Abstract
Background:
Attention-deficit/hyperactivity disorder (ADHD) and bipolar disorder (BPD) are frequently co-occurring and highly heritable mental health conditions. We hypothesized that BPD cases with an early age of onset (≤21 years) would be particularly likely to show genetic covariation with ADHD.
Methods:
GWAS data were available for 4,609 individuals with ADHD, 9,650 individuals with BPD (5,167 thereof with early-onset BPD) and 21,363 typically developing controls. We conducted a cross-disorder GWAS meta-analysis to identify whether the observed comorbidity between ADHD and BPD could be due to shared genetic risks.
Results:
We found a significant SNP-based genetic correlation between ADHD and BPD in the full and age-restricted samples (rGfull = 0.64, p = 3.13×10−14; rGrestricted = 0.71, p = 4.09×10−16). The meta-analysis between the full BPD sample identified two genome-wide significant (prs7089973 = 2.47×10−8; Prs11756438 = 4.36×10−8) regions located on chromosomes 6 (CEP85L) and 10 (TAF9BP2). Restricting the analyses to BPD cases with an early onset yielded one genome-wide significant association (prs58502974 = 2.11×10−8) on chromosome 5 in the ADCY2 gene. Additional nominally significant regions identified contained known eQTLs with putative functional consequences for NT5DC1, NT5DC2 and CACNB3 expression, while functional predictions implicated ABLIM1 as an allele-specifically expressed gene in neuronal tissue.
Conclusions:
The SNP-based genetic correlation between ADHD and BPD is substantial, significant, and consistent with the existence of genetic overlap between ADHD and BPD, with potential differential genetic mechanisms involved in early and later BPD onset.
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is the most frequent neuropsychiatric disorder in childhood and frequently persists into adulthood. Bipolar disorder (BPD) is amongst the most prevalent mental diseases in adulthood. Both disorders are highly heritable (1,2). However, in both cases the mode of inheritance is complex and polygenic (3). Although differing from one another with regard to core signs and symptoms, age of onset, presentation and treatment response, the two disorders share several clinical features. This is especially the case for the manic phase of BPD, which is associated with irritability, increased impulsivity, distractibility, and restlessness (4). Furthermore, ADHD often co-presents with depression (5), a core feature of BPD. In adulthood, when BPD is most commonly diagnosed, co-occurrence of the two disorders occurs more often than would be expected by chance (6). For patients with BPD rates of ADHD vary between 9.5% and 28%, depending on study characteristics (7, 8). The rate of BPD in adult ADHD has been estimated at around 20% (7). Meta-analyses of family studies confirm elevated rates of BPD in first-degree relatives of ADHD patients and vice versa (8).
Although a shared genetic basis for ADHD and BPD seems plausible given the above, molecular genetic studies thus far provide limited evidence for this (9-11). For instance, risk allele frequencies of candidate genes, identified through prior ADHD genome-wide association studies, are not increased in BPD. This failure to find a shared genetic signal may be due to prior studies’ lack of statistical power, and the limited set of polymorphisms examined. To address these shortcomings, a genome-wide cross-disorder meta-analysis of BPD and ADHD was conducted in a large sample of individuals with BPD, with ADHD and typically developing controls from the Psychiatric Genomics Consortium. Because ADHD is a childhood onset disorder (prior to 12 years), we hypothesized that the overlap would be most obvious in BPD cases with a relatively early onset (age of onset ≤21 years), as this group could be assumed to have a more neuro-developmental etiology (8). Restricting the age range of the sample to those with an onset ≤21 years may also increase the power for gene-finding since it could reduce heterogeneity (12). We thus performed analyses with the total number of BPD cases as well as the age-restricted set. GWAS meta-analysis top-findings were further characterized using eQTL analysis to investigate potential functional consequences for gene expression.
Methods and Materials
Samples
Cases, controls, and family-based samples assembled for previous genome-wide PGC analyses of individual-level data were included in the current analysis (13,14). A description of individual study data contributions and genotyping platforms is included in the Supplemental Tables S1a and S1b. The ADHD sample comprised 4,609 cases and 8,519 controls. The full BPD sample comprised 9,650 cases and 12,844 controls. For tests of the age of onset hypothesis we restricted the BPD sample to cases with an age of onset ≤21 years of age. This reduced the number of cases to 5,167 (restricted sample). All available controls from the BPD samples were included in the age restricted sample to maximize power. Control individuals that featured in both the ADHD and BPD samples were identified and removed prior to analysis.
Genetic Analysis
Raw genotype and phenotype data for each study was uploaded to a central server and processed through the same quality control, imputation, and analysis process to ensure comparability between the samples. The quality control and analysis pipeline is described elsewhere (3).
Statistical analysis
Linkage disequilibrium score regression (LDSR) was used to estimate the SNP-based genetic correlation (rG) between ADHD and both BPD samples. For LDSR, each data set underwent additional filtering. Only markers overlapping with HapMap Project Phase 3 SNPs and passing the following filters were included: INFO score > 0.9, study missingness of 0 and MAF > 1% (where available). Indels and strand-ambiguous SNPs were removed.
The analysis was conducted using a two-step procedure with the LD-scoring analysis package (41). An unconstrained regression was run to estimate the regression intercepts for each phenotype, followed by an analysis with regression intercepts constrained to those estimated in the first step and an unconstrained covariance intercept (we took steps to exclude overlapping samples). Standard errors were estimated using a block jackknife procedure and used to calculate P-values.
GWAS was initially performed for each ADHD study separately (n=8). Four multidimensional scaling components were included to account for potential population stratification. GWAS was then also performed for each BPD study separately (n=12). In this case a total of seven multidimensional scaling components (both total and restricted samples). These GWAS were free from genomic inflation as judged by quantile-quantile plots (data not shown). For each disorder, results were then combined in a disorder-specific meta-analysis. Finally, results from the disorder-specific meta-analyses were combined in cross-disorder meta-analyses for both the primary and the age-restricted samples. For all meta-analyses, we applied a weighted Z-score approach using Plink 1.07, in which weights equaled the inverse of the regression coefficient’s standard error (15). This strategy assumed a fixed-effects model, in which all studies/disorders had the same direction of effect, with weights indicating the sample size and imputation accuracy of the disease-specific studies. The fixed effects model was compared to a genome-wide random-effects model, in which studies/disorders were allowed to have a different direction of effect.
Prediction of allele-specific effects on transcription
The overlap between polymorphic loci with miRNA binding sites was examined suing the PolymiRTS 3.0 database (16). To check for the known influence of identified SNPs on gene expression, we searched a database of cis-acting eQTLs defined with RNA-seq data of lymphoblastoid cell lines from 462 individuals, most of which were also examined in the 1000 Genomes Phase I dataset (17). Allele-specific transcription factor binding sites (TFBS) were predicted with the web-based tool MatInspector version 2.1 (18). TFBS searches were performed for all promoter regions (40 kb upstream) of the top-100 SNPs identified in the full and the age-restricted meta-analyses (i.e. each allele flanked by 10 bp up- and downstream sequence). Sequences with a transcription factor-specific matrix core similarity of at least 0.75 were defined as potentially containing the respective TFBS.
Results
SNP-Based Genetic Correlation
The SNP-based rG between ADHD and BPD was substantial and significant for both the full and age-restricted samples. Interestingly, the rG was higher for the age restricted as compared to the fUll sample (rGfull = 0.64, SE= 0.02, p = 3.13×10−14; rGrestricted = 0.71, SE= 0.02, p = 4.09×10−16; Table 1).
Table 1:
Results from the univariate heritability analysis (h2g [SE]), and genetic correlation between data from ADHD and BPD complete and age restricted samples.
| BPD cohort | N (cases/controls) | Heritability | SE | Genetic correlation with ADHD | SE | Z-score | P-value |
|---|---|---|---|---|---|---|---|
| Age restricted | 5,167/12,844 | 0.119 | 0.023 | 0.7062 | 0.087 | 7.593 | 4.09 × 10−16 |
| Full sample | 9,650/12,844 | 0.068 | 0.024 | 0.6392 | 0.084 | 8.136 | 3.13 × 10−14 |
ADHD: Ncases=4,609 and Ncontrols=8,519, SNP-based h2 (observed)= 0.0652 (SE=0.0279)
Full sample
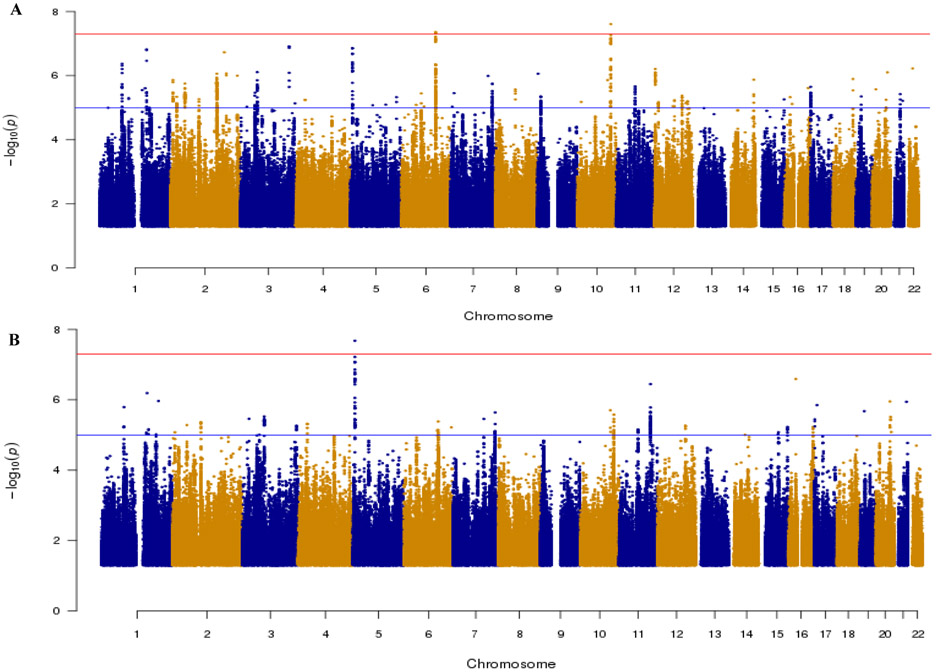
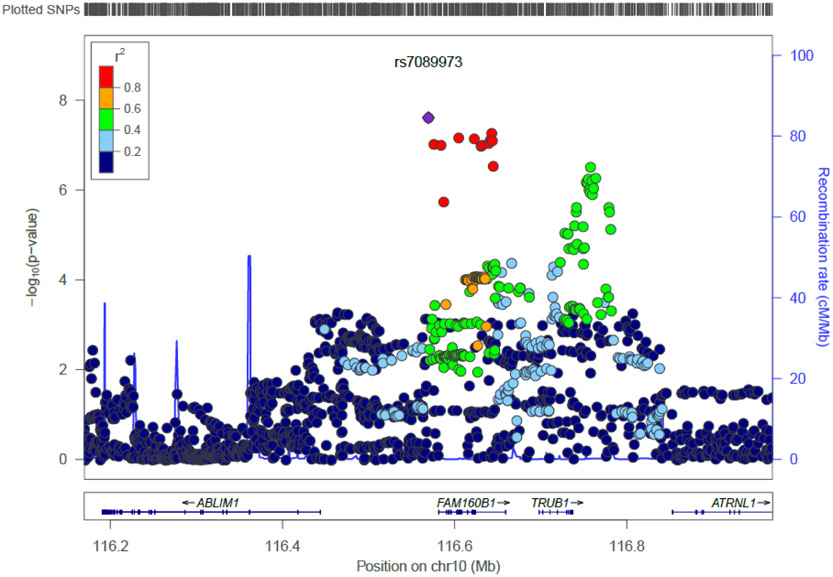
Figure 1A shows the Manhattan plot of the primary cross-disorder meta-analysis and Figure S1A shows the corresponding QQ plot. Two independent loci, located on chromosomes 6 and 10, reached genome-wide significance (p<5×10−8). The strongest signal (p = 2.47×10−8) was for SNP rs7089973, located on chromosome 10 in an intronic region of the TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa pseudogene 2 gene (TAF9BP2; Figure 2A) which is a pseudogene of yet unknown function. SNP rs7089973 is in linkage disequilibrium (LD) with SNPs in the TruB Pseudouridine (Psi) Synthase Homolog 1 gene (TRUB1; best p = 8.89×10−6) and with the family with sequence similarity 160 member B1 gene (FAM160B1; best p = 5.45×10−8).
Figure 1.
Manhattan plot of the primary (a) and restricted (b) cross-disorder meta-analyses. Only SNPs with p values ≤ 0.05 are shown; Horizontal lines show threshold for genome-wide significance (p<5×10−8 in red) and suggestive association (p<1×10−6 in blue).
Figure 2A.
Regional plot for genome-wide significant locus on chromosome 10 for the primary meta-analysis.
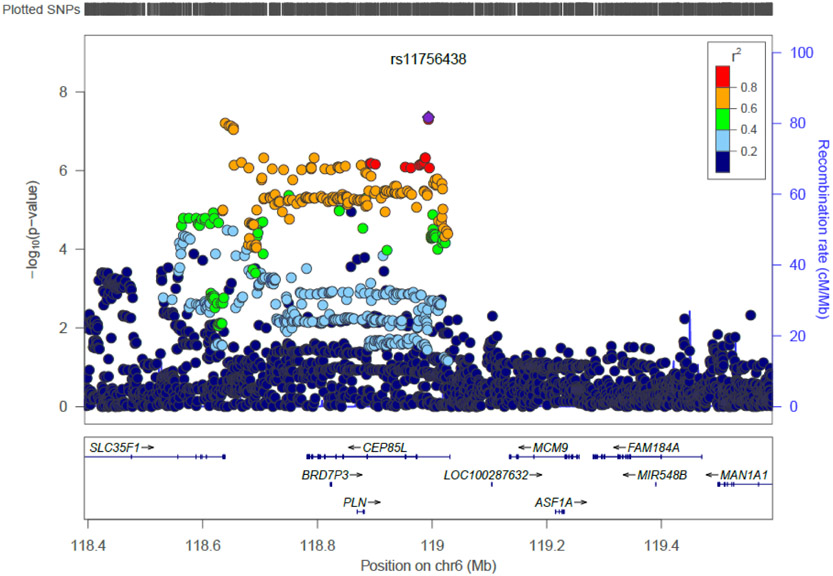
On chromosome 6, the strongest signal was for SNP rs11756438, located in an intronic region of the centrosomal protein 85kDa-like gene (CEP85L; p = 4.36×10−8; Figure 2B) encoding a protein of unknown function. Furthermore, rs11756438 is in LD with SNPs in the Bromodomain Containing 7 Pseudogene 3 gene (BRD7P3; best p = 9.39× 10−7), the phospholamban gene (PLN; best p = 7.25×10−7), and the Solute Carrier Family 35, Member F1 gene (SLC35F1; best p = 1.00×10−5). Both associated regions showed the same direction of effect in ADHD and BPD. The disorder-specific contribution to each genome-wide significant locus can be found in Table 2, and forest plots from individual GWAS are shown in Figure S5. Top-ranked SNPs for this analysis (p < 10−6) are summarized in Table S3.
Figure 2B.
Regional plot for genome-wide significant locus on chromosome 6 for the primary meta-analysis.
Table 2.
Association results showing effect size and p for genome-wide significant loci by disorder for the primary and restricted meta-analyses.
| Primary Meta-Analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Location (bp:chr) |
A1/A2 | MAF | ORADHD | PADHD | SEADHD | ORBPD | PBPD | SEBPD | ORMA | PMA | Gene |
| rs7089973 | 10:116569565 | A/C | 0.36 | 0.90 | 1.56× 10−3 | 0.03 | 0.90 | 4.39×l0−6 | 0.02 | 0.90 | 2.47× 10−8 | TAF9BP2 |
| rs11756438 | 6: 118993632 | A/C | 0.34 | 1.11 | 6.14×10−4 | 0.03 | 1.11 | 1.85×l0−5 | 0.02 | 1.10 | 4.36× 10−8 | CEP85L |
| Age Restricted Meta-Analysis | ||||||||||||
| SNP | Location (bp:chr) |
A1/A2 | MAF | ORADHD | PADHD | SEADHD | ORBPD | PBPD | SEBPD | ORMA | PMA | Gene |
| rs58502974 | 5:7755900 | A/T | 0.44 | 1.14 | 1.84× 10−5 | 0.03 | 1.11 | 2.33×l0−4 | 0.03 | 1.13 | 2.11×l0−8 | ADCY2 |
bp=base pairs; Chr= chromosome; MAF=minor allele frequency; OR=odds ratio; P=p value; SE=standard=standard error of the OR; ADHD=Attention- Deficit/Hyperactivity Disorder; BPD=Bipolar Disorder.
Age-Restricted BPD sample
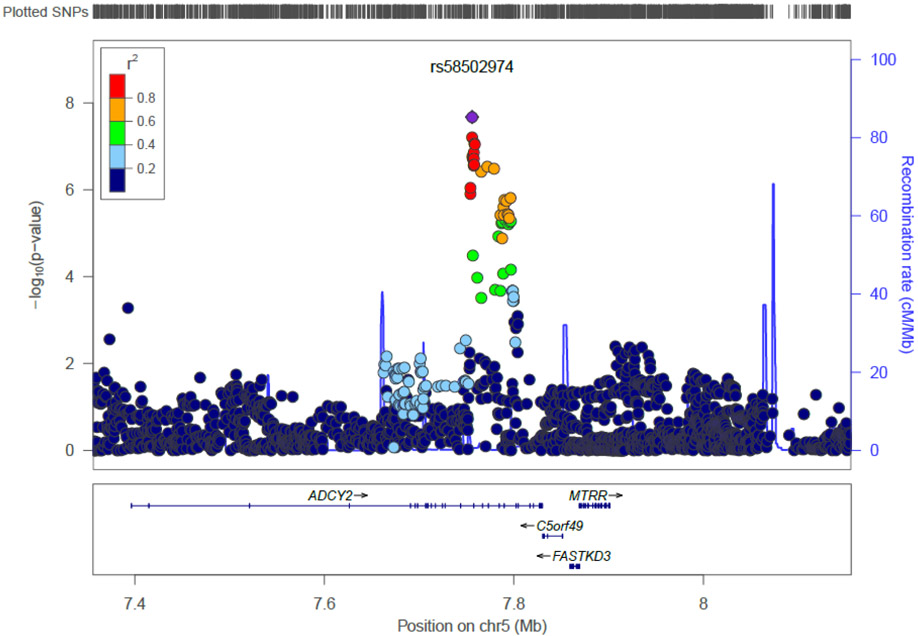
The Manhattan plot for the age-restricted analysis is shown in Figure 1B; the respective QQ plot is provided in Figure S1B. One SNP on chromosome 5 reached genome-wide significance; forest plots of results from individual GWAS are shown in Figure S5. The strongest signal (p = 2.47×10−8) was observed for SNP rs58502974, located in an intron of the Adenylate Cyclase 2 (Brain) (ADCY2) gene (Figure 2C). The product of this gene is a member of the family of adenylate cyclases, which are membrane-associated enzymes that catalyze the formation of the secondary messenger cyclic adenosine monophosphate (cAMP). Top-ranked SNPs for this analysis (p < 10−4) are summarized in Table S3.
Figure 2C.
Regional plot for genome-wide significant locus on chromosome 5 for the restricted meta-analysis.
Results are shown as –log(p-value) for genotyped and imputed SNPs. The SNP showing strongest association is shown in the purple circle. The color of the remaining markers reflects r2 of the strongest associated SNP. The recombination rate is plotted in blue.
Allele-specific transcriptional activity
None of the top-100 associated SNPs in the primary and restricted analyses (Supplemental Table S3) were listed in the Geuvadis database of cis-eQTLs defined in lymphoblastoid cell lines that were mainly derived from the 1000 Genomes sample (17). To estimate the potential eQTL function in other tissues, direct bioinformatics prediction was thus used and found differential binding of transcription factors for 13 markers in the primary and four in the restricted GWAS, which involved neuro-relevant transcription factors at seven and one sites, respectively (Supplemental Table S5). A notable finding among those was for the gene ABLIM1. Allele-specific binding sites of miRNAs at the 3’-UTR of family with sequence similarity 160, member B1 (FAM160B1) transcripts were also observed (Supplemental Table S5).
Extending this analysis to evaluate top-ranked SNPs (p < 10−4) for effects on transcriptional activity, we queried the Geuvadis database (17) for known eQTLs. Of the 4,806 SNPs displaying a trend towards association (p<1×104) in one of the two analyses, 192 indeed were known eQTLs (Supplemental Table S4). Of those, 74 markers influenced the expression of 5’-nucleotidase domain containing 2 (NT5DC2); the expression of its homolog NT5DC1 was modulated by 17 SNPs. Two SNPs were found to influence the expression of the calcium channel, voltage-dependent, beta 3 subunit (CACNB3). The accumulation of eQTLs at the NT5DC loci especially may be explained by LD. Indeed, when we tested the total number of eQTLs, it did not exceed expectation in either the full or the restricted analysis (ORprimary=0.91, pprimary=0.27; ORrestricted=0.97, pprestricted=0.84).
Discussion
In this study, we set out to identify shared genetic risk factor for ADHD and BPD through cross-disorder meta-analysis of the existing GWAS samples from the Psychiatric Genomics Consortium. We hypothesized that overlap between the disorders might be most pronounced in BPD cases with an early age of onset, and that restricting the analysis to samples with an onset ≤21 would increase the power of gene-finding by reducing heterogeneity (8). Our findings clearly show the substantial and significant genetic correlation between ADHD and BPD which supports the existence of genome-wide significant, shared genetic risk variants for ADHD and BPD (rGfull = 0.64, SE= 0.08, p = 3.13×10−14; rGrestricted = 0.71, SE= 0.09, p = 4.09×10−16; Table 1). We contrasted these results by estimating the genetic correlation between ADHD and BPD using previously published data sets and found the same correlation pattern (Supplemental Table S2).
In the full sample, we found two genome-wide significant loci and 10 with suggestive evidence of association (p<10−6; Supplemental Table S3). Although the statistical power was potentially lower in the age-restricted sample, one significant finding was observed. The region identified was different from the top-findings in the primary analysis, raising the possibility of development-specific gene activity in these two conditions. When specifically considering the BPD analyses (Table S3, also compare Figure S2A with S2B), it becomes clear that almost all top cross-disorder SNPs have lower p values in the age-restricted BPD sample as compared to the full BPD sample, despite its lower power. This might point to a stronger genetic component of early-onset BPD and consequently a different etiology as compared to late-onset BPD (19). However, these findings are still preliminary addressing this issue was not the major aim of our analysis and further work is clearly needed to adequately answer such questions.
Our top hit in the full analysis was present on chromosome 10 in the gene TAF9BP2. This being a pseudogene of unknown function, we cannot speculate about its potential involvement in the ADHD-BPD covariation. TRUB1, which includes variants in LD with the top SNP may constitute a potential candidate gene; its product is a member of the pseudouridine synthase gene family and may function as RNA chaperone, altering aspects of mRNA metabolism known to be affected by RNA structure (20). The protein encoded by CEP85L, suggested to be involved by the hit on chromosome 6, was identified as a breast cancer antigen (21); the gene was also associated in a meta-analysis with the myocardial repolarization (22,23), the latter lending some support to the notion that the gene’s product is involved in neural conduction. This is the first time that genes in these loci are associated with psychiatric diseases, so the mechanisms by which variants in those might affect disease risk remain elusive. Our eQTL analyses did not provide evidence for a direct effect of the variation on the expression of the implicated genes or surrounding genes in LD with the hits. Furthermore, the functions of these genes are not well described, highlighting the need for further research.
The best hit found in the age-restricted sample is more obviously functionally significant. ADCY2, which codes for adenylate cyclase 2, is a key regulator of cyclic AMP metabolism and thus of second messaging by activating PKA, thereby triggering CREB phosphorylation (24). Both the PKA and CREB pathways (25,26) have been implicated in BPD, and CREB1 is a well-documented candidate gene for BPD (27). Recently, a SNP in ADCY2 was also found to be genome-wide significantly associated with BPD (28). There is a vast literature on disturbances of serotonin and dopamine signaling in ADHD; both monoamines target G-protein coupled receptors (GPCRs) that use ADCY2 in their signal transduction cascades, so it is conceivable that this protein is also relevant to ADHD. As this association became significant (p=4.5×10−8) when restricting the BPD sample to an age of onset ≤21 years, the signal likely may be associated with a more neuro-developmental form of BPD. One hypothesis for future research is that adenylate cyclase signaling is a core mechanism mediating the comorbidity between ADHD and BPD.
Our primary meta-analysis yielded 10 suggestive loci (p<10−6). In addition to the aforementioned genes, these loci comprise the following candidate genes, each supported by at least three suggestive associations: On chromosome 1, we observed a suggestive association with the retinoid X receptor, gamma gene (RXRG). RXRG codes for retinoid receptor gamma - a regulator of dopamine signaling, cocaine response, and affective behaviors in mice (29,30). This makes it an attractive candidate for both BPD and ADHD. The gene has been suggested to have an effect on hippocampal volume by QTL analysis in mice (31) and has been associated with sensation seeking (32), a personality trait associated with combined-type ADHD (33). On chromosome 3, a signal for the neuroligin 1 (NLGN1) gene was also found. The neuroligin protein family has already been implicated in a wide range of neuropsychiatric disorders including autism, schizophrenia, and BPD (34).
Presence of eQTLs was evaluated in SNPs with association p-values <10−4 in either of the cross-disorder GWAS meta-analyses (Supplemental Table S5). Several interesting candidate genes for ADHD-BPD covariation were found to be regulated by multiple SNPs in our list serving as eQTLs. Of greatest interest are the findings implicating two members of the NT5DC family, a family of haloacid dehalogenase (HAD-type phosphatases (35). NT5DC1 has been associated with BPD in several studies, showing the strongest association with the disorder in NIMH Genetics Initiative bipolar pedigrees (36). Importantly, we recently observed an association between rare variants in this gene and adult ADHD (under review). NT5DC2 has previously been found associated with schizophrenia; furthermore, NT5DC2 is a target of miR-137 and is differentially methylated as a function of childhood maltreatment in BPD patients (37). Another interesting finding was for CACNB3, which codes for a voltage-gated calcium channel involved in neuronal morphology and differentiation; its transcript is targeted by miR-34a, which is upregulated in cerebellum of bipolar patients (38). Allelic variation in ABLIM1, which has previously been associated with novelty seeking, harm avoidance, reward and alcohol dependence (39), also appears to be a plausible finding.
Our finding that putative promoter regions containing the top-ranked associated SNPs do not contain known eQTLs was against our predictions of allele-specific effects on transcription. This discordance may, on the one hand, be attributed to the known high false positive rate of pattern searches with position-specific scoring matrices (18). On the other hand, the eQTL dataset was derived from analyses of lymphoblastoid cell lines (17), and it is therefore conceivable that the allele-specific transcription factor binding sites may serve as functional eQTLs in a different tissue context.
The findings described here need to be interpreted in the light of several caveats: The differential diagnosis especially between BPD and adult ADHD can be challenging. While this does not affect the ADHD sample, which was ascertained in childhood, it might pose a problem for the BPD sample: some shared signals might be due to patients with adult ADHD having falsely received a diagnosis of BPD. Given that all studies included here relied on DSM inclusion criteria, we are confident that misdiagnosis is at least not common enough to account for genome-wide significant hits. Ethnic heterogeneity might influence the results as well, although we aimed to control for this by including MDS components in every GWAS included in the meta-analyses. Finally, sample sizes in our cross-disorder analyses were still rather small for complex genetic traits. This was particularly damaging in the restricted analysis, where the reduction in sample size masked the potential increase in power due to the reduction in etiological heterogeneity (discussed above).
In conclusion, we provide evidence for the genetic overlap between ADHD and BPD. This genome-wide SNP-based genetic overlap (40) confirms the involvement of pathways such as G-protein-coupled signaling already known for their role in hyperactivity and/or emotional behaviors, and implicate a new candidate pathway (i.e. mRNA stability) in the pathophysiology of both ADHD and BPD.
Supplementary Material
Acknowledgments
Core funding for the PGC is provided by the NIMH (U01 MH094421). Work was also supported by grants to JRK from the NIMH (MH081804, MH078151, MH59567). The composition of PGC ADHD and BPD workgroups is detailed in the Supplementary Material (section 4).
This work was carried out on the Dutch national e-infrastructure with the support of SURF Foundation. The authors especially thank Willem Vermin (SURF Foundation, Amsterdam, The Netherlands) for his valuable support in the creating computer codes that fulfilled our needs for an efficient analysis of the data.
SVF is supported by the K.G. Jebsen Centre for Research on Neuropsychiatric Disorders, University of Bergen, Bergen, Norway, the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement n°602805 and NIMH grants R13MH059126 and R01MH094469. JRK is supported by NIH grants MH078151, MH081804, MH59567 and MH094483. AR is supported by the Deutsche Forschungsgemeinschaft (KFO 125, TRR 58/B06 and Z02 to AR, RE1632/5-1, RTG 1256 AR) and the European Community‘s Seventh Framework Programme (FP7/2007–2013) under grant agreement n° 602805 (“AGGRESSOTYPE”).
CJS and HW are supported by Interdisziplinäres Zentrum für Klinische Forschung (IZKF) grant Z-6.
KvH, BF, and AAV are supported by grants from the Netherlands Organization for Scientific Research (NWO), i.e. the NWO Brain & Cognition Excellence Program (grant 433-09-229) and a Vici grant to BF (grant 016-130-669), and by grants from the Netherlands Brain Foundation (grant 15F07[2]27) and BBMRI-NL (grant CP2010-33). The research leading to these results also received funding from the European Community’s Seventh Framework Programme (FP7/2007 – 2013) under grant agreements n° 602805 (AGGRESSOTYPE), n° 278948 (TACTICS), and n° 602450 (IMAGEMEND), and from the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreement n° 643051 (MiND) and n° 667302 (CoCA). In addition, their work is supported by the ECNP for the Research Network ‘ADHD across the Lifespan’.
Footnotes
Financial Disclosures
Kimm J.E. van Hulzen, Claus J. Scholz, Stephan Ripke, Marieke Klein, Andrew McQuillin, John R. Kelsoe, Klaus-Peter Lesch, Heike Weber, Alejandro Arias-Vasquez, Andreas Reif, Richard J.L. Anney, Alejandro Arias Vasquez, Maria J. Arranz, Philip Asherson, Tobias J. Banaschewski, Mònica Bayés, Joseph Biederman, Jan K. Buitelaar, Miguel Casas, Alice Charach, Bru Cormand, Jennifer Crosbie, Soeren Dalsgaard, Mark J. Daly, Alysa E. Doyle, Richard P. Ebstein, Josephine Elia, Christine Freitag, Michael Gill, Hakon Hakonarson, Amaia Hervas, Peter Holmans, Lindsey Kent, Jonna Kuntsi, Nanda Lambregts-Rommelse, Kate Langley, Sandra K. Loo, Joanna Martin, James J. McGough, Sarah E. Medland, Jobst Meyer, Eric Mick, Ana Miranda, Fernando Mulas, Benjamin M. Neale, Stan F. Nelson, Michael C. O'Donovan, Robert D. Oades, Michael J. Owen, Haukur Palmason, Josep A. Ramos-Quiroga, Marta Ribasés, Herbert Roeyers, Jasmin Romanos, Marcel Romanos, Aribert Rothenberger, Cristina Sánchez-Mora, Russell Schachar, Joseph Sergeant, Susan L. Smalley, Edmund J.S. Sonuga-Barke, Hans-Christoph Steinhausen, Anita Thapar, Alexandre Todorov, Susanne Walitza, Yufeng Wang, Andreas Warnke, Nigel Williams, Yanli Zhang-James, Devin Absher, Huda Akil, Adebayo Anjorin, Lena Backlund, Judith A. Badner, Jack D. Barchas, Thomas B. Barrett, Nick Bass, Michael Bauer, Frank Bellivier, Sarah E. Bergen, Wade Berrettini, Douglas Blackwood, Cinnamon S. Bloss, Michael Boehnke, Gerome Breen, René Breuer, William E. Bunney, Margit Burmeister, William Byerley, Sian Caesar, Kim Chambert, Sven Cichon, David A Collier, Aiden Corvin, William Coryell, Nick Craddock, David W. Craig, Mark Daly, Richard Day, Franziska Degenhardt, Srdjan Djurovic, Frank Dudbridge, Howard J. Edenberg, Amanda Elkin, Bruno Etain, Anne Farmer, Manuel Ferreira, I. Nicol Ferrier, Matthew Flickinger, Tatiana Foroud, Josef Frank, Christine Fraser, Louise Frisén, Janice Fullerton, Elliot S. Gershon, Katherine Gordon-Smith, Elaine K. Green, Tiffany A. Greenwood, Detelina Grozeva, Weihua Guan, Hugh Gurling, Ómar Gustafsson, Marian L. Hamshere, Martin Hautzinger, Chantal Henry, Stefan Herms, Maria Hipolito, Peter A. Holmans, Christina Hultman, Stéphane Jamain, Edward G. Jones, Ian R. Jones, Lisa Jones, Jean-Pierre Kahn, Radhika Kandaswamy, James L. Kennedy, George Kirov, Daniel L. Koller, Phoenix Kwan, Niklas Langstrom, Mark Lathrop, Jacob Lawrence, William B. Lawson, Marion Leboyer, Phil H. Lee, Jun Li, Paul Lichtenstein, Danyu Lin, Chunyu Liu, Falk W. Lohoff, Susanne Lucae, Pamela B. Mahon, Sandra Maier, Wolfgang Maier, Nick Martin, Manuel Mattheisen, Keith Matthews, Morten Mattingsdal, Kevin McGhee, Peter McGuffin, Melvin G. McInnis, Andrew McIntosh, Rebecca McKinney, Alan W. McLean, Francis J. McMahon, Ingrid Melle, Fan Guo Meng, Philip B. Mitchell, Grant W. Montgomery, Jennifer Moran, Gunnar Morken, Derek Morris, Valentina Moskvina, Pierandrea Muglia, Thomas W. Mühleisen, Walter J. Muir, Bertram Müller-Mysok, Richard M. Myers, Caroline M. Nievergelt, Ivan Nikolov, Vishwajit Nimgaonkar, Markus M. Nöthen, John I. Nurnberger, Jr., Evaristus A. Nwulia, Colm O'Dushlaine, Urban Osby, Högni Óskarsson, Roy Perlis, Hannes Petursson, Benjamin S. Pickard, James B. Potash, Peter Propping, Shaun Purcell, Emma Quinn, Soumya Raychaudhuri, John Rice, Marcella Rietschel, Douglas Ruderfer, Martin Schalling, Alan F. Schatzberg, William A. Scheftner, Peter R. Schofield, Nicholas J. Schork, Thomas G. Schulze, Johannes Schumacher, Markus Schwarz, Ed Scolnick, Laura J. Scott, Paul D. Shilling, Engilbert Sigurðsson, Pamela Sklar, Erin N. Smith, Jordan Smoller, David St. Clair, Hreinn Stefansson, Kari Stefansson, Michael Steffens, Stacy Steinberg, John Strauss, Jana Strohmaier, Patrick Sullivan, Szabolcs Szelinger, Robert C. Thompson, Þorgeir Þorgeirsson, Federica Tozzi, Jens Treutlein, John B. Vincent, Stanley J. Watson, Thomas F. Wienker, Richard Williamson, Stephanie H. Witt, Adam Wright, Wei Xu, Allan H Young, Peter P. Zandi, Peng Zhang and Sebastian Zöllner report no competing interests.
Barbara Franke received an educational speaking fee from Merz.
In the past year, Steve V. Faraone received income, travel expenses, potential income and/or research support from Pfizer, Ironshore, Shire, Akili Interactive Labs, CogCubed, Alcobra, VAYA Pharma, Neurovance, Impax, NeuroLifeSciences. With his institution, he has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. He receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health, Oxford University Press: Schizophrenia: The Facts and Elsevier, ADHD: Non-Pharmacologic Interventions.
Edmund J. Sonuga-Barke received fees for speaking, consultancy, research funding and conference support from Shire Pharma, speaker fees from Janssen Cilag, Medice, book royalties from OUP and Jessica Kingsley and consultancy from Neurotech solutions.
Mikael Landén declares that, over the past 36 months, he has received lecture honoraria from Biophausia Sweden, Servier Sweden, AstraZeneca, and served at advisory board for Lundbeck pharmaceuticals. No other equity ownership, profit-sharing agreements, royalties, or patent.
Ole Andreassen received speakers’ honoraria from Lundbeck and GSK.
References
- 1.Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, et al. (2005): Molecular genetics of attention-deficit/hyperactivity disorder. Biological psychiatry. 57:1313–1323. [DOI] [PubMed] [Google Scholar]
- 2.Shih RA, Belmonte PL, Zandi PP (2004): A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. International review of psychiatry. 16:260–283. [DOI] [PubMed] [Google Scholar]
- 3.Cross-Disorder Group of the Psychiatric Genomics Consortium (2013): Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klassen LJ, Katzman MA, Chokka P (2010): Adult ADHD and its comorbidities, with a focus on bipolar disorder. J Affect Disord. 124:1–8. [DOI] [PubMed] [Google Scholar]
- 5.Gross-Lesch S, Dempfle A, Reichert S, Jans T, Geissler J, Kittel-Schneider S, et al. (2013): Sex- and Subtype-Related Differences in the Comorbidity of Adult ADHDs. J Atten Disord. [DOI] [PubMed] [Google Scholar]
- 6.Faraone SV, Asherson P, Banaschewski T, Biederman J, Buitelaar JK, Ramos-Quiroga JA, et al. (2015): Attention-deficit/hyperactivity disorder. Nature Reviews Disease Primers. 15020. [DOI] [PubMed] [Google Scholar]
- 7.Wingo AP, Ghaemi SN (2007): A systematic review of rates and diagnostic validity of comorbid adult attention-deficit/hyperactivity disorder and bipolar disorder. J Clin Psychiatry. 68:1776–1784. [DOI] [PubMed] [Google Scholar]
- 8.Ryden E, Thase ME, Straht D, Aberg-Wistedt A, Bejerot S, Landen M (2009): A history of childhood attention-deficit hyperactivity disorder (ADHD) impacts clinical outcome in adult bipolar patients regardless of current ADHD. Acta Psychiatr Scand. 120:239–246. [DOI] [PubMed] [Google Scholar]
- 9.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. (2013): Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schimmelmann BG, Hinney A, Scherag A, Putter C, Pechlivanis S, Cichon S, et al. (2013): Bipolar disorder risk alleles in children with ADHD. J Neural Transm. [DOI] [PubMed] [Google Scholar]
- 11.Landaas ET, Johansson S, Halmoy A, Oedegaard KJ, Fasmer OB, Haavik J (2011): Bipolar disorder risk alleles in adult ADHD patients. Genes Brain Behav. 10:418–423. [DOI] [PubMed] [Google Scholar]
- 12.Manchia M, Adli M, Akula N, Ardau R, Aubry JM, Backlund L, et al. (2013): Assessment of Response to Lithium Maintenance Treatment in Bipolar Disorder: A Consortium on Lithium Genetics (ConLiGen) Report. PLoS One. 8:e65636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neale BM, Medland S, Ripke S, Anney RJ, Asherson P, Buitelaar J, et al. (2010): Case-control genome-wide association study of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 49:906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. (2011): Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 43:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. (2007): PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya A, Ziebarth JD, Cui Y (2014): PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 42:D86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. (2013): Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 501:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. (2005): MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 21:2933–2942. [DOI] [PubMed] [Google Scholar]
- 19.Faraone SV, Glatt SJ, Tsuang MT (2003): The genetics of pediatric-onset bipolar disorder. Biological psychiatry. 53:970–977. [DOI] [PubMed] [Google Scholar]
- 20.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV (2014): Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 515:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Scelo G, Purdue MP, Rothman N, Johansson M, Ye Y, et al. (2012): A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 21:456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, et al. (2013): Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 127:1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. (2009): Common variants at ten loci influence QT interval duration in the QTGEN Study. Nature genetics. 41:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delghandi MP, Johannessen M, Moens U (2005): The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal. 17:1343–1351. [DOI] [PubMed] [Google Scholar]
- 25.Nurnberger JI Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, et al. (2014): Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 71:657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerner B, Rao AR, Christensen B, Dandekar S, Yourshaw M, Nelson SF (2013): Rare Genomic Variants Link Bipolar Disorder with Anxiety Disorders to CREB-Regulated Intracellular Signaling Pathways. Front Psychiatry. 4:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Luo XJ, Rietschel M, Lewis CM, Mattheisen M, Muller-Myhsok B, et al. (2014): Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Molecular psychiatry. 19:452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. (2014): Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 5:3339. [DOI] [PubMed] [Google Scholar]
- 29.Krzyzosiak A, Szyszka-Niagolov M, Wietrzych M, Gobaille S, Muramatsu S, Krezel W (2010): Retinoid x receptor gamma control of affective behaviors involves dopaminergic signaling in mice. Neuron. 66:908–920. [DOI] [PubMed] [Google Scholar]
- 30.Krezel W, Ghyselinck N, Samad TA, Dupe V, Kastner P, Borrelli E, et al. (1998): Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. Science. 279:863–867. [DOI] [PubMed] [Google Scholar]
- 31.Lu L, Airey DC, Williams RW (2001): Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J Neurosci. 21:3503–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alliey-Rodriguez N, Zhang D, Badner JA, Lahey BB, Zhang X, Dinwiddie S, et al. (2011): Genome-wide association study of personality traits in bipolar patients. Psychiatr Genet. 21:190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob CP, Gross-Lesch S, Reichert S, Geissler J, Jans T, Kittel-Schneider S, et al. (2014): Sex-and Subtype-Related Differences of Personality Disorders (Axis II) and Personality Traits in Persistent ADHD. J Atten Disord. [DOI] [PubMed] [Google Scholar]
- 34.Carroll LS, Owen MJ (2009): Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 1:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seifried A, Schultz J, Gohla A (2013): Human HAD phosphatases: structure, mechanism, and roles in health and disease. The FEBS journal. 280:549–571. [DOI] [PubMed] [Google Scholar]
- 36.Bigdeli TB, Maher BS, Zhao Z, Sun J, Medeiros H, Akula N, et al. (2013): Association study of 83 candidate genes for bipolar disorder in chromosome 6q selected using an evidence-based prioritization algorithm. Am J Med Genet B Neuropsychiatr Genet. 162B:898–906. [DOI] [PubMed] [Google Scholar]
- 37.Prados J, Stenz L, Courtet P, Prada P, Nicastro R, Adouan W, et al. (2015): Borderline personality disorder and childhood maltreatment: a genome-wide methylation analysis. Genes Brain Behav. 14:177–188. [DOI] [PubMed] [Google Scholar]
- 38.Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, et al. (2015): Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Molecular psychiatry. 20:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang KS, Liu X, Aragam N, Mullersman JE, Jian X, Pan Y, et al. (2012): Polymorphisms in ABLIM1 are associated with personality traits and alcohol dependence. Journal of molecular neuroscience : MN. 46:265–271. [DOI] [PubMed] [Google Scholar]
- 40.Larsson H, Ryden E, Boman M, Langstrom N, Lichtenstein P, Landen M (2013): Risk of bipolar disorder and schizophrenia in relatives of people with attention-deficit hyperactivity disorder. Br J Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics, C., et al. , (2015): LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.