ABSTRACT
Recycling of wood ash from energy production may counteract soil acidification and return essential nutrients to soils. However, wood ash amendment affects soil physicochemical parameters that control composition and functional expression of the soil microbial community. Here, we applied total RNA sequencing to simultaneously assess the impact of wood ash amendment on the active soil microbial communities and the expression of functional genes from all microbial taxa. Wood ash significantly affected the taxonomic (rRNA) as well as functional (mRNA) profiles of both agricultural and forest soil. Increase in pH, electrical conductivity, dissolved organic carbon and phosphate were the most important physicochemical drivers for the observed changes. Wood ash amendment increased the relative abundance of the copiotrophic groups Chitinonophagaceae (Bacteroidetes) and Rhizobiales (Alphaproteobacteria) and resulted in higher expression of genes involved in metabolism and cell growth. Finally, total RNA sequencing allowed us to show that some groups of bacterial feeding protozoa increased concomitantly to the enhanced bacterial growth, which shows their pivotal role in the regulation of bacterial abundance in soil.
Keywords: metatranscriptomics, total RNA, wood ash, biodiversity, soil biota, protozoa
We applied total RNA sequencing to simultaneously assess the impact of wood ash amendment on the active soil microbial communities and the expression of functional genes from all microbial taxa.
INTRODUCTION
Wood ash from energy production is often considered a waste product (Vance 1996; Demeyer, Voundi Nkana and Verloo 2001) despite that recycling of wood ash may have beneficial effects as it counteracts acidification and returns essential nutrients to soil (Demeyer, Voundi Nkana and Verloo 2001; Augusto, Bakker and Meredieu 2008). Wood combustion is becoming more popular in several countries and increased reuse of wood ash as soil amendment holds the potential to improve the sustainability of this practice (Karltun et al. 2008; Huotari et al. 2015). However, wood ash application affects several soil physicochemical parameters important to the structure and function of microbial communities, e.g. pH, electrical conductivity and dissolved organic carbon (DOC; Ohno and Susan Erich 1990; Demeyer, Voundi Nkana and Verloo 2001; Pitman 2006; Augusto, Bakker and Meredieu 2008; Hansen et al. 2017; Maresca, Hyks and Astrup 2017). As the soil microbiota carries out an array of key biochemical processes (Blagodatskaya and Kuzyakov 2013), knowledge of its response to disturbance is important, not least in production soils due to potential impact on soil fertility.
The soil microbiome, which includes prokaryotes as well as microeukaryotes, is one of the most diverse and complex biomes on Earth. It has a pivotal role in nutrient cycling and carbon sequestration and is a key component in the maintenance of soil fertility of managed ecosystems (Wall et al. 2012; Fierer 2017). Wood ash amendment causes changes in soil microbiome composition, activity and quantity (Perkiömäki and Fritze 2002; Aronsson and Ekelund 2004; Huotari et al. 2015). Ash amendment induces changes in community structure followed by increased microbial activity and growth, which is usually explained by the increased soil pH brought about by the alkaline oxides in the ash (Cruz-Paredes et al. 2017; Vestergård et al. 2018). Still, some studies show no or only minor microbial response to wood ash application (Aronsson and Ekelund 2004; Huotari et al. 2015).
Only few studies have concomitantly analyzed microorganisms from all domains of life (i.e. Archaea, Bacteria and Eukaryotes) and most of these rely on cultivation, model organisms or molecular fingerprinting, which provide only limited resolution of taxonomical and functional responses. Total RNA sequencing, or metatranscriptomics, makes it possible to investigate active soil microbial communities from all domains of life, including their transcriptional activity, simultaneously. By targeting RNA—and not DNA—most of the biases associated with relic DNA are avoided. Relic DNA can, because of its relative slow degradation in soil, result in biases with delayed functional and community responses (Carini et al. 2016). Total RNA sequencing allows for the study of immediate regulatory responses to environmental changes (Carvalhais et al. 2012), and it has proven useful in the assessment of active microbial communities’ functional roles in soil (Urich et al. 2008; Epelde et al. 2015; Geisen et al. 2015; Hultman et al. 2015; Schostag et al. 2019).
We therefore aimed to investigate how the active soil prokaryotic and microeukaryotic communities in agricultural and forest soil responded structurally and functionally (transcriptional) to wood ash application. Both soil types are relevant for large-scale application of wood ash. We applied wood ash in concentrations corresponding to field application of 0, 3, 12 and 90 t ha−1, where 3 t ha−1 is the currently allowed dose in Scandinavian countries. We expected wood ash to increase soil pH, electrical conductivity and DOC and therefore hypothesized that (i) the pH increase would favor bacteria more than fungi, (ii) the nutrients in the wood ash would benefit the copiotrophic microbial groups, (iii) multitrophic responses would appear gradually over time after wood ash application and (iv) microbial stress responses would be observable in the transcriptome.
MATERIALS AND METHODS
Soils and wood ash
We used two contrasting soils for the experiment. The first was a loamy sand (Typic Hapludult) from the plough layer (0–10 cm) of an agricultural field (Research Center Foulum, DK; 56°29′42′N 9°33′36′E). The other was from the O-horizon (0–10 cm) of a forest (Gedhus, DK; 56°16′38′N 09°05′12′E). The forest is a second-generation Norway spruce stand [Picea abies (L.) Karst.] on Podzol heathland. Qin et al. (2017) provide soil characteristics for both soils. On both sites, we removed visible plant parts before taking ten 100 g soil samples within a 30 m2 area. The 10 samples from each site were sieved (4 mm), pooled and stored in the dark for 14 days at 4°C until further processing.
Wood ash was a mixture of bottom and fly ash from a heating plant (Brande, Denmark) produced by combustion of wood chips from predominantly coniferous trees. We homogenized the ash by sieving (2 mm). Maresca, Hyks and Astrup (2017) provide a list of mineral nutrients and heavy metals in the ash.
Microcosm setup and incubation
We prepared microcosms in triplicates of 50 g soil in 250 ml sterilized airtight glass jars. We mixed the ash thoroughly with soil-to-ash concentrations corresponding to field application of 0, 3, 12 and 90 t ash ha−1. The water content was adjusted to 50% of the water holding capacity of the two soils. We prepared 12 microcosms for each soil-ash combination to allow four destructive samplings, i.e. a total of 96 microcosms. Samples were also collected at the start of the experiment. Microcosms were incubated at 10°C in the dark and all microcosms were opened once a week inside an LAF bench to maintain aerobic conditions.
Physicochemical soil parameters
At destructive sampling, after 3, 10, 30 and 100 days of incubation, we prepared soil extracts from 15 g soil and 75 ml sterile ddH2O followed by 1 h shaking and settling for 0.5 h. In the supernatant, we measured electrical conductivity using a TetraCon 325 electrode adapted to a conductivity meter Cond 340i (WTW, Weilheim, Germany) and pH using a pH electrode (Sentix Mic, WTW, Weilheim, Germany) connected to pH meter Multi 9310 (WTW, Weilheim, Germany). The remaining supernatant was filtered (5C filters; Advantec, Tokyo, Japan; 1 μm pore size) and analyzed for DOC, nitrate (NO3−), ammonium (NH4+) and phosphate (PO43−). DOC concentrations were determined on a TOC-5000A (Shimadzu, Kyoto, Japan). Nitrate, ammonium and phosphate concentrations were determined by flow injection analysis (FIAstar 5000, FOSS, Hillerød, Denmark), following the manufacturer's instructions.
Nucleic acid extraction, qPCR and library preparation for sequencing
RNA and DNA were co-extracted from 2 g soil samples using the RNA PowerSoil Total RNA Isolation Kit (MOBIO, Carlsbad, CA, USA) in combination with DNA Elution Accessory Kit (MOBIO), following the manufacturer's protocol. The soil for nucleic acid extraction were immediately frozen in liquid nitrogen (N) after collection (to preserve RNA) and subsequently stored at −80°C until extraction. Agricultural soil amended with the highest ash concentration had an RNA yield below detection limit and was not sequenced.
We quantified 16S rRNA and ITS2 gene copies (DNA level) using qPCR. 16S rRNA genes were amplified in technical duplicates using a CFX Connect (Bio-Rad, Richmond, VA, USA). We used a dilution series of genomic DNA from Escherichia coli K-12 (with seven copies of 16S rRNA genes) as a standard (Blattner et al. 1997). The master mix consisted of 2 µl bovine serum albumin (BSA; 20 mg/ml; BIORON, Ludwigshafen, Germany), 10 µl SsoFast EvaGreen Supermix (Bio-Rad), 0.8 μl of primer 341f (5′-CCTAYGGGRBGCASCAG-3′), 0.8 μl of primer 806r (5′-GGACTACNNGGGTATCTAAT-3′; Hansen et al. 2012), 1 μl of 10× diluted template and 5.4 µl of sterile DEPC-treated water. PCR conditions for 16S rRNA gene amplification were 98°C for 15 min, followed by 35 cycles of 98°C for 30 s, 56°C for 30 s and 72°C for 30 s (with fluorescence measurements) and ending with 72°C for 7 min and production of melt curves. The PCR efficiencies for the 16S assays were 96.1 ± 1.0% (SEM, n = 3) with R2 = 0.99 ± 0.001. ITS gene copies were quantified as described for the 16S rRNA above with minor modifications: Vector cloned ITS2 DNA regions from Aureobasidium pullulans were included as standards, primers used were gITS7 (5′-GTGARTCATCGARTCTTTG-3′; Ihrmark et al. 2012) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′; White et al. 1990), annealing temperature was 60°C and 40 amplification cycles were used. The PCR efficiencies for the ITS assays were 106.0 ± 4.6% with R2 = 0.99 ± 0.003.
Prior to total RNA library building, we removed potential DNA carryovers using the DNase Max Kit (MOBIO), following the manufacturer's protocol. Successful DNA removal of RNA extracts was tested with the 16S qPCR protocol described above but with 50 amplification cycles: All DNase-treated RNA extracts had Cq values higher than or equal to those of the negative samples (sterile DEPC-treated water as template) and DNA was thereby not present.
Quality of the DNase-treated RNA was tested using RNA 6000 Nano Kit (Agilent, Santa Clara, CA, USA) on a 2100 Bioanalyzer System (Agilent), following the manufacturer's protocol [average RIN number was 7.85 ± 0.13 (SEM, n = 69)].
Subsequently, DNase-treated RNA extracts from time points 0, 3, 30 and 100 days were fragmented into ∼150 bp segments and prepared for sequencing using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina in combination with the NEBNext Multiplex Oligos for Illumina (New England BioLabs, Ipswich, MA, USA), according to the manufacturer's protocol. We sequenced the resulting metatranscriptome libraries using HiSeq 2500 (Illumina Inc., San Diego, CA, USA) in high-output mode (8 HiSeq lanes, 125 bp, paired-end reads) at the National High-throughput DNA Sequencing Centre (Copenhagen, Denmark).
Bioinformatic processing
We obtained a total of 3.3 billion paired sequences (SRA accession number: PRJNA512608) and processed them through the following bioinformatic pipeline (see Table S1, Supporting Information, for numbers of sequences and contigs during the processing steps). Adapters, poly-A tails, sequences shorter than 60 nt and nucleotides with Phred score below 20 at the 5′ and 3′ end of sequences were removed using Cutadapt v.1.9.1 (Martin 2011). Five samples were removed prior to subsequent processing due to low quality of reads (one replicate of 3 t ha−1, day 100 from the agricultural soil; two replicates of 0 t ha−1, day 0; and two replicates of 0 t ha−1, day 100 from the forest soil). Sequences were then sorted into small subunit (SSU) rRNA, large subunit (LSU) rRNA and non-rRNA sequences using SortMeRNA v.2.1 (Kopylova, Noé and Touzet 2012).
rRNA
A subset of 1.5 million randomly chosen SSU rRNA sequences per sample was assembled into longer SSU rRNA contigs using EMIRGE (Miller et al. 2011). The subset of sequences was done partly to normalize the number of sequences per sample (to deal with unequal sequencing depth, i.e. different numbers of sequences per sample after HiSeq sequencing), partly due to computational constraints. Contigs were taxonomically classified using CREST (Lanzén et al. 2012) and rRNA reads were mapped to resulting EMIRGE contigs using BWA (Li and Durbin 2009), as in Epelde et al. (2015), resulting in a table of taxonomically annotated read abundance across samples (Datasheet S1, Supporting Information).
mRNA
A combined pool of non-ribosomal sequences from all samples was assembled using trinity v.2.0.6 (Grabherr et al. 2011). From the resulting assembled contigs, non-coding RNA contigs were filtered away by aligning contigs to the Rfam database v.12.0 (Nawrocki et al. 2015) using cmsearch v.1.1.1 with a significant e-value threshold of <10−3. Input sequences used for non-ribosomal RNA assembly were then mapped to coding mRNA contigs. We normalized the contigs by removing those with relative expression lower than 1 out of the number of sequences in the dataset with least number of sequences. EMBOSS (Rice, Longden and Bleasby 2000) was used to search six possible open reading frames (ORFs) of the contigs. SWORD (Vaser, Pavlović and Šikić 2016) was used to align ORFs against the Md5nr protein database (Wilke et al. 2012). The output was then parsed with custom Python scripts and filtered hits with minimum e-value of 10−5 as threshold. Best hit for each contig was then selected based on alignment statistics and annotated against the eggnog hierarchical database v.4.5 (Jensen et al. 2008). The output was an abundance table of numbers of sequences assigned to groups of different functional genes (COGs; Datasheet S2, Supporting Information). The mRNA processing has been validated by Anwar et al. (2019).
Statistical analysis and data processing
Statistical validation for both taxonomy and functional abundance was done in R v.3.4.0 (R Core Team 2015) using vegan (Oksanen et al. 2008). The rRNA abundance was converted into relative abundance and collapsed taxonomically into Archaea, Bacteria and Eukaryota. We further grouped Eukaryota into Fungi, Metazoa, and protists (with main focus on bacterivorous protozoa). We calculated Richness (number of rRNA contigs) and Shannon diversity on the total number of rRNA contigs and abundance of sequence reads mapped to them. Non-metric multidimensional scaling (NMDS) was carried out using Bray–Curtis dissimilarities of community composition (rRNA contigs and abundance of sequence reads mapped to them) between samples. Soil physicochemical parameters were fitted to the resulting NMDS using the function envfit. Variables explaining overall differences in community composition were evaluated using the function Adonis, which performs permutational analysis of variance (PERMANOVA; 10 000 permutations) using Bray–Curtis dissimilarities as response variable. A forward selection strategy was carried out to only include explanatory variables with significant P-values in Adonis models.
Significant effects of wood ash amendment and incubation time on taxonomic groups were determined using non-parametric Kruskal–Wallis tests (due to the non-normal distribution of taxon abundances). To separate the pronounced changes in community responses observed at the 90 t ha−1 amendment in the forest soil from the less pronounced changes observed at 0–12 t ha−1, we performed Kruskal–Wallis tests with wood ash concentration as independent variable for both the ranges of 0–12 and 0–90 t ha−1. We also used Kruskal–Wallis to test the effect of time on differential abundances of taxa within the wood ash concentrations separately. P-values were adjusted for false discovery rate (FDR) using the Benjamini–Hochberg method in all tests.
NMDS on Bray–Curtis dissimilarities of functional gene compositions (mRNA) and Adonis testing were carried out as described above. Likewise were soil physicochemical parameters fitted to the resulting NMDS, as described above. mRNA gene counts between samples were normalized using the DESeq2 algorithm (Love, Huber and Anders 2014). Significantly differentially expressed genes (mRNA) were analyzed using the DESeq2 module of SARTools (Varet et al. 2016). These analyses were conducted by pairwise comparisons of gene transcription (mRNA) levels between samples of increasing wood ash concentration to control samples (0 t ha−1) at different incubation times. For the forest samples at time 100 days, only one replicate remained for the 0 t ha−1 treatment. Therefore, we compared instead the 12 and 90 t ha−1 to the 3 t ha−1.
We used linear Pearson regression to test for significant correlations between wood ash concentration and time against measured physicochemical parameters. Additionally, we performed two-way ANOVAs with Tukey's post-hoc tests using wood ash concentration and time as explanatory variables, with all physicochemical parameters as dependent variables. Variance homogeneity was tested using Levene's test and normal distribution of data was tested using the Shapiro–Wilk test in combination with QQ-plots prior to ANOVA tests.
We used a significance level of 0.05, unless otherwise explicitly mentioned, and the Results section provide descriptions at this significance level.
RESULTS
Physicochemical parameters
Soil pH, electrical conductivity and DOC correlated positively with wood ash concentration for both soils (Table 1). For the 90 t ha−1 ash amendment, soil pH increased from 6.4 to 11.5 in the agricultural and from 4.1 to 8.5 in the forest soil (Figure S1, Supporting Information). Similarly, the 90 t ha−1 resulted in 15- and 19-fold increases in electrical conductivity for the agricultural and forest soil, respectively. In the agricultural soil, ammonium increased with time in samples both with and without ash amendment, while nitrate showed no significant changes. In the forest soil, ammonium and nitrate increased after 3 days in the 90 t ha−1 amendment, followed by a decrease after 30 days. In the other treatments, increased concentrations were observed during the entire incubation period. In both soils, concentrations of dissolved phosphate increased up to 12 t ash ha−1 followed by a decrease at 90 t ha−1.
Table 1.
Pearson correlation values (r) and associated significance levels between ash dose (field equivalents 0, 3, 12 and 90 t ha−1) and incubation time, and soil physicochemical parameters.
| Agricultural soil | Forest soil | |||
|---|---|---|---|---|
| Explanatory variable | Ash dose (t ha−1) | Time (days) | Ash dose (t ha−1) | Time (days) |
| pH | 0.76*** | 0.15 | 0.98*** | 0.07 |
| Conductivity (µS cm−1) | 0.82*** | 0.14 | 0.99*** | 0.07 |
| DOC (mg g−1 DW soil) | 0.74*** | 0.33* | 0.91*** | 0.05 |
| Ammonium (µg g−1 DW soil) | 0.05 | 0.57*** | 0.40** | 0.36 |
| Nitrate (µg g−1 DW soil) | −0.45*** | 0.28* | 0.63*** | −0.15 |
| Phosphate (µg g−1 DW soil) | −0.61* | −0.07 | 0.26 | −0.04 |
*P< 0.05, **P< 0.01, ***P< 0.001.
Quantitative PCR
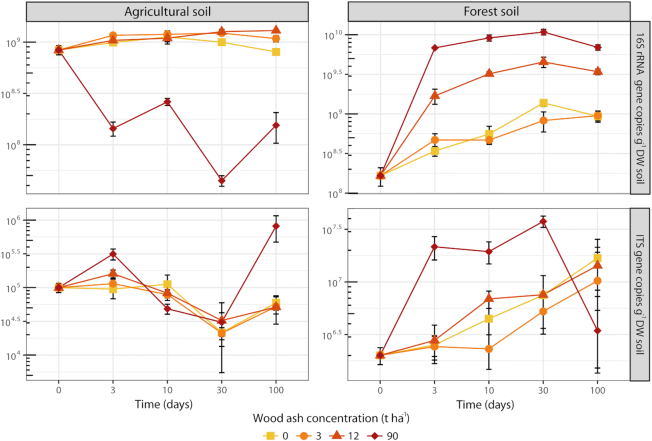
Prokaryotic abundance (number of 16S rRNA gene copies) increased in the agricultural soil after the wood ash application of 12 t ha−1, but decreased after application of 90 t ha−1 (Fig. 1). Fungal abundance (number of ITS copies) remained fairly unchanged over time regardless of ash application with the exception of an increase after 100 days at 90 t ha−1. In the forest soil, prokaryotic abundance increased over time for all treatments (Fig. 1); however, addition of 12 and 90 t ha−1 resulted in a stronger increase. The fungal abundance in the forest soil showed higher abundance for most of the period with wood ash concentrations of 90 t ha−1.
Figure 1.
Numbers of 16S rRNA gene copies (top row) and ITS gene copies (bottom row) g−1 DW of the agricultural soil (left panel) and the forest soil (right panel) across wood ash concentrations and incubation times. Symbols represent averages with SEM (n = 3). The presented data are results from qPCR on DNA. Note logarithmic y-axes and different ranges of values on y-axes.
rRNA—community composition
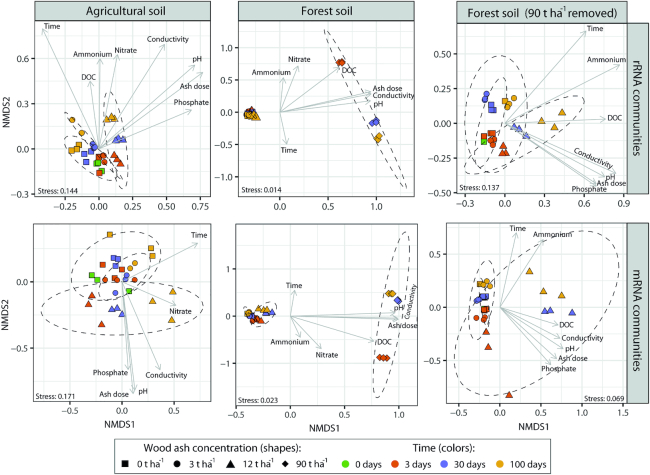
The number of unique rRNA contigs ranged from 1216 to 5931 per sample and originated from all domains of life. Community composition differed significantly (P < 0.001; R2 = 0.86; Adonis) between the two soil types. For forest soil, amendment with 90 t ha−1 resulted in highly altered community composition (Fig. 2) compared to 0–12 t ha−1. Though less pronounced, changes from 0–3 to 12 t ha−1 were also clearly visible for both soil types (Fig. 2). Moreover, microcosms for particular soil type/ash dose combinations were clearly separated by sampling times (Fig. 2).
Figure 2.
NMDS plots based on Bray–Curtis dissimilarities of the taxonomic (rRNA; top row) and functional (mRNA; bottom row) profiles of an agricultural soil and a forest soil amended with wood ash. Dashed lines represent 95% confidence ellipses around samples with same wood ash concentration. Arrows indicate the direction of fitted physicochemical parameters (using envfit function; only significant parameters shown) onto the NMDS ordination space (longer arrows indicate better fit). To improve the resolution of the forest soil at wood ash concentrations 0–12 t ha−1, we removed the 90 t ha−1 samples and repeated the analysis (rightmost two panels).
In both soils, wood ash dose, incubation time, pH and electrical conductivity correlated to the transformed NMDS community space (Fig. 2). Optimized Adonis models (Table 2) supported that wood ash concentration, time, pH and electrical conductivity together significantly explained the variation in microbial communities after ash application in both soils. Additionally, dissolved phosphate significantly explained the variation in microbial communities in both soils up to 12 t ha−1 ash amendments and DOC, ammonium and nitrate in the forest soil.
Table 2.
Explanatory strength of physicochemical variables on rRNA and mRNA dissimilarity profiles of the two soils after ash amendment testing using permutational multivariate analysis of variance (Adonis).
| rRNA | mRNA | |||||
|---|---|---|---|---|---|---|
| Explanatory variable | Agriculture (0–12 t ha−1) | Forest (0–90 t ha−1) | Forest (0–12 t ha−1) | Agriculture (0–12 t ha−1) | Forest (0–90 t ha−1) | Forest (0–12 t ha−1) |
| pH | 0.184*** | 0.536*** | 0.216*** | 0.079* | 0.386*** | 0.224*** |
| Conductivity (µS cm−1) | 0.081*** | 0.056*** | 0.108*** | 0.140* | 0.061*** | 0.100*** |
| Wood ash concentration (t ha−1) | 0.113*** | 0.044*** | 0.041* | 0.063* | 0.049*** | 0.051** |
| Time (days) | 0.089*** | 0.068*** | 0.173*** | 0.092* | 0.086*** | 0.258*** |
| Phosphate (µg g−1 DW soil) | 0.039* | NS | 0.076*** | 0.065* | NS | 0.118*** |
| DOC (mg g−1 DW soil) | NS | 0.094*** | 0.038* | NS | 0.162*** | 0.033** |
| Ammonium (µg g−1 DW soil) | NS | 0.034** | 0.029* | NS | 0.066*** | 0.040** |
| Nitrate (µg g−1 DW soil) | NS | 0.027** | 0.036* | NS | 0.038*** | 0.028* |
| Wood ash concentration:time | 0.064*** | 0.015* | 0.043* | NS | 0.025** | 0.039** |
| Residuals (unexplained variance) | 0.430 | 0.127 | 0.239 | 0.560 | 0.126 | 0.109 |
Values refer to R2 values of the Adonis test on Bray–Curtis dissimilarities between samples.
Asterisks refers to significance level (*0.01 < P< 0.05, **0.001 < P< 0.01, ***P < 0.001).
Non-significant (P> 0.05) parameters are written as 'NS'.
rRNA—taxonomic distribution and diversity
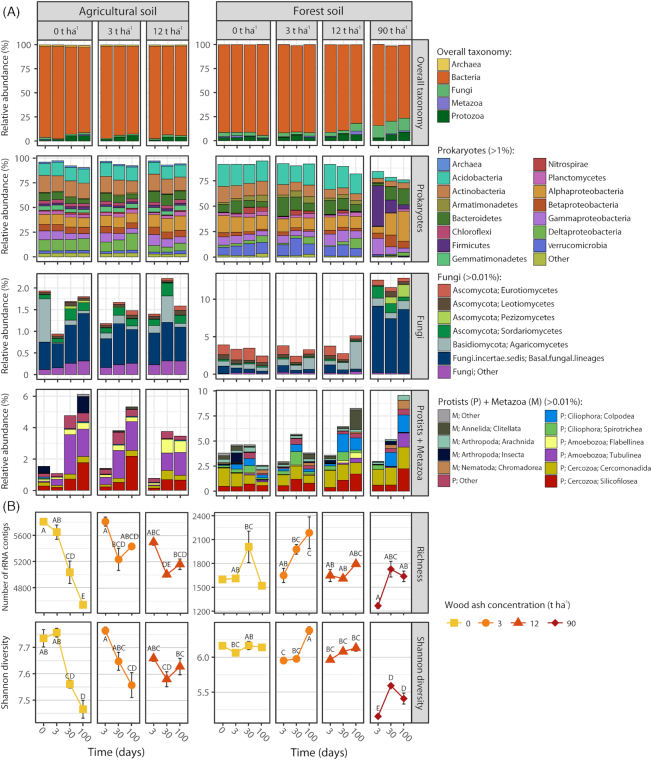
A majority (85%) of rRNA sequence reads, mapped to rRNA contigs, could be annotated to order rank (99% to phylum and 97% to class rank; Fig. 3A). Fewer sequences could be assigned lower taxonomic ranks (60 and 27% to family and genus level, respectively). Therefore, to include sufficient community information at the lowest taxonomic rank possible, we evaluated possible significant differences in abundance of taxa at order rank as the lowest taxonomic rank (see Datasheets S3 and S4, Supporting Information, for P-values and averages of relative abundances, respectively). Furthermore, for taxonomic groups that significantly changed in relative abundance, we examined the community data at lower taxonomic ranks (family and genus) to determine if specific groups were the main contributors for the observed response, as described below. Richness and Shannon diversity decreased in the unamended agricultural soil over time, while ash amendments of 3 and 12 t ha−1 counteracted this decrease (Fig. 3B). In the forest soil, these measures generally remained unchanged up to 12 t ha−1 amendments (with a single exception of increased richness at 3 t ha−1 after 100 days of incubation), while the 90 t ha−1 amendment caused reduction of Shannon diversity.
Figure 3.
Community composition and diversity across the two soils at increasing wood ash amendment and incubation times based on PCR-free, total RNA-seq. (A) The most abundant taxonomic groups (cutoff levels of average relative abundances are shown in legend header) are presented in upper panel (overall taxonomy), i.e. Archaea, Bacteria, Fungi, Protists and Metazoa. Bars represent averages of triplicates [excluding agricultural soil 3 t ha−1 at 100 days (n = 2), forest soil 0 t ha−1 at 0 days (n = 1) and forest soil 0 t ha−1 at 100 days (n = 1)]. (B) Richness and Shannon diversity. Statistically significant different richness and diversity measures (P < 0.05) between samples within each measure and soil are indicated by different letters. Symbols represent averages, as described for the bar plots.
Prokaryotic community
In both soil types, the relative abundance of Chitinophagaceae (Bacteroidetes) increased with wood ash application (Fig. 3A). In the agricultural soil, ash amendment also caused increases in Alphaproteobacteria and Betaproteobacteria. In the forest soil, the 3 and 12 t ha−1 ash amendments increased Myxococcales (Deltaproteobacteria), while Acidimicrobiia (Actinobacteria) decreased.
In the forest soil, the 90 t ha−1 ash amendment resulted in major prokaryotic community changes. Actinobacteria, Acidobacteria, Armatimonadetes and Verrucomicrobia decreased, with Acidobateria having the strongest decrease with an initial relative abundance of 21.7% with no ash amendment to 6.7% 3 days after the 90 t ha−1 ash amendment. On the contrary, Firmicutes, Bacteroidetes and Proteobacteria increased after the 90 t ha−1 ash amendment. Firmicutes dominated after 3 days, with Paenibacillus as most abundant with relative abundance of 21.3%, followed by a gradual decrease toward 1.1% after 100 days. Similarly, Gammaproteobacteria decreased during incubation after an initial increase. Chitinophagaceae (Bacteroidetes) and Rhizobiales (Alphaproteobacteria) showed the opposite temporal trend after 90 t ha−1 ash amendment and were most abundant after 100 days; Chitinophagaceae increased in relative abundance from 0.9% after 3 days to 9.8% after 100 days and Rhizobiales increased from 2.5% after 3 days to 16.7% after 100 days.
Fungal community
The 3 and 12 t ha−1 ash amendments did not affect fungal community composition in the agricultural soil (Fig. 3A). In the forest soil, no major changes were found at low amendments, while application of 90 t ha−1 resulted in increase in members of the genus Mortierella (incertae sedis) that 3 days after ash amendment became the dominant fungal group with a relative abundance of the total community of 6.5%. Also, the order Hypocreales (Sordariomycetes) and the genus Peziza (Pezizomycetes) increased with the 90 t ha−1 ash amendment.
Microeukaryotic community
In the agricultural soil, the relative abundances of Tubulinea (Amoebozoa), Thaumatomonadida (Cercozoa) and Silicofilosea (Cercozoa) increased over time in all treatments (Fig. 3A). In the forest soil, members belonging to the genus Colpoda (Ciliophora) increased with time in all treatments, though more pronouncedly at higher wood ash amendments. Further, Tubulinea (Amoebozoa), Heteromitidae (Cercozoa) and Silicofilosea (Cercozoa) increased in the 12 and 90 t ha−1 amendments.
mRNA—functional genes
A total of 0.9 million sequences were mapped to 463 mRNA contigs. The two soils possessed distinct pools of expressed genes (P < 0.001; R2 = 0.82; Adonis; Figure S2 and Datasheet S2, Supporting Information). Accordingly, scaling of Bray–Curtis dissimilarities of mRNA pools between samples on NMDS plots revealed clear clustering patterns and the Bray–Curtis dissimilarities and fitting of physicochemical parameters to these revealed similar trends as for rRNA taxonomic communities (Fig. 2 and Table 2).
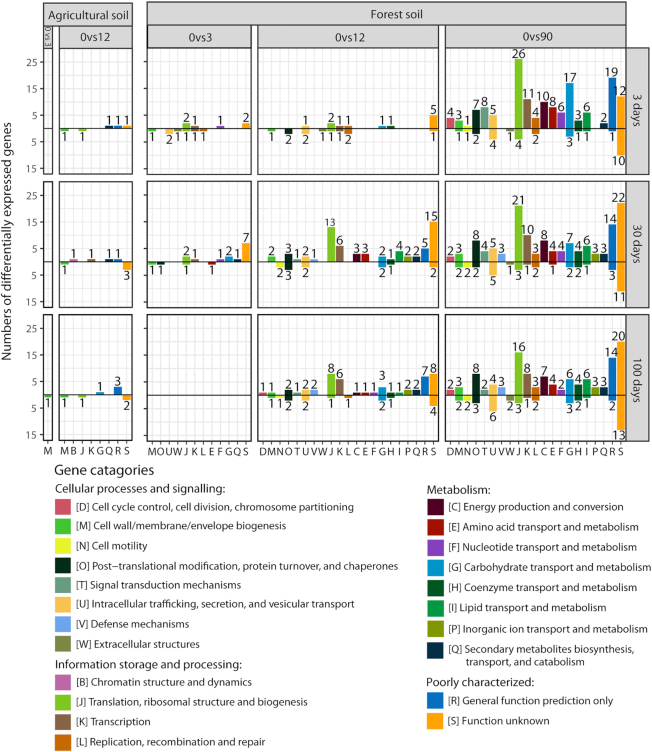
In the agricultural soil, we observed only minor functional gene responses to time and ash amendment, while more genes were differentially expressed in the forest soil (Fig. 4; see full list of differential expressed genes in Datasheet S5, Supporting Information). Overall, the number of differential expressed functional genes increased with higher wood ash amendments and the ash amendments resulted in more functional genes being upregulated than downregulated. Of the well characterized genes, four functional categories contained most of the differentially expressed genes, i.e. ‘Post-translation modification, protein turnover and chaperones’; ‘Transcription’; ‘Replication, recombination and repair’; and ‘Carbohydrate transport and metabolism’. Furthermore, genes related to stress responses such as chaperones (e.g. ‘COG0443 Molecular Chaperone’), sporulation (e.g. ‘NOG08151 Stage III sporulation protein D’), transmembrane transporters (e.g. ‘COG1744 ABC-type transport system’) and general stress response genes (e.g. ‘COG1825 Ribosomal protein L25–general stress protein Ctc’) increased mainly in the forest soil at 90 t ha−1 ash amendments (Figure S3, Supporting Information).
Figure 4.
Numbers of differentially expressed genes within functional categories across agricultural and forest soil by pairwise comparisons of gene transcription levels between samples of increasing wood ash concentration to reference samples without ash amendment at different incubation times. ‘0vs3, ‘0vs12’ and ‘0vs90’ denote the wood ash doses compared, i.e. wood ash dose 0 t ha−1 compared to 3 t ha−1 is written as ‘0vs3’. Increasing and decreasing gene transcription levels are presented above and below the black horizontal zero line, respectively. The pairwise comparisons for forest soil, 100 days, were carried out using 3 t ha−1, 100 days, as reference samples because only one replicate was acquired from the 0 t ha−1, 100 days, samples (hence the empty plot in 0vs3, 100 days, forest plot). Digits above/below bars represent the number of differentially expressed genes within a gene category.
DISCUSSION
Here, we present the first detailed analysis of changes in soil microbial prokaryotic and eukaryotic communities after amendment with ash using the total RNA sequencing procedure.
Bacterial responses to wood ash application
The general copiotrophic groups of bacteria, i.e. Bacteroidetes, Alphaproteobacteria and Betaproteobacteria, were stimulated by wood ash application. Members of Bacteroidetes benefit from wood ash application (Noyce et al. 2016; Bang-Andreasen et al. 2017); they are initial metabolizers of labile carbon and respond positively to increased soil pH and electrical conductivity (Fierer, Bradford and Jackson 2007; Lauber et al. 2009; Kim et al. 2016). Alpha- and Betaproteobacteria are also generally copiotrophic (Cleveland et al. 2007; Fierer, Bradford and Jackson 2007) and Betaproteobacteria thrive in soils with higher pH (Kim et al. 2016), whereas Alphaproteobacteria are favored at high N availability (Nemergut et al. 2010; Fierer et al. 2012).
Acidobacteria and Verrucomicrobia declined after the 90 t ha−1 amendment to the forest soil. These phyla are considered oligotrophic (Fierer, Bradford and Jackson 2007; Bergmann et al. 2011; Ramirez, Craine and Fierer 2012; Cederlund et al. 2014; Kielak et al. 2016) and Acidobacteria are generally most abundant under acidic conditions (Rousk et al. 2010; Kielak et al. 2016). Likewise are many members of the class Acidimicrobiia acidophilic that likely explain the observed decrease in this group after wood ash amendment—the acidophilic members are probably not able to cope with the wood ash-induced increases in soil pH (Johnson et al. 2009; Itoh et al. 2011). Thus, increases in pH, bioavailable DOC and nutrients induced by wood ash allow copiotrophic groups to thrive at the expense of oligotrophic groups. The shift toward a more copiotrophic-dominated community after ash amendment was further supported by the mRNA profile of the soil. Here, an increasing number of functional genes involved in metabolism and cell growth (‘Translation’, ‘Transcription’ and ‘Replication’) showed significant higher transcription levels. Moreover, the observed increase in 16S rRNA gene copies, as analyzed by qPCR, supports a shift toward a more copiotrophic community with higher average 16S rRNA gene number per genome as well as increased prokaryotic growth (Klappenbach, Dunbar and Schmidt 2000; Roller, Stoddard and Schmidt 2016).
Of the Bacteroidetes, Chitinonophagaceae showed the strongest positive response to wood ash application. Members of this family can degrade a broad spectrum of carbon compounds (Kämpfer et al. 2006; Hanada et al. 2014). Thus, they are well suited for the ash-induced increased DOC availability. Rhizobiales dominated the increasing Alphaproteobacterial fraction of the forest soil after ash amendment. They are copiotrophs (Starke et al. 2016; Lladó and Baldrian 2017) and can degrade organic pollutants and cope with heavy metals (Teng et al. 2015). They probably have advantageous properties, as the wood ash induces increase of heavy metals and nutrients in the soils. Deltaproteobacterial Myxococcales responded positively to wood ash amendment in the forest soil. Myxococcales are a group with known fungal-like behaviors including the production of extracellular enzymes involved in carbon degradation and ability to produce spores when nutrients are scarce (Sozinova et al. 2005). These traits might give Myxococcales an advantage after wood ash application. Noteworthy, the increase in Myxococcales occurred late in the incubation where especially Chitinophagaceae and Alphaproteobacteria decreased. Myxococcales are ‘micropredators’ and attack and lyse other bacteria, which might explain the increased dominance of this group at the expense of other bacterial groups (Reichenbach 1999).
The increase in 16S rRNA gene copy numbers after ash amendment (up to 12 and 90 t ha−1 for the agricultural and forest soil, respectively) is consistent with other reports of increasing bacterial numbers after wood ash application (Bååth and Arnebrant 1994; Fritze et al. 2000; Perkiömäki and Fritze 2002; Bang-Andreasen et al. 2017; Vestergård et al. 2018). The large increase in the forest soil is further consistent with the increased pH as most bacteria thrive better at pH around 7 (Rousk, Brookes and Bååth 2009). Increased prokaryotic growth and a shift toward a copiotrophic-dominated community with higher average 16S rRNA gene number per genome, as described above, are likely causing the 16S rRNA gene copy increase.
The 90 t ha−1 ash amendment to the forest soil caused immediate dominance of Firmicutes and Gammaproteobacteria. Both groups are copiotrophs that thrive upon addition of easily degradable carbon and nitrogen to soil, which probably partly explain their success upon ash application (Cleveland et al. 2007; Fierer, Bradford and Jackson 2007; Nemergut et al. 2010; Fierer et al. 2012; Ramirez, Craine and Fierer 2012). However, bacteria from these phyla are also known to be tolerant to heavy metals (Jacquiod et al. 2017). Moreover, within Firmicutes the endospore-forming genus Paenibacillus dominated (de Hoon, Eichenberger and Vitkup 2010), and we found increased transcription of genes involved in sporulation in these samples. Combined, these capabilities probably enable members of these groups to withstand the initial wood ash-induced changes to the soil, including increased heavy metal concentrations, thereby allowing them to be initial utilizers of newly available labile resources. Reduced diversity at this ash dose further indicates that less organisms can cope with the ash-induced changes to the soil system
Fungal responses to wood ash application
In both soil types, fungal response to ash amendment was slight compared to the prokaryotic response. Likewise, Högberg, Högberg and Myrold (2007), Rousk, Brookes and Bååth (2009, 2011) and Cruz-Paredes et al. (2017) found bacteria to be more stimulated by nutrient addition and increases in pH than fungi. Similarly, effects of ash amendment have been reported by Mahmood et al. (2003) and Noyce et al. (2016). The 90 t ha−1 amendment in the forest soil caused increased ITS gene copy numbers and a fungal community shift with increased dominance of Mortierella, Peziza and Hypocreales. These fungi are opportunistic saprotrophs with high growth rates and can exploit readily available nutrients before other fungi arrive (Carlile, Watkinson and Gooday 2001; Tedersoo et al. 2006; Druzhinina, Shelest and Kubicek 2012). Further, some Peziza spp. are early post-fire colonizers adapted to ash conditions (Egger 1986; Rincón et al. 2014). The increase in these groups further supports that copiotrophic-like lifestyles are favored by wood ash application.
Microeukaryote responses to wood ash application
The microeukaryotes also responded to wood ash application in the forest soil, probably because the stimulation of copiotrophic bacteria and fungi provided more food for nematodes and protozoa (Rønn, Vestergård and Ekelund 2012). Ciliates (Colpoda), amoebae (Tubulinea) and small heterotrophic flagellates (Heteromitidae and Silicofilosea) increased with more pronounced responses at the later incubation times. Protozoa generally have longer generation times than prokaryotes, and thus need longer time to increase in population size. Further, they cannot start growth before a reasonable bacterial population has been formed (Fenchel 1987; Ekelund, Frederiksen and Rønn 2002). The protozoan increase may explain the small decrease in prokaryotic 16S rRNA gene copies at day 100, where we observed the largest fraction of protozoa. The positively responding protozoa were likely primarily bacterivorous (Ekelund and Rønn 1994; Ekelund 1998), consistent with the decreasing relative fraction of bacterial rRNA sequences and the increasing relative fraction of fungal and protozoan rRNA sequences in the later incubation times after the application of 12 and 90 t ha−1 ash. Thus, preferential protozoan grazing on bacteria can explain the relative larger rRNA fraction of fungi and protozoa at day 100. We found no significant effect of ash amendment on microeukaryotes in the agricultural soil, which is consistent with the relative minor effects on prokaryotes and fungi in this soil.
Stress responses at high wood ash amendments
We recorded increased transcription of stress-response genes at the 90 t ha−1 amendments, which supports that this high dose exerts harmful effects on many members of the microbiome. For example, we found increased transcription of genes involved in sporulation. Sporulation is a known survival mechanism to unfavorable conditions (de Hoon, Eichenberger and Vitkup 2010). Also, transmembrane transporter proteins balance osmotic pressure of cells, regulate cytosolic pH and can export toxins such as metals from the cell (Alberts et al. 2002; Ma, Jacobsen and Giedroc 2009; Wilkens 2015). Increased activity of transmembrane transporters is probably a response to wood ash-induced osmotic changes to the soil system, increased pH, metal concentration and other toxic compounds. Moreover, chaperones ensure correct folding of proteins and are involved in cellular coping with stress-induced denaturation of proteins (Feder and Hofmann 1999) and the observed increase in transcription level of these probably is a stress response.
The changes in the microbial communities are linked to physicochemical soil parameters
We found that ash amendment strongly increased soil pH, which is a strong driver of microbial community composition and functioning (Fierer and Jackson 2006; Rousk et al. 2010) also after wood ash application (Frostegård et al. 1993; Zimmermann and Frey 2002; Högberg, Högberg and Myrold 2007; Peltoniemi et al. 2016; Bang-Andreasen et al. 2017). DOC and phosphate concomitantly increased. Several factors may contribute to this: (i) pH dependent changes in solubility (Evans et al. 2012; Maresca, Hyks and Astrup 2017), (ii) release from dead organisms incapable of coping with the wood ash or wood ash-induced changes to the soil system, (iii) increased mineralization rates after wood ash application (Bååth and Arnebrant 1994; Vestergård et al. 2018) and (iv) the phosphorus in the bio-ash (Pitman 2006; Maresca, Hyks and Astrup 2017).
Since pH, conductivity, DOC and phosphate all correlated positively to wood ash concentrations, it is difficult to disentangle the direct effect of these components as they might all be covariates of the wood ash amendments. pH changes induce a cascade of effects in soil parameters and therefore affect mineral nutrient availability, salinity, metal solubility and organic C (Lauber et al. 2009). Many of the wood ash-induced changes were likely caused directly or indirectly by the pH increase, which is probably the major reason that pH is an essential driver of taxonomic and functional soil characteristics (Lauber et al. 2009; Rousk et al. 2010; Fierer 2017; Vestergård et al. 2018).
Wood ash contains virtually no nitrogen; hence, measurable effects on soil nitrate and ammonium are probably caused by pH effects on microbial N mineralization (Vestergård et al. 2018) and ion solubility (Pitman 2006). Changes in nitrate and ammonium were significant as explanatory variables on the observed rRNA and mRNA dissimilarity profiles of the forest soil but not in the agricultural soil. Forest soil is generally more N limited than agricultural soil, where N is kept at a high level through fertilization.
Conclusions
We used detailed total RNA sequencing to demonstrate drastic taxonomic and functional changes in the active prokaryotic and eukaryotic microbiomes of agricultural and forest soil after wood ash amendment. Our analyses suggested that increase in pH, electrical conductivity, DOC and phosphate were the main drivers of the observed changes. Wood ash amendment of 3 and 12 t ha−1 resulted in increased prokaryotic abundance and dominance of copiotrophic groups and elevated expression of genes involved in metabolism and cell growth. Amendment of 90 t ha−1 caused collapse of the microbiome in the agricultural soil, while in the forest soil the copiotrophic microbiome, also including fast-growing saprotrophic fungi, was further stimulated. However, diversity was reduced, and expression of stress response genes increased. Bacterivorous protozoan groups increased as a response to enhanced bacterial growth, which supports that the protozoa have a pivotal role in controlling bacterial abundance in soil following wood ash application. Overall, prokaryotic community and quantity responded more pronouncedly to wood ash amendment than fungi in both forest and agricultural soil.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pia Bach Jakobsen for laboratory assistance.
FUNDING
This work was supported by the ‘Center for Bioenergy Recycling (ASHBACK)’ project, funded by the Danish Council for Strategic Research (grant no. 0603-00587B) and Danish Geocenter (grant no. 5298507). AL was supported by a Juan de la Cierva scholarship from the Spanish Government (FJCI-2014-19572). MZA was supported by the European Union's Horizon 2020 - Research and Innovation Framework Programme under the Marie Skłodowska-Curie project MicroArctic (grant no. 675546). FE and RR were supported by the Danish Council for Independent Research (DFF-4002-00274).
Conflicts of interest. None declared.
REFERENCES
- Alberts B, Johnson A, Lewis J et al.. Molecular Biology of the Cell. 4th edn New York: Garland Science, 2002. [Google Scholar]
- Anwar MZ, Lanzen A, Bang-Andreasen T et al.. To assemble or not to resemble—a validated comparative metatranscriptomics workflow (CoMW). GigaScience. 2019;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson KA, Ekelund NGA. Biological effects of wood ash application to forest and aquatic ecosystems. J Environ Qual. 2004;33:1595–605. [DOI] [PubMed] [Google Scholar]
- Augusto L, Bakker MR, Meredieu C. Wood ash applications to temperate forest ecosystems—potential benefits and drawbacks. Plant Soil. 2008;306:181–98. [Google Scholar]
- Bååth E, Arnebrant K. Growth rate and response of bacterial communities to pH in limed and ash treated forest soils. Soil Biol Biochem. 1994;26:995–1001. [Google Scholar]
- Bang-Andreasen T, Nielsen JT, Voriskova J et al.. Wood ash induced pH changes strongly affect soil bacterial numbers and community composition. Front Microbiol. 2017;8:1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG et al.. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagodatskaya E, Kuzyakov Y. Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol Biochem. 2013;67:192–211. [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA et al.. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1453–62. [DOI] [PubMed] [Google Scholar]
- Carini P, Marsden PJ, Leff JW et al.. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol. 2016;2:16242. [DOI] [PubMed] [Google Scholar]
- Carlile MJ, Watkinson SC, Gooday GW. 2001. The Fungi. 2nd edn San Diego, CA: Academic Press. [Google Scholar]
- Carvalhais LC, Dennis PG, Tyson GW et al.. Application of metatranscriptomics to soil environments. J Microbiol Methods. 2012;91:246–51. [DOI] [PubMed] [Google Scholar]
- Cederlund H, Wessén E, Enwall K et al.. Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl Soil Ecol. 2014;84:62–8. [Google Scholar]
- Cleveland CC, Nemergut DR, Schmidt SK et al.. Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry. 2007;82:229–40. [Google Scholar]
- Cruz-Paredes C, Wallander H, Kjøller R et al.. Using community trait-distributions to assign microbial responses to pH changes and Cd in forest soils treated with wood ash. Soil Biol Biochem. 2017;112:153–64. [Google Scholar]
- De Hoon MJL, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr Biol. 2010;20:R735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeyer A, Voundi Nkana J., Verloo M. Characteristics of wood ash and influence on soil properties and nutrient uptake: an overview. Bioresour Technol. 2001;77:287–95. [DOI] [PubMed] [Google Scholar]
- Druzhinina IS, Shelest E, Kubicek CP. Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol Lett. 2012;337:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger KN. Substrate hydrolysis patterns of post-fire ascomycetes (Pezizales). Mycologia. 1986;78:771. [Google Scholar]
- Ekelund F. Enumeration and abundance of mycophagous protozoa in soil, with special emphasis on heterotrophic flagellates. Soil Biol Biochem. 1998;30:1343–7. [Google Scholar]
- Ekelund F, Frederiksen HB, Rønn R. Population dynamics of active and total ciliate populations in arable soil amended with wheat. Appl Environ Microbiol. 2002;68:1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekelund F, Rønn R. Notes on protozoa in agricultural soil with emphasis on heterotrophic flagellates and naked amoebae and their ecology. FEMS Microbiol Rev. 1994;15:321–53. [DOI] [PubMed] [Google Scholar]
- Epelde L, Lanzén A, Blanco F et al.. Adaptation of soil microbial community structure and function to chronic metal contamination at an abandoned Pb–Zn mine. FEMS Microbiol Ecol. 2015;91:1–11. [DOI] [PubMed] [Google Scholar]
- Evans CD, Jones TG, Burden A et al.. Acidity controls on dissolved organic carbon mobility in organic soils. Glob Change Biol. 2012;18:3317–31. [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82. [DOI] [PubMed] [Google Scholar]
- Fenchel T. Ecology of Protozoa. 1st edn Berlin, Heidelberg: Springer, 1987. [Google Scholar]
- Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90. [DOI] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–64. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Ramirez KS et al.. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze H, Perkiömäki J, Saarela U et al.. Effect of Cd-containing wood ash on the microflora of coniferous forest humus. FEMS Microbiol Ecol. 2000;32:43–51. [DOI] [PubMed] [Google Scholar]
- Frostegård A, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol. 1993;59:3605–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisen S, Tveit AT, Clark IM et al.. Metatranscriptomic census of active protists in soils. ISME J. 2015;9:2178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M et al.. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada S, Tamaki H, Nakamura K et al.. Crenotalea thermophila gen. nov., sp. nov., a member of the family Chitinophagaceae isolated from a hot spring. Int J Syst Evol Microbiol. 2014;64:1359–64. [DOI] [PubMed] [Google Scholar]
- Hansen M, Bang-Andreasen T, Sørensen H et al.. Micro vertical changes in soil pH and base cations over time after application of wood ash on forest soil. Forest Ecol Manag. 2017;406:274–80. [Google Scholar]
- Hansen CHF, Krych L, Nielsen DS et al.. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55:2285–94. [DOI] [PubMed] [Google Scholar]
- Högberg MN, Högberg P, Myrold DD. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia. 2007;150:590–601. [DOI] [PubMed] [Google Scholar]
- Hultman J, Waldrop MP, Mackelprang R et al.. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 2015;521:208–12. [DOI] [PubMed] [Google Scholar]
- Huotari N, Tillman-Sutela E, Moilanen M et al.. Recycling of ash—for the good of the environment? Forest Ecol Manag. 2015;348:226–40. [Google Scholar]
- Ihrmark K, Bödeker ITM, Cruz-Martinez K et al.. New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77. [DOI] [PubMed] [Google Scholar]
- Itoh T, Yamanoi K, Kudo T et al.. Aciditerrimonas ferrireducens gen. nov., sp. nov., an iron-reducing thermoacidophilic actinobacterium isolated from a solfataric field. Int J Syst Evol Microbiol. 2011;61:1281–5. [DOI] [PubMed] [Google Scholar]
- Jacquiod S, Cyriaque V, Riber L et al.. Long-term industrial metal contamination unexpectedly shaped diversity and activity response of sediment microbiome. J Hazard Mater. 2017;344:299–307. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M et al.. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Bacelar-Nicolau P, Okibe N et al.. Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic actinobacteria. Int J Syst Evol Microbiol. 2009;59:1082–9. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, Young C-C, Sridhar KR et al.. Transfer of [Flexibacter] sancti, [Flexibacter] filiformis, [Flexibacter] japonensis and [Cytophaga] arvensicola to the genus Chitinophaga and description ofChitinophaga skermaniisp. nov. Int J Syst Evol Microbiol. 2006;56:2223–8. [DOI] [PubMed] [Google Scholar]
- Karltun E, Saarsalmi A, Ingerslev M et al.. Wood ash recycling—possibilities and risks. In: Röser D, Asikainen A, Raulund-Rasmussen K, Stupak I(eds). PCR Protocols—A Guide to Methods and Applications. Dordrecht: Springer Netherlands, 2008, 79–108. [Google Scholar]
- Kielak AM, Barreto CC, Kowalchuk GA et al.. The ecology of acidobacteria: moving beyond genes and genomes. Front Microbiol. 2016;7:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Roh A-S, Choi S-C et al.. Soil pH and electrical conductivity are key edaphic factors shaping bacterial communities of greenhouse soils in Korea. J Microbiol. 2016;54:838–45. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. [DOI] [PubMed] [Google Scholar]
- Lanzén A, Jørgensen SL, Huson DH et al.. CREST—classification resources for environmental sequence tags. PLoS One. 2012;7:e49334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R et al.. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó S, Baldrian P. Community-level physiological profiling analyses show potential to identify the copiotrophic bacteria present in soil environments. PLoS One. 2017;12:e0171638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jacobsen FE, Giedroc DP. Metal transporters and metal sensors: how coordination chemistry controls bacterial metal homeostasis. Chem Rev. 2009;109:4644–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood S, Finlay RD, Fransson A-M et al.. Effects of hardened wood ash on microbial activity, plant growth and nutrient uptake by ectomycorrhizal spruce seedlings. FEMS Microbiol Ecol. 2003;43:121–31. [DOI] [PubMed] [Google Scholar]
- Maresca A, Hyks J, Astrup TF. Recirculation of biomass ashes onto forest soils: ash composition, mineralogy and leaching properties. Waste Manag. 2017;70:127–38. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetJl. 2011;17:10. [Google Scholar]
- Miller CS, Baker BJ, Thomas BC et al.. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 2011;12:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Burge SW, Bateman A et al.. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Cleveland CC, Wieder WR et al.. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem. 2010;42:2153–60. [Google Scholar]
- Noyce GL, Fulthorpe R, Gorgolewski A et al.. Soil microbial responses to wood ash addition and forest fire in managed Ontario forests. Appl Soil Ecol. 2016;107:368–80. [Google Scholar]
- Ohno T, Susan Erich M. Effect of wood ash application on soil pH and soil test nutrient levels. Agric Ecosyst Environ. 1990;32:223–39. [Google Scholar]
- Oksanen J, Kindt R, Legendre P et al.. vegan: Community Ecology Package https://cran.r-project.org/web/packages/vegan/17.6.2019;
- Peltoniemi K, Pyrhönen M, Laiho R et al.. Microbial communities after wood ash fertilization in a boreal drained peatland forest. Eur J Soil Biol. 2016;76:95–102. [Google Scholar]
- Perkiömäki J, Fritze H. Short and long-term effects of wood ash on the boreal forest humus microbial community. Soil Biol Biochem. 2002;34:1343–53. [Google Scholar]
- Pitman RM. Wood ash use in forestry—a review of the environmental impacts. Forestry. 2006;79:563–88. [Google Scholar]
- Qin J, Hovmand MF, Ekelund F et al.. Wood ash application increases pH but does not harm the soil mesofauna. Environ Pollut. 2017;224:581–9. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing http://www.R-project.org/17.6.2019.
- Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Change Biol. 2012;18:1918–27. [Google Scholar]
- Reichenbach H. The ecology of the myxobacteria. Environ Microbiol. 1999;1:15–21. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the european molecular biology open software suite. Trends Genet. 2000;16:276–7. [DOI] [PubMed] [Google Scholar]
- Rincón A, Santamaría BP, Ocaña L et al.. Structure and phylogenetic diversity of post-fire ectomycorrhizal communities of maritime pine. Mycorrhiza. 2014;24:131–41. [DOI] [PubMed] [Google Scholar]
- Roller BRK, Stoddard SF, Schmidt TM. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat Microbiol. 2016;1:16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønn R, Vestergård M, Ekelund F. Interactions between bacteria, protozoa and nematodes in soil. Acta Protozool. 2012;51:223–35. [Google Scholar]
- Rousk J, Bååth E, Brookes PC et al.. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–51. [DOI] [PubMed] [Google Scholar]
- Rousk J, Brookes PC, Bååth E. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol. 2009;75:1589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Brookes PC, Bååth E. Fungal and bacterial growth responses to N fertilization and pH in the 150-year “Park Grass” UK grassland experiment. FEMS Microbiol Ecol. 2011;76:89–99. [DOI] [PubMed] [Google Scholar]
- Schostag M, Priemé A, Jacquiod R et al.. Bacterial and protozoan dynamics upon thawing and freezing of an active layer permafrost soil. ISME J. 2019;13:1345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozinova O, Jiang Y, Kaiser D et al.. A three-dimensional model of myxobacterial aggregation by contact-mediated interactions. Proc Natl Acad Sci. 2005;102:11308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke R, Kermer R, Ullmann-Zeunert L et al.. Bacteria dominate the short-term assimilation of plant-derived N in soil. Soil Biol Biochem. 2016;96:30–8. [Google Scholar]
- Tedersoo L, Hansen K, Perry BA et al.. Molecular and morphological diversity of pezizalean ectomycorrhiza. New Phytol. 2006;170:581–96. [DOI] [PubMed] [Google Scholar]
- Teng Y, Wang X, Li L et al.. Rhizobia and their bio-partners as novel drivers for functional remediation in contaminated soils. Front Plant Sci. 2015;6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T, Lanzén A, Qi J et al.. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2008;3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance ED. Land application of wood-fired and combination boiler ashes: an overview. J Environ Qual. 1996;25:937. [Google Scholar]
- Varet H, Brillet-Guéguen L, Coppée J-Y et al.. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS One. 2016;11:e0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Pavlović D, Šikić M. SWORD—a highly efficient protein database search. Bioinformatics. 2016;32:i680–4. [DOI] [PubMed] [Google Scholar]
- Vestergård M, Bang-Andreasen T, Buss SM et al.. The relative importance of the bacterial pathway and soil inorganic nitrogen increase across an extreme wood-ash application gradient. GCB Bioenergy. 2018;10:320–34. [Google Scholar]
- Wall DH, Bardgett RD, Behan-Pelletier V et al.. Soil Ecology And Ecosystem Services. 1st edn USA: Oxford University Press, 2012. [Google Scholar]
- White T, Bruns R, Lee S et al.. Amplification and direct sequencing of fungal ribosomalrna genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T(eds). PCR Protocols—A Guide to Methods and Applications. New York, US: Academic Press, 1990, 315–22. [Google Scholar]
- Wilke A, Harrison T, Wilkening J et al.. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinformatics. 2012;13:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S, Frey B. Soil respiration and microbial properties in an acid forest soil: effects of wood ash. Soil Biol Biochem. 2002;34:1727–37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.