Summary
The PE and PPE proteins of Mycobacterium tuberculosis have been studied with great interest since their discovery. Named after the conserved proline (P) and glutamic acid (E) residues in their N‐terminal domains, these proteins are postulated to perform wide‐ranging roles in virulence and immune modulation. However, technical challenges in studying these proteins and their encoding genes have hampered the elucidation of molecular mechanisms and leave many open questions regarding the biological functions mediated by these proteins. Here, I review the shared and unique characteristics of PE and PPE proteins from a molecular perspective linking this information to their functions in mycobacterial virulence. I discuss how the different subgroups (PE_PGRS, PPE‐PPW, PPE‐SVP and PPE‐MPTR) are defined and why this classification of paramount importance to understand the PE and PPE proteins as individuals and or groups. The goal of this MicroReview is to summarize and structure the existing information on this gene family into a simplified framework of thinking about PE and PPE proteins and genes. Thereby, I hope to provide helpful starting points in studying these genes and proteins for researchers with different backgrounds. This has particular implications for the design and monitoring of novel vaccine candidates and in understanding the evolution of the M. tuberculosis complex.
Structure of the EspG5‐PPE41‐PE25 complex (4KXR). The EspG chaperone (Gold) binds to the EspG‐binding domain of the PPE protein (Light blue), thereby conferring TypeVII secretion system specificity (Daleke et al., 2012b; Korotkova et al., 2014; Phan et al., 2017). The rest of the PPE‐protein interacts with its PE partner (Teal) via hydrophobic interactions. The conserved WxG residues of the PPE (Pink) and YxxxD/E of the PE (Red) are closely associated and may form a composite TypeVII secretion signal. The C‐terminus of the PPE (Green) extends toward the C‐terminus of the PE (Red), suggesting that PE/PPE protein pairs with C‐terminal extensions may have further PE/PPE interactions between those specific domains.

Introduction
One of the most striking findings of the Mycobacterium tuberculosis genome sequencing project was the identification of a family of genes, called pe and ppe genes, covering up to 10% of the M. tuberculosis genome (Cole et al., 1998; Fishbein et al., 2015). The PE and PPE proteins are named after the conserved proline (P) and glutamic acid (E) residues in their N‐terminal sequences and have intrigued researchers since their discovery. However, although significant advances have been made in understanding these proteins, progress has been hampered by technological challenges associated with studying PE and PPE proteins and genes. These challenges include the high gc‐content of pe and ppe genes, which can range up to 80%, making sequencing, alignment and cloning of these genes an ordeal (Hermans et al., 1992; Poulet and Cole, 1995; Cole et al., 1998). This problem is aggravated by many gene duplications and repetitive sequences that distinguish certain subgroups of pe and ppe genes (Gey van Pittius et al., 2006; McEvoy et al., 2012; Copin et al., 2014). Because of the challenges associated with mapping pe/ppe reads to mycobacterial reference genomes, these genes have generally been excluded from frequently used bioinformatic pipelines and have remained relatively understudied with genome‐based techniques (Coll et al., 2014; Phelan et al., 2016; Meehan et al., 2019). Similarly, mass spectrometry of PE and PPE proteins is hampered by a paucity of trypsin cleavage sites and high homology between and within PE and PPE proteins (Banu et al., 2002; Schubert et al., 2013; Ates et al., 2015). However, emerging techniques, such as long‐read sequencing methods (e.g. Nanopore and Pacific Biosciences technologies) and novel biochemical insights into these proteins, may pave the way for new discoveries and improve understanding of the biology of these proteins (Ates et al., 2015; 2018b; Rodríguez et al., 2015; Pandey et al., 2018).
When discussing PE and PPE proteins and their functions, it is paramount to consider their subcellular localization. Some PPE proteins are thought to function as outer membrane nutrient transport proteins and would therefore need to be localized in the mycobacterial outer membrane to perform their function (Ates et al., 2015; Tufariello et al., 2016; Mitra et al., 2017). Other PE and PPE proteins are described to play roles in host–pathogen interaction or immune evasion (Sampson, 2011; Saini et al., 2016). To interact directly with the host cell, the protein needs to be either surface associated and/or secreted in soluble form. In all these cases, the PE and PPE proteins need to be first transported over the bacterial inner membranes, by their cognate TypeVII secretion systems ESX‐1, ESX‐3 and ESX‐5 (Bitter et al., 2009; Houben et al., 2012b; Gröschel et al., 2016; Beckham et al., 2017). For clarity, in this review ‘secretion’ is defined as protein transport over the inner membrane by a TypeVII secretion system.
Due to the methodological obstacles, the precise subcellular localization of PE and PPE proteins is understudied and remains uncertain in many cases. Despite some progress to isolate mycobacterial outer membrane proteins, no single method has been truly standardized or widely adopted (Abdallah et al., 2006; 2009; Sani et al., 2010; van der Woude et al., 2013; Danilchanka et al., 2014; Ates et al., 2015; Fishbein et al., 2015; Mitra et al., 2017). Similarly, different methods of secretion analysis can come to different results depending on timescale, growth medium and the detection method used. Finally, many studies investigating PE and PPE proteins use Mycobacterium smegmatis as a model organism, which has no functional ESX‐5 system and is therefore unable to solubly express or translocate the majority of PE and PPE proteins (Abdallah et al., 2007; Sampson, 2011; Gröschel et al., 2016; Beckham et al., 2017).
In spite of all these methodological challenges, perhaps the biggest hurdle for researchers attempting to study PE and PPE proteins is to make sense of the vast body of literature. In this review, I aim to provide a framework to study PE and PPE proteins from a molecular biology perspective by establishing their common characteristics while highlighting the importance of classifying PE and PPE proteins within subgroups (Gey van Pittius et al., 2006). This framework is used to discuss recent progress in understanding of the role of PE and PPE proteins in the pathogenesis, immune recognition and evolution of the M. tuberculosis complex (MTBC).
General features of PE and PPE proteins
To study any protein of interest, it is important to understand the features that define its function. A first step in studying PE and PPE proteins is, therefore, to divide the protein of interest into their conserved PE or PPE domains and the highly variable C‐terminal domains. Although the function of PE and PPE proteins remains mostly unknown, molecular and biochemical studies have greatly improved our understanding of the PE and PPE domains (Strong et al., 2006; Abdallah et al., 2009; Sayes et al., 2012; Ekiert and Cox, 2014; Korotkova et al., 2014). In this section, I will first discuss these general features of the PE and PPE domains before discussing the more variable C‐terminal domains.
The N‐termini of all PE proteins are highly conserved and consist of approximately 100 amino acids that form a helix‐turn‐helix structure (PFAM: PF00934 [Finn et al., 2014]) (Cole et al., 1998; Gey van Pittius et al., 2006; Strong et al., 2006). The pro‐glu residues responsible for naming the protein are generally situated in the ten N‐terminal amino acids, while the C‐terminus of the PE‐domain contains the conserved YxxxD/E TypeVII secretion signal (Fig. 1) (Champion et al., 2006; Daleke et al., 2012a). This secretion signal is required for secretion of these proteins and may be involved in substrate recognition by the TypeVII secretion systems (Champion et al., 2006; Daleke et al., 2012a; Rosenberg et al., 2015; Solomonson et al., 2015). Similarly, the PPE proteins contain highly conserved features. The PPE domain (PFAM: PF00823 [Finn et al., 2014]) is typically around 180 amino acids long and contains the pro‐pro‐glu motif near its N‐terminus. In some cases, both PE and PPE amino acid sequences may not be fully conserved, which should therefore not be used as the sole criterion to recognize PPE proteins (Gey van Pittius et al., 2006). A conserved WxG motif is found between the second and third alpha helix of all PPE proteins, but PPE proteins do not contain the YxxxD/E secretion signal (Fig. 1; Daleke et al., 2012a; Phan and Houben, 2018).
Figure 1.
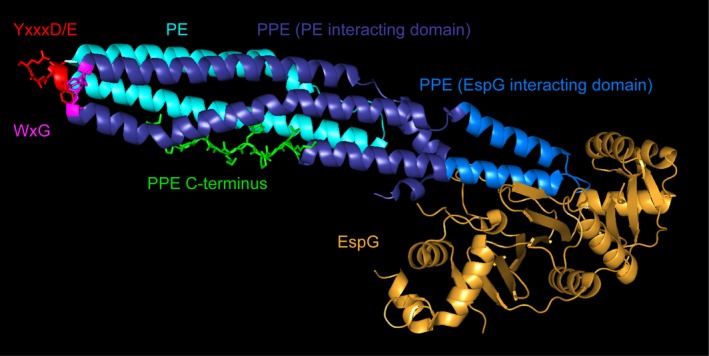
Structure of the EspG5‐PPE41‐PE25 complex (4KXR). The EspG chaperone (Gold) binds to the EspG‐binding domain of the PPE protein (Light blue), thereby conferring TypeVII secretion system specificity (Daleke et al., 2012b; Korotkova et al., 2014; Phan et al., 2017). The rest of the PPE‐protein interacts with its PE partner (Teal) via hydrophobic interactions. The conserved WxG residues of the PPE (Pink) and YxxxD/E of the PE (Red) are closely associated and may form a composite TypeVII secretion signal. The C‐terminus of the PPE (Green) extends toward the C‐terminus of the PE (Red), suggesting that PE/PPE protein pairs with C‐terminal extensions may have further PE/PPE interactions between those specific domains.
In all studies with sufficient biochemical characterization, PE and PPE proteins were found to be secreted as heterodimers. The PPE protein binds to its cognate PE protein via hydrophobic interactions with the three N‐terminal alpha‐helices of the PPE (Strong et al., 2006; Ekiert and Cox, 2014; Korotkova et al., 2014; Chen et al., 2017). This heterodimeric structure places the conserved WxG of the PPE protein and the YxxxD/E sequence of the PE protein in close proximity and these sequences likely form a composite recognition structure for TypeVII secretion (Fig. 1; Daleke et al., 2012a; Poulsen et al., 2014; Ates et al., 2016a). This PE and PPE complex structure highly resembles the structure of the heterodimers formed by Esx proteins, such as EsxA‐EsxB (ESAT‐6/CFP10) (Renshaw et al., 2005; Poulsen et al., 2014), or even the Staphylococcus aureus EsxA or EsxC homodimers (Sundaramoorthy et al., 2008; Anderson et al., 2013; Ates et al., 2016a). The structure of PE and PPE proteins is also highly reminiscent of that of a number of ESX‐1 secretion‐associated proteins (esp) proteins such as the EspA/C and EspE/F heterodimers and EspB (Bitter et al., 2009; Solomonson et al., 2015; Lou et al., 2017; Phan et al., 2018; Phan and Houben, 2018). Together, these data suggest that the helix‐bundle structure and composite secretion signal are among the defining features of PE and PPE heterodimers and other TypeVII‐associated substrates (Daleke et al., 2012a).
A unique characteristic of the Esp and PPE proteins is that helices 4 and 5 do not directly interact with the PE secretion partner. Instead, they bind to cytosolic EspG chaperones and thereby confer system specificity to these substrates (Fig. 1; Daleke et al., 2012b; Ekiert and Cox, 2014; Korotkova et al., 2014; Phan et al., 2017; Phan and Houben, 2018). Intriguingly, exchanging the EspG interacting domain of the ESX‐1 secreted PPE68_1 with that of the ESX‐5 substrate PPE18 was sufficient to reroute this protein from ESX‐1 to ESX‐5 in M. marinum (Phan et al., 2017). Therefore, the sequence of the EspG‐binding domain of the PPE protein may be sufficient to predict system specificity of PPE proteins, although experimental validation of this hypothesis remains incomplete beyond the ESX‐1 and ESX‐5 systems (Ekiert and Cox, 2014; Korotkova et al., 2014; Chen et al., 2017; Phan and Houben, 2018). Many of these studies have been performed in the model organism M. marinum, but most results have been verified in M. tuberculosis. However, some mechanistic differences also remain between M. marinum and M. tuberculosis, which is understandable when considering the different host ranges and physiological characteristics such as optimal growth temperature. For instance, EspG5 was found to be required for all ESX‐5‐dependent phenotypes in M. marinum (Abdallah et al., 2009), but dispensable for ESX‐5‐mediated secretion of selected ESX‐5 substrates or virulence in M. tuberculosis (Bottai et al., 2012). Similarly, while essentiality of the ESX‐5 system of M. marinum can be complemented by introduction of M. tuberculosis esx‐5, this complementation did not fully restore PE‐PPE secretion (Ates et al., 2015). A new report pinpoints this species specificity to EccC5 and specifically to the linker 2 domain, which is suggested to mediate secretion of specific PE and PPE proteins secreted by ESX‐5 (Bunduc et al., 2019). This illustrates that even when differences between model organisms and M. tuberculosis are found, such species specificity can be used to gain more insight into the mechanisms of TypeVII secretion (Bunduc et al., 2019).
More insight into PE and PPE proteins may be obtained by comparing them with other closely related TypeVII secretion substrates. Recent studies have postulated that the EspB protein and the EspA/C heterodimer may multimerize and may form large pore‐ or needle‐like structures that could span the mycobacterial outer membrane (Solomonson et al., 2015; Lou et al., 2017). It remains to be determined if similar structures are also formed by PE and PPE dimers to carry out channel‐like functions, which would align with other experimental data discussed below. An alternative hypothesis is that TypeVII secretion substrates function as autotransporter‐like molecules, which require PE and PPE heteromultimeric complexes for transport over the mycobacterial outer membrane. The structure of the M. xenopi ESX‐5 membrane complex was recently visualized by negative stain cryo‐electron microscopy and revealed that the conserved membrane components (EccBCDE) are all present in the inner membrane and their dimensions do not allow for a one‐step secretion process over both the inner and outer mycobacterial membrane (Beckham et al., 2017). Even more recently, high‐resolution cryo‐electron microscopy structures have been resolved of the M. smegmatis ESX‐3 secretion systems (Famelis et al., 2019; Poweleit et al., 2019). These structures mostly confirm what was found for M. xenopi ESX‐5, but do show in more detail that EccB3 protrudes into the periplasmic space, where it could perhaps interact with other periplasmic, or outer membrane, proteins. Yet, no outer membrane components for TypeVII secretion machinery have been identified to date. It is tempting to speculate that such a mechanism for transport over the outer membrane would be encoded in the esx‐loci together with the inner membrane components. This function is not performed by other widely present genes in the esx‐loci, such as the inner membrane‐localized mycosin proteases (MycP), the EspG chaperones or the cytosolic EccA ATPases, which have clearly characterized other functions and localization (Daleke et al., 2012b; Wagner et al., 2014; Van Winden et al., 2016). There are therefore no clear candidates encoded in the esx‐loci that could be involved in outer membrane translocation, apart from the ESX/PE/PPE/Esp substrates. This suggests that either these substrates, or an individually encoded locus, is responsible for outer membrane transport of TypeVII secretion substrates. Interestingly, TypeVII‐like secretion systems have also been identified in monoderm gram‐positive bacteria such as S. aureus and S. intermedius. These secretion systems secrete bacterial toxins belonging to the WxG (PF06013) and LxG (PF04740) family of TypeVII secretion substrates (Finn et al., 2014; Cao et al., 2016; Whitney et al., 2017). However, since no PE and PPE orthologues have been found in these monoderm bacteria it is tempting to speculate on a role in the secretion process for the PE and PPE proteins, but this is currently nothing more than a highly speculative hypothesis. The fundamental question of how TypeVII secretion substrates cross the mycobacterial outer membrane is clearly of great interest and identification of such mechanisms could even provide leads toward the discovery of novel antimycobacterial compounds (Rybniker et al., 2014).
In summary, PE and PPE proteins share general features in the form of a helix‐bundle structure that allows PE and PPE proteins to interact. This interaction creates a composite secretion signal of the WxG sequence on the PPE and YxxxD/E on the PE, which are likely important for substrate recognition. The PPE proteins contain a two‐helix domain that interacts with EspG chaperones to confer system specificity.
Subclassification of PE and PPE proteins
Most of the diversity and arguably the most interesting features of the PE and PPE proteins can be found in the highly variable C‐terminal domains. These C‐terminal domains can also be used to classify the PE and PPE proteins in different subgroups, which may greatly aid researchers to formulate testable research hypotheses and provide functional or evolutionary insight (Gordon et al., 1999; Brennan and Delogu, 2002; Gey van Pittius et al., 2006; Delogu et al., 2017).
The largest subgroup that can be distinguished are the polymorphic gc‐rich sequences (PGRS), which define 65 of the 99 PE proteins present in the M. tuberculosis reference genome H37Rv (Supplemental Table 1; Poulet and Cole, 1995; Cole et al., 1998; Gey van Pittius et al., 2006). With gc‐contents reaching over 80% in some cases, hydrophobic glycine‐rich repeats and large sizes (up to 1400 amino acids), the PE_PGRS subgroup is the most difficult to study. The rest of the PE proteins cannot be reliably subgrouped in further functional categories based on C‐terminal sequences. However, more information can be obtained from the sequences of the PE domains and their genetic environment (Supplemental Table 1). Gey van Pittius et al. have used sequence phylogeny of the PE proteins to thus classify these into five sublineages (Gey van Pittius et al., 2006). The ancestral pe‐genes encoded within ESX‐1 (PE35) and ESX‐3 (PE5) are defined as sublineage I and II, respectively. Sublineage III may be the most loosely defined sublineage and includes the esx2‐localized PE36 as well as the ESX‐5 substrate PE25. Sublineage IV is a group of ESX‐5 secreted PE proteins, whose genes are often transcribed as bicistronic transcripts together with PPE‐SVP’s (discussed below) (Gey Van Pittius et al., 2001; 2006). Sublineage V includes all PE_PGRS proteins, but also a number of proteins with unique C‐terminal domains, including putative hydrolases, lipases and cutinases, with LipY as a best‐known example (Supplemental Table 1; Deb et al., 2006; Daleke et al., 2011; Burggraaf et al., 2019). A number of other PE proteins are putative hydrolases (PE1, PE3, PE4 & PE16) based on structural homology searches by Phyre2 software (Three closest hits with 99% confidence: PDB 3AJA, 1QOZ & 3HC7; Berman et al., 2000; Payne et al., 2009). This hydrolase‐like fold is annotated as the PF08237 domain, which was originally named the ‘PE/PPE domain’ by Adindla and Guruprasad (Adindla and Guruprasad, 2003; Gey van Pittius et al., 2006; Kelley and Sternberg, 2009; Finn et al., 2014). The PFAM08237 domain is also found in the PPE proteins PPE28, PPE42 and PPE63, as well as non‐TypeVII substrate proteins (Gey van Pittius et al., 2006; Finn et al., 2014), indicating that these putative enzymatic domains may have become TypeVII substrates by genetic recombinations with pe and ppe genes. It should be noted that PE2, which also contains PFAM08237, is likely not an actual PE protein, since it does not contain any of the PE, YxxxD/E or helix‐turn‐helix hallmarks (Supplemental Table 1; Cole et al., 1998; Kelley and Sternberg, 2009). Interestingly, the lipase LipY is a PE protein in M. tuberculosis, while it is a PPE protein in M. marinum. Together, the identification of these ESX‐5‐secreted lipases and putative hydrolases and cutinases suggests that such TypeVII‐secreted PE and PPE proteins may be involved in the biogenesis and homeostasis of the cell envelope, or in the uptake of lipid‐based nutrients (Ates et al., 2015; Bosserman and Champion, 2017).
The PPE protein can be subdivided in even clearer subgroups (Fig. 3). Besides the ESX‐1‐specific (PPE68) and ESX‐2‐specific (PPE69) PPE’s, the first clearly defined subgroup is that of the PPE‐PPW proteins. These can be distinguished by a PxxPxxW amino acid motif generally placed between 10 and 30 amino acids from their C‐terminus. M. tuberculosis H37Rv contains 10 PPW proteins (Supplemental Table 2), although PPE48 and PPE67 are truncated proteins that may not be functional (Cole et al., 1998; Gey van Pittius et al., 2006; Kapopoulou et al., 2011).
Figure 3.

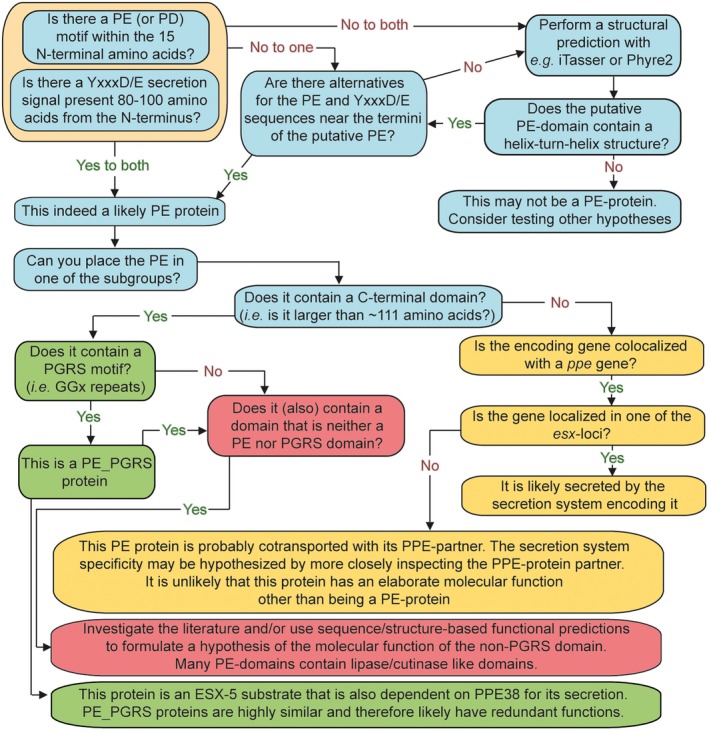
A proposed flow chart to aid researchers to identify and classify PPE proteins in order to formulate testable hypotheses. When interested in a PPE gene or protein, a first step can be to verify it is indeed a functional PPE protein, by investigating presence of the defining characteristics (Start at the left top of the flow chart). A following step is to classify the PPE‐protein as a PPE‐PPW (Green), PPE‐SVP (Red), PPE‐MPTR (Yellow), or other (blue), based on its C‐terminal domains (or lack thereof).
With 26 members, the largest subgroup of PPE proteins are the PPE‐SVP proteins, which are named for the conserved SVP (Serine‐Valine‐Proline) amino acid sequence present in their C‐terminal domain (PFAM: PF12484). PPE50 is likely a truncated protein and PPE9 is not a true PPE‐SVP, because it consists of only a PPE domain of 180 amino acids, but was classified as a PPE‐SVP protein based on phylogenetic analysis (Gey van Pittius et al., 2006). The remaining 24 PPE's are all between 350 and 468 amino acids in size.
Finally, the most recently evolved subgroup of PPE proteins is that of the Major Polymorphic Tandem Repeat (MPTR)‐containing PPE‐MPTR proteins (Hermans et al., 1992; Cole et al., 1998; Gey van Pittius et al., 2006). The MPTR‐repeat consists of repeats that vary around a NxGxGNxG motif. While some PPE‐MPTR proteins are moderately sized, members of this group have unusually large sizes for secreted proteins, ranging up to 3716 amino acids (PPE56; Cole et al., 1998; Gey van Pittius et al., 2006; Kapopoulou et al., 2011).
Secretion and functions of specific PE and PPE protein subgroups
Subdividing PE and PPE proteins as described above (Figs 2 and 3) can help researchers to systematically formulate hypotheses and approaches for experiments. First of all, while generally all pe and ppe genes are excluded from bioinformatic datasets, this approach is overly stringent in most cases. Even short‐read sequencing techniques can reliably map almost all pe and ppe genes thanks to paired‐end technologies and increased read lengths, if only pe_pgrs and ppe‐mptr genes/transcripts are excluded (Miran and Farhat – personal communication, Holt et al., 2018; Ates et al., 2018a; Walter et al., 2019). Furthermore, knowing the subgroup of a PE/PPE can be an excellent starting point to hypothesize the most‐likely route of secretion and may even suggest functions, or redundancy. It should be noted that even within the subgroups discussed below, individual proteins may behave markedly different from the other members of their groups. Therefore, the subgrouping may help to provide testable hypotheses, but these should always be validated experimentally before strong conclusions can be drawn.
Figure 2.
A proposed flow chart to aid researchers to identify and classify PE‐proteins and to formulate testable hypotheses. When interested in a PE gene or protein, a first step can be to verify that it is indeed a functional PE protein, by investigating presence of the defining characteristics (Start at the left top of the flow chart). A following step is to classify the PE‐protein based on its C‐terminal domains (or lack thereof) and genomic localization.
The best‐studied member of the PPE‐PPW proteins is PPE4, which is genetically encoded within the ESX‐3 genetic locus and is likely secreted by this secretion system, together with its secretion partner PE5 (Korotkova et al., 2014; Tufariello et al., 2016). These proteins are important for mycobactin‐mediated iron acquisition (Siegrist et al., 2009; Tufariello et al., 2016). The PPE‐PPW PPE20 and its cognate partner PE15 were experimentally identified as ESX‐3 substrates (Tufariello et al., 2016). Two other PPW proteins, PPE36 and PPE37, are also described to play a role in iron homeostasis through functions in heme‐iron acquisition (Mitra et al., 2017; Tullius et al., 2019). Together with sequence analysis of the EspG‐binding domain, these data render the hypothesis that PPE‐PPW proteins may be generally dependent on ESX‐3, rather than ESX‐5 for their translocation, increasingly likely (Abdallah et al., 2009; Korotkova et al., 2014; Tufariello et al., 2016).
There is strong experimental evidence that all PPE‐SVP proteins are secreted by ESX‐5 (Abdallah et al., 2009; Bottai et al., 2012; Sayes et al., 2012; Ates et al., 2015). PPE‐SVP proteins are often transcribed as bicistronic transcripts with a sublineage IV PE‐protein and in many cases also two Esx‐proteins (Gey Van Pittius et al., 2001; 2006). These operonic clusters are likely to be the first pe and ppe genes duplicated from the esx genetic loci after introduction of the ESX‐5 system into the most‐recent common ancestor of the slow‐growing mycobacteria (Gey van Pittius et al., 2006; Fishbein et al., 2015; Dumas et al., 2016). The PE and PPE proteins encoded by such operons are thought to be exclusively secreted as PE and PPE heterodimers that may not be promiscuous to other secretion partners (Ekiert and Cox, 2014; Korotkova et al., 2014; Chen et al., 2017; Phan et al., 2017; Phan and Houben, 2018). A notable example of such operonic organization is the PE8‐PPE15‐EsxI‐EsxJ operon, which was shown to facilitate protein secretion of other TypeVII and non‐TypeVII substrates (Shah et al., 2015). Similarly, the PE13‐PPE18‐EsxK‐EsxL is another well‐studied operon, of which PPE18 represents an important antigen component of the M72/AS01E subunit vaccine candidate (Meeren et al., 2018). Another important PPE‐SVP protein is PPE38, which is required for the secretion of all detected PE_PGRS and PPE‐MPTR proteins, which will be discussed in detail below (Ates et al., 2018b; 2018c).
The PPE‐MPTR proteins are secreted by ESX‐5 (Abdallah et al., 2009) and are furthermore dependent on PPE38 for their secretion (Ates et al., 2018b). Loss‐of‐function mutations in the ppe38 locus render many clinical M. tuberculosis isolates and other MTBC members unable to secrete these proteins and may therefore provide information on these groups as a whole (Ates et al., 2018b; 2018c). An important function in capsule integrity of M. marinum was shown for the ancestral PPE‐MPTR protein PPE10 (Gey van Pittius et al., 2006; Ates et al., 2016b), although this phenotype was not apparent in M. tuberculosis (Ates et al., 2018c). PPE62 is reported to be a surface accessible heme‐binding protein, which is not strictly required for virulence of M. tuberculosis (Mitra et al., 2017). The PPE‐MPTR protein PPE42 is an immunogenic protein that is part of the ID93 subunit vaccine candidate (Bertholet et al., 2008; 2010; Baldwin et al., 2015).
The secretion of all PE‐proteins with available data, except those encoded within the ESX‐1, ESX‐2 and ESX‐3 loci and those associated with PPE‐PPW proteins, is thought to be mediated by the ESX‐5 secretion system (Abdallah et al., 2009; Sayes et al., 2012, 2016; 2018; Houben et al., 2012b; Ates et al., 2015; 2018c). The esx5‐encoded PE19 is of general interest since it has been postulated to play a role in phosphate homeostasis and stress resistance (Ramakrishnan et al., 2015), and it is part of the ppe25‐pe19 gene cluster that attenuates M. tuberculosis when deleted in a preclinical vaccine candidate (Bottai et al., 2012; Sayes et al., 2012).
A significant amount of research has been dedicated to the largest group of the PE_PGRS proteins, in part because of the availability of a monoclonal antibody that recognizes the repetitive PGRS domain (Abdallah et al., 2009). Similar to the PPE‐MPTR proteins, PE_PGRS are not only dependent on ESX‐5 for their secretion, but also on the presence of a functional PPE38 protein (Ates et al., 2018b). It is not yet known whether this is true only for PE_PGRS proteins, or also for other sublineage‐V PE‐proteins, that is, PPE38 required for secretion of the PGRS/MPTR domains specifically? Or, is it required for all sublineage‐V PE and PPE’s by for instance functioning as the partner in heterodimeric secretion complexes that can interact with the chaperone EspG5 (Daleke et al., 2012b; Phan et al., 2017)? While this latter possibility was the major hypothesis upon identification of PPE38 as an essential factor for PE_PGRS secretion, it is hard to reconcile this hypothesis with the finding that PPE38 is also required for PPE‐MPTR secretion (Ates et al., 2018b).
Many virulent M. tuberculosis strains, such as the modern L2 lineage, do not secrete PE_PGRS and PPE‐MPTR proteins, because of loss‐of‐function mutations in the ppe38 genetic locus (Ates et al., 2018b; 2018c; Fig. 4). From these findings it can be postulated that secretion of neither PE_PGRS nor PPE‐MPTR proteins is essential for virulence of M. tuberculosis in humans. In fact, loss of secretion of these proteins actually increases virulence of M. tuberculosis (Ates et al., 2018b). The latter observation is bolstered by the finding that a M. marinum mutant with a disrupted espG5 gene is hypervirulent in adult zebrafish (Weerdenburg et al., 2012). Intriguingly, the increased virulence of M. tuberculosis ppe38 mutants in mice is only apparent in later stages of infection (3–6 weeks post infection) and M. marinum espG5::tn was similarly not hypervirulent in zebrafish embryos (Weerdenburg et al., 2012; Ates et al., 2018b). Therefore, this hypervirulence phenotype could be related to interactions with the host’s adaptive immunity or bacterial adaptation at later stages of infection. Importantly, genetic perturbations that completely abrogate ESX‐5 secretion are lethal to slow growing mycobacteria (Di Luca et al., 2012; Ates et al., 2015). The absence of in vitro growth defects and the increased virulence of M. tuberculosis with ppe38 deletions suggests that substrates that are responsible for the essentiality of ESX‐5 likely do not belong to the PE_PGRS or PPE‐MPTR subgroups (Ates et al., 2018b). Finally, it should be noted that only one particular transposon insertion site in the 5′ of espG5 was tolerated by M. marinum. Likely, because this results in a truncated EspG5 that allows some residual ESX‐5 secretion (Ates et al., 2015). Full deletions, or transposon insertion in the middle of espG5, were only possible in the context of expression of MspA, which circumvents essentiality of ESX‐5 in M. marinum (Ates et al., 2015). Care should be taken to extrapolate these findings to M. tuberculosis, because EspG5 may have a more redundant role in M. tuberculosis compared to M. marinum (Bottai et al., 2012).
Figure 4.
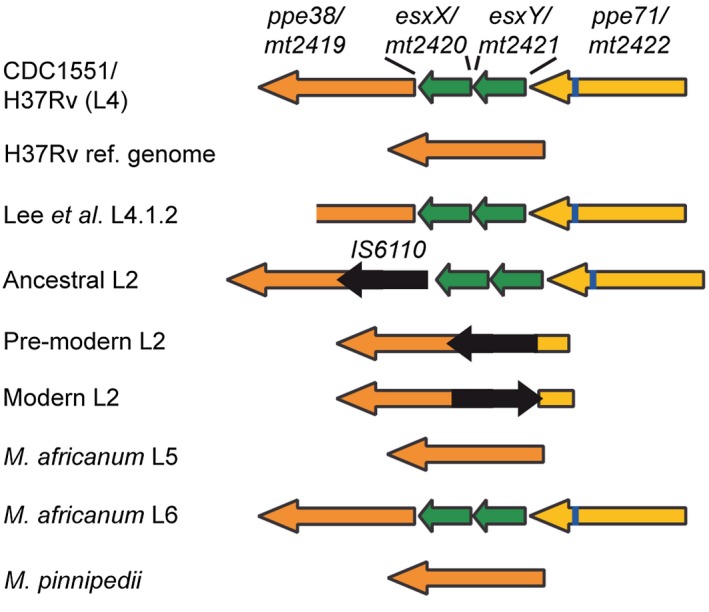
A selection of existing configurations of the ppe38‐71 locus. Gene numbers correspond to the M. tuberculosis CDC1551 reference genome. ppe38 (orange) and ppe71 (yellow) are genetically identical except for a 21bp deletion (blue) in ppe71. The two esx‐genes (green) have been named esxX and esxY based on standardized nomenclature (Bitter et al., 2009; Ates et al., 2018b). The most common configuration of the locus is depicted at the top (McEvoy et al., 2009a). The H37Rv reference genome (second line) is misannotated as having only one copy of ppe38, but multiple H37Rv strains were shown to possess the CDC1551‐like locus (Box 1; McEvoy et al., 2009a; Ates et al., 2018b). However, the configuration of a single copy of either ppe38, or ppe71, does occur due to recombination in diverse clinical isolates and is referred to as RvD7 (McEvoy et al., 2009a). An RD5‐like deletion that truncates ppe38, but keeps ppe71 intact, was found in certain L4 isolates. This polymorphism did not negatively affect PPE38‐dependent secretion (Lee et al., 2015; Ates et al., 2018c). Similarly, in an ancestral L2‐isolate (SAWC_2088) an IS6110 (black) insertion truncation ppe38 does not negatively affect PPE‐38‐dependent secretion. Subsequent recombination that occurred at the branching point of the modern L2‐lineages, have also truncated ppe71 and deleted esxXY, resulting in a loss of PPE38‐dependent secretion (McEvoy et al., 2009a; Ates et al., 2018b). It should be emphasized that many more configurations of the ppe38‐71 locus, including different RD5‐like deletions have been more thoroughly described in McEvoy et al., 2009b (McEvoy et al., 2009a).
Confusingly, the recent data reporting increased virulence in strains that do not secrete PE_PGRS and PPE‐MPTR proteins are in stark contrast with numerous previous studies reporting PE_PGRS as important virulence factors that perform a plethora of functions related to host–pathogen interaction. These proposed functions include inhibition of apoptosis or antigen presentation, induction of host‐cell death and modulation of immune responses by binding to Toll‐like receptors (reviewed in: Sampson, 2011; Fishbein et al., 2015; Delogu et al., 2017). Many studies on PE_PGRS proteins used expression of these proteins in M. smegmatis as a model, which usually lead to increased persistence of the bacteria in macrophage or mouse models. It is hard to envisage the biological relevance of this model, since M. smegmatis does not possess the appropriate chaperones, secretion partners or secretion system to express PE_PGRS proteins in soluble form and secrete them. A promising new approach that could overcome this obstacle is the recent success in expression of a functional ESX‐5 system from M. xenopi in M. smegmatis, which enabled resolution of the first TypeVII secretion system structure by cryo‐electron microscopy. This heterologously expressed ESX‐5 system was able to facilitate secretion of the PPE‐SVP protein PPE18 as well as EsxN in M. smegmatis (Beckham et al., 2017). No secretion of PE_PGRS proteins has been reported in this model, which is also not expected to occur, since no functional PPE38 is present in this system. However, such a model has great potential to build a ‘bottom‐up’ model of the secretion requirements of different classes of PE and PPE proteins.
Other studies, performed in arguably more appropriate models, such as M. tuberculosis or M. marinum, have also reported roles for PE_PGRS proteins as virulence factors. For instance, Saini et al. reported that PE_PGRS47 suppresses autophagy and impairs antigen presentation (Saini et al., 2016). Similarly, PE_PGRS30 was reported to be important for M. tuberculosis virulence in mice, especially in chronic infection (Iantomasi et al., 2012). Another interesting study identified the M. marinum‐specific PE_PGRS protein MMAR_0242 as an important virulence factor that was dependent on the unique C‐terminal domain and not the PGRS domain itself (Singh et al., 2016). These high‐quality studies represent only a fraction of the available literature on the various roles of PE_PGRS (and also PPE‐MPTR) proteins in virulence, which is reviewed more extensively by Delogu et al., (2017), Fishbein et al. (2015) and Sampson (2011). In contrast with the available literature, we have not found differences in dendritic cell activation, antigen presentation or cytokine production in response to short‐term infection with M. tuberculosis or M. bovis BCG strains with or without functional PPE38‐dependent PE_PGRS/PPE‐MPTR secretion (Ates et al., 2018c). This may however be dependent on the dynamics of infections in different infection models, which may vary in their response depending on host species and cell type utilized. Furthermore, the finding that many virulent clinical isolates do not secrete these proteins does not directly suggest an important role as virulence factors at the host–pathogen interface for these proteins (Ates et al., 2018a; 2018b; 2018c). However, a major difference between these studies is the contrast between blocking secretion of the complete PE_PGRS/PPE‐MPTR groups, while leaving expression of the proteins untouched, versus creating genetic knock‐outs of single pe_pgrs genes. These conflicting data from different approaches could suggest roles for the PE_PGRS proteins that are not dependent on their protein secretion and are therefore likely independent of their direct interaction with the host. Alternatively, deletion of single PE_PGRS proteins could have indirect effects on other ESX‐5 substrates that may explain certain virulence effects. Together, these exciting and seemingly contradictory findings create a puzzling tension in the field that begs for a unifying explanation which integrates these findings. Together, recent work has provided new roads forward that have the potential to finally bring more insight into the actual function of PE_PGRS and PPE‐MPTR proteins (Ates et al., 2015; 2018b; Beckham et al., 2017).
The potential and pitfalls of PE and PPE proteins as vaccine components
PE and PPE proteins contain many immunogenic epitopes and are therefore of great interest for the development of new tuberculosis vaccines (Brennan, 2017). The PGRS domains of PE_PGRS proteins possess only a few predicted immunogenic epitopes, especially compared to their more antigenic PE domains (McEvoy et al., 2012; Copin et al., 2014). However, especially the PPE proteins contain many predicted and experimentally validated antigenic epitopes. Important examples of immunogenic PPE proteins include the PPE‐SVP protein PPE18, which is part of M72/AS01E subunit vaccine candidate (Meeren et al., 2018) and the PPE‐MPTR protein PPE42, which is a part of the subunit vaccine ID93 (Bertholet et al., 2008; 2010; Baldwin et al., 2015). Furthermore, immune responses to PE and PPE proteins also play a large role in designing whole‐cell vaccines. For instance, the preclinical vaccine candidate M. tuberculosis Δppe25‐pe19 is attenuated by a partial deletion of the esx‐5 genetic locus (Bottai et al., 2012; Sayes et al., 2012). Despite this deletion, ESX‐5 secretion remains functional, leading to cross immune recognition of PE and PPE domains from orthologous proteins that are still secreted, thereby minimizing the reduction in the antigenic repertoire of this vaccine strain compared to wildtype (Sayes et al., 2012; 2016). In contrast, this immune recognition is completely lost when blocking ESX‐5 secretion by deleting the ESX‐5 membrane components eccC5 or eccD5 (Sayes et al., 2012; 2016), because secretion through the cognate TypeVII protein secretion system is essential for antigen processing, presentation and recognition by immune cells (Sayes et al., 2018).
Importantly, the BCG vaccine strain does not secrete PE_PGRS and PPE‐MPTR proteins due to deletion of the ppe38‐containing RD5‐genetic region in the ancestral M. bovis isolate (McEvoy et al., 2009a; Ates et al., 2018c). This secretion defect can likely be generalized to an inability of BCG to induce immune responses against all PE‐PGRS and PPE‐MPTR proteins, which was experimentally confirmed for the ancestral PPE‐MPTR PPE10. Although complementation of BCG with the ppe38‐71 genetic region restored PE_PGRS secretion and immune recognition of PPE10, no improved (or reduced) protection of this recombinant strain was seen in two different mouse models (Ates et al., 2018c). In spite of this disappointing effect, when faced with a choice it would seem preferable to induce the widest antigenic recognition possible. In that light, attenuated M. tuberculosis vaccine candidates, such as the promising MTBVAC (Arbues et al., 2013; Tameris et al., 2019), have the advantage over recombinant BCG that they will usually contain functional PPE38‐dependent secretion.
It is also important to consider the genetic and phenotypic variability within M. tuberculosis that can lead to differing PE and PPE recognition. The pe and ppe genes are among the most genetically variable genes within the M. tuberculosis genome and this variation should therefore be taken into account, especially when subunit vaccines are rolled out in patient populations (McEvoy et al., 2009a; 2012; Copin et al., 2014; Phelan et al., 2016). A prime example for this is the PPE18 component of the M72/AS01E vaccine candidate, for which Homolka et al. described 28 non‐synonymous single‐nucleotide polymorphisms, three deletions and one insertion in a panel of 71 MTBC strains (Homolka et al., 2016). Furthermore, expression patterns of PE and PPE proteins may not be constant during infection, which may influence T cell responses and therefore vaccine efficacy (Dheenadhayalan et al., 2006; Goldstone et al., 2009; Moguche et al., 2017).
However, only genetic investigation of epitopes of interest may still underestimate variability, as these proteins need to be secreted to be efficiently processed and presented as antigenic epitopes (Sayes et al., 2018). For instance, the modern L2 (modern Beijing) strains with ppe38 deletions are not expected to be recognized by T cell or antibody immune responses aimed at epitopes from PE_PGRS or PPE‐MPTR proteins (Ates et al., 2018c). Using such epitopes in subunit vaccines could infer increased protection to strains that also secrete these proteins, and conversely protect less against strains that do not secrete these proteins. This could be doubly disadvantageous, since such strains also seem to be more virulent (Hanekom et al., 2011; Ates et al., 2018b). This could be of particular relevance for PPE42, a PPE‐MPTR protein that is part of the ID93 vaccine candidate, which is currently being tested in phase II clinical trials (Johnson et al., 2008; Phase 2a ID93 + GLA‐SE Vaccine Trial in TB Patients After Treatment Completion – Full Text View – ClinicalTrials.gov, n.d.). Although these scenarios are presently purely hypothetical, it is important to design trials and preclinical investigation in ways that take into account the existing diversity of M. tuberculosis. Such measures could include bioinformatic analyses of the expected diversity of epitopes of vaccine candidate proteins within M. tuberculosis strains, preferably supplemented by laboratory tests. For instance, immunogenicity and protection of individual subunit components should be tested against multiple divergent strains of M. tuberculosis. Finally, epidemiologic monitoring of the circulating strain genotypes after introduction of novel vaccines, as is done for other bacterial vaccines, could help to observe bacterial adaptation once a vaccine is implemented (McIntyre et al., 2012; Azarian et al., 2018).
Implications of pe and ppe gene variation for the evolution of Mycobacteria
The finding that pe and ppe genes cover 7–10% of the M. tuberculosis genome suggests an important role for these proteins in the evolution of M. tuberculosis (Cole et al., 1998; Fishbein et al., 2015). This is further substantiated by the finding that the fast‐growing mycobacteria, which include predominantly saprophytic species, only possess the PE and PPE proteins localized in the esx‐loci ESX‐1 and ESX‐3 (Gey van Pittius et al., 2006). Since the slow‐growing mycobacteria, which include most species pathogenic to humans and other animals, have seen remarkable expansion of pe and ppe genes, it is tempting to suggest that this expansion has been part of adaptation to an intracellular and pathogenic lifestyle (Gey van Pittius et al., 2006; Fishbein et al., 2015). In contrast, the major mycobacterial human pathogen, Mycobacterium leprae, only has a small number of remaining pe and ppe genes that have not been subject to reductive evolution (Cole et al., 2001; Gey van Pittius et al., 2006). The only exceptions of PE/PPE proteins present in both M. leprae and M. tuberculosis are PE15‐PPE20 (ML_0538‐9), PE13‐PPE18 (ML_1053 and ML_1182 (an M. leprae specific PPE18 paralog)), PE5 (ML_2534c), PPE1 (ML_1991), PPE2 (ML_1828c) and PPE68 (ML_0051c; Cole et al., 1998; 2001; Gey van Pittius et al., 2006). It could be argued that these widely conserved genes may be most relevant to study conserved aspects of mycobacterial pathogenesis and these may include the substrates that make the ESX‐3 and ESX‐5 systems essential for slow‐growing Mycobacteria (Di Luca et al., 2012; Serafini et al., 2013; 2014; Ates et al., 2015; Tufariello et al., 2016).
Neither the PE_PGRS or PPE‐MPTR proteins, nor PPE38 are present as potentially functional ORFs in the M. leprae genome, thereby indicating that these proteins are not strictly required for a pathogenic lifestyle in humans. This is concordant with the finding that natural ppe38 deletion strains that do not secrete PPE‐MPTR and PE_PGRS proteins are highly virulent in humans. However, a role for these proteins in the co‐evolution with human hosts should not be dismissed based on these data. For instance, the majority of pathogenic M. tuberculosis strains possess functional copies of PPE38 and corresponding PE_PGRS and PPE‐MPTR secretion. This observation, in combination with that the ppe38 locus is among the most variable regions of the M. tuberculosis genome, suggests that there is an evolutionary pressure to retain PPE38‐dependent secretion (McEvoy et al., 2009a; Ates et al., 2018a; 2018b; 2018c). In contrast, a signature of convergent evolution toward loss of the ppe38 locus seems especially apparent in the animal adapted isolates of the MTBC (Fig. 5).
Figure 5.
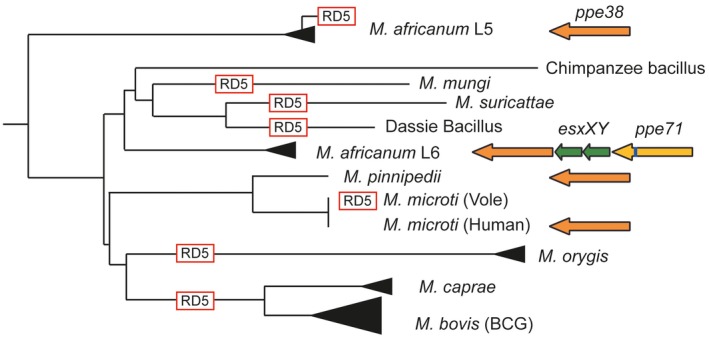
RD5 and ppe38 polymorphisms in M. africanum and the animal‐adapted species of the M. tuberculosis complex. The phylogeny is adapted from Brites et al. with permission of the author (Brites et al., 2018). Independent RD5 deletions, with unique remaining flanking sequences, are depicted in the phylogenetic tree. The organization of the ppe38 locus in M. africanum L5, L6 and M. microti and M. pinnipedii is indicated with arrows, similar as in Fig. 4. Only one out of 18 M. africanum L5 isolates was found to have an RD5‐like deletion (Ates et al., 2018a). In a study by Brodin et al. three M. microti isolates from humans had an intact RD5‐locus, while four strains isolated from voles had RD5‐deletions (Brodin et al., 2002).
These deletions, known as region of difference (RD)5, have occurred independently in M. bovis/M. caprae, M. orygis, M. suricattae and M. mungi, the Dassie Bacillus and a subset of M. microti strains isolated from voles (Brodin et al., 2002; Garnier et al., 2003; Mostowy et al., 2004; Dippenaar et al., 2015; Brites et al., 2018). In fact, the only known animal adapted MTBC members that have retained the RD5 locus are the Chimpanzee bacillus, M. pinnipedii and a subset of M. microti isolates isolated from humans (Brodin et al., 2002; Coscolla et al., 2013; Brites et al., 2018). Similarly, also the closely related human‐adapted M. africanum Lineage six seems to have mostly intact RD5 regions (Fig. 5; Brosch et al., 2002; Brites et al., 2018; Ates et al., 2018a). Together, these evolutionary observations suggest an advantage for the loss of the ppe38 locus only under specific conditions. An interesting hypothesis is that increased virulence resulting from ppe38 deletion may confer an advantage under population‐dense situations, allowing fast spread between individuals and relatively quick progression to disease as has been reported for the modern Beijing isolates (Hanekom et al., 2011; Merker et al., 2015; Ates et al., 2018b). However, in most human populations, these traits may actually be disadvantageous and a more chronic disease may be evolutionary favorable (Brites and Gagneux, 2015). This hypothesis would translate well to the animal adapted isolates, where species infecting small animals with relatively short lifespans such as voles, mongooses and meerkats have developed RD5 deletions (Brodin et al., 2002; Dippenaar et al., 2015; Alexander et al., 2016a). It is also particularly intriguing that these mycobacterial species (but also M. pinnipedii with an intact RD5) have all obtained independent RD1‐like deletions inactivating the ESX‐1 secretion system, which should theoretically lead to complete attenuation (Brodin et al., 2002; Dippenaar et al., 2015; Alexander et al., 2016a; 2016b). Perhaps the route of transmission in these animals is different from the aerosol transmission between humans and other larger animals. This was suggested for M. mungi, which seems to be transmitted via direct contact of mucous tissue in particularly in the nasal and anal areas (Alexander et al., 2016b). The RD1/ESX‐1‐dependent rupture of phagosomal membranes that leads to cellular necrosis and extracellular bacilli may be particularly important for aerosol transmission and may be less important when direct contact between infected and noninfected tissues occurs regularly (van der Wel et al., 2007; Simeone et al., 2012; Houben et al., 2012a; Gröschel et al., 2016; Dallenga et al., 2017). Since the ESX‐1 substrates are highly immunogenic, the potential reduction of virulence could be offset by a reduction in antigenic recognition of the pathogen, especially in this context of reduced importance for transmission. The RD5 deletions in the larger livestock animals such as cattle and goats could potentially be explained by the artificially shortened lifespans due to consumption of these animals. However, it is unclear to what extend this is true for the time in which the most‐recent common ancestor of M. bovis and M. caprae emerged (Loiseau et al., 2019). In line with the hypothesis that RD5‐deletions make M. bovis more virulent, there is experimental evidence that M. bovis is more aggressive in cattle compared to M. tuberculosis (Whelan et al., 2010). Chimpanzees, seals and sea lions may have longer expected lifespans, and this may therefore have formed an advantage for the chimpanzee bacillus and M. pinnipedii to retain ppe38. As intriguing as this hypothesis is, it is purely speculative and based on circumstantial evidence. Although the evidence for increased virulence of modern lineage two isolates and ppe38 deletion mutants is strong in experimental animals (Aguilar et al., 2010; Hanekom et al., 2011; Weerdenburg et al., 2012; Gopal et al., 2014; Ates et al., 2018b), differences are much harder to substantiate in human populations in spite of mounting evidence (Hanekom et al., 2011; Huyen et al., 2013; Merker et al., 2015; Holt et al., 2018). Furthermore, analysis of the full spectrum of genetic variation of ppe38 locus has been hampered by sequencing and mapping difficulties, due to the misannotated reference genome (Box 1, Fig. 4; McEvoy et al., 2009a).
Box 1. Methodological difficulties to identify ppe38 polymorphisms.
The ppe38‐containing genomic locus is one of the most variable regions in the M. tuberculosis genome (McEvoy et al., 2009a). Unfortunately, a large part of this diversity remains currently uncharted, because of an annotation error in the M. tuberculosis H37Rv reference genome (Genbank: AL123456). While the reference genome only contains a single ppe38 gene, most M. tuberculosis genomes contain two copies of the ppe (ppe38 and ppe71, which are identical or differ only by a seven‐amino acid insertion) that together flank two esx genes, esxXY (Fig. 4; McEvoy et al., 2009a; Ates et al., 2018b). Therefore, it may be more informative to use a different reference genome (e.g. the M. tuberculosis CDC1551 reference genome (Genbank: AE000516 – mt2422‐19)) when assessing polymorphisms in this region (McEvoy et al., 2009a; Lee et al., 2015; Ates et al., 2018b). Even then, it may be needed to perform PCR amplification and Sanger sequencing, or long‐read sequencing as a verification, because IS6110 insertions may remain difficult to detect (McEvoy et al., 2009a; 2009b).
The potential of new sequencing techniques and refined sequence analyses
Increased understanding of the actual role of the PE and PPE proteins have played in the evolution of the MTBC may come from more detailed molecular studies identifying the mechanisms of their secretion and their interacting partners. Another approach may be to more accurately map the diversity in pe and ppe genes present in the global M. tuberculosis isolates with emerging new long read sequencing techniques, but also by more careful analyses. These may include only discarding those pe and ppe genes which are not reliably sequenced (e.g. a curated subset of the pe_pgrs and ppe/mptr genes), but may also be aided by using more refined reference genomes and mapping algorithms (Maciuca et al., 2016). Refining bioinformatic analysis methods to include pe and ppe genes may allow to re‐analyze thousands of published genome datasets and therefore may uncover exciting new information on M. tuberculosis (Bradley et al., 2015; The CRyPTIC Consortium and the 100.00 Genomes Project, 2018). For instance, it may be plausible to uncover polymorphisms in pe and ppe genes that affect antibiotic sensitivity, since PE and PPE proteins are involved in membrane permeability and nutrient uptake (Ates et al., 2015; Tufariello et al., 2016; Mitra et al., 2017). Furthermore, such technical advances are needed to perform more large‐scale studies on the evolutionary pressures exerted on pe and ppe genes, such as by studying dN/dS rates in different domains/subgroups of pe and ppe genes or in predicted antigenic epitope sequences (McEvoy et al., 2012; Copin et al., 2014). A very interesting open question is whether loss of ppe38 leads to increased/reduced selection pressure on pe_pgrs/ppe‐mptr genes, since these protein products are then no longer expected to be under selection pressure from immune recognition.
Such approaches may be especially powerful when combined with laboratory and clinical parameters to link genotypes to particular phenotypes. For instance, we recently found a new sublineage of M. africanum lineage 5 that was deficient in PE_PGRS and PPE‐MPTR secretion while have a seemingly intact ppe38 locus (Ates et al., 2018a). This phenotype was not complemented by introduction of the ppe38‐71 locus, suggesting that another variation is responsible for this phenotype. However, the development of novel tools such as specific antibodies to PPE38 and PPE‐MPTR proteins may be required to gain more biochemical insight in such cases.
Conclusions
The PE and PPE proteins have puzzled researchers for decades and it is unlikely that their biological roles will be completely elucidated in the coming decade. However, big advances in our knowledge on these proteins have still been achieved recently and ongoing methodological advances may provide further progress. It is of paramount importance to be aware of the associated methodological difficulties when studying these proteins, such as redundancy, sequence complexity, interdependency of substrates and essentiality of secretion systems. Similarly, being aware of the different subgroups of PE/PPE proteins is an essential step toward elucidation of the functions of these proteins, since the most diverse and unique molecular features of individual proteins may be found in the variable C‐terminal domains. Some PE/PPE proteins may be best studies as individuals, for example, single gene knock‐out and complementation studies. In contrast, the PE_PGRS and PPE‐MPTR may in cases be more aptly studied as a subgroup, because of their high similarity and putative redundancy. Such groupings may be especially valuable for bioinformatic studies, since variation and selection pressures may differ remarkably between different subgroups of pe and ppe genes and grouping all of these genes together may lead to a loss of such signals. In summary, big advances in our knowledge on these proteins have been achieved recently and ongoing methodological advances may provide further progress. If the molecular biology of these intriguing, but sometimes confusing proteins is taken into account, this will lead to a better understanding of the biology of the most successful human pathogen of our times.
Conflict of interest
The author declares no financial conflict of interest.
Funding information
Louis S Ates is supported by a PostDoc stipend of the Amsterdam Infection and Immunity Institute and by the Netherlands Organisation for Scientific Research (VIDI grant 91717305 to Jeroen WJ van Heijst).
Supporting information
Acknowledgements
The author would like to thank Matthias Gröschel for proofreading and providing feedback on the manuscript. I am grateful to Coen Kuijl, Edith Houben, Roland Brosch and Wilbert Bitter for insightful discussions regarding the topics discussed herein, as well as for their continued mentoring support.
References
- Abdallah, A.M. , Verboom, T. , Hannes, F. , Safi, M. , Strong, M. , Eisenberg, D. , et al (2006) A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Molecular Microbiology, 62, 667–679. [DOI] [PubMed] [Google Scholar]
- Abdallah, A.M. , Gey van Pittius, N.C. , Champion, P.A.D. , Cox, J. , Luirink, J. , Vandenbroucke‐Grauls, C.M.J.E. , et al (2007) Type VII secretion – mycobacteria show the way. Nature Reviews Microbiology, 5, 883–891. [DOI] [PubMed] [Google Scholar]
- Abdallah, A.M. , Verboom, T. , Weerdenburg, E.M. , Gey van Pittius, N.C. , Mahasha, P.W. , Jiménez, C. , et al (2009) PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX‐5. Molecular Microbiology, 73, 329–340. [DOI] [PubMed] [Google Scholar]
- Adindla, S. and Guruprasad, L. (2003) Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. Journal of Biosciences, 28, 169–179. [DOI] [PubMed] [Google Scholar]
- Aguilar, D. , Hanekom, M. , Mata, D. , Gey van Pittius, N.C. , van Helden, P.D. , Warren, R.M. , et al (2010) Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis (Edinb), 90, 319–325. [DOI] [PubMed] [Google Scholar]
- Alexander, K.A. , Larsen, M.H. , Robbe‐Austerman, S. , Stuber, T.P. and Camp, P.M. (2016a) Draft genome sequence of the Mycobacterium tuberculosis complex pathogen M. mungi, identified in a banded mongoose (Mungos mungo) in Northern botswana. Genome Announcements, 4, e00471–e00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, K.A. , Sanderson, C.E. , Larsen, M.H. , Robbe‐Austerman, S. , Williams, M.C. and Palmer, M.V. (2016b) Emerging tuberculosis pathogen hijacks social communication behavior in the group‐living banded mongoose (Mungos mungo ). MBio, 7, e00281–e00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. , Aly, K.A. , Chen, Y.‐H. and Missiakas, D. (2013) Secretion of atypical protein subtrates by the ESAT‐6 secretion system of Staphylococcus aureus . Molecular Microbiology, 90, 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbues, A. , Aguilo, J.I. , Gonzalo‐Asensio, J. , Marinova, D. , Uranga, S. , Puentes, E. , et al (2013) Construction, characterization and preclinical evaluation of MTBVAC, the first live‐attenuated M. tuberculosis‐based vaccine to enter clinical trials. Vaccine, 31, 4867–4873. [DOI] [PubMed] [Google Scholar]
- Ates, L.S. , Ummels, R. , Commandeur, S. , van der Weerd, R. , Sparrius, M. , Weerdenburg, E. , et al (2015) Essential role of the ESX‐5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genetics, 11, e1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates, L.S. , Houben, E.N.G. and Bitter, W . (2016a) Type VII secretion: A highly versatile secretion system. Microbiolspec, 4 10.1128/microbiolspec.VMBF-0011-2015. [DOI] [PubMed] [Google Scholar]
- Ates, L.S. , van der Woude, A.D. , Bestebroer, J. , van Stempvoort, G. , Musters, R.J.P. , Garcia‐Vallejo, J.J. , et al (2016b) The ESX‐5 system of pathogenic mycobacteria is involved in capsule integrity and virulence through its substrate PPE10. PLoS Pathogens, 12, e1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates, L.S. , Dippenaar, A. , Sayes, F. , Pawlik, A. , Bouchier, C. , Ma, L. , et al (2018a) Unexpected genomic and phenotypic diversity of mycobacterium africanum lineage 5 affects drug resistance, protein secretion, and immunogenicity. Genome Biology and Evolution, 10, 1858–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates, L.S. , Dippenaar, A. , Ummels, R. , Piersma, S.R. , van der Woude, A.D. , van der Kuij, K. , et al (2018b) Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis . Nature Microbiology, 3, 181–188. [DOI] [PubMed] [Google Scholar]
- Ates, L.S. , Sayes, F. , Frigui, W. , Ummels, R. , Damen, M.P.M. , Bottai, D. , et al (2018c) RD5‐mediated lack of PE_PGRS and PPE‐MPTR export in BCG vaccine strains results in strong reduction of antigenic repertoire but little impact on protection. PLoS Pathogens, 14, e1007139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarian, T. , Mitchell, P.K. , Georgieva, M. , Thompson, C.M. , Ghouila, A. , Pollard, A.J. , et al (2018) Global emergence and population dynamics of divergent serotype 3 CC180 pneumococci. PLoS Pathogens, 14, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, S.L. , Reese, V.A. , Huang, P.D. , Beebe, E.A. , Podell, B.K. , Reed, S.G. , et al (2015) Protection and long‐lived immunity induced by the ID93/GLA‐SE vaccine candidate against a clinical Mycobacterium tuberculosis isolate. Clinical and Vaccine Immunology, 23, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banu, S. , Honoré, N. , Saint‐Joanis, B. , Philpott, D. , Prévost, M.‐C. and Cole, S.T. (2002) Are the PE‐PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Molecular Microbiology, 44, 9–19. [DOI] [PubMed] [Google Scholar]
- Beckham, K.S.H. , Ciccarelli, L. , Bunduc, C.M. , Mertens, H.D.T. , Ummels, R. , Lugmayr, W. , et al (2017) Structure of the mycobacterial ESX‐5 type VII secretion system membrane complex by single particle analysis. Nature Microbiology, 2, 1–8. [DOI] [PubMed] [Google Scholar]
- Berman, H.M. , Westbrook, J.D. , Feng, Z. , Gilliland, G.L. , Bhat, T.N. , Weissig, H. , et al (2000) The protein data bank. Nucleic Acids Research, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet, S. , Ireton, G.C. , Kahn, M. , Guderian, J. , Mohamath, R. , Stride, N. , et al (2008) Identification of human T cell antigens for the development of vaccines against Mycobacterium tuberculosis . The Journal of Immunology, 181, 7948–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet, S. , Ireton, G.C. , Ordway, D.J. , Windish, H.P. , Pine, S.O. , Kahn, M. , et al (2010) A defined tuberculosis vaccine candidate boosts BCG and protects against multidrug‐resistant Mycobacterium tuberculosis . Science Translational Medicine, 2, 53–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter, W. , Houben, E.N.G. , Bottai, D. , Brodin, P. , Brown, E.J. , Cox, J.S. , et al (2009) Systematic genetic nomenclature for type VII secretion systems. PLoS Pathogens, 5, e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosserman, R.E. and Champion, P.A. (2017) Esx systems and the mycobacterial cell envelope: what’s the connection? Journal of Bacteriology, 199, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottai, D. , Luca, M.Di , Majlessi, L. , Frigui, W. , Simeone, R. , Sayes, F. , et al (2012) Disruption of the ESX‐5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Molecular Microbiology, 83, 1195–1209. [DOI] [PubMed] [Google Scholar]
- Bradley, P. , Gordon, N.C. , Walker, T.M. , Dunn, L. , Heys, S. , Huang, B. , et al (2015) Rapid antibiotic‐resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis . Nature Communications, 6, 10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, M.J. (2017) The enigmatic PE/PPE multi‐gene family of Mycobacteria and TB vaccination. Infection and Immunity, 85, e00969‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, M.J. and Delogu, G. (2002) The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends in Microbiology, 10, 246–249. [DOI] [PubMed] [Google Scholar]
- Brites, D. and Gagneux, S. (2015) Co‐evolution of Mycobacterium tuberculosis and Homo sapiens . Immunological Reviews, 264, 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites, D. , Loiseau, C. , Menardo, F. , Borrell, S. , Boniotti, M.B. , Warren, R. , et al (2018) A new phylogenetic framework for the animal‐adapted Mycobacterium tuberculosis complex. Frontiers in Microbiology, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin, P. , Eiglmeier, K. , Marmiesse, M. , Billault, A. , Garnier, T. , Niemann, S. , et al (2002) Bacterial artificial chromosome‐based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT‐6 deletion mutant. Infection and Immunity, 70, 5568–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch, R. , Gordon, S.V. , Marmiesse, M. , Brodin, P. , Buchrieser, C. , Eiglmeier, K. , et al (2002) A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proceedings of the National Academy of Sciences, 99, 3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunduc, C.M. , Ummels, R. , Bitter, W. , and Houben, E.N.G . (2019) Species‐specific secretion of ESX‐5 type VII substrates is determined by the linker 2 of EccC5. bioRxiv 765206. 10.1101/765206v1. [DOI] [PMC free article] [PubMed]
- Burggraaf, M.J. , Ates, L.S. , Speer, A. , van der Kuij, K. , Kuijl, C. and Bitter, W. (2019) Optimization of secretion and surface localization of heterologous OVA protein in mycobacteria by using LipY as a carrier. Microbial Cell Factories, 18, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Z. , Casabona, M.G. , Kneuper, H. , Chalmers, J.D. and Palmer, T. (2016) The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nature Microbiology, 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion, P.A.D. , Stanley, S.A. , Champion, M.M. , Brown, E.J. and Cox, J.S. (2006) C‐terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis . Science, 313, 1632–1636. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Cheng, H.‐F. , Zhou, J. , Chan, C.‐Y. , Lau, K.‐F. , Tsui, S.K.‐W. , et al (2017) Structural basis of the PE‐PPE protein interaction in Mycobacterium tuberculosis . Journal of Biological Chemistry, 292, 16880–16890. 10.1074/jbc.M117.802645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S.T. , Brosch, R. , Parkhill, J. , Garnier, T. , Churcher, C. , Harris, D. et al (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Cole, S.T. , Eiglmeier, K. , Parkhill, J. , James, K.D. , Thomson, N.R. , Wheeler, P.R. , et al (2001) Massive gene decay in the leprosy bacillus. Nature, 409, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Coll, F. , McNerney, R. , Guerra‐Assunção, J.A. , Glynn, J.R. , Perdigão, J. , Viveiros, M. , et al (2014) A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nature Communications, 5, 4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin, R. , Coscollá, M. , Seiffert, S.N. , Bothamley, G. , Sutherland, J. , Mbayo, G. , et al (2014) Sequence diversity in the pe_pgrs genes of Mycobacterium tuberculosis is independent of human T cell recognition. MBio, 5, e00960‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscolla, M. , Lewin, A. , Metzger, S. , Maetz‐Rennsing, K. , Calvignac‐Spencer, S. , Nitsche, A. , et al (2013) Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerging Infectious Diseases, 19, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, M.H. , Cascioferro, A. , de Punder, K. , Ummels, R. , Abdallah, A.M. , van der Wel, N. , et al (2011) Conserved Pro‐Glu (PE) and Pro‐Pro‐Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX‐5 pathway. Journal of Biological Chemistry, 286, 19024–19034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, M.H. , Ummels, R. , Bawono, P. , Heringa, J. , Vandenbroucke‐Grauls, C.M.J.E. , Luirink, J. , et al (2012a) General secretion signal for the mycobacterial type VII secretion pathway. Proceedings of the National Academy of Sciences of the United States of America, 109, 11342–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, M.H. , Van Der Woude, A.D. , Parret, A.H.A. , Ummels, R. , De Groot, A.M. , Watson, D. , et al (2012b) Specific chaperones for the type VII protein secretion pathway. Journal of Biological Chemistry, 287, 31939–31947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallenga, T. , Repnik, U. , Corleis, B. , Eich, J. , Reimer, R. , Griffiths, G.W. , et al (2017) M. tuberculosis‐induced necrosis of infected neutrophils promotes bacterial growth following phagocytosis by macrophages. Cell Host & Microbe, 22, 519–530.e3. [DOI] [PubMed] [Google Scholar]
- Danilchanka, O. , Sun, J. , Pavlenok, M. , Maueröder, C. , Speer, A. , Siroy, A. , et al (2014) An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proceedings of the National Academy of Sciences of the United States of America, 111, 6750–6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb, C. , Daniel, J. , Sirakova, T.D. , Abomoelak, B. , Dubey, V.S. and Kolattukudy, P.E. (2006) A novel lipase belonging to the hormone‐sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis . Journal of Biological Chemistry, 281, 3866–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu, G. , Brennan, M.J. and Manganelli, R. (2017) PE and PPE genes: a tale of conservation and diversity. Advances in Experimental Medicine and Biology, 1019, 191–207. [DOI] [PubMed] [Google Scholar]
- Dheenadhayalan, V. , Delogu, G. , Sanguinetti, M. , Fadda, G. and Brennan, M.J. (2006) Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. Journal of Bacteriology, 188, 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippenaar, A. , Parsons, S.D.C. , Sampson, S.L. , van der Merwe, R.G. , Drewe, J.A. , Abdallah, A.M. , et al (2015) Whole genome sequence analysis of Mycobacterium suricattae . Tuberculosis (Edinb), 95, 682–688. [DOI] [PubMed] [Google Scholar]
- Dumas, E. , Boritsch, E.C. , Vandenbogaert, M. , De La Vega, R.C.R. , Thiberge, J.M. , Caro, V. , et al (2016) Mycobacterial pan‐genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biology and Evolution, 8, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert, D.C. and Cox, J.S. (2014) Structure of a PE‐PPE‐EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proceedings of the National Academy of Sciences of the United States of America, 111, 14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famelis, N. , Rivera‐Calzada, A. , Degliesposti, G. , Wingender, M. , Mietrach, N. , Skehel, J.M. , et al (2019) Architecture of the mycobacterial type VII secretion system. Nature, 1, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D. , Bateman, A. , Clements, J. , Coggill, P. , Eberhardt, R.Y. , Eddy, S.R. , et al (2014) Pfam: the protein families database. Nucleic Acids Research, 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein, S. , Wyk, N. , Warren, R.M. and Sampson, S.L. (2015) Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Molecular Microbiology, 96, 901–916. [DOI] [PubMed] [Google Scholar]
- Garnier, T. , Eiglmeier, K. , Camus, J.‐C.J.‐C. , Medina, N. , Mansoor, H. , Pryor, M. , et al (2003) The complete genome sequence of Mycobacterium bovis . Proceedings of the National Academy of Sciences of the United States of America, 100, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey Van Pittius, N.C. , Gamieldien, J. , Hide, W. , Brown, G.D. , Siezen, R.J. and Beyers, A.D. (2001) The ESAT‐6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram‐positive bacteria. Genome Biology, 2, RESEARCH0044. https://genomebiology.biomedcentral.com/articles/10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius, N.C. , Sampson, S.L. , Lee, H. , Kim, Y. , van Helden, P.D. and Warren, R.M. (2006) Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT‐6 (esx) gene cluster regions. BMC Evolutionary Biology, 15, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone, R.M. , Goonesekera, S.D. , Bloom, B.R. and Sampson, S.L. (2009) The transcriptional regulator Rv0485 modulates the expression of a pe and ppe gene pair and is required for Mycobacterium tuberculosis virulence. Infection and Immunity, 77, 4654–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, R. , Monin, L. , Slight, S. , Uche, U. , Blanchard, E. , Fallert Junecko, A. , et al (2014) Unexpected role for IL‐17 in protective immunity against hypervirulent Mycobacterium tuberculosis HN878 infection. PLoS Pathogens, 10, e1004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S.V. , Brosch, R. , Billault, A. , Garnier, T. , Eiglmeier, K. and Cole, S.T. (1999) Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Molecular Microbiology, 32, 643–655. [DOI] [PubMed] [Google Scholar]
- Gröschel, M.I. , Sayes, F. , Simeone, R. , Majlessi, L. and Brosch, R. (2016) ESX secretion systems: mycobacterial evolution to counter host immunity. Nature Reviews Microbiology, 14, 677–691. [DOI] [PubMed] [Google Scholar]
- Hanekom, M. , Gey van Pittius, N.C. , McEvoy, C. , Victor, T.C. , Van Helden, P.D. and Warren, R.M. (2011) Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb), 91, 510–523. [DOI] [PubMed] [Google Scholar]
- Hermans, P.W. , van Soolingen, D. and van Embden, J.D. (1992) Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae . Journal of Bacteriology, 174, 4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, K.E. , McAdam, P. , Thai, P.V.K. , Thuong, N.T.T. , Ha, D.T.M. , Lan, N.N. , et al (2018) Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nature Genetics, 50, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka, S. , Ubben, T. and Niemann, S. (2016) High sequence variability of the PPE18 gene of clinical Mycobacterium tuberculosis complex strains potentially impacts effectivity of vaccine candidate M72/AS01E. PLoS ONE, 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, D. , Demangel, C. , van Ingen, J. , Perez, J. , Baldeón, L. , Abdallah, A.M. , et al (2012a) ESX‐1‐mediated translocation to the cytosol controls virulence of mycobacteria. Cellular Microbiology, 14, 1287–1298. [DOI] [PubMed] [Google Scholar]
- Houben, E.N.G. , Bestebroer, J. , Ummels, R. , Wilson, L. , Piersma, S.R. , Jiménez, C.R. , et al (2012b) Composition of the type VII secretion system membrane complex. Molecular Microbiology, 86, 472–484. [DOI] [PubMed] [Google Scholar]
- Huyen, M.N.T. , Buu, T.N. , Tiemersma, E. , Lan, N.T.N. , Dung, N.H. , Kremer, K. , et al (2013) Tuberculosis relapse in vietnam is significantly associated with Mycobacterium tuberculosis beijing genotype infections. Journal of Infectious Diseases, 207, 1516–1524. [DOI] [PubMed] [Google Scholar]
- Iantomasi, R. , Sali, M. , Cascioferro, A. , Palucci, I. , Zumbo, A. , Soldini, S. , et al (2012) PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis . Cellular Microbiology, 14, 356–367. [DOI] [PubMed] [Google Scholar]
- Johnson, M. , Zaretskaya, I. , Raytselis, Y. , Merezhuk, Y. , McGinnis, S. and Madden, T.L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Research, 36, W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapopoulou, A. , Lew, J.M. and Cole, S.T. (2011) The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb), 91, 8–13. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J.E. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols, 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Korotkova, N. , Freire, D. , Phan, T.H. , Ummels, R. , Creekmore, C.C. , Evans, T.J. , et al (2014) Structure of the Mycobacterium tuberculosis type VII secretion system chaperone EspG5 in complex with PE25‐PPE41 dimer. Molecular Microbiology, 94, 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.S. , Radomski, N. , Proulx, J.‐F. , Levade, I. , Shapiro, B.J. , McIntosh, F. et al (2015) Population genomics of Mycobacterium tuberculosis in the Inuit. Proceedings of the National Academy of Sciences, 2000, 201507071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau, C. , Menardo, F. , Aseffa, A. , Hailu, E. , Gumi, B. , Ameni, G. , et al (2019) An African origin for Mycobacterium bovis . bioRxiv. https://www.biorxiv.org/content/10.1101/773192v1. [DOI] [PMC free article] [PubMed]
- Lou, Y. , Rybniker, J. , Sala, C. and Cole, S.T. (2017) EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX‐1 secretion. Molecular Microbiology, 103, 26–38. [DOI] [PubMed] [Google Scholar]
- Luca, M.Di , Bottai, D. , Batoni, G. , Orgeur, M. , Aulicino, A. , Counoupas, C. , et al (2012) The ESX‐5 associated eccB‐EccC locus is essential for Mycobacterium tuberculosis viability. PLoS ONE, 7, e52059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciuca, S. , Elias, C.D.O. , McVean, G. and Iqbal, Z. (2016) A natural encoding of genetic variation in a burrows‐wheeler transform to enable mapping and genome inference In: Frith M., Storm Pedersen C. (Eds.) Algorithms in Bioinformatics. WABI 2016. Lecture Notes in Computer Science. Cham: Springer, Vol. 9838. [Google Scholar]
- McEvoy, C.R.E. , van Helden, P.D. , Warren, R.M. and Gey van Pittius, N.C. (2009a) Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evolutionary Biology, 9, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy, C.R.E. , Warren, R.M. , van Helden, P.D. and Gey van Pittius, N.C. (2009b) Multiple, independent, identical IS6110 insertions in Mycobacterium tuberculosis PPE genes. Tuberculosis (Edinb), 89, 439–442. [DOI] [PubMed] [Google Scholar]
- McEvoy, C.R.E. , Cloete, R. , Müller, B. , Schürch, A.C. , van Helden, P.D. , Gagneux, S. , et al (2012) Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints. PLoS ONE, 7, e30593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre, P.B. , O’Brien, K.L. , Greenwood, B. and Van De Beek, D . (2012) Effect of vaccines on bacterial meningitis worldwide. Lancet, 380, 1703–1711. [DOI] [PubMed] [Google Scholar]
- Meehan, C.J. , Goig, G.A. , Kohl, T.A. , Verboven, L. , Dippenaar, A. , Ezewudo, M. , et al (2019) Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nature Reviews Microbiology, 17, 533–545. [DOI] [PubMed] [Google Scholar]
- Merker, M. , Blin, C. , Mona, S. , Duforet‐Frebourg, N. , Lecher, S. , Willery, E. , et al (2015) Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nature Genetics, 47, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A. , Speer, A. , Lin, K. , Ehrt, S. and Niederweis, M. (2017) PPE surface proteins are required for heme utilization by Mycobacterium tuberculosis . MBio, 8, e01720‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moguche, A.O. , Musvosvi, M. , Penn‐Nicholson, A. , Plumlee, C.R. , Mearns, H. , Geldenhuys, H. , et al (2017) Antigen availability shapes T cell differentiation and function during tuberculosis. Cell Host & Microbe, 21, 695–706.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy, S. , Cousins, D. and Behr, M.A. (2004) Genomic interrogation of the dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. Journal of Bacteriology, 186, 104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S. , Lavu, E. , Coulter, C. , Duarte, T. , Bainomugisa, A. , Marais, B.J. , et al (2018) A complete high‐quality MinION nanopore assembly of an extensively drug‐resistant Mycobacterium tuberculosis Beijing lineage strain identifies novel variation in repetitive PE/PPE gene regions. Microbial Genomics, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, K. , Sun, Q. , Sacchettini, J. and Hatfull, G.F. (2009) Mycobacteriophage Lysin B is a novel mycolylarabinogalactan esterase. Molecular Microbiology, 73, 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T.H. and Houben, E.N.G. (2018) Bacterial secretion chaperones: the mycobacterial type VII case. FEMS Microbiology Letters, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T.H. , Ummels, R. , Bitter, W. and Houben, E.N.G. (2017) Identification of a substrate domain that determines system specificity in mycobacterial type VII secretion systems. Scientific Reports, 7, 42704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T.H. , van Leeuwen, L.M. , Kuijl, C. , Ummels, R. , van Stempvoort, G. , Rubio‐Canalejas, A. , et al (2018) EspH is a hypervirulence factor for Mycobacterium marinum and essential for the secretion of the ESX‐1 substrates EspE and EspF. PLoS Pathogens, 14, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phase 2a ID93 + GLA‐SE Vaccine Trial in TB Patients After Treatment Completion – Full Text View – ClinicalTrials.gov . https://clinicaltrials.gov/ct2/show/NCT02465216. [Accessed 21 September, 2017].
- Phelan, J.E. , Coll, F. , Bergval, I. , Anthony, R.M. , Warren, R. , Sampson, S.L. , et al (2016) Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages. BMC Genomics, 17, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet, S. and Cole, S.T. (1995) Characterization of the highly abundant polymorphic GC‐rich‐repetitive sequence (PGRS) present in Mycobacterium tuberculosis . Archives of Microbiology, 163, 87–95. [DOI] [PubMed] [Google Scholar]
- Poulsen, C. , Panjikar, S. , Holton, S.J. , Wilmanns, M. and Song, Y.‐H. (2014) WXG100 protein superfamily consists of three subfamilies and exhibits an α‐helical C‐terminal conserved residue pattern. PLoS ONE, 9, e89313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poweleit, N. , Czudnochowski, N. , Nakagawa, R. , Murphy, K. , Sassetti, C. and Rosenberg, O.S . (2019) A large inner membrane pore defines the ESX translocon. bioRxiv https://www.biorxiv.org/content/10.1101/800169v1.
- Ramakrishnan, P. , Aagesen, A.M. , McKinney, J.D. and Tischler, A.D. (2015) Mycobacterium tuberculosis resists stress by regulating PE19 expression. Infection and Immunity, 84, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw, P.S. , Lightbody, K.L. , Veverka, V. , Muskett, F.W. , Kelly, G. , Frenkiel, T.A. , et al (2005) Structure and function of the complex formed by the tuberculosis virulence factors CFP‐10 and ESAT‐6. EMBO Journal, 24, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, J.G. , Pino, C. , Tauch, A. and Murcia, M.I. (2015) Complete genome sequence of the clinical Beijing‐like strain Mycobacterium tuberculosis 323 using the PacBio real‐time sequencing platform. Genome Announcements, 3, 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, O.S. , Dovala, D. , Li, X. , Connolly, L. , Bendebury, A. , Finer‐Moore, J. , et al (2015) Substrates control multimerization and activation of the multi‐domain ATPase motor of Type VII secretion. Cell, 161, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybniker, J. , Chen, J.M. , Sala, C. , Hartkoorn, R.C. , Vocat, A. , Benjak, A. , et al (2014) Anticytolytic screen identifies inhibitors of mycobacterial virulence protein secretion. Cell Host & Microbe, 16, 538–548. [DOI] [PubMed] [Google Scholar]
- Saini, N.K. , Baena, A. , Ng, T.W. , Venkataswamy, M.M. , Kennedy, S.C. , Kunnath‐Velayudhan, S. , et al (2016) Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nature Microbiology, 1, 16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, S.L. (2011) Mycobacterial PE/PPE proteins at the host‐pathogen interface. Clinical and Developmental Immunology, 2011, 497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani, M. , Houben, E.N.G. , Geurtsen, J. , Pierson, J. , de Punder, K. , van Zon, M. , et al (2010) Direct visualization by cryo‐EM of the mycobacterial capsular layer: a labile structure containing ESX‐1‐secreted proteins. PLoS Pathogens, 6, e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayes, F. , Sun, L. , Luca, M.Di , Simeone, R. , Degaiffier, N. , Fiette, L. et al (2012) Strong immunogenicity and cross‐reactivity of Mycobacterium tuberculosis ESX‐5 type VII secretion: encoded PE‐PPE proteins predicts vaccine potential. Cell Host & Microbe, 11, 352–363. [DOI] [PubMed] [Google Scholar]
- Sayes, F. , Pawlik, A. , Frigui, W. , Gröschel, M.I. , Crommelynck, S. , Fayolle, C. , et al (2016) CD4+ T cells recognizing PE/PPE antigens directly or via cross reactivity are protective against pulmonary Mycobacterium tuberculosis infection. PLoS Pathogens, 12, e1005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayes, F. , Blanc, C. , Ates, L.S.L.S. , Deboosere, N. , Orgeur, M. , Le Chevalier, F. , et al (2018) Multiplexed quantitation of intraphagocyte Mycobacterium tuberculosis secreted protein effectors. Cell Reports, 23, 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, O.T. , Mouritsen, J. , Ludwig, C. , Röst, H.L. , Rosenberger, G. , Arthur, P.K. , et al (2013) The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis . Cell Host & Microbe, 13, 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini, A. , Pisu, D. , Palù, G. , Rodriguez, G.M. and Manganelli, R. (2013) The ESX‐3 secretion system is necessary for iron and Zinc homeostasis in Mycobacterium tuberculosis . PLoS ONE, 8, e78351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, S. , Cannon, J.R. , Fenselau, C. and Briken, V. (2015) A duplicated ESAT‐6 region of ESX‐5 is involved in protein export and virulence of mycobacteria. Infection and Immunity, 83, 4349–4361. 10.1128/IAI.00827-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist, M.S. , Unnikrishnan, M. , McConnell, M.J. , Borowsky, M. , Cheng, T.‐Y. , Siddiqi, N. , et al (2009) Mycobacterial Esx‐3 is required for mycobactin‐mediated iron acquisition. Proceedings of the National Academy of Sciences of the United States of America, 106, 18792–18797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist, M.S. , Steigedal, M. , Ahmad, R. , Mehra, A. , Dragset, M.S. , Schuster, B.M. , et al (2014) Mycobacterial Esx‐3 requires multiple components for iron acquisition. MBio, 5, e01073‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, R. , Bobard, A. , Lippmann, J. , Bitter, W. , Majlessi, L. , Brosch, R. and Enninga, J. (2012) Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathogens, 8, e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V.K. , Berry, L. , Bernut, A. , Singh, S. , Carrère‐Kremer, S. , Viljoen, A. , et al (2016) A unique PE_PGRS protein inhibiting host cell cytosolic defenses and sustaining full virulence of Mycobacterium marinum in multiple hosts. Cellular Microbiology, 18, 1489–1507. [DOI] [PubMed] [Google Scholar]
- Solomonson, M. , Setiaputra, D. , Makepeace, K.A.T. , Lameignere, E. , Petrotchenko, E.V. , Conrady, D.G. , et al (2015) Structure of EspB from the ESX‐1 Type VII secretion system and insights into its export mechanism. Structure, 23, 571–583. [DOI] [PubMed] [Google Scholar]
- Strong, M. , Sawaya, M.R. , Wang, S. , Phillips, M. , Cascio, D. and Eisenberg, D. (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis . Proceedings of the National Academy of Sciences of the United States of America, 103, 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy, R. , Fyfe, P.K. and Hunter, W.N. (2008) Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. Journal of Molecular Biology, 383, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameris, M. , Mearns, H. , Penn‐nicholson, A. , Gregg, Y. , Bilek, N. , Mabwe, S. , et al (2019) Articles Live‐attenuated Mycobacterium tuberculosis vaccine MTBVAC versus BCG in adults and neonates: a randomised controlled, double‐blind dose‐escalation trial. Lancet Respiratory Medicine, 2600, 1–14. [DOI] [PubMed] [Google Scholar]
- The CRyPTIC Consortium and the 100.000 Genomes Project . (2018) Prediction of susceptibility to first‐line tuberculosis drugs by DNA sequencing. New England Journal of Medicine, 379, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufariello, J.M. , Chapman, J.R. , Kerantzas, C.A. , Wong, K.‐W. , Vilchèze, C. , Jones, C.M. , et al (2016) Separable roles for Mycobacterium tuberculosis ESX‐3 effectors in iron acquisition and virulence. Proceedings of the National Academy of Sciences of the United States of America, 113, E348–E357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius, M.V. , Nava, S. and Horwitz, M.A. (2019) PPE37 is essential for Mycobacterium tuberculosis heme‐iron acquisition (HIA), and a defective PPE37 in Mycobacterium bovis BCG prevents HIA. Infection and Immunity, 87, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Meeren, O. , Hatherill, M. , Nduba, V. , Wilkinson, R.J. , Muyoyeta, M. , Van Brakel, E. , et al (2018) Phase 2b controlled trial of M72/AS01E vaccine to prevent tuberculosis. New England Journal of Medicine, 379, 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, J.M. , Evans, T.J. and Korotkov, K.V. (2014) Crystal structure of the N‐terminal domain of EccA₁ ATPase from the ESX‐1 secretion system of Mycobacterium tuberculosis . Proteins, 82, 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, K.S. , Colijn, C. , Cohen, T. , Mathema, B. , Liu, Q. , Bowers, J. ,et al (2019) Genomic variant identification methods alter Mycobacterium tuberculosis transmission inference. bioRxiv, 10.1101/733642. [DOI] [PMC free article] [PubMed]
- Weerdenburg, E.M. , Abdallah, A.M. , Mitra, S. , de Punder, K. , van der Wel, N.N. , Bird, S. , et al (2012) ESX‐5‐deficient Mycobacterium marinum is hypervirulent in adult zebrafish. Cellular Microbiology, 14, 728–739. [DOI] [PubMed] [Google Scholar]
- van der Wel, N. , Hava, D. , Houben, D. , Fluitsma, D. , van Zon, M. , Pierson, J. , et al (2007) M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell, 129, 1287–1298. [DOI] [PubMed] [Google Scholar]
- van der Woude, A.D. , Mahendran, K.R. , Ummels, R. , Piersma, S.R. , Pham, T.V. , Jiménez, C.R. , et al (2013) Differential detergent extraction of Mycobacterium marinum cell envelope proteins identifies an extensively modified threonine‐rich outer membrane protein with channel activity. Journal of Bacteriology, 195, 2050–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, A.O. , Coad, M. , Cockle, P.J. , Hewinson, G. , Vordermeier, M. and Gordon, S.V. (2010) Revisiting host preference in the Mycobacterium tuberculosis complex: experimental infection shows M. tuberculosis H37Rv to be avirulent in cattle. PLoS ONE, 5, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J.C. , Peterson, S.B. , Kim, J. , Pazos, M. , Verster, A.J. , Radey, M.C. , et al (2017) A broadly distributed toxin family mediates contact‐dependent antagonism between gram‐positive bacteria. elife, 6, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Winden, V.J.C. , Ummels, R. , Piersma, S.R. , Jiménez, C.R. , Korotkov, K.V. , Bitter, W. , et al (2016) Mycosins are required for the stabilization of the ESX‐1 and ESX‐5 type VII secretion membrane complexes. MBio, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


