Abstract
Interstrand DNA-DNA cross-links (ICLs) are generated by endogenous processes, drugs, and environmental toxins. Understanding the cellular pathways by which various ICLs are repaired is critical to understanding their biological effects. Recent studies showed that replication-dependent repair of an ICL derived from the reaction of an abasic (AP) site with an adenine residue (dA) on the opposing strand of duplex DNA proceeds via a novel mechanism in which the DNA glycosylase NEIL3 unhooks the ICL. Here we examined the ability of the glycosylase domain of murine NEIL3 (MmuNEIL3-GD) to unhook dA-AP ICLs. The enzyme selectively unhooks the dA-AP ICL located at the duplex/single-strand junction of splayed duplexes that model the strand-separated DNA at the leading edge of a replication fork. We show that the ability to unhook the dA-AP ICL is a specialized function of NEIL3 as this activity is not observed in other BER enzymes. Importantly, NEIL3 only unhooks the dA-AP ICL when the AP residue is located on what would be the leading template strand of a model replication fork. The same specificity for the leading template strand was observed with a 5,6-dihydrothymine monoadduct, demonstrating that this preference is a general feature of the glycosylase and independent of the type of DNA damage. Overall, the results show that the glycosylase domain of NEIL3, lacking the C-terminal NPL4 and GRF zinc finger motifs, is competent to unhook the dA-AP ICL in splayed substrates and independently enforces important substrate preferences on the repair process.
Keywords: DNA-DNA interstrand cross-link; ICL; replication-dependent DNA repair; unhooking; NEIL3, DNA glycosylase; abasic site, AP-derived cross-link
Graphical Abstract
1. Introduction
The generation of interstrand covalent cross-links (ICLs) in cellular DNA has profound biological consequences because these lesions block the strand separation that is required for most DNA transactions.1, 2 ICLs are generated by endogenous processes, clinically-used anticancer drugs, and environmental toxins.3-8 Failure to repair ICLs can lead to cell death, genomic instability, and tissue dysfunction.2, 6, 9-13 As a result, genetic defects in ICL repair factors are associated with cancer, neurodegeneration, and accelerated aging.2, 6, 9-12, 14
Mechanisms of cellular ICL repair are complex and not well understood. Nonetheless, it is clear that a critical early step in the classical model for ICL repair involves “unhooking” of the cross-link via dual incisions generated by endonucleases such as XPF-ERCC1 (Scheme 1A).12, 15-18 The resulting double-strand break is repaired by homologous recombination,19 while the gapped duplex on the other side of the cross-link is reconstituted by error-prone translesion synthesis (Scheme 1A).17, 20, 21
Scheme 1.
Mechanisms for interstrand cross-link unhooking and repair. Panel A: The classical model for ICL unhooking involves dual incisions that generate a double-strand break.The upper strand is repaired by homologous recombination (HR) and the lower strand reconstituted by translesion synthesis polymerases (bypass). Panel B: Unhooking the dA-AP ICL by the glycosylase NEIL3 evades double-strand break formation.
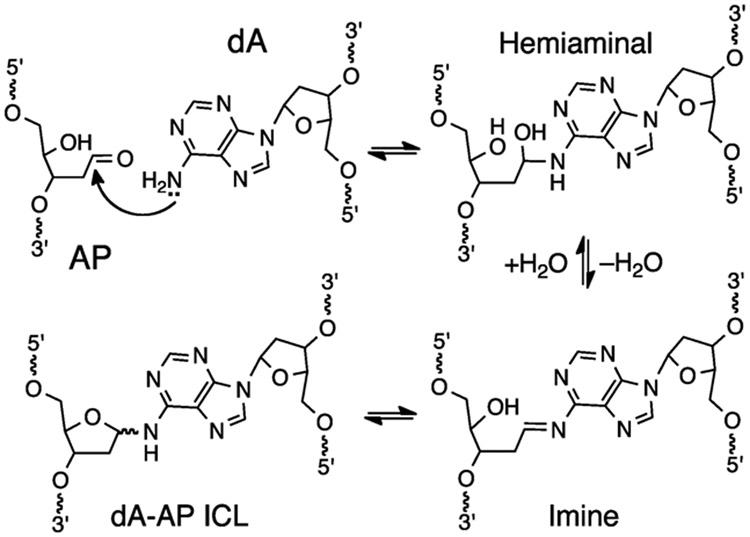
Recent studies of replication-dependent ICL repair in Xenopus egg extracts revealed an unprecedented cross-link unhooking mechanism that evades the double strand break generated by dual incisions (Scheme 1B).22 These studies examined the unhooking and bypass of a recently-characterized ICL derived from reaction of an abasic (AP) site in duplex DNA with an adenine residue (dA) on the opposing strand (Scheme 2).23-25 AP sites are common in cellular DNA.26, 27 Most AP sites are repaired by components of the base excision pathway,28-30 but ICLs derived from these lesions may contribute to the load of unavoidable endogenous cross-links6, 31 that drive the evolution and retention of cross-link repair pathways across most walks of life.32, 33 The dA-AP ICL is chemically stable for days in neutral aqueous solution24, 25 and further was shown to completely block DNA replication by the highly processive, strand-displacing ϕ29 polymerase.34
Scheme 2.
Formation of dA-AP ICL via the reaction of an AP site with an adenine residue on the opposing strand of duplex DNA.
The studies of Semlow et al. provided evidence that the base excision repair (BER) DNA glycosylase NEIL3 unhooks the dA-AP ICL and also psoralen-derived ICLs (Figure 1B).22, 35 This observation was surprising because DNA glycosylases are known for the removal of single damaged, mispaired, or misincorporated nucleobases from duplex DNA as the first step in the BER pathway.29, 36, 37 In humans, there are at least eleven BER glycosylases, each with distinct substrate preferences.28 Typically, the action of glycosylases proceeds via extrusion (“flipping out”) of the target base from the double-helix into the active site of the enzyme, followed by hydrolysis of the glycosidic bond to release the inappropriate base from the DNA backbone.36, 37 Such a base flipping mechanism is not compatible with ICL lesions.
Figure 1.
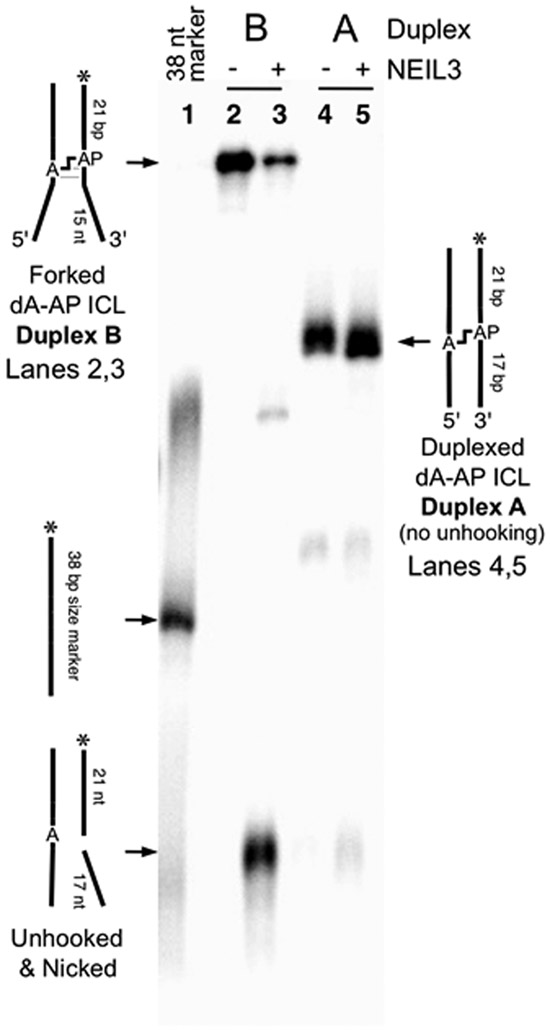
NEIL3-GD selectively unhooks the dA-AP cross-link in a splayed duplex. The 5’-32P-labeled cross-linked duplexes A and B were incubated with NEIL3 in HEPES (20 mM, pH 7.4) containing NaCl (100 mM), EDTA (1 mM), dithiothreitol (1 mM) and bovine serum albumin (100 μg/mL) at 37 °C for 2 h. Formamide loading buffer was added and the DNA in the samples resolved by electrophoresis on a denaturing 20% polyacrylamide gel. Following separation, the 32P-labeled oligonucleotides in the gel were visualized by phosphorimager analysis.
NEIL3 is one of the most recently discovered DNA glycosylases and its mechanism of action, substrate specificity, and cellular roles are not yet well defined.38, 39 There is evidence that the α-amino group of the N-terminal valine residue of NEIL3 is required for catalytic activity.38, 40, 41 Consistent with unusual roles for this enzyme in DNA repair, NEIL3 is expressed in S-phase, is associated with the replication fork and nascent DNA,38, 42, 43 and prefers to excise oxidized nucleobase lesions in non-canonical DNA structures such as bubbles, single-stranded, and telomeric structures.38, 39, 41, 44-47
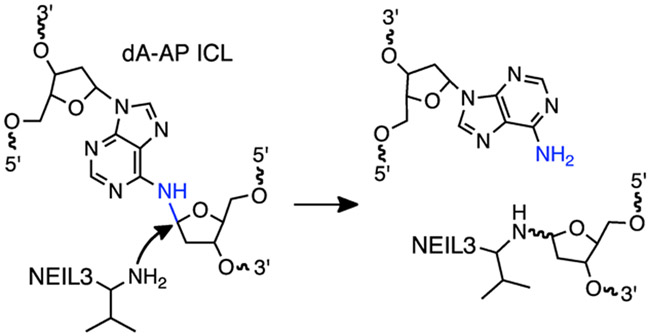
The results of cross-link repair in Xenopus egg extracts suggested a mechanism whereby NEIL3 hydrolyzed the non-native glycosidic bond in the dA-AP ICL to release the AP-containing strand and the complementary strand containing the native adenine residue (Scheme 3).22 Biochemical experiments further showed that full length Xenopus NEIL3 did not unhook a dA-AP ICL that was centrally located within a 24 base pair (bp) DNA-DNA duplex and showed modest unhooking activity on the ICL located in DNA-RNA heteroduplexes.22 On the other hand, the enzyme completely unhooked a small, trinucleotide ICL remnant attached to a 24 nucleotide (nt) DNA oligomer via the dA-AP linkage.22 Given the significance of ICL repair in biology and medicine it is important to understand the biochemical and structural mechanisms of this novel glycosylase-dependent unhooking process. In the work described here, we characterized the ability of NEIL3 to selectively unhook the dA-AP ICL located at the duplex/single strand junction of splayed structures that model the strand-separated DNA at the leading edge of a replication fork.16, 48-51
Scheme 3.
NEIL3 glycosylase unhooks the dA-AP ICL by cleaving the non-native glycosidic bond in the cross-linkage between N6 of the adenine residue and C1 of the AP site. Val2 of NEIL3 acts as a nucleophile in its base excision repair reactions.41
2. Materials and methods
2.1. Materials and general procedures.
Reagents were purchased from the following suppliers and were of the highest purity available. Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA) or, in the case of DHT-containing oligonucleotides, from Midland Certified Reagent Co. (Midland, TX). Uracil DNA glycosylase (UDG), and T4 DNA polynucleotide kinase (T4 PNK), formamidopyrimidine DNA glycosylase (Fpg) and Endonuclease III (Nth) were from New England Biolabs (Ipswich, MA). [γ-32P]-ATP (6000 Ci/mmol) was purchased from PerkinElmer. C-18 Sep-Pak cartridges were purchased from Waters (Milford, MA), and BS Poly-prep columns were obtained from BioRad (Hercules, CA). Acrylamide/bis-acrylamide 19:1 (40% Solution/Electrophoresis) was purchased from Fisher Scientific (Waltham, MA), and other reagents were purchased from Sigma-Aldrich (St. Louis, MO). Quantification of radioactivity in polyacrylamide gels was carried out using a Personal Molecular Imager (BIORAD) with Quantity One software (v.4.6.5).
2.2. MmuNEIL3-GD purification.
MmuNEIL3-GD (residues 1-282) was expressed in E. coli and purified by nickel affinity and heparin-sepharose chromatography as previously described 40, with the following modifications. The cell lysis buffer was supplemented with 1 μM leupeptin and 1 μM pepstatin A and did not contain detergent. Protein was stored at −80°C in 40 mM HEPES-NaOH pH 7.0, 150 mM NaCl, 20% glycerol, and 1 mM DTT. Catalytically inactive E3Q and K82A mutants were expressed and purified the same as wild-type.
2.3. Preparation of cross-linked DNA substrates.
A single-stranded, uracil-containing 2’-deoxy-oligonucleotide was 5’-32P-labeled using standard procedures,52 annealed to its complementary strand, and the resulting duplex treated with the enzyme UDG (50 units/mL, final concentration) to generate an AP-containing duplex. The UDG enzyme was removed by phenol-chloroform extraction and the DNA ethanol precipitated. For ICL generation, AP-containing duplexes were incubated in 50 mM HEPES pH 7 and 100 NaCl at 37 °C for 120 h. The DNA was ethanol precipitated, resuspended in formamide loading buffer and the slowly migrating cross-linked material was separated from uncross-linked material by electrophoresis for 10 h at 200 V on a 2 mm thick, 20% denaturing polyacrylamide gel. The splayed ICLs used in these studies were generated in approximately 30-50% yields. The DNA bands in the gel were visualized by autoradiography and the slow-moving band corresponding to cross-linked duplex was excised from the gel. The gel slice was crushed and agitated in an elution buffer composed of aqueous 200 mM NaCl and 1 mM EDTA pH 8 at 24 °C for 1 h. The gel fragments were removed by filtering through a Poly-Prep column and the filtrate was ethanol precipitated. The resulting residue was briefly dried in a Speed-Vac concentrator and stored at −20 °C. ICLs labeled with 5(6)-carboxyfluorescein (FAM) were prepared in a similar manner using a FAM-labeled dU-containing strand prepared by treatment of 5’ or 3’-amino modified oligonucleotides (/5AmMC6/ or /3AmMO/ from IDT) with 5(6)-carboxyfluorescein N-hydroxysuccinimidyl ester,53 followed by gel purification. Sequences of all DNA substrates used in these studies are shown in Figure S1.
2.4. Glycosylase activity assays.
Unhooking reactions typically contained 10,000-30,000 cpm (~1-10 nM) of labeled dA-AP DNA substrate and 25 nM of NEIL3-GD in 10 μL of a solution containing 20 mM HEPES-NaOH pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, and 100 μg/mL BSA. Following incubation at 37 °C for the specified time, formamide loading buffer was added directly to the assay and the DNA in the samples resolved by electrophoresis for 4 h at 1600 V on a denaturing 20% polyacrylamide gel. The 32P-labeled oligonucleotides in the gel were visualized by phosphorimager analysis.
To assay DHT excision from splayed substrates, a 25-mer oligodeoxynucleotide containing a central DHT and a 5’-FAM label was annealed to a 2-fold molar excess of the appropriate complementary strand to generate splayed substrates. NEIL3-GD (10 μM) was incubated with 100 nM FAM-DNA in 20 mM HEPES-NaOH pH 7.0, 100 mM NaCl, 10 μg/mL BSA, 1 mM DTT, and 5% glycerol at 25 °C. At various timepoints, 12 μL aliquots were removed, spiked with 0.1 N NaOH (final concentration) and incubated at 70 °C for 5 minutes. An equal volume of formamide loading buffer [80% (w/v) formamide, 10 mM EDTA] was added and samples were stored at −20 °C. Samples were electrophoresed on 20% polyacrylamide/8 M urea gels in 0.5X TBE buffer (45 mM Tris-base, 45 mM boric acid, 1 mM EDTA pH 8.0). Band fluorescence intensities were quantified on a Typhoon Trio Variable Mode Imager (GE Healthcare).
In the unhooking assays shown in Figure 3, the conditions were as follows: 25 nM NEIL3 in 20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 100 μg/mL BSA; 25 nM hNEIL1 in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA and 100 μg/mL BSA; 10 units Fpg in 10 mM Bis-Tris-propane-HCl pH 7, 10 mM MgCl2, 1 mM DTT, 100 μg/mL BSA; 10 units Endo III in 20 mM Tris-HCl pH 8, 1 mM EDTA, 1 mM DTT; and 10 units APE in 50 mM potassium acetate, 20 mM Tris-acetate pH 7.9, 10 mM magnesium acetate, 1 mM DTT. Control reactions showed that all enzymes displayed robust activity on an (uncross-linked) AP-containing substrate under the assay conditions. The reactions were incubated for 24 h at 37 °C. Following incubation at 37 C, formamide loading buffer was added, and DNA fragments in the samples resolved by electrophoresis for 4 h at 1600 V on a denaturing 20% polyacrylamide gel.
Figure 3.
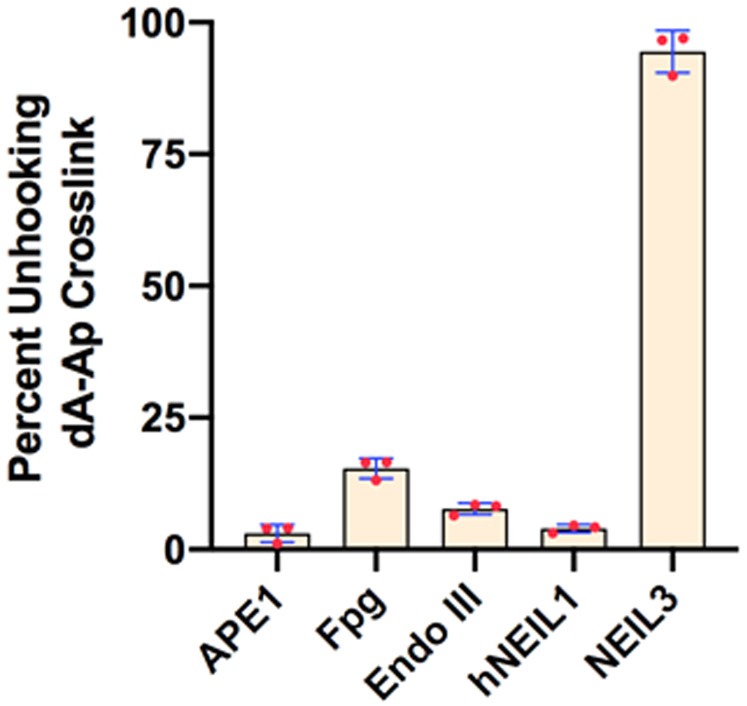
Base excision repair enzymes other than NEIL3-GD do not effectively unhook the dA-AP ICL in the splayed duplex B. The splayed duplex was incubated with various enzymes in their preferred buffers at 37 °C for 24 h. The bar graph depicts average ± S.D. (n=3) for gel electrophoretic analyses of the reactions.
2.5. DNA binding assays.
Fluorescence anisotropy was monitored as a catalytically inactive E3Q mutant of MmuNEIL3-GD was added to FAM-labeled fork structures (Figure S1). Reactions (25 μL) were incubated for 30 min at 4 °C and contained 25 nM DNA, 25 mM HEPES-NaOH pH 7.0, 50 mM NaCl, 3% glycerol, and 1 mM DTT. Measurements were recorded in 384-well plates (Corning 4514) using a BioTek Synergy H1 plate reader with 485 nm excitation and 528 nm emission filters. Equilibrium dissociation constants (Kd) were derived by fitting a one-site binding model to the data. We did not see any quenching of total fluorescence intensity that would indicate an interaction between the protein and the FAM label.
3. Results
3.1. NEIL3 glycosylase preferentially unhooks the dA-AP ICL in a DNA fork structure
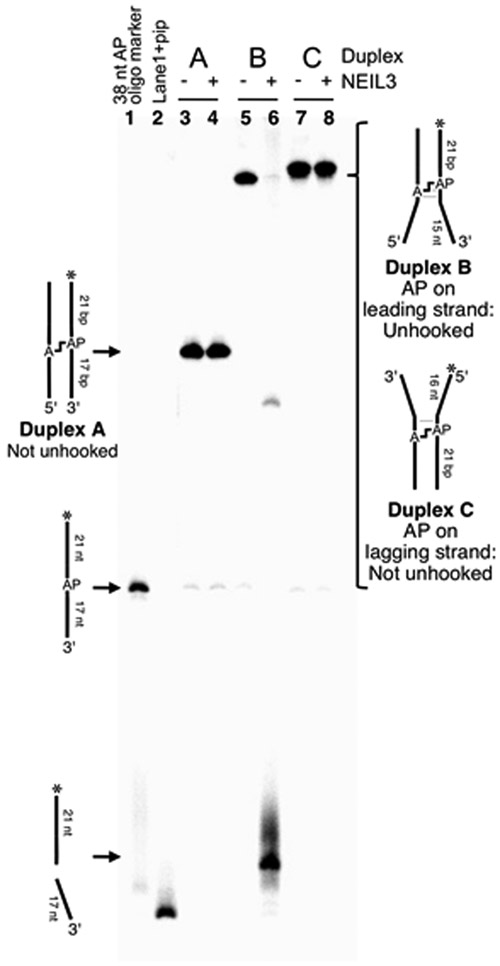
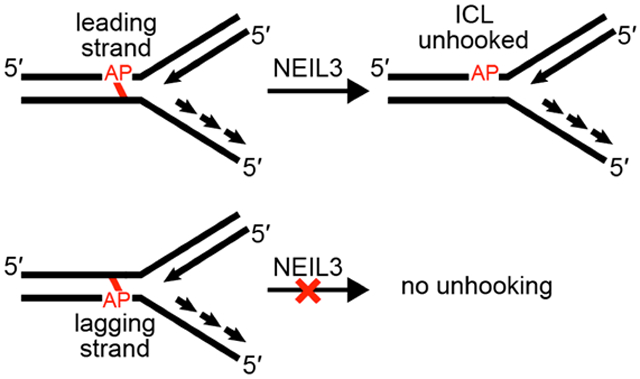
We set out to examine the ability of the glycosylase domain of murine NEIL3 (MmuNEIL3-GD)41 to unhook dA-AP cross-links in splayed DNA substrates that mimic the type of structures present at a stalled replication fork. Using previously described methods, we prepared and isolated a series of fully base-paired and splayed duplexes containing the dA-AP ICL.23, 24, 34, 54 We found that NEIL3-GD did not unhook a dA-AP ICL embedded in the center of a 35 bp duplex A (Figure 1, lanes 4 and 5). In contrast, NEIL3-GD readily unhooked the dA-AP cross-link located at the duplex/single-strand junction of the splayed duplex B, composed of a 21 bp duplex and two 15 nt unpaired arms (Figure 1, lanes 2 and 3). In experiments where the 5’-32P-label was located on the AP-containing strand of duplex B (Figure 1), we observed that unhooking was accompanied by complete cleavage at the AP site, with no full length AP strand observed during the reaction time-course (Figure S2). A complementary experiment with the 5’-32P-label placed on the opposite strand of duplex B showed that the unhooking action of NEIL3-GD released the full-length dA-containing strand (Figure S3). Taken together, these results are consistent with the proposed22 unhooking mechanism involving hydrolysis of the non-native glycosidic bond of the dA-AP ICL (blue bond in Scheme 3).
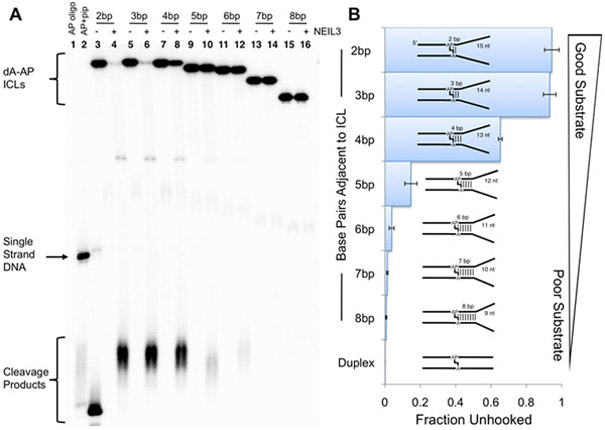
3.2. Five or more base pairs adjacent to the ICL inhibits unhooking by NEIL3-GD
To determine how far away the AP-ICL could reside from the fork and still be recognized by NEIL3-GD, we prepared a series of duplexes that systematically introduced base pairing into the splayed region adjacent to the cross-link. We found that a clamp of 5 bp significantly inhibits unhooking and clamps of ≥6 bp preclude unhooking of the dA-AP ICL by NEIL3-GD (Figure 2). Clearly, Watson-Crick base pairing adjacent to the cross-link disrupts the ability of NEIL3-GD to bind or process the dA-AP ICL.
Figure 2.
Base pairing adjacent to the dA-AP cross-link dramatically inhibits unhooking by NEIL3-GD. Substrates consist of 21 nt of paired duplex region on the 5’-side of the AP site and 17 nt, with varying numbers of base pairs, on the 3’-side of the AP as shown in the duplex cartoons. Sequences of substrates are shown in Figure S1. Panel A: the gel-purified, 5’-32P-labeled, cross-linked duplexes were incubated with NEIL3-GD in HEPES (20 mM, pH 7.4) containing NaCl (100 mM), EDTA (1 mM), dithiothreitol (1 mM) and bovine serum albumin (100 μg/mL) at 37 °C for 2 h. Formamide loading buffer was added and the DNA in the samples resolved by electrophoresis on a denaturing 20% polyacrylamide gel. Following separation, the 32P-labeled oligonucleotides in the gel were visualized by phosphorimager analysis. Panel B: Average ± S.D. for gel electrophoretic analyses of unhooking reactions.
3.3. Other base excision repair enzymes do not display activity against the dA-AP ICL
We next examined the ability of other base excision repair enzymes – NEIL1, Fpg, EndoIII glycosylases, and the apurinic endonuclease APE1 – to unhook the dA-AP ICL. Importantly, none of these enzymes was able to generate significant amounts of unhooked product from the splayed ICL-containing duplex B, even with extended (24 h) incubation times (Figures 3 and S4). This provides evidence that AP-lyase or AP-endonuclease activities alone are not sufficient for unhooking the dA-AP ICL and that NEIL3-GD has a specialized ability to recognize and process this cross-linkage in splayed duplexes.
3.4. NEIL3-GD selectively targets lesions on the leading template strand of a model replication fork
If we view the splayed substrates as models for the strand-separated DNA at the leading edge of a replication fork, the AP residue of the dA-AP ICL in duplex B (Figure 4) resides on the leading template strand. For comparison, we constructed a cross-linked duplex C in which the AP residue of the dA-AP ICL was placed on the lagging strand template (Figure 4). Interestingly, we found that NEIL3-GD was unable to unhook the dA-AP lagging strand ICL in duplex C (Figure 4).
Figure 4.
NEIL3-GD selectively unhooks the dA-AP ICL located at the duplex/single-strand junction of a splayed duplex in which the AP site resides on the leading strand template of a model replication fork.
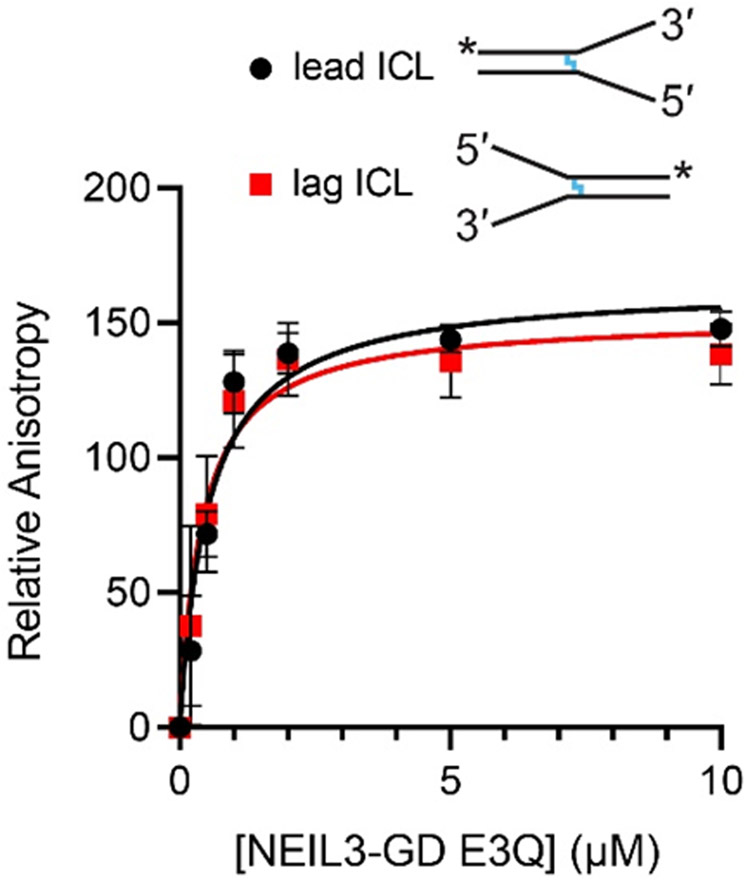
We used fluorescence anisotropy to examine the binding affinity of NEIL3-GD for the cross-linked duplexes containing the AP site on the leading (fork B) and lagging strand template (fork C). For these experiments, we employed analogs of the splayed duplexes B and C containing fluorescent labels in the duplex region and the catalytically-inactive E3Q mutant of NEIL3-GD. The enzyme binds to the leading and lagging ICL forks with the same affinity (leading, 0.5 ± 0.1 μM; lagging, 0.4 ± 0.2 μM) (Figure 5). The results suggest that the difference in catalytic activity of NEIL3-GD for leading and lagging substrates cannot be explained by a simple binding preference.
Figure 5.
Fluorescence anisotropy experiments provide evidence that NEIL3-GD has the same affinity for leading and lagging strand ICL substrates. Solutions containing FAM-labeled AP-ICL substrate were titrated with the catalytically-inactive E3Q mutant of NEIL3-GD. The plot shows average ± S.D. for three independent measurements. The calculated Kd for the binding of NEIL3-GD E3Q to the leading strand ICL is 0.5±0.1 μM and that for the lagging strand ICL is 0.4 ± 0.2 μM. In the fork schematics, the blue line indicates the position of the ICL and the asterisk denotes the position of the FAM label.
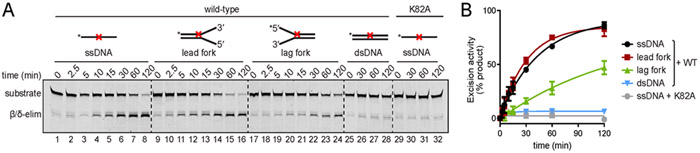
We further explored this aspect of NEIL3-GD substrate specificity by examining the rates at which the enzyme excised a 5,6-dihydrothymine (DHT) monoadduct located on either the leading or lagging strand templates of model forks. We found that the enzyme is more effective at removing the DHT lesion located on the leading strand template of a splayed substrate than from the lagging strand template (Figure 6). Removal of the DHT from the leading strand template was comparable to that from a single-stranded substrate. The enzyme is ineffective at the removal of DHT from the center of a double-stranded substrate (Figure 6). Overall, these results provide evidence that NEIL3-GD selectively targets the deoxyribose residue of lesions located on the leading strand template of a model replication fork.
Figure 6.
NEIL3-GD preferentially removes a dihydrothymine (DHT) monoadduct from the leading template strand of a splayed substrate. A. Representative polyacrylamide gel showing a time-course for NEIL3-GD activity against DHT (represented by red X)-containing structures. Lanes 1-28, wild type MmuNEIL3-GD; lanes 29-32, K82A inactive mutant control. B. Quantification of data from three independent experiments (average ± S.D.).
4. Discussion and conclusions
Recent studies provided evidence that NEIL3 is at the center of a previously unrecognized, front-line system for unhooking some ICLs at stalled replication forks.22 This pathway evades double-strand break formation and the need to engage potentially error prone55 homologous recombination. Our results show that the glycosylase domain of NEIL3 selectively unhooks the dA-AP ICL in splayed duplexes and that this is a specialized feature of NEIL3-GD, not shared by other BER enzymes examined here. The unhooking of the ICL by NEIL3-GD in our experiments, occurring with a half-time of approximately 2 h, (Figure S2) is slow compared to the processing of favored substrates such as spiroiminohydantoin in single stranded DNA which are reported to take place with a half-times of one minute or less.41, 56 In our experiments using the glycosylase domain of the mouse ortholog, unhooking of the dA-AP ICL is accompanied by nicking of the AP-containing strand. In contrast, NEIL3-mediated unhooking of the dA-AP cross-link in Xenopus egg extracts takes place without concomitant cleavage of the AP strand, thereby avoiding formation of a double-strand break.22 It is uncertain how the AP lyase activity of NEIL3 might be regulated at the replication fork. The difference in lyase activity may result from differences in the proteins used although, in general, a consistent picture has not emerged in the literature regarding NEIL3 lyase activity on simple, single-stranded substrates.41, 56
We find that NEIL3-GD can unhook the dA-AP ICL only when the AP residue of the cross-link is located on the leading strand template of a model replication fork (single-stranded DNA on the 3’-side of the AP site) but not when the AP residue is located on the lagging strand template (single-stranded DNA on the 5’-side of the AP site). We find that the enzyme displays approximately 40-fold higher affinity for the leading strand ICL over the lagging strand ICL. These results are in agreement with the unhooking of the dA-AP observed in Xenopus egg extracts, where stalling of the “rightward fork” was not resolved by NEIL3, but led to a small amount of CMG helicase unloading and FANC-dependent unhooking via the incision pathway.22 Crystallographic studies have shown that NEIL3-GD has a positively-charged channel that is incompatible with duplex DNA on one side of the lesion.57 The single-stranded regions of the splayed substrates may associate with this DNA-binding channel in a manner that aligns the dA-AP ICL with the catalytic machinery of the enzyme. Indeed, modeling the interaction of a forked ICL with NEIL3 based on the protein-DNA complexes of other Nei family members places the single-stranded DNA on the 3’-side of the AP site against the single strand-binding channel, thus positioning the enzyme for attack on the deoxyribose residue of the lesion on the leading strand template.
NEIL3 contains several zinc finger (ZF) motifs C-terminal to the glycosylase domain. Recent work showed that the NPL4-like (NZF, a.k.a. RanBP-ZF) in full-length NEIL3 is required for unhooking the dA-AP ICL in Xenopus egg extracts, presumably by recruiting NEIL3 to the stalled fork through interaction with ubiquitylated MCM subunits of the CMG helicase.58 The two GRF-ZFs at the extreme C-terminus bind DNA and may play an additional role in orienting NEIL3 at the fork.58 The N-terminal glycosylase domain employed in our work lacks these C-terminal domains but clearly possesses the capacity to unhook the dA-AP ICL. Our data further suggest that the glycosylase domain of NEIL3 independently enforces important substrate preferences on the unhooking process. Additional studies will be required to characterize the atypical glycosylase activity involved in ICL unhooking by NEIL3.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH grants R01ES021007 (K.S.G.) and R01GM131071 (B.F.E.). A.A.R. was funded by the Molecular Biophysics Training Program (T32GM008320) and an NSF Graduate Research Fellowship (DGE-1445197).
Footnotes
Conflict of interest
The authors declare no conflicts of interest
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https//doi.org/10.1016/j.dnarap.2019 DNA sequences used in this study, time-course for the unhooking of the dA-AP ICL in splayed duplex B, unhooking of the dA-AP ICL in a splayed duplex containing a 5’-32P-label on the dA strand of the cross-link, gel electrophoretic data showing that base pairing adjacent to the dA-AP ICL inhibits unhooking by NEIL3 glycosylase, and gel electrophoretic data for the unhooking of the dA-AP ICL by various base excision repair enzymes.
References
- 1.Brookes P; Lawley PD The reaction of mono and di-functional alkylating agents with nucleic acids. Biochem. J 1961, 80, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawley PD; Phillips DH DNA adducts from chemotherapeutic agents. Mutation Res. 1996, 355, 13–40. [DOI] [PubMed] [Google Scholar]
- 3.Rajski SR; Williams RM DNA cross-linking agents as antitumor drugs. Chem. Rev 1998, 98, 2723–2795. [DOI] [PubMed] [Google Scholar]
- 4.Schärer OD DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem 2005, 6, 27–32. [DOI] [PubMed] [Google Scholar]
- 5.Rycenga HB; Long DT The evolving role of DNA inter-strand crosslinks in chemotherapy. Curr. Opin. Pharmacol 2018, 41, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duxin JP; Walter JC What is the DNA repair defect underlying Fanconi anemia. Curr. Opin. Cell. Biol 2015, 37, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imani-Nejad M; Johnson KM; Price NE; Gates KS A new cross-link for an old cross-linking drug: the nitrogen mustard anticancer agent mechlorethamine generates cross-links derived from abasic sites in addition to the expected drug-bridged cross-links. Biochemistry 2016, 55, 7033–7041. [DOI] [PubMed] [Google Scholar]
- 8.Langevin F; Crossan GP; Rosado IV; Arends MJ; Patel KJ The Fanconi Anaemia DNA repair pathway counteracts the toxic effects of naturally produced aldehydes. Nature 2011, 375, 53–58. [DOI] [PubMed] [Google Scholar]
- 9.Niedernhofer LJ; Lalai AS; Hoeijmakers JHJ Fanconi anemia (cross)linked to DNA repair. Cell 2005, 123, 1191–1198. [DOI] [PubMed] [Google Scholar]
- 10.Osawa T; Davies D; Hartley JA Mechanism of cell death resulting from DNA interstrand cross-linking in mammalian cells. Cell Death Dis. 2011, 2, e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeij WP; Hoeijmakers JHJ; Pothof J Aging: not all DNA damage is equal. Curr. Opin. Gen. Devel 2014, 26, 124–130. [DOI] [PubMed] [Google Scholar]
- 12.Kottemann MC; Smogorzewska A Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 2013, 493, 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto S; Anai H; Hanada K Mechanisms of interstrand DNA crosslink repair and human disorders. Genes and Environment 2016, 38, DOI: 10.1186/s41021-016-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto M; Greenberg MM; Kow YW; Hwang J-T; Cunningham RP The 2-deoxyribonolactone lesion produced in DNA by neocarzinostatin and other damaging agents forms cross-links with the base-excision repair enzyme endonuclease III. J. Am. Chem. Soc 2001, 123, 3161–3162. [DOI] [PubMed] [Google Scholar]
- 15.Ceccaldi R; Sarangi P; D'Andrea AD The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol 2016, 17, 337–349. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J; Walter JC Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair 2014, 19, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clauson C; Schärer OD; Niedernhofer LJ Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspectives in Biology 2013, 5, a012732/1–a012732/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douwel DK; Boonen RACM; Long DT; Szypowska AA; Räschle M; Walter JC; Knipscheer P XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 2014, 54, 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long DT; Räschle M; Joukov V; Walter JC Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 2011, 333, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Räschle M; Knipscheer P; Enoiu M; Angelov T; Sun J; Griffith JD; Ellenberger TE; Schärer OD; Walter JC Mechanism of replication-coupled DNA interstrand cross-link repair. Cell 2008, 134, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TV; Schärer OD Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Env. Mol. Mutagenesis 2010, 51, 552–566. [DOI] [PubMed] [Google Scholar]
- 22.Semlow DR; Zhang J; Budzowska M; Drohat AC; Walter JC Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 2016, 167, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price NE; Johnson KM; Wang J; Fekry MI; Wang Y; Gates KS Interstrand DNA–DNA Cross-Link Formation Between Adenine Residues and Abasic Sites in Duplex DNA. J. Am. Chem. Soc 2014, 136, 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price NE; Catalano MJ; Liu S; Wang Y; Gates KS Chemical and structural characterization of interstrand cross-links formed between abasic sites and adenine residue in duplex DNA. Nucleic Acids Res. 2015, 43, 3434–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catalano MJ; Price NE; Gates KS Effective molarity in a nucleic acid controlled reaction. Bioorg. Med. Chem. Lett 2016, 26, 2627–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gates KS An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol 2009, 22, 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura J; Swenberg JA Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999, 59, 2522–2526. [PubMed] [Google Scholar]
- 28.Krokan HE; Bjørås M Base Excision Repair. Cold Spring Harb. Perspect. Biol 2013, 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson III DM; Bohr VA The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair 2007, 6, 544–559. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava DK; Vande Berg BJ; Prasad R; Molina JT; Beard WA; Tomkinson AE; Wilson SH Mammalian abasic site base excision repair. Identification of the reaction sequence and rate determining steps. J. Biol. Chem. 1998, 273, 21203–21209. [DOI] [PubMed] [Google Scholar]
- 31.Chen H-JC; Liu C-T; Li Y-J Correlation between glyoxal-induced DNA cross-links and hemoglobin modifications in human blood measured by mass spectrometry. Chem. Res. Toxicol 2019, 32, 179–189. [DOI] [PubMed] [Google Scholar]
- 32.Wilson III DM; Rieckher M; Williams AB; Schumacher B Systematic analysis of DNA crosslink repair pathways during development and aging in Caenorhabditis elegans. Nucleic Acids Res. 2017, 45, 9467–9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McVey M Strategies for DNA interstrand cross-link repair: Insights from worms, flies, frogs, and slime molds. Env. Mol. Mutagenesis 2010, 51, 646–658. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z; Johnson KM; Price NE; Gates KS Characterization of interstrand DNA-DNA cross-links derived from abasic sites using bacteriophage φ29 DNA polymerase. Biochemistry 2015, 54, 4259–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z; Nejad MI; Gamboa Varela J; Price NE; Wang Y; Gates KS A role for the base excision repair enzyme NEIL3 in replication-dependent repair of interstrand cross-links derived from psoralen and abasic sites. DNA Repair 2017, 52, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks SC; Adhikary S; Rubinson EH; Eichman BF Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta 2013, 1834, 247–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stivers JT; Jiang YJ A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev 2003, 103, 2729–2759. [DOI] [PubMed] [Google Scholar]
- 38.Liu M; Doublié S; Wallace SS Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutation Res. 2013, 743-744, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming AM; Burrows CJ Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Rad. Biol. Med 2017, 107, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M; Bandaru V; Holmes A; Averill AM; Cannan W; Wallace SS Expression and purification of active mouse and human NEIL3 proteins. Protein Expr. Purif 2012, 84, 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M; Bandaru V; Bond JP; Jaruga P; Zhao XS; Christov PP; Burrows CJ; Rizzo CJ; Dizdaroglu M; Wallace SS The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Nat. Acad. Sci. USA 2010, 107, 4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjoras KO; Sousa MML; Sharma A; Fonseca DM; Sogaard CK; Bjoras M; Otterlei M Monitoring the spatial and temporal dynamics of BER/SSBR pathway proteins, including MYH, UNG2, MPG, NTH1, and NEIL1-3, during DNA replication. Nucleic Acids Res. 2017, 45, 8291–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larson NB; Gao AO; Sparks JL; Gallina I; Wu RA; Mann M; Räschle M; Walter JC; Duxin JP Replication-coupled DNA-protein crosslink repair by SPRTN and the proteasome in Xenopus egg extracts. Mol. Cell 2019, 73, 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J; Fleming AM; Averill AM; Burrows CJ; Wallace SS The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015, 43, 4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin PR; Couvé S; Zutterling C; Albelazi MS; Groisman R; Matkarimov BT; Parsons JL; Elder RH; Saparvaev MK The human DNA glycosylases NEIL1 and NEIL3 excise psoralen-induced cross-links in a four-stranded DNA structure. Sci. Rep 2017, 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albelazi MS; Martin PR; Mohammed S; Mutti L; Parsons JL; Elder RH The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes 2019, 10 doi: 10.3390/genes10040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J; Chan J; Lambelé M; Stumpff J; Opresco PL; Thali M; Wallace SS NEIL3 repairs telomere damage during S phase to secure chromosome segregation at mitosis. Cell Rep. 2017, 20, 2044–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgescu R; Yuan Z; Bai L; de Luna Almeida Santos R; Sun J; Zhang DD; Yurieva O; Li H; O’Donnell ME Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Nat. Acad. Sci. USA 2017, doi: 10.1073/pnas.1620500114, E697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amunugama R; Willcox S; Wu RA; Abdullah UB; El-Sagheer AH; Brown T; McHugh PJ; Griffith JD; Walter JC Replication fork reversal during DNA interstrand cross-link repair requires CMG unloading. Cell 2018, 23, 3419–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J; Shi Y; Georgescu RE; Yuan Z; Chait BT; Li H; O’Donnell ME The architecture of a eucarytoic replisome. Nat. Struct. Mol. Biol 2015, 22, 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langston L; O’Donnell M Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. eLife 2017, 6, e23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J; Fritsch EF; Maniatis T Molecular Cloning: A Lab Manual. Cold Spring Harbor Press: Cold Spring Harbor, NY, 1989. [Google Scholar]
- 53.Giusti WG; Adriano T Synthesis and characterization of 5’-fluorescent-dye-labeled oligonucleotides. Genome Res. 1993, 2, 223–227. [DOI] [PubMed] [Google Scholar]
- 54.Gamboa Varela J; Gates KS Simple, high-yield syntheses of DNA duplexes containing interstrand DNA-DNA cross-links between an N4-aminocytidine residue and an abasic site. Curr. Protoc. Nucleic Acid Chem 2016, 65, 5.16.1–5.16.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonnalagadda VS; Matsuguchi T; Engelward BP Interstrand crosslink-induced homologous recombination carries an increased risk of deletions and insertions. DNA Repair 2005, 4, 594–605. [DOI] [PubMed] [Google Scholar]
- 56.Krokeide SZ; Laerhahl JK; Salah M; Luna L; Cederkvist FH; Fleming AM; Burrows CJ; Dalhus B; Bjoras M Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroiminohydantoin and guanidinohydantoin. DNA Repair 2013, 12, 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu M; Imamura K; Averill AM; Wallace SS; Doublié S Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure 2013, 21, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu RA; Semlow DR; Kamimae-Lanning AN; Kochenova OV; Chistol G; Hodskinson MRG; Amunugama R; Sparks JL; Wang M; Deng L; Mimoso CA; Low E; Patel KJ; Walter JC TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 2019, 567–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.