Abstract
Cascade-specific termination of G protein signaling is catalyzed by the Regulator of G protein Signaling (RGS) family members, including RGS2. Angiotensin, vasopressin, and endothelin are implicated in preeclampsia, and RGS2 is known to inhibit G protein cascades activated by these hormones. Mutations in RGS2 are associated with human hypertension and increased risk of developing preeclampsia and its sequelae. RGS family members are known to influence maternal vascular function, but the role of RGS2 within the placenta has not been explored. Here, we hypothesized that reduced expression of RGS2 within the placenta represents a risk factor for the development of preeclampsia. Although cAMP/CREB signaling was enriched in placentas from human pregnancies affected by preeclampsia compared to clinically-matched controls and RGS2 is known to be a CREB-responsive gene, RGS2 mRNA was reduced in placentas from pregnancies affected by preeclampsia. Experimentally reducing Rgs2 expression within the feto-placental unit was sufficient to induce preeclampsia-like phenotypes in pregnant wildtype C57BL/6J mice. Stimulation of RGS2 transcription within immortalized human HTR8/SVneo trophoblasts by cAMP/CREB signaling was discovered to be dependent upon the activity of histone deacetylase activity, and more specifically, histone deacetylase-9 (HDAC9), and HDAC9 expression was reduced in placentas from human pregnancies affected by preeclampsia. We conclude that reduced expression of RGS2 within the placenta may mechanistically contribute to preeclampsia. More generally, this work identifies RGS2 as an HDAC9-dependent CREB-responsive gene, which may contribute to reduced RGS2 expression in placenta during preeclampsia.
Keywords: Pre-Eclampsia, Pregnancy, Hypertension, Placenta, G protein signaling
Graphical Abstract
Summary
Reduced expression of RGS2 within the placenta is a risk factor for the development of PreE. Understanding the causes and consequences of reduced placental expression of RGS2 may identify novel diagnostic and therapeutic approaches to PreE.
Introduction
The pregnancy-specific hypertensive disorder, preeclampsia (PreE), remains a dominant worldwide cause of maternal and infant mortality, and a wealth of evidence supports a primary role for the placenta in PreE development and maintenance.1 Increased G Protein coupled receptor (GPCR) signaling in response to various hormones including angiotensin, endothelin, and vasopressin is implicated in human PreE and animal models of the condition.2–6
Termination of G protein signaling after activation by GPCRs involves hydrolysis of a phosphate from the guanosine triphosphate (GTP) bound to the Gα subunit, and this is accomplished via enzymatic activity of the Gα subunit. Regulator of G protein Signaling (RGS) family members accelerate this GTPase activity. The B/R4 family of RGS proteins is increasingly recognized as a potential contributor to cardiovascular control during pregnancy.7 Holobotovskyy demonstrated that loss of RGS5 in maternal myometrium is associated with hypertensive disorders of pregnancy, including PreE.8 The Staff group has also demonstrated that a specific single nucleotide polymorphism (SNP) in the 3’ untranslated region of the RGS2 gene (rs4606) in mothers is associated with PreE.9–11 Mutations in RGS2 are associated with human hypertension,12, 13 and reduced RGS2 expression has been demonstrated in hypertensive populations.14 Deletion of Rgs2 has also been shown to increase resistance and reduce flow in uterine arteries of non-pregnant mice.15
Given the role of the placenta in the pathogenesis and maintenance of PreE, it follows that B/R4 family members such as RGS2 expressed within the placenta may contribute to this disorder. Here we demonstrate that RGS2 expression is suppressed in human PreE placenta, and that feto-placental disruption of Rgs2 is sufficient to initiate selected characteristics of PreE in pregnant C57BL/6J mice. Finally, we document that RGS2 transcription in immortalized HTR8/SVneo human trophoblasts is dependent upon histone deacetylase (HDAC9) activity. This discovery supports the concept that reduced HDAC9 activity may account for the observed reduction in RGS2 expression in PreE placenta.
Methods
Please see associated Supplemental Methods and Data file for details; data and materials are available from the corresponding authors upon reasonable request.
De-identified placental tissue samples and clinical data were obtained from the University of Iowa Maternal Fetal Tissue Bank. Mice harboring a null allele for Rgs2 were obtained from the MMRRC (B6.129P2-Rgs2tm1Dgen/Mmnc, 011639-UNC). All studies were approved by the University of Iowa Institutional Animal Care & Use Committee. HTR8/SVneo cells were purchased from American Type Culture Collection (CRL-3271).
Transcriptomes of embryonic/gestational day (GD) 12.5 placentas were assessed by RNA-sequencing; data are deposited in the NCBI Gene Expression Omnibus (GSE122503).
Analytical comparisons were performed using t-test, Mann-Whitney U, or ANOVA followed by Tukey or Bonferroni multiple-comparison procedures, as indicated. Differences were considered significant at p<0.05 using two-tailed tests.
Results
RGS2 is expressed in placenta, and this expression is reduced during PreE
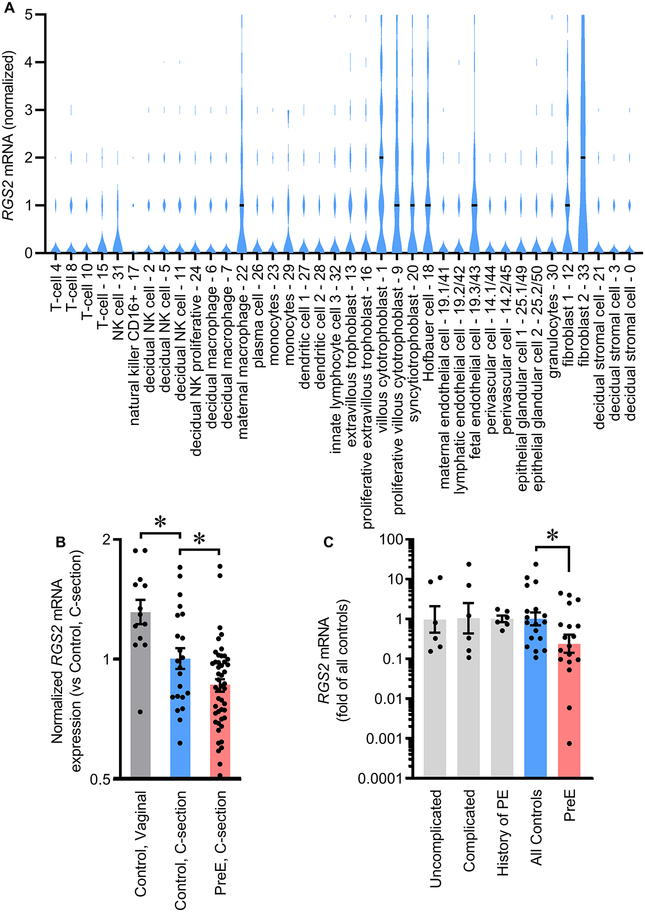
We previously reported that RGS B/R4 family members are expressed in multiple layers of human placenta throughout gestation, and that RGS2 is among the most strongly expressed members.7 Reanalysis of single-cell RNAseq data from human placentas at 6–14 weeks of gestation16 demonstrates that in RGS2 is expressed in many cell types (Figure 1A). Similar results are reported at 8 weeks of gestation17 and in third-trimester (Figures S1A–B).18 Single-cell sequencing similarly demonstrates that Rgs2 is expressed in essentially all cells of mouse placenta at GD14.5 (Figure S1C).19 Fluorescent in situ hybridization (RNAscope) also confirms Rgs2 expression across various layers of placenta in wildtype C57BL/6J mice (Figure S2).
Figure 1. RGS2 mRNA is reduced in placenta during PreE in humans.
(A) RGS2 expression in individual cell types of human placenta at 6–14 weeks of gestation, assessed by droplet single-cell sequencing (data from Vento-Tormo et al.16). Horizonal black lines represent median expression level within each cell type. (B) RGS2 expression in preterm human placenta during PreE (control, Vaginal n = 13 ; control, C-section n = 22, PreE, C-section n = 47; data from GSE75010). *p<0.05 by t-test. (C) RGS2 expression in human placental tissue from clinically diverse normal pregnancies and PreE pregnancies (Uncomplicated n = 6, Complicated n = 6, History of PreE n = 6, All controls n = 18, PreE n =18). *p<0.05 by Mann-Whitney U. (B-C) Summary data presented as mean±SEM.
Comparison of gestational-age matched human placentas (GSE7501020) revealed RGS2 expression is significantly reduced in preterm placentas from PreE pregnancies compared to preterm physiological pregnancies (Figures 1B, S3). RGS2 was also sensitive to delivery method, as expression is lower in placentas delivered by Cesarean section than vaginal birth. Confirming these in silico studies, the expression of RGS2 was suppressed in placentas from pregnancies complicated by PreE obtained via the University of Iowa Maternal Fetal Tissue Bank (MFTB21) (Figures 1C. Reanalysis of GSE9383918 and qPCR analysis of MFTB samples (Figure S3C) also support differential expression of RGS2 across layers of the placenta.
Commercially-available antibodies against human RGS2 failed quality-control experiments for Western blotting applications (Figure S4), and therefore rigorous quantification of RGS2 protein is not possible at this time. In contrast, analysis of mouse RGS2 protein in placentas from mice confirms both the quality of antibodies against mouse RGS2, and cytoplasmic localization of RGS2 in the cytoplasm of cells in specific layers of the placenta (Figure S5). These findings bolster the concept that RGS2 protein is expressed by trophoblasts, and its subcellular localization within the cytoplasm is expected to permit RGS2 to perform its canonical second-messenger modulating function.
Disruption of Rgs2 in the feto-placental unit is sufficient to model some clinical and many molecular features of PreE in mice
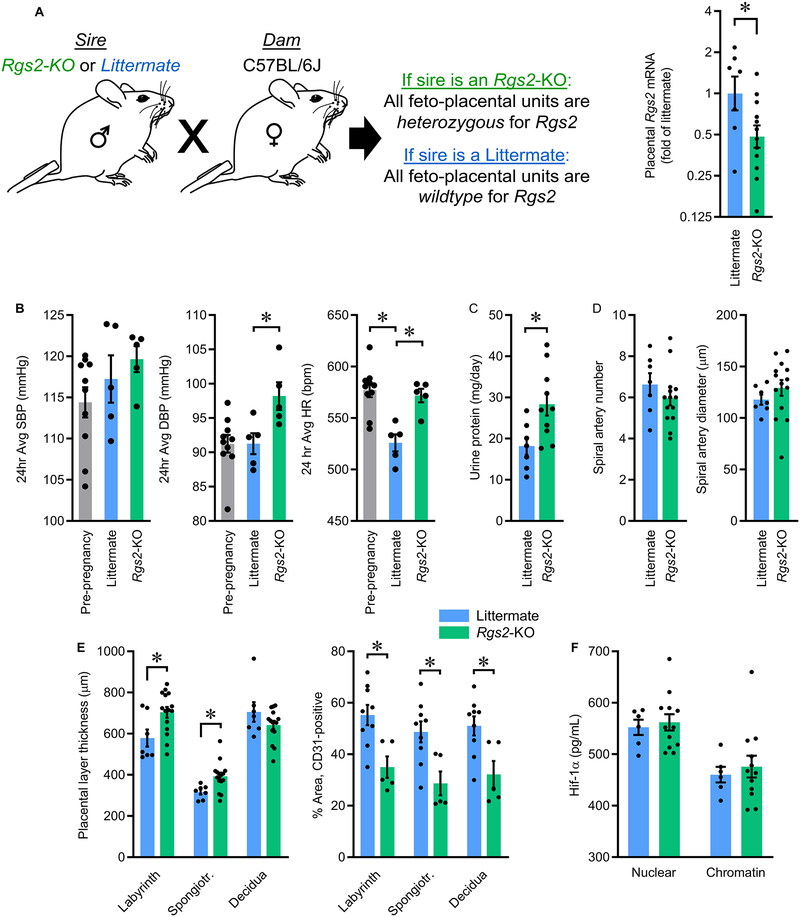
To dissociate the effects of disrupting Rgs2 in the dam as well as in the placenta, we used a selective breeding strategy in which wildtype C57BL/6J dams were mated with Rgs2-deficient (Rgs2-KO) sires or with wildtype littermates of these Rgs2-KO sires. These matings resulted in pregnant wildtype C57BL/6J dams carrying feto-placental units that were either heterozygous or wildtype for Rgs2, respectively (Figure 2A). This breeding approach was selected instead of attempting to generate homozygous-null feto-placental units, as generating such feto-placental units would have required a breeding strategy in which the dam was either homozygous-null (ie - Rgs2-KO) or heterozygous-null for Rgs2, and it has been previously demonstrated that adult mice with either heterozygous or homozygous disruption of Rgs2 exhibit hypertension in the non-pregnant state.22 Future studies examining the effect of homozygous Rgs2 disruption in the placenta, perhaps as a model of preeclampsia superimposed upon chronic hypertension, are warranted but beyond the objectives of the current study. Heterozygous disruption of Rgs2 resulted in an approximate 50% reduction in Rgs2 mRNA in the placenta (Figure 2A), confirming that Rgs2 expression is sensitive to gene copy number, and that use of this breeding scheme results in suppression of Rgs2 mRNA within the feto-placental unit during pregnancy in an otherwise wildtype C57BL/6J dam.
Figure 2. Selective breeding paradigm to reduce Rgs2 in the feto-placental unit of pregnant C57BL/6J mice.
(A) Schematic illustrating breeding scheme, and Rgs2 mRNA in GD17.5 placentas from dams mated with wildtype littermate (Littermate) or Rgs2-deficient (Rgs2-KO) males (Littermate n=7, Rgs2-KO n=12, each from an independent pregnancy). (B) Effects of reduced feto-placental Rgs2 on maternal systolic (SBP) and diastolic (DBP) blood pressure, and heart rate (HR) (Littermate n=5, Rgs2-KO n=5). (C) Maternal urine protein excretion (Littermate n=7, Rgs2-KO n=10). (D) Spiral artery number and diameter (Littermate n=7, Rgs2-KO n=14). (E) Thicknesses of placental layers, and percentage of area in placental layers staining positive for CD31 as an index of total vascularization (Littermate n=7, Rgs2-KO n=15). (F) Hif-1α localization in nuclear and chromatin-precipitated fractions of placentas at GD17.5 (Littermate n=6, Rgs2-KO n=12). Summary data presented as mean±SEM. *p<0.05 by two-tailed t-test (A, C), or Bonferroni correction for multiple comparisons (B, E).
Blood pressure was assessed in dams in the last week of gestation using previously-implanted radiotelemetric probes. Systolic blood pressure (SBP) was not modified by sire genotype, but diastolic blood pressure (DBP) and heart rate (HR) were significantly increased throughout the light cycle in dams mated with an Rgs2-KO sire (Figures 2B, S6). Protein loss in urine was significantly increased for dams mated with an Rgs2-KO sire (Figure 2C) and tissue damage was suggested by increased plasma alanine transferase concentrations, though no major structural abnormalities (such as glomerular endotheliosis) were observed in kidneys from either group by electron microscopy (Figures S7, S8A).
At GD12.5 and at GD17.5 the number of total feto-placental units and rate of spontaneous resorption were indistinguishable between groups (Table S1). Placental masses and fetal masses were increased at GD12.5 but normalized by GD17.5 in dams mated with Rgs2-KO sires (Figures S8B–C). No significant differences in number of spiral arteries or diameter of spiral arteries were observed between groups at GD12.5 (Figure 2D). No differences were observed in maximum invasion depth by individual cytokeratin-8 (CK8)-positive trophoblasts into the decidua layer, nor the maximum depth of CK8-positive, remodeled spiral arteries (Figure S8D–E). No differences in angiogenic factors were observed in placenta at GD17.5, including placental growth factor (PGF) or Fms-related tyrosine kinase 1 (FLT1) (Figure S9A–B). The labyrinth and spongiotrophoblast layers were thickened at GD12.5, but no difference in decidual thickness was noted (Figure 2E). Immunohistochemical detection of platelet endothelial cell adhesion molecule (CD31) was also used to assess total vascularization of the placental layers at GD12.5. In contrast to assessments of trophoblast invasion, the fraction of CD31-positive cells was significantly reduced in each layer of the placenta from dams mated with Rgs2-KO sires (Figure 2E), consistent with a reduction in total vascularization of the placenta despite normal morphology of the spiral arteries.
Hypoxia is frequently documented in human PreE placenta, and commonly-used animal models of PreE such as the reduced uterine perfusion pressure (RUPP) model specifically rely upon mid-gestational placental hypoxia/ischemia as a primary etiology,23 but no evidence for hypoxia was observed in placentas from dams mated with Rgs2-KO sires. Subcellular localization of HIF1α to the nucleus or to precipitated chromatin were both unchanged (Figure 2F), and expression levels of hypoxia-responsive genes were unchanged in placenta (Figure S9C–D). Further, transcriptomes of GD12.5 placentas (including decidua) were assessed using RNA sequencing, and Gene Set Enrichment Analysis (GSEA) failed to provide evidence for enrichment of canonical hypoxia target genes (GRD gene set: FDR q-value=0.22, normalized enrichment score (NES)=1.25; and Harris gene set: MSigDB: M10508, FDR q-value=0.16, NES=1.30).
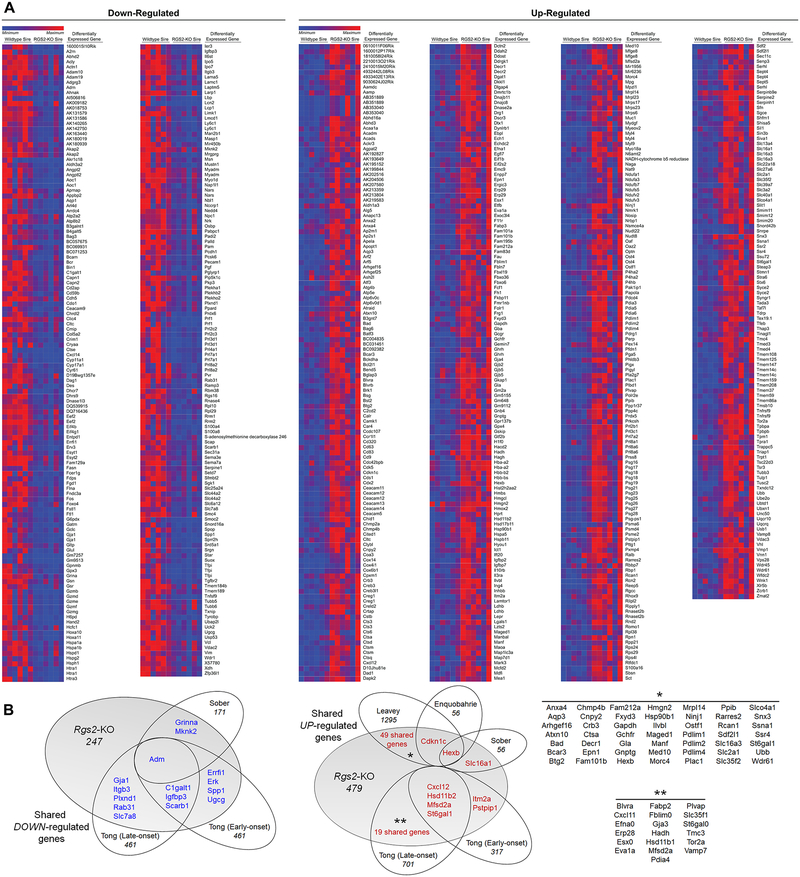
Differential gene expression analyses identified increased expression of 479 genes and decreased expression of 247 genes in the GD12.5 placentas from dams mated with Rgs2-KO sires (Figure 3A, S10). Many transcriptomic changes observed in these placentas were similar to expression changes previously reported in human PreE placentas (Figure 3B). Ingenuity Pathway Analysis (IPA) was then used to identify enriched pathways and networks, and many gene expression patterns previously identified as altered in placenta from humans with PreE were identified as significantly enriched in GD12.5 placentas from dams mated with Rgs2-KO sires, including mitochondrial dysfunction, unfolded protein responses, cell death & survival, cell movement, cell growth & proliferation, oxidative stress, increased levels of ALT, and red blood cell production (Tables S2–S5).
Figure 3. Reduced feto-placental expression of Rgs2 induces changes in GD12.5 placental gene expression consistent with PreE.
(A) Heat map illustrating relative expression of differentially upregulated (479) and downregulated (247) genes identified by DeSeq2 (FDR < 0.1) (Littermate n=6, Rgs2-KO n=6 placentas, each from an independent pregnancy). (B) Venn diagrams demonstrating comparisons of the up- and downregulated genes in Rgs2-heterozygous placentas to genes similarly changed in human placentas from pregnancies affected by PreE.20, 36–38 Numbers indicate the total number of genes significantly changed in the individual datasets, or shared between datasets.
Transcriptional control of RGS2 by CREB is altered in placenta during PreE
Previously, the rs4606 SNP “G” allele for RGS2 in mothers was associated with PreE, and the same allele is suspected to reduce RGS2 transcript stability.9–11, 14 In the current cohort, we determined that expression of RGS2 in placenta did not correlate with rs4606 genotype (Figure S11). This suggests that other mechanisms also contribute to reduced RGS2 in the placenta during PreE.
Methylation of the RGS2 promoter has also been implicated in control of RGS2 expression,24 but little methylation was observed in any placental sample by the bisulfite conversion method, and no change in methylation of the RGS2 promoter was observed during PreE (Figure S12).
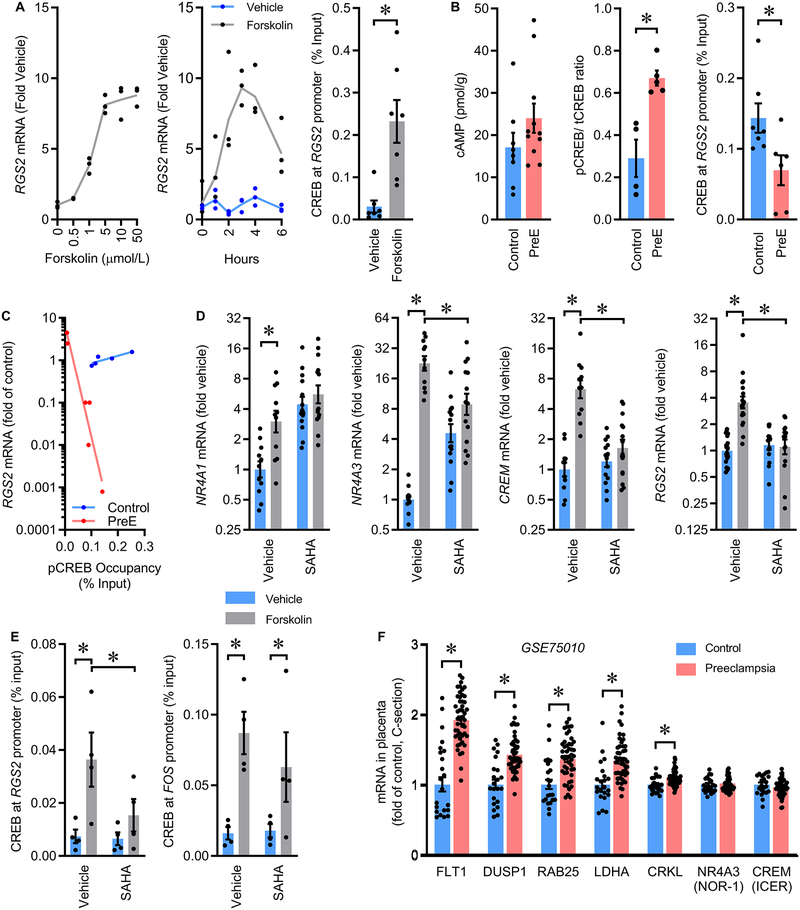
Expression of Rgs2 is stimulated in vascular smooth muscle by cAMP/CREB through serine-133 phosphorylated CREB (pCREB) binding to a specific cAMP response element (CRE) sequence within the Rgs2 promoter.25 We examined whether this pathway was functional in immortalized HTR-8/SVneo (HTR8) first-trimester human trophoblasts. Increasing cAMP via forskolin (FSK) resulted in dose- and time-dependent increases in expression of RGS2, and chromatin immunoprecipitation (ChIP) confirmed FSK-stimulated binding of pCREB to the RGS2 promoter in these cells (Figure 4A).
Figure 4. Transcriptional regulation of RGS2 by the cAMP/CREB signaling pathway is impaired in human PreE.
(A) Dose- and time-dependent responses of RGS2 mRNA in HTR8/SVneo cells treated with Forskolin (n=3 each dose / time), and serine-133 phosphorylated CREB (pCREB) presence at the RGS2 promoter in HTR8 cells after treatment with FSK (n=7 each). (B) cAMP concentrations (Control n=8, PreE n=11), ratio of serine-133 phosphorylated CREB to total CREB (Control n=4, PreE n=5), and serine-133 phosphorylated CREB presence at the RGS2 promoter (Control n=7, PreE n=6) in placenta from humans with or without PreE. (C) Correlation of RGS2 mRNA content to pCREB occupancy at the RGS2 promoter in human placental tissue (Control n=5, PreE n=6); comparison of curves by extra sum-of-squares: F=56.65 (2,7), p<0.0001. (D) NR4A1, NR4A3, CREM, and RGS2 mRNA in HTR8/SVneo cells after stimulation by forskolin in the presence or absence of the non-selective HDAC inhibitor, SAHA (Vehicle n=11, SAHA n=14). (E) Serine-133 phosphorylated CREB occupancy at the RGS2 and FOS promoters of HTR8/SVneo cells (n=4 each). (F) Canonical CREB target gene expression in human placentas by in silico reanalysis of GSE75010 (Control n=22, PreE n=47). Summary data presented as mean±SEM. *p<0.05 by t-test (A, B, F) or Tukey multiple-comparison procedure (D, E).
We examined the status of the cAMP/CREB/RGS2 mechanism in PreE placenta, as others have previously documented increased cAMP/CREB activities in independent cohorts of PreE placenta.26, 27 In the present cohort, the concentration of cAMP in placental tissue exhibited a non-significant increase during PreE, and CREB phosphorylation at serine residue 133 was significantly increased during PreE (Figures 4B, S13). Additional in silico reanalysis of GSE7501020 by GSEA also uncovered significant enrichment of CREB target gene expression during PreE (FDR q-value=0.037, NES=1.54). Despite these multiple lines of evidence supporting normal or increased cAMP/CREB signaling in the placenta during PreE, pCREB binding to the RGS2 promoter was significantly reduced in PreE placenta, consistent with an RGS2 locus-specific effect (Figure 4B). Correlational analyses of the expression of RGS2 versus pCREB binding to the RGS2 promoter in individual placenta samples illustrates a complex alteration in the relationship between pCREB binding and RGS2 expression during PreE, as both the slope and intercept of the lines of regression are changed during PreE (Figure 4C).
Previously, Fass demonstrated that inhibition of histone deacetylases (HDACs) increases expression of multiple CREB target genes in PC12 cells with FSK application, including FOS and NR4A1.28 In contrast, HDAC inhibition antagonized FSK-induced expression of a subset of CREB targets including NR4A3 and CREM (ICER). Consistent with the findings of Fass, we determined that FSK application increased expression of NR4A1, NR4A3, and CREM in cultured HTR8 cells, and the non-specific HDAC inhibitor, suberanilohydroxamic acid (SAHA) interfered with induction of NR4A3 and CREM (Figure 4D). We next discovered that the induction of RGS2 expression by FSK is dependent upon HDAC activity (Figure 4D). SAHA co-treatment also attenuated pCREB binding to the RGS2 promoter (Figures 4E, S14). Importantly, pCREB binding to another (HDAC-independent) cAMP responsive gene, FOS, was unaffected by SAHA treatment. Consistent with the concept that HDAC activity may be impaired in PreE placenta, mRNA levels of canonical CREB responsive genes such as FLT1, DUSP1, RAB25, LDHA and CRKL were increased in placenta during PreE, while the HDAC-dependent CREB responsive genes NR4A3 and CREM were unchanged (Figure 4F, S15).
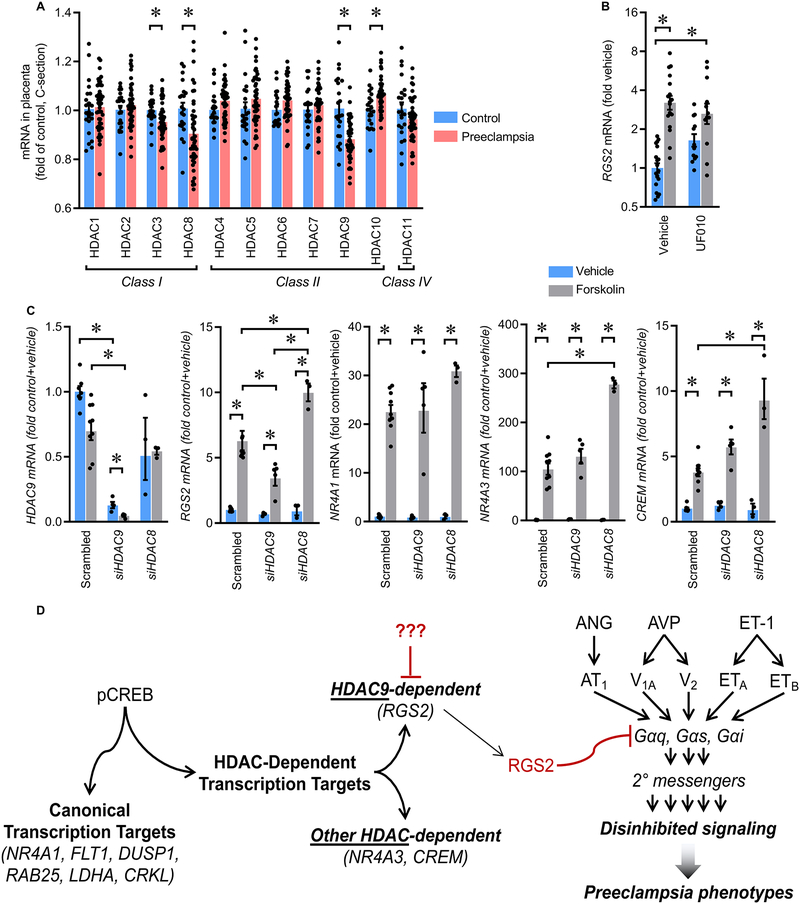
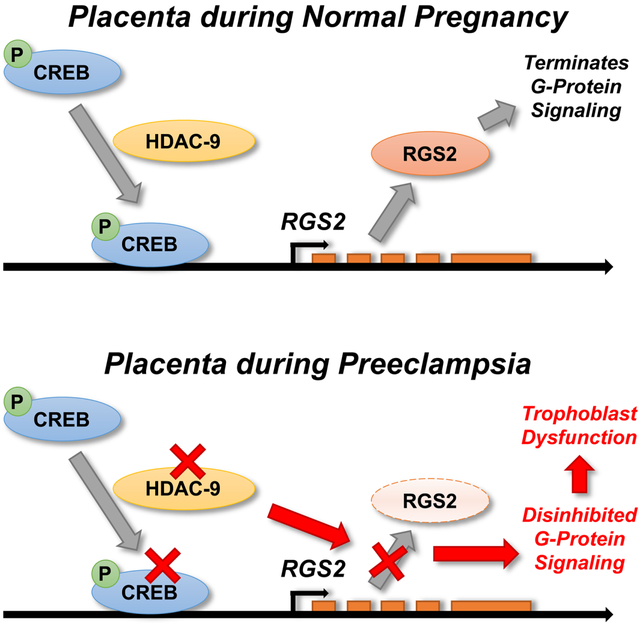
Reductions in HDAC3, HDAC8, and HDAC9, and increased HDAC10 mRNA were identified in PreE placenta via in silico reanalysis of GSE75010 (Figure 5A). These findings are consistent with the recent report that HDAC9 expression is suppressed in PreE placenta, and that HDAC9 knockdown inhibits proliferation, migration and invasion of HTR8/svneo cells.29 Activities of HDAC3, HDAC8, and HDAC10 are inhibited by benzoylhydrazide scaffold compound UF010, but this compound has no effect on HDAC9.30 UF010 had no inhibitory effect upon FSK-mediated stimulation of RGS2 in HTR8 cells (Figure 5B), consistent with a role for HDAC9 in RGS2 control. Further, siRNA-mediated selective knockdown of HDAC9 but not HDAC8 in HTR8 cells resulted in reduced FSK-mediated stimulation of RGS2 but not the SAHA-insensitive NR4A1 (Figure 5C). Interestingly, HDAC9 knockdown had no inhibitory effect upon FSK-mediated stimulation of other SAHA-sensitive genes NR4A3 and CREM, indicating that subsets of HDAC-dependent CREB-responsive genes are regulated by distinct HDAC enzymes. These results lead us to the working model that reduced HDAC9 activity in results in reduced CREB-mediated induction of RGS2 in the trophoblast, which would be expected to disinhibit GPCR signaling and promote placental dysfunction and PreE (Figure 5D).
Figure 5. Transcriptional control of RGS2 by CREB is dependent upon HDAC9 activity.
(A) HDAC gene mRNA in human placentas by in silico reanalysis of GSE75010 (Control n=22, PreE n=47). (B) RGS2 mRNA in HTR8/SVneo cells after stimulation with forskolin (FSK) in the presence or absence of the selective HDAC inhibitor, UF010, which inhibits HDAC3, HDAC8, and HDAC10 but not HDAC9 (Vehicle n=19, UF010 n=18, FSK n=18, FSK+UF010 n=12). (C) HDAC9, RGS2, NR4A1, NR4A3, and CREM mRNA in HTR8/SVneo cells after stimulation with FSK following selective knockdown of HDAC9 or HDAC8 by siRNA (scrambled siRNA control: n=7 vs n=8; siRNA against HDAC9: n=4 vs n=5; siRNA against HDAC8: n=3 vs n=3). Summary data presented as mean±SEM. *p<0.05 by Tukey multiple-comparison procedure. (D) Working model.
Discussion
A major role for the placenta in the development and maintenance of PreE is widely accepted, and the current study demonstrates an association between PreE with reduced expression of RGS2 in the human placenta, and the sufficiency of reduced feto-placental expression of Rgs2 to elicit several physiological and molecular PreE-like phenotypes in wildtype C57BL/6J dams.
Previously we reported the sufficiency of chronic low-dose infusion of arginine vasopressin to induce all of the clinical features of PreE in C57BL/6J dams without inducing placental hypoxia.6 Similarly, here we report that reduced expression of Rgs2 in the feto-placental unit is sufficient to induce PreE-like clinical features without inducing placental hypoxia. Interestingly, both models induced other molecular signatures within the placenta that parallel PreE, including mitochondrial dysfunction, oxidative stress and unfolded protein responses. Others have demonstrated that surgical induction of placental hypoxia/ischemia (eg – the RUPP model) is sufficient to induce PreE-like phenotypes without modifying AVP secretion.31 These findings lead to the concept that aberrant GPCR signaling and hypoxia – which are both documented in human PreE – exist as independent, and perhaps synergistic, contributors to the pathogenesis of PreE.6 The variable contributions of multiple independent mechanisms to the pathogenesis of PreE may help explain the heterogeneity of clinical presentations of this disorder (early/late, mild/severe, etc.) and the growing appreciation for multiple independent ‘molecular subclasses’ of PreE.20
RGS2 has previously been associated with hypertensive disorders of pregnancy such as PreE, though not specifically within the placenta. Mothers carrying the G allele for the rs4606 SNP in RGS2 exhibit increased risk of PreE.9–11 This association is modified by relevant co-variates such as BMI,9 which may reflect obesity-associated increases in circulating hormones that activate RGS2-sensitive G protein signaling, such as angiotensin II32 or arginine vasopressin33. Although HUNT2 studies demonstrated that the maternal genotype for rs4606 was associated with increased risk of PreE, it is notable that RGS2 mRNA expression levels were not assessed, and that the placental rs4606 genotypes were also not assessed. The rs4606 genotype has been studied outside the context of PreE, however, as Semplicini et al. demonstrated that rs4606 is associated with human hypertension and is associated with decreased RGS2 mRNA and protein in peripheral blood mononuclear cells.14 Our data that placental rs4606 genotype did not correlate with RGS2 mRNA align with findings from Mendelova, in which no association between maternal rs4606 genotype and PreE status was observed.34 We conclude that rs4606 may confer moderate, perhaps tissue-specific, changes in control of RGS2 but that this mechanism is unlikely to account for the large reductions in RGS2 observed in placental tissue during PreE – and may indicate that RGS2 plays multiple independent roles in mother and placenta that each independently contribute to PreE.
Impaired transcriptional regulation of RGS2 could also occur due to mutations within the promoter, or deficiencies in critical transcriptional regulators. More than 30 polymorphisms have been identified within the RGS2 gene in hypertensive humans, and three of these SNP’s flank the functional CRE element within the RGS2 promoter.25 It is possible that mutations in these locations could interfere with CREB binding to the CRE sequence,25 however their low prevalence is unlikely to account for the reductions in RGS2 expression observed in PreE at the population level. The mechanism by which HDAC9 modulates CREB-mediated induction of transcription remains unclear but likely involves a complex of interacting factors at the RGS2 promoter and upstream enhancer (Tables S8, S9), warranting future studies.
One major limitation facing the current study is the lack of tools available to assess RGS2 protein levels or to assess its activity in human tissues. Another major limitation facing the current study is the lack of data addressing cell- or layer-specific changes in gene expression in PreE. Ongoing work by our group and others to explore cell-type specific changes in transcriptomes of PreE versus control placentas using single-cell RNAsequencing techniques promises to provide this type of analysis. A third major limitation stems from the lack of an animal harboring a conditional Rgs2 allele (eg – Rgs2flox) to permit layer-specific disruptions of the gene in mouse placenta. Our team is currently working to regenerate such a model, as the only previously-reported animal of this type was lost in 2012.35 The current study provides clear rationale for the pursuit of such tools and studies.
Supplementary Material
Perspectives.
Reduced RGS2 expression in the placenta is associated with PreE, and reduced Rgs2 expression in the feto-placental unit is sufficient to initiate PreE-like phenotypes in wildtype C57BL/6J dams. Our data identify RGS2 as an HDAC9-dependent CREB-responsive gene, and the suppression of RGS2 expression in human placenta during PreE is associated with suppressed HDAC9 activity. These findings identify RGS2 and HDAC9-modulated CREB signaling within the placenta as potential diagnostic and therapeutic targets for PreE.
Novelty & Significance.
What is new?
RGS2 mRNA is reduced in human placenta during PreE despite enhanced CREB signaling
Reduced Rgs2 expression in mouse placenta is sufficient to cause PreE-like phenotypes
RGS2 expression is stimulated by CREB via an HDAC9-dependent mechanism
HDAC9 expression is reduced in human placenta during PreE
What is Relevant?
Placental RGS2 expression, independent of the rs4606 genotype, modifies PreE risk
Paternal and placental genetics importantly contribute to PreE risk
RGS2 is an HDAC9-depenedent CREB-responsive gene
Acknowledgments
The authors gratefully acknowledge the participation of mothers in the University of Iowa MFTB. Multiple core facilities supported the project, including the University of Iowa Office of Animal Resources, Genome Editing Facility, and DNA Core Facility.
Sources of Funding
This work was supported by the American Heart Association (15SFRN23730000, 16PRE30980043, 16POST30960016, 17PRE33660633, 18PRE33960377, 18EIA33890055, 19POST34380239), National Institutes of Health (HL134850, HL084207, HD089940, UL1TR002537, P30CA086862), Roy Carver Trust, American Physiological Society, and University of Iowa Medical Student and Summer Undergraduate MSTP Research programs.
Footnotes
Disclosures
JLG, DAS, and MKS hold patents related to AVP for the prediction and treatment of PreE: US #9,937,182 (April 10, 2018), EU #2,954,324 (July 31, 2019), and PCT/US2018/027152.
References
- 1.Roberts JM. The perplexing pregnancy disorder preeclampsia: What next? Physiological genomics. 2018;50:459–467 [DOI] [PubMed] [Google Scholar]
- 2.Saleh L, Danser JA, van den Meiracker AH. Role of endothelin in preeclampsia and hypertension following antiangiogenesis treatment. Current opinion in nephrology and hypertension. 2016;25:94–99 [DOI] [PubMed] [Google Scholar]
- 3.Santillan MK, Santillan DA, Scroggins SM, Min JY, Sandgren JA, Pearson NA, Leslie KK, Hunter SK, Zamba GK, Gibson-Corley KN, Grobe JL. Vasopressin in preeclampsia: A novel very early human pregnancy biomarker and clinically relevant mouse model. Hypertension. 2014;64:852–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gant NF, Daley GL, Chand S, Whalley PJ, MacDonald PC. A study of angiotensin ii pressor response throughout primigravid pregnancy. The Journal of clinical investigation. 1973;52:2682–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou CC, Ahmad S, Mi T, Abbasi S, Xia L, Day MC, Ramin SM, Ahmed A, Kellems RE, Xia Y. Autoantibody from women with preeclampsia induces soluble fms-like tyrosine kinase-1 production via angiotensin type 1 receptor and calcineurin/nuclear factor of activated t-cells signaling. Hypertension. 2008;51:1010–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandgren JA, Deng G, Linggonegoro DW, Scroggins SM, Perschbacher KJ, Nair AR, Nishimura TE, Zhang SY, Agbor LN, Wu J, Keen HL, Naber MC, Pearson NA, Zimmerman KA, Weiss RM, Bowdler NC, Usachev YM, Santillan DA, Potthoff MJ, Pierce GL, Gibson-Corley KN, Sigmund CD, Santillan MK, Grobe JL. Arginine vasopressin infusion is sufficient to model clinical features of preeclampsia in mice. JCI insight. 2018;3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perschbacher KJ, Deng G, Fisher RA, Gibson-Corley KN, Santillan MK, Grobe JL. Regulators of g protein signaling in cardiovascular function during pregnancy. Physiological genomics. 2018;50:590–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holobotovskyy V, Chong YS, Burchell J, He B, Phillips M, Leader L, Murphy TV, Sandow SL, McKitrick DJ, Charles AK, Tare M, Arnolda LF, Ganss R. Regulator of g protein signaling 5 is a determinant of gestational hypertension and preeclampsia. Science translational medicine. 2015;7:290ra288. [DOI] [PubMed] [Google Scholar]
- 9.Karppanen T, Kaartokallio T, Klemetti MM, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Staff AC, Laivuori H. An rgs2 3’utr polymorphism is associated with preeclampsia in overweight women. BMC genetics. 2016;17:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kvehaugen AS, Melien O, Holmen OL, Laivuori H, Dechend R, Staff AC. Hypertension after preeclampsia and relation to the c1114g polymorphism (rs4606) in rgs2: Data from the norwegian hunt2 study. BMC medical genetics. 2014;15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kvehaugen AS, Melien O, Holmen OL, Laivuori H, Oian P, Andersgaard AB, Dechend R, Staff AC. Single nucleotide polymorphisms in g protein signaling pathway genes in preeclampsia. Hypertension. 2013;61:655–661 [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Kamide K, Kokubo Y, Takiuchi S, Tanaka C, Banno M, Miwa Y, Yoshii M, Horio T, Okayama A, Tomoike H, Kawano Y, Miyata T. Genetic variations of regulator of g-protein signaling 2 in hypertensive patients and in the general population. Journal of hypertension. 2005;23:1497–1505 [DOI] [PubMed] [Google Scholar]
- 13.He F, Luo J, Zhang Z, Luo Z, Fan L, He Y, Wen J, Zhu D, Gao J, Wang Y, Qian Y, Zhou H, Chen X, Zhang W. The rgs2 (−391, c>g) genetic variation correlates to antihypertensive drug responses in chinese patients with essential hypertension. PloS one. 2015;10:e0121483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semplicini A, Lenzini L, Sartori M, Papparella I, Calo LA, Pagnin E, Strapazzon G, Benna C, Costa R, Avogaro A, Ceolotto G, Pessina AC. Reduced expression of regulator of g-protein signaling 2 (rgs2) in hypertensive patients increases calcium mobilization and erk1/2 phosphorylation induced by angiotensin ii. Journal of hypertension. 2006;24:1115–1124 [DOI] [PubMed] [Google Scholar]
- 15.Jie L, Owens EA, Plante LA, Fang Z, Rensing DT, Moeller KD, Osei-Owusu P. Rgs2 squelches vascular gi/o and gq signaling to modulate myogenic tone and promote uterine blood flow. Physiological reports. 2016;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, Lin HY, Zhu C, Ge H, Cross JC, Wang H. Single-cell rna-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell research. 2018;28:819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gormley M, Ona K, Kapidzic M, Garrido-Gomez T, Zdravkovic T, Fisher SJ. Preeclampsia: Novel insights from global rna profiling of trophoblast subpopulations. American journal of obstetrics and gynecology. 2017;217:200 e201–200 e217 [DOI] [PubMed] [Google Scholar]
- 19.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan GC, Chen M, Guo G. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107.e1017 [DOI] [PubMed] [Google Scholar]
- 20.Leavey K, Benton SJ, Grynspan D, Kingdom JC, Bainbridge SA, Cox BJ. Unsupervised placental gene expression profiling identifies clinically relevant subclasses of human preeclampsia. Hypertension. 2016;68:137–147 [DOI] [PubMed] [Google Scholar]
- 21.Santillan MK, Leslie KK, Hamilton WS, Boese BJ, Ahuja M, Hunter SK, Santillan DA. “Collection of a lifetime: A practical approach to developing a longitudinal collection of women’s healthcare biological samples”. European journal of obstetrics, gynecology, and reproductive biology. 2014;179:94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heximer SP, Knutsen RH, Sun X, Kaltenbronn KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin AJ, Steinberg TH, Wyss JM, Mecham RP, Blumer KJ. Hypertension and prolonged vasoconstrictor signaling in rgs2-deficient mice. The Journal of clinical investigation. 2003;111:445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaMarca B, Cornelius D, Wallace K. Elucidating immune mechanisms causing hypertension during pregnancy. Physiology (Bethesda, Md.). 2013;28:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff DW, Xie Y, Deng C, Gatalica Z, Yang M, Wang B, Wang J, Lin MF, Abel PW, Tu Y. Epigenetic repression of regulator of g-protein signaling 2 promotes androgen-independent prostate cancer cell growth. International journal of cancer. 2012;130:1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Liu D, Liu S, Calderon L, Zhao G, Turk J, Guo Z. Identification of a camp-response element in the regulator of g-protein signaling-2 (rgs2) promoter as a key cis-regulatory element for rgs2 transcriptional regulation by angiotensin ii in cultured vascular smooth muscles. The Journal of biological chemistry. 2011;286:44646–44658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaiman D, Calicchio R, Miralles F. Landscape of transcriptional deregulations in the preeclamptic placenta. PLoS One. 2013;8:e65498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moslehi R, Mills JL, Signore C, Kumar A, Ambroggio X, Dzutsev A. Integrative transcriptome analysis reveals dysregulation of canonical cancer molecular pathways in placenta leading to preeclampsia. Sci Rep. 2013;3:2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for camp activation of a subset of creb target genes. The Journal of biological chemistry. 2003;278:43014–43019 [DOI] [PubMed] [Google Scholar]
- 29.Xie D, Zhu J, Liu Q, Li J, Song M, Wang K, Zhou Q, Jia Y, Li T. Dysregulation of hdac9 represses trophoblast cell migration and invasion through timp3 activation in preeclampsia. American journal of hypertension. 2019;32:515–523 [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Stowe RL, Pinello CE, Tian G, Madoux F, Li D, Zhao LY, Li JL, Wang Y, Wang Y, Ma H, Hodder P, Roush WR, Liao D. Identification of histone deacetylase inhibitors with benzoylhydrazide scaffold that selectively inhibit class i histone deacetylases. Chemistry & biology. 2015;22:273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palei AC, Warrington JP, Granger JP. The effect of placental ischemia-induced hypertension on circulating copeptin levels of pregnant rats. Faseb Journal. 2016;30:lb765 [Google Scholar]
- 32.Littlejohn NK, Grobe JL. Opposing tissue-specific roles of angiotensin in the pathogenesis of obesity, and implications for obesity-related hypertension. American journal of physiology. Regulatory, integrative and comparative physiology. 2015;309:R1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asferg CL, Andersen UB, Linneberg A, Goetze JP, Jeppesen JL. Copeptin, a surrogate marker for arginine vasopressin secretion, is associated with higher glucose and insulin concentrations but not higher blood pressure in obese men. Diabetic medicine : a journal of the British Diabetic Association. 2014;31:728–732 [DOI] [PubMed] [Google Scholar]
- 34.Mendelova A, Holubekova V, Grendar M, Zubor P, Svecova I, Loderer D, Snahnicanova Z, Biringer K, Danko J, Lasabova Z. Association between 3’utr polymorphisms in genes acvr2a, agtr1 and rgs2 and preeclampsia. Gen Physiol Biophys. 2018;37:185–192 [DOI] [PubMed] [Google Scholar]
- 35.Osei-Owusu P, Sabharwal R, Kaltenbronn KM, Rhee MH, Chapleau MW, Dietrich HH, Blumer KJ. Regulator of g protein signaling 2 deficiency causes endothelial dysfunction and impaired endothelium-derived hyperpolarizing factor-mediated relaxation by dysregulating gi/o signaling. The Journal of biological chemistry. 2012;287:12541–12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong J, Zhao W, Lv H, Li WP, Chen ZJ, Zhang C. Transcriptomic profiling in human decidua of severe preeclampsia detected by rna sequencing. Journal of cellular biochemistry. 2018;119:607–615 [DOI] [PubMed] [Google Scholar]
- 37.Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. American journal of obstetrics and gynecology. 2008;199:566.e561–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sober S, Reiman M, Kikas T, Rull K, Inno R, Vaas P, Teesalu P, Marti JM, Mattila P, Laan M. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci Rep. 2015;5:13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.