S. , & Numata, H. (2009). Roles of PER immunoreactive neurons in circadian rhythms and photoperiodism in the blow fly, Protophormia terraenovae . Journal of Experimental Biology, 212, 867–877. 10.1242/jeb.027003 [DOI] [PubMed] [Google Scholar]
- Smith, P. (1987). Naturally occurring arrhythmicity in eclosion and activity in Lucilla cuprina: Its genetic basis. Physiological Entomology, 12, 99–107. 10.1111/j.1365-3032.1987.tb00728.x [DOI] [Google Scholar]
- Stanewsky, R. , Kaneko, M. , Emery, P. , Beretta, B. , Wager‐Smith, K. , Kay, S. A. , … Hall, J. C. (1998). The cryb mutation identifies Cryptochrome as a circadian photoreceptor in Drosophila . Cell, 95, 681–692. 10.1016/S0092-8674(00)81638-4 [DOI] [PubMed] [Google Scholar]
- Stehlík, J. , Závodská, R. , Shimada, K. , Sauman, I. , & Kostál, V. (2008). Photoperiodic induction of diapause requires regulated transcription of timeless in the larval brain of Chymomyza costata . Journal of Biological Rhythms, 23, 129–139. 10.1177/0748730407313364 [DOI] [PubMed] [Google Scholar]
- Tauber, E. , Zordan, M. , Sandrelli, F. , Pegoraro, M. , Osterwalder, N. , Breda, C. , … Costa, R. (2007). Natural selection favors a newly derived timeless allele in Drosophila melanogaster . Science, 316, 1895–1898. 10.1126/science.1138412 [DOI] [PubMed] [Google Scholar]
- Throckmorton, L. (1975). The phylogeny, ecology, and geography of Drosophila In King R. C. (Ed.), Invertebrates of genetic interest (pp. 421–469). New York, NY: Plenum Press; 10.1007/978-1-4615-7145-2 [DOI] [Google Scholar]
- Vanlalhriatpuia, K. , Chhakchhuak, V. , Moses, S. K. , Iyyer, S. B. , Kasture, M. S. , Shivagaje, A. J. , … Joshi, D. S. (2007). Effects of altitude on circadian rhythm of adult locomotor activity in Himalayan strains of Drosophila helvetica . Journal of Circadian Rhythms, 5, 1 10.1186/1740-3391-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak, P. , Coupar, J. , Hughes, S. E. , Fozdar, P. , Kilby, J. , Garren, E. , … Hirsh, J. (2013). Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genetics, 9, e1003615 10.1371/journal.pgen.1003615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. L. , & Hey, J. (1996). The speciation history of Drosophila pseudoobscura and close relatives: Inferences from DNA sequence variation at the period locus. Genetics, 144, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warman, G. R. , Newcomb, R. D. , Lewis, R. D. , & Evans, C. W. (2000). Analysis of the circadian clock gene period in the sheep blow fly Lucilia cuprina . Genetical Research, 75, 257–267. 10.1017/S0016672399004425 [DOI] [PubMed] [Google Scholar]
- Weeks, A. R. , McKechnie, S. W. , & Hoffmann, A. A. (2006). In search of clinal variation in the period and clock timing genes in Australian Drosophila melanogaster populations. Journal of Evolutionary Biology, 19, 551–557. 10.1111/j.1420-9101.2005.01013.x [DOI] [PubMed] [Google Scholar]
- Wei, H. , Yasar, H. , Funk, N. W. , Giese, M. , Baz, E.‐S. , & Stengl, M. (2014). Signaling of pigment‐dispersing factor (PDF) in the Madeira cockroach Rhyparobia maderae . PLoS ONE, 9, e108757 10.1371/journal.pone.0108757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, D. A. , Hamblen‐Coyle, M. J. , Dushay, M. S. , & Hall, J. C. (1993). Behavior in light‐dark cycles of Drosophila mutants that are arrhythmic, blind, or both. Journal of Biological Rhythms, 8, 67–94. 10.1177/074873049300800106 [DOI] [PubMed] [Google Scholar]
- Wheeler, D. A. , Kyriacou, C. P. , Greenacre, M. L. , Yu, Q. , Rutila, J. E. , Rosbash, M. , & Hall, J. C. (1991). Molecular transfer of a species‐specific behavior from Drosophila simulans to Drosophila melanogaster . Science, 251, 1082–1085. 10.1126/science.1900131 [DOI] [PubMed] [Google Scholar]
- Wülbeck, C. , Grieshaber, E. , & Helfrich‐Förster, C. (2008). Pigment‐dispersing factor (PDF) has different effects on Drosophila's circadian clocks in the accessory medulla and in the dorsal brain. Journal of Biological Rhythms, 23, 409–424. 10.1177/0748730408322699 [DOI] [PubMed] [Google Scholar]
- Yamada, H. , & Yamamoto, M.‐T. (2011). Association between circadian clock genes and diapause incidence in Drosophila triauraria . PLoS ONE, 6, e27493 10.1371/journal.pone.0027493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M. , Nishimura, K. , & Shiga, S. (2017). Clock and hormonal controls of an eclosion gate in the flesh fly Sarcophaga crassipalpis . Zoological Science, 34, 151–160. 10.2108/zs160153 [DOI] [PubMed] [Google Scholar]
- Yamamoto, M. , Shiga, S. , & Goto, S. G. (2017). Distribution of PERIOD‐immunoreactive neurons and temporal change of the immunoreactivity under long‐day and short‐day conditions in the larval brain of the flesh fly Sarcophaga similis . Chronobiology International, 34(6), 819–825. 10.1080/07420528.2017.1310736 [DOI] [PubMed] [Google Scholar]
- Yassin, A. (2013). Phylogenetic classification of the Drosophilidae Rodani (Diptera): The role of morphology in the postgenomic era. Systematic Entomology, 38, 349–364. 10.1111/j.1365-3113.2012.00665.x [DOI] [Google Scholar]
- Yasuyama, K. , Hase, H. , & Shiga, S. (2015). Neuroanatomy of pars intercerebralis neurons with special reference to their connections with neurons immunoreactive for pigment‐dispersing factor in the blow fly Protophormia terraenovae . Cell and Tissue Research, 362, 33–43. 10.1007/s00441-015-2192-x [DOI] [PubMed] [Google Scholar]
- Yoshii, T. , Hermann‐Luibl, C. , & Helfrich‐Förster, C. (2016). Circadian light‐input pathways in Drosophila . Communicative & Integrative Biology, 9, e1102805 10.1080/19420889.2015.1102805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii, T. , Rieger, D. , & Helfrich‐Förster, C. (2012). Two clocks in the brain: An update of the morning and evening oscillator model in Drosophila . Progress in Brain Research, 199, 59–82. 10.1016/B978-0-444-59427-3.00027-7 [DOI] [PubMed] [Google Scholar]
- Yoshii, T. , Wülbeck, C. , Sehadova, H. , Veleri, S. , Bichler, D. , Stanewsky, R. , & Helfrich‐Förster, C. (2009). The neuropeptide pigment‐dispersing factor adjusts period and phase of Drosophila's clock. Journal of Neuroscience, 29, 2597–2610. 10.1523/JNEUROSCI.5439-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q. , Colot, H. V. , Kyriacou, C. P. , Hall, J. C. , & Rosbash, M. (1987). Behaviour modification by in vitro mutagenesis of a variable region within the period gene of Drosophila . Nature, 326, 765–769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This review does not report unpublished primary data.


