Abstract
Incremental proofs demonstrate that miRNAs, the essential regulators of gene expression, are implicated in various biological procedures, including mammary development and milk synthesis. Here, the role of miR‐574‐5p in milk synthesis, apoptosis, and proliferation of goat mammary epithelial cells (GMECs) are explored without precedent, and the molecular mechanisms for the impacts are elucidated. Small RNA libraries were constructed using GMECs transfected with miR‐574‐5p mimics and negative control followed by sequencing via Solexa technology. Overall, 332 genes were distinguishingly expressed entre two libraries, with 74 genes upregulated and 258 genes downregulated. This approach revealed mitogen‐activated protein kinase kinase kinase 9 (MAP3K9), an upstream activator of MAPK signaling, as a differentially expressed unigene. miR‐574‐5p targeted seed sequences of the MAP3K9 3′‐untranslated region and suppressed its messenger RNA (mRNA) and protein levels, correspondingly. GMECs with miR‐574‐5p overexpression and MAP3K9 inhibition showed increased cell apoptosis and decreased cell proliferation resulting from sustained suppression of MAPK pathways, while MAP3K9 elevation manifested the opposite results. miR‐574‐5p repressed the phosphorylation of members of protein kinase B (AKT)–mammalian target of rapamycin pathway via downregulating MAP3K9 and AKT3, resulting in reducing the secretion of β‐casein and triglycerides in GMECs. Finally, according to the constructed circular RNA (circRNA) libraries and bioinformatics prediction approach, we selected circ‐016910 and found it acted as a sponge for miR‐574‐5p and blocked its relevant behaviors to undertake biological effects in GMECs. The circRNA–miRNA–mRNA network facilitates further probes on the function of miR‐574‐5p in mammary development and milk synthesis.
Keywords: AKT3, circRNA, MAPK, miRNA, transcriptome
miR‐574‐5p inactive c‐Jun N‐terminal kinase/extracellular‐signal‐regulated kinase pathways resulting in decreased cell proliferation, enhanced cell apoptosis and weakens milk synthesis by protein kinase B–mammalian target of rapamycin pathway via mitogen‐activated protein kinase kinase kinase 9 and AKT3 in goat mammary epithelial cells, meanwhile, circ‐016910 acts as a sponge for miR‐574‐5p and shows a circRNA–miRNA–mRNA network to undertake biological effects.
1. INTRODUCTION
miRNAs are a series of noncoding single‐stranded endogenous RNAs with 22 nucleotides (nts) in length approximately, identified as important posttranscriptional regulators (Shukla, Singh, & Barik, 2011). Generally, miRNAs modulate gene expression via binding to the 3′‐untranslated region (3′‐UTR) of their target genes (Eulalio, Huntzinger, & Izaurralde, 2008), eliciting translation blockage and/or messenger RNA (mRNA) degradation (Liang, 2009). miRNAs have been reported to function in diversiform vital biological processes, including cell development, differentiation (Martin et al., 2016), proliferation (Shiah et al., 2014), apoptosis (Ambros, 2003), and milk performance (H. M. Li, Wang, Li, & Gao, 2012). Due to our previous work, we selected miR‐574‐5p for further research based on its significant effect on mammary gland development and lactation performance after a comparative analysis of our sequence data with published mammary gland transcriptome data during colostrum and common milk stages (Hou et al., 2017). Previous studies about miR‐574‐5p have specialized in its antioncogenic and antimetastatic activities, (Cui, Tang, Chen, & Wang, 2014; Ji et al., 2013; Q. Li et al., 2012; Zhou et al., 2016)), whereas, the effect of miR‐574‐5p remains relatively uncharacterized in goat mammary epithelial cells (GMECs). Circular RNAs (circRNAs) are new noncoding transcripts that are observed in eukaryotic cells by electron microscopy 40 years ago (Jeck & Sharpless, 2014). According to bioinformatics and high‐throughput sequencing, circRNAs have been sequentially identified in numerous cell lines and varieties of species (Memczak et al., 2013). Vary from unidimensional RNAs, circRNAs are basically formed by exon or intron back‐splicing (Salzman, Gawad, Wang, Lacayo, & Brown, 2012). Exonic circRNAs are structured in a heat‐to‐tail closed continuous loop with the 3′ RNA and 5′ RNA joined covalently. Recently studies report circRNAs as “miRNA sponges” that take suppressive effects on miRNA regulation (Guo, Agarwal, Guo, & Bartel, 2014). However, the function of circRNAs as “miRNA sponges” are not extensively understood in GMECs.
Mitogen‐activated protein kinases (MAPKs) are a cluster of serine/threonine kinases protein and modulate signal transduction cascades. They regulate cell biology on account of their involvement in important cell physiology, such as cell proliferation, cell apoptosis (Slattery, Lundgreen, & Wolff, 2012), and milk synthesis (Lu, Li, Huang, & Gao, 2013). Each MAPK pathway is a three‐tiered cascade that includes MAP kinase (MAPK), MAPK kinase (MAP2K), and MAPKK kinase (MAP3K; Cargnello & Roux, 2011). Many research have furnished important evidence revealing that MAP3K9 are connected with noteworthy cell biology. MAP3K9 suppresses proliferation and apoptosis in renal cancer cells by MAPK/c‐Jun N‐terminal kinase (JNK) signaling (Nie et al., 2016). Interference of MAP3K9 inhibits the motivation of extracellular‐signal‐regulated kinase (ERK) and p38 MAPK pathways, which further inhibits proliferation and promotes apoptosis of NP cells (Cai et al., 2017). Attenuation of MAP3K9 inhibits OS cell proliferation and metastasis, which shows stem cell properties (Zhao et al., 2015). Overexpression of MAP3K9 mutant depresses phosphorylation of downstream MAPKs in HEK293T cells, while knockdown MAP3K9 function using siRNA brings about enhanced melanoma cell viability (Stark et al., 2011). Whereas, it remains indistinct whether MAP3K9 is capable of taking effects on GMECs. AKT3/PKBγ is a member of the AKT/PKB family in mammalian cells, the other two closely correlative isoforms are AKT1/PKBα and AKT2/PKBβ (Stahl et al., 2004). AKTs are second messenger‐regulated serine/threonine protein kinases and refer to many vital biological processes, such as cell apoptosis, cell proliferation, and milk synthesis (LoPiccolo, Blumenthal, Bernstein, & Dennis, 2008).
The RNA sequencing (RNA‐Seq) technology provides us a high‐throughput deep‐sequencing platform that detects the functional complexity of transcriptomes with low abundance, verifies and uncovers the discrepant expression of novel transcriptional regions among tested samples (Grabherr et al., 2011). The integral observation of transcriptomes and its community furnished using the sequencing technology manifests numerous newfangled transcript units, single‐nucleotide polymorphisms, and splice isoforms and allows purification of gene mechanics (He & Liu, 2013). Compared with a microarray procedure, which is restricted by the number of probes on the array, the RNA‐Seq method analyzes the expression of genes in tested samples (Trapnell, Pachter, & Salzberg, 2009).
Herein, we framed the transcribed outline of GMECs at an overall level and made a comparison of the genes expressed in GMECs transfected with miR‐574‐5p mimics and NC. The use of this strategy allowed us to select MAP3K9 for further studies, owing to its involvement in the lactation mechanism. Then we selected a circRNA to explore the “miRNA sponges” function based on constructed circRNA libraries in our laboratory via bioinformatics prediction method and built a circRNA–miRNA–mRNA network. The purpose of the hodiernal study was devised to examine the effects and molecular mechanisms of miR‐574‐5p in GMECs.
2. MATERIALS AND METHODS
2.1. Ethics statement
The animal use and care protocol were confirmed by the Review Committee for the Use of Animal Subjects of Northwest A&F University, Yang Ling, China. All surgeries were framed with a struggle to minimize martyrdom to animals.
2.2. Animals and cell culture
The elite herd of dairy goats used in the study was from a local farm in Northwest A&F University of China. Mammary glands were collected and stored in phosphate‐buffered saline (PBS) supplemented with 100 μg/ml streptomycin and 100 μg/ml penicillin and then were transferred to the laboratory within 1 hr. The GMECs were seeded in Dulbecco's modified Eagle media/F12 medium (Gibco, CA) containing 10% fetal bovine serum, 10 ng/ml epidermal growth factor 1 (Gibco), 5 mg/ml insulin, 100 U/ml streptomycin/ml penicillin, and 0.3 mmol/L hydrocortisone at 37°C in a humidified atmosphere with 5% CO2. GMECs were fractionated and cultured according to previous reports (Chen et al., 2017).
2.3. pcDNA3.1‐MAP3K9/AKT3 vector construction
Goat MAP3K9 CDS sequence (XM_018054019.1) and AKT3 CDS sequence (XM_018060259.1) were extended by polymerase chain reaction (PCR) using total RNA extracted from GMECs and cloned into pMD™19‐T vector (TaKaRa, Beijing, China) and entirely sequenced. The MAP3K9 and AKT3 fragments were then subcloned into the multiple cloning sites of the pcDNA3.1 vector (Thermo Fisher Scientific, Shanghai, China) between the Xho I and Kpn I sites, and the mechanics were appraised by DNA sequencing.
The forward primer of MAP3K9: Kpn I 5′‐CGGGTACCATGGAGCCCTCCAGAGCACTT‐3′ and reverse primer: Xho I 5′‐CGCTCGAGCTAAGACCAGAACTCCTGCTGGA‐3′. The forward primer of AKT3: Kpn I 5′‐CTGGGTACCATGTCGGAGAAAAAATATGTATC GGAAAAGATTGC‐3′, and reverse primer: Xho I 5′‐CGCTCGAGTTATTCTCGTCCACTT GCAGAGTAGG‐3′.
2.4. pcD2.1‐circ‐016910 vector construction
Goat circ‐016910 sequence was extended by PCR and cloned into pMD™19‐T vector then subcloned into the multiple cloning sites of the pcD2.1‐cir vector (Geneseed Biotech, Guangzhou, China) between the Kpn I and Bamh I sites. The forward primer of circ‐016910: Kpn I 5′‐GGGGTACCATGATGACTTTGACCAGTTTGATAAGCAG‐3′ and reverse primer: Bamh I 5′‐CGGGATCCCGACGTAAGGCAGAGAGCAAC‐3′.
2.5. Transfection and RNA extraction
GMECs were seeded in six‐well plates at 3 × 105 cells/cm2 until cell density reached 80–90%. GMECs were transfected with NC, inhNC, miR‐574‐5p mimics/inhibitors, siRNA‐MAP3K9/AKT3/circ‐016910 (GenePharma, Shanghai, China), pcDNA3.1, pcDNA3.1‐MAP3K9/AKT3 and pcD2.1‐circ‐016910 plasmids using Lipofectamine™ RNAiMAX Reagent (Invitrogen, CA) based on the manufacturer's specifications. Briefly, 2 μl miR‐574‐5p mimics/inhibitors or siRNA‐MAP3K9/AKT3/circ‐016910 with 5 μl RNAiMAX, 4 μg pcDNA3.1 or pcDNA3.1‐MAP3K9/AKT3 or pcD2.1‐ciR or pcD2.1‐circ‐016910 plasmids with 7.5 μl RNAiMAX, 2 μl miR‐574‐5p mimics and 2 μg pcDNA3.1‐MAP3K9/AKT3 or pcD2.1‐circ‐016910 with 7.5 μl RNAiMAX were diluted using 250 μl Gibco Opti‐MEM I medium and the complexes were cultivated at room temperature for 20 min. Cells were harvested for further examination after 24 and 48 hr transfection. Total RNA was isolated using TRIzol reagent (Invitrogen). The RNA concentration and purity were evaluated using Agilent 2100 Bioanalyzer (Agilent Technologies, CA) and the RNA was stored at −80°C before use. Figure S1 exhibited expressions of miR‐574‐5p/circ‐016910 and MAP3K9/AKT3 in mRNA and protein levels after transfection.
2.6. Transcriptome sequencing
2.6.1. Library construction and sequencing
Generation sequencing library preparations of MC and NC groups were constructed on the basis of the manufacturer's specifications (NEBNext® Ultra™ RNA Library Prep Kit for Illumina®) after RNA purity and integrity were measured (Table S1). Briefly, the poly(A) mRNA extraction was implemented via NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB). The mRNA was interrupted to short sequences (~300 bp) after adding the fragmentation buffer, then the first‐strand cDNA was synthesized by NEBNext Random Primers and ProtoScript II Reverse Transcriptase by the mRNA fragments as templates. The second‐strand synthesis was conducted by incubation with dNTPs, RNase H and DNA polymerase I while second‐strand primer and strand synthesis enzyme Mix were added. The resulting double‐stranded cDNA fragments were subsequently treated with End Prep Enzyme Mix to end repair after purified with AxyPrep Mag PCR Clean‐up (Axygen). Then a dA‐tailing and T‐A ligation were added to both ends, followed by sequencing adapters roped to the sequences. The essential fragments were purified using AxyPrep Mag PCR Clean‐up and enriched by PCR. The library production was used for the ultimate sequencing reaction.
2.7. Mapping reads on the goat reference genome and data analysis
After the initial image was taken, the data were transformed into sequence data. High‐throughput sequencing of the pooled RNA was processed and analyzed in GENEWIZ, Beijing, China. Genome websites (UCSC, ENSEMBL, and NCBI) provided reference genome sequences and gene model commentary records of proportional species. The reference genome sequence was indexed and aligned clean data by Hisat2 (v2.0.1). Asprofile v1.0, a range of procedures for isolating, quantifying, and contrasting alternative splicing items of sequencing data, took a GTF transcript file created by Cufflinks as its input. SAMtools v0.1.18 with command mpileup and BCFtools v0.1.19 were used to do SNV calling. Cuffcompare (Dai et al., 2013) assembly was used to know transcripts and predict novel transcripts after assembling a transcriptome from one or more samples.
2.8. Analysis of differentially expressed genes
The mapped reads counting for each gene were standardized for RNA length and for the entire read number in the lane draw on reads per kilobase of exon model per million mapped reads (RPKM), which accelerates the parabole of transcript levels among samples (Smyth, 2004). The expression abundance for gene i was calculated by the RPKM approach: RPKMi = 109 × Ci/(Ni × Li), where i denotes the gene index, Ci represents the sum of short read calculates mapped to exons and exon–exon junctions, Ni is whole mapped read calculates in the lane, and Li refers to the sum of exon lengths (Tarazona et al., 2015). HTSeq (v0.6.1) was used to assess gene and isoform expression levels from pair‐end clean data with the file as a reference gene file. The differentially expressed genes (DEGs) were determined using DESeq Bioconductor package, a model on the basis of the passive binomial distribution. p Value of genes was set less than .05 to explore diverse expressed genes after corrected by Benjamini and Hochberg's measure for possessing the inaccurate discovery rate (Grabherr et al., 2011).
2.9. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis of DEGs
Gene ontology (GO) is a comprehensive criterion gene functional category scheme (Tweedie et al., 2009). The DEGs were itemized into the categories of biological process, cellular component and molecular function by the GO annotation. The hypergeometric detection was demanded to match all DEGs to terms in the GO database (http://www.geneontology.org/) (Camon et al., 2004) and to inquiry for remarkably enriched GO terms in DEGs via in comparison them of the genome background. GO terms were identified using GO‐Term Finder that note on a series of enriched genes with a remarkable p < .05. The p value formula was as follows:
where N represents the number of all genes with GO annotation; n refers to the number of DEGs in N; M is the number of all genes annotated to certain GO terms; and m denotes the number of DEGs in M. The counted p value was exposed to Bonferroni adjustment (Benjamini & Yekutieli, 2001).
Next, we used a primary public pathway‐related database called the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) to fulfill a pathway enrichment assay of DEGs (Kanehisa & Goto, 2000). The calculation formula was uniform with that in the GO annotation. The pathway enrichment approach offers a farther comprehending of the biological effects on genes. Using the calculated p less than .05 as a threshold, we found notably enriched KEGG terms in the input list of DEGs in comparison with their genomic background and determined significantly enriched signal transduction pathways or metabolic pathways (Kanehisa et al., 2014).
2.10. Luciferase reporter assay
To produce reporter structures for the luciferase assay, approximately 210, 204, and 427 bp fragments incorporating with predicted miRNA target sites in the 3′‐UTR of MAP3K9, AKT3 and circ‐016910 were composited and inlet into the psiCHECK‐2 vectors (Addgene, CA). Primers were designed with particular restriction enzyme sites between Xho I and Not I (Table S2). All constructs were subjected to sequencing for identification. GMECs were cultured in 48‐well plates at a density of 50,000 cells/well before transfection. Then cells were cotransfected with 0.33 mg psiCHECK‐2 luciferase reporter gene constructs and 10 pmol miR‐574‐5p mimics or inhibitors using Lipofectamine™ RNAiMAX Reagent. After 24 hr, renilla and firefly luciferase activities were measured using Thermo Scientific Varioskan Flash (Thermo Fisher Scientific) by the Dual‐Glo luciferase assay system (Promega).
2.11. Quantitative real‐time polymerase chain reaction
The total RNA was reverse‐transcribed into cDNA via the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa). In brief, reverse transcription was exercised as follows: a 10 μl mixture consists of a total of 800 ng of total RNA, 2 μl of 5X gDNA eraser buffer, 1 μl of gDNA Eraser, and RNase‐free dH2O and was cultivated at 42°C for 2 min, then a total of 4 μl of 5X prime script buffer 2, 1 μl of PrimeScript RT Enzyme Mix 1, 1 μl of RT Primer Mix and RNase‐Free dH2O was added into a final volume of 20 μl, and the resulting reaction was incubated at 42°C for 15 min therewith 85°C for 5 s. The mRNA standards of disparate genes were quantified via SYBR Premix Ex Taq II (TaKaRa), and analysis was achieved through the CFX Connect Real‐Time PCR Detection System (Bio‐Rad, CA). The validated primers used for real‐time polymerase chain reaction (RT‐PCR) were listed in Table S3. The RT‐PCR conditions were: 95°C for 10 min and then 40 cycles at 94°C for 15 s, 60 °C for 30 s, followed by 72 °C for 30 s. β‐Actin or U6 mRNA level was used for normalization. Three independent experimental repeats were performed. Relative expression was calculated by the method.
2.12. Analysis of cell apoptosis
Apoptosis of GMECs was measured using the flow cytometry method (FCM) and an Annexin V‐FITC propidium iodide (PI) staining apoptosis assay kit (SeaBioTech, Shanghai, China) was used for the detection of apoptotic impact. After 24 hr transfection, the number of apoptotic cells was calculated using flow cytometry within 1 hr behind staining of a cell population (4 × 105) with annexin V‐FITC and PI, on account of the manufacturer's protocol.
2.13. Analysis of cell viability and cell proliferation
The viability of GMECs was measured using the cell counting kit‐8 (CCK‐8) assay. GMECs were cultured into 96‐well plates at a density of 2 × 104 cells per well in a 200 μl volume and incubated overnight. Then the cells were transfected for 24, 48, 72, and 96 hr. Subsequently, 20 μl of CCK‐8 solution (ZETATM life) was added into each well and in incubation at 37°C for 2 hr. The absorbance was measured at 450 nm using Epoch microplate reader (Biotek, Winooski, VT), after the compound oscillated for 15 min.
The cell proliferation assay was performed with EdU assay after cells treated for 24 hr in 96‐well plates. Briefly, cells were dyed with EdU (RiboBio, Guangzhou, China) with a final concentration of 50 μM for 2 hr and then with DAPI for 15 min in 37 ℃ after washed in PBS three times. Fluorescence microscopy resolved the images.
2.14. Determination of β‐casein and triglycerides
GMECs were posttransfection for 24 hr and the obtained cell‐free supernatants and cell lysate were used to estimate the concentrations of β‐casein by goat β‐casein enzyme‐linked immunosorbent assay kit (Tongwei, Shanghai, China) and triglycerides using triglycerides quantitative assay kit (Applygen, Beijing, China). According to the manufacturer's instructions, 50 μl supernatants and 10 μl cell lysate were used, and absorbance at 450 and 550 nm was measured by an Epoch microplate reader (Biotek). Concentrations were calculated after protracting the standard curve. The sensitivity of kits was 1.0 μg/ml, and mean intra‐ and interassay variable coefficient values were less than 15% and 10%, respectively.
2.15. Western blot
As to western blot analysis, cells were collected and extracted in ice‐cold radioimmunoprecipitation lysis buffer (BioTeke, Beijing, China) inclusive of phenylmethanesulfonyl fluoride (Solarbio, Beijing, China) at 0.1 mg/ml after 48 hr posttransfection. BCA protein assay kit (Vazyme Biotech, Nanjing, China) was used to verify the concentration of proteins. Approximately 30 μg total protein was subjected to a 12% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis. The separated proteins were then transferred to polyvinylidene difluoride (Merck Millipore, MA) membranes, followed by the membrane blocked with 10% skim milk powder in Tris‐buffered saline inclusive of 0.1% Tween 20 (pH 7.6) at room temperature for 1 hr. Membranes were then probed with the indicated primary antibodies overnight at 4°C, washed and whereafter in incubation with the suitable horseradish peroxidase‐conjugated secondary antibodies against mouse, rabbit or goat at 4°C for 2 hr. The primary antibodies were shown in Table S4. Proteins were detected by facilitated chemiluminescence (Advansta). The Quantity One program (Bio‐Rad) was used to perform quantification.
2.16. Statistical analysis
The SPSS software package, SPSS 19.0 (Beijing, China) was used to calculate the analysis of statistics. Data were shown as means ± standard error of three independent experiments. Differences between the groups were detected using one‐way and two‐way analysis of variance, followed by a Bonferroni post hoc correction for all group comparisons, considering statistically significant at *p < .05 and **p < .01.
3. RESULTS
3.1. Massively sequencing to transcriptome
To acquire a global view of the GMECs transcriptome, total RNA from GMECs transfected with miR‐574‐5p mimics and NC were utilized to construct RNA libraries for high‐throughput sequencing. Behind screening the unique adapter sequences, these containing low‐quality sequences and N sequences, we obtained 70,944,975 and 47,056,906 clean reads in each RNA sequencing library yet. Respectively, 94.294% and 92.852% were mapped to the reference genome for MC and NC groups in the overall sequenced reads. Of these, more than 85% or 7% mapped either to unique or to multiple genomic locations of respective samples while the residuals were unmapped (Table 1), owing to only reads aligning completely inside exonic regions can be mapped. The distribution of reads in distinct regions of the reference genome was evaluated using the following standard metrics: Exon, intron, and intergenic reads (Figure 1a).
Table 1.
Summary of sequence read alignments to the reference genome
| Category | miRNA‐574‐5p (MC) | Negative control (NC) | ||
|---|---|---|---|---|
| Reads number | Percentage | Reads number | Percentage | |
| Total reads | 70,944,975 | 100% | 47,056,906 | 100% |
| Total mapped reads | 66,896,622 | 94.294% | 43,693,024 | 92.852% |
| Multiple mapped reads | 5,681,987 | 8.009% | 3,418,700 | 7.265% |
| Uniquely mapped reads | 61,214,635 | 86.285% | 40,274,324 | 85.586% |
| Reads mapped in proper pairs | 58,013,589 | 81.773% | 37,666,159 | 80.048% |
Note: total reads, total number of sequencing reads; total mapped reads, the reads that can be aligned to reference sequence and the ratio of it; multiple mapped reads, in total mapped reads, reads aligned to two or more places; uniquely mapped reads, in total mapped reads, reads aligned to only one position.
Figure 1.
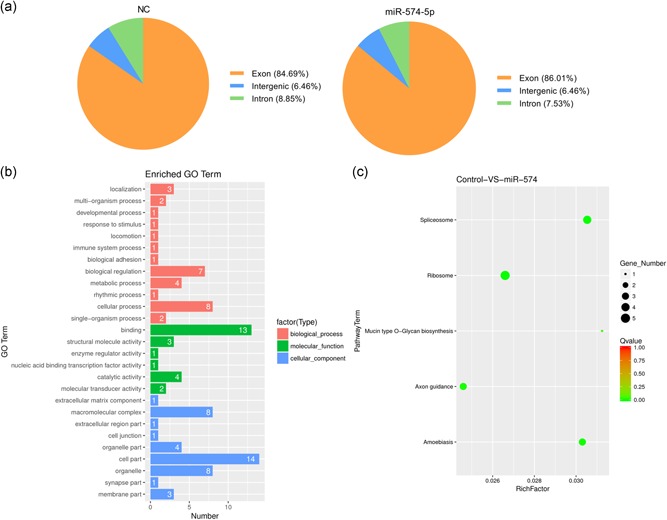
Transcriptome sequencing. (a) The distribution of reads in the reference genome in different areas. (b) Histogram presentation of the top 30 gene ontology (GO) functional annotations for the differentially expressed genes (DEGs). The x‐axis indicates the number of DEGs, the y‐axis indicates the GO terms. (c) The scatter plot of the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of differential genes. The x‐axis indicates the rich factor; the y‐axis indicates the KEGG pathway. The size of the dot indicates the number of differentially expressed genes in the pathway, and the color of the dot corresponds to the different Q value range
3.2. Identification of DEGs
On behalf of identifying the promising candidates regulated by miR‐574‐5p, we investigated the DEGs after miR‐574‐5p overexpressed. Although abundant genes were expressed differentially between the two libraries, the present study focused on genes meeting the appointed standard of p < .05 and |log2FoldChange| > 1 (Table S5). An amount of 332 genes were distinguishingly expressed range from the MC and NC libraries (Table S5), with 74 genes upregulated and 258 genes downregulated. Compared with the NC group, noticeably, DGKI, DGKH, MAP3K9, MAP3K2, SVEP1, RGS9BP, and IGFBP5 were downregulated in the MC group. ISG15, ISG20, RAB38, GAPDHS, and LIPH were upregulated in the MC group than the NC group.
3.3. GO analysis of DEGs
GO is extensively used to standardize the representation of genes and provides information related to three independent ontology categories cellular component, biological process, and molecular function (Camon et al., 2004; Tweedie et al., 2009). Therefore, we executed a GO analysis using running queries for respective DEGs specific to the GO database and explored the associated biological processes. Figure 1b and Table S6 showed the results of GO analysis in functional annotations. Overall, 97 GO terms were distributed to the DEGs, and these were itemized into three ontologies: (a) 32 (33.0%) terms in accordance with biological processes, (b) 41 (42.3%) terms in accordance with cellular components, and (c) 24 (23.7%) terms in accordance with molecular functions. In the molecular function category, the three most imperative enriched terms were associated with binding, catalytic activity, and structural molecular activity. Within the biological processes category, the GO terms with the high level of significances were a cellular process, biological regulation, and metabolic process. Cell part accounted for the most of terms in the molecular function category, accompanied by organelle, macromolecular complex, and organelle part.
3.4. KEGG pathway analysis of DEGs
KEGG analysis offers us a further understanding of the biological effects on DEGs, according to manifold genes collaborated with each other to perform their biological regulatory functions (Kanehisa & Goto, 2000; Kanehisa et al., 2014). In total, the DEGs were remarkably enriched in five KEGG pathways, meeting the standard of Q values less than .05 (Figure 1c and Table S7), and implicated that these pathways may take vital impacts on the development of GMECs. The most significantly enriched pathway was ribosomal with five DEGs enriched. Spliceosome, axon guidance, amebiasis, and mucin‐type O‐glycan biosynthesis were also among the significantly enriched pathways.
3.5. miR‐574‐5p specifically targeted MAP3K9 in GMECs
To identify new potential molecular targets of the miR‐574‐5p during transformation, we used TargetScan (http://www.targetscan.org/) for predicted identification. Among them, we prioritized and focused on MAP3K9 for the following reasons: (a) TargetScan prediction revealed that there was a miR‐574‐5p binding site at nucleotides 10714–10720 of MAP3K9‐3′‐UTR; (b) MAP3K9 was a differentially expressed unigenes according to transcriptome sequencing; and (c) MAP3K9 was considered as an imperative regulator for various metabolic processes (Durkin et al., 2004). Afterward, to ascertain MAP3K9 as a target of miR‐574‐5p, we set up a two‐tier luciferase assay by the psiCHECK‐2 (psC) vector and constructed luciferase reporters with the wild‐type (wt)‐MAP3K9‐3′‐UTR or mutant (mut)‐MAP3K9‐3′‐UTR, separately. As shown in Figure 2a, cotransfection of psC‐MAP3K9‐3′‐UTR‐wt luciferase reporter with miR‐574‐5p mimics into GMECs produced attenuated relative luciferase activities compared with the negative control, and the opposite results were exhibited in the miR‐574‐5p inhibitors transfected GMECs. This repressive effect was eliminated by psC‐MAP3K9‐3′‐UTR‐mut (Figure 2a).
Figure 2.
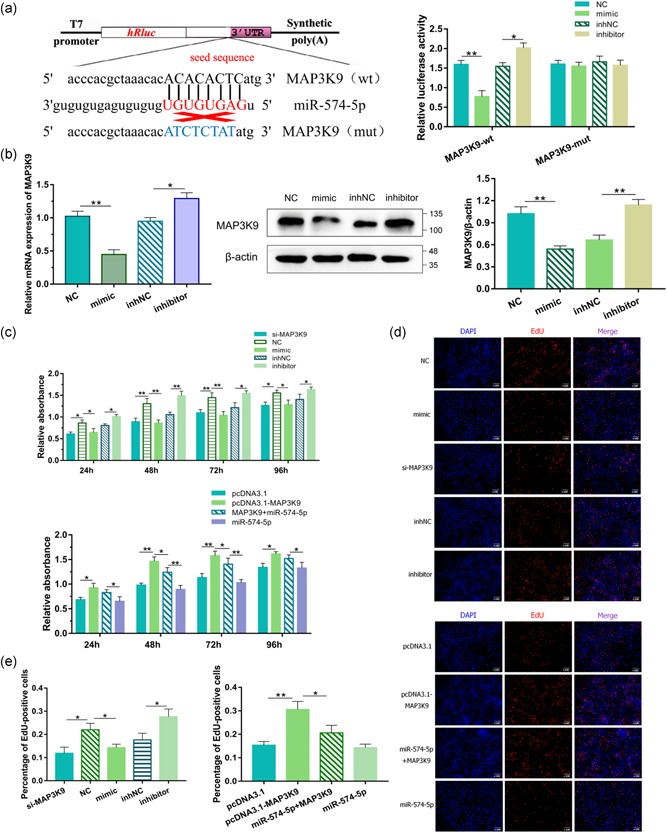
miR‐574‐5p regulated the proliferation of goat mammary epithelial cells (GMECs) via mitogen‐activated protein kinase kinase kinase 9 (MAP3K9). The target site for miR‐574‐5p in the MAP3K9 3′‐untranslated region (3′‐UTR) and the construction of the luciferase expression vector (Luc) fused with the MAP3K9 3′‐UTR (a). wt represents the Luc reporter vector with the wt MAP3K9 3′‐UTR 10714–10720); mut represents the Luc reporter vector with the mutation at the miR‐574‐5p site in MAP3K9 3′‐UTR. After 24 hr, the luciferase activities were measured. (b) Messenger RNA and protein expressions of MAP3K9 in GMECs transfected with miR‐574‐5p mimics or inhibitors for 24 and 48 hr. Expression was quantified by quantitative real‐time polymerase chain reaction and western blot, β‐actin was used as an internal control. (c) Cell viability was determined using the CCK‐8 assay for 24, 48, 72, and 96 hr. GMECs were transfected with miR‐574‐5p mimics, inhibitors, siRNA‐MAP3K9 or pcDNA3.1‐MAP3K9 expression vectors or cotransfected MAP3K9 with miR‐574‐5p. (d, e) EdU staining assay. After 24 hr transfection, GMECs in the S phase were stained with EdU in red, while cell nuclei were dyed with DAPI in blue. *p < .05; **p < .01
Then we assessed whether miR‐574‐5p induced consistent diversities in the mRNA and protein expressions of MAP3K9. MAP3K9 mRNA levels were significantly suppressed after miR‐574‐5p overexpressed, as demonstrated in Figure 2b, while they enhanced when miR‐574‐5p interfered with antisense molecules. Consistent with these results, we found a conspicuous decrease or increase of endogenous MAP3K9 protein level in GMECs transfected with miR‐574‐5p mimics or inhibitors compared with control groups (Figure 2b). Taken together, these results revealed that miR‐574‐5p acted as a demotivated regulator of MAP3K9 expression by directly binding to the 3′‐UTR in GMECs.
3.6. miR‐574‐5p inhibited the proliferation and promoted the apoptosis of GMECs via MAP3K9
We next screened for miR‐574‐5p that were involved in regulating the proliferation of GMECs. A CCK‐8 assay was conducted to investigate any changes in cell viability following cells posttransfected with miR‐574 mimics or inhibitors for 24, 48, 72, and 96 hr. The data evinced that miR‐574‐5p motivation or blockade inhibited or promoted the cell viability compared with control groups (Figure 2c). The EdU staining assay was consistent with the result, with decreased S‐phage cells after miR‐574‐5p treatment (Figure 2d,e). Subsequently, we explored whether miR‐574‐5p functioned in the proliferation of GMECs via its negativity to MAP3K9. Therefore, siRNA‐MAP3K9 and a vector carrying MAP3K9 coding sequence (CDS), were introduced into GMECs for the inhibition and overexpression of MAP3K9. The proliferation of cells transfected with si‐MAP3K9 was notably decreased in comparison with negative control and on contrast, the proliferation of cells transfected with pcDNA3.1‐MAP3K9 vectors was significantly elevated. As expected, MAP3K9 weakened miR‐574‐5p‐attenuated impacts on GMECs proliferation by reducing cell viability and EdU positive cells (Figure 2c–e), indicating that miR‐574‐5p could inhibit the proliferation of GMECs via MAP3K9.
To authenticate the impact of miR‐574‐5p and MAP3K9 on GMECs apoptosis, the degree of apoptosis was analyzed by Annexin V‐FITC/PI staining and the protein levels of critical apoptotic genes. FCM was executed to evaluate the count of apoptotic cells incorporating early and late apoptosis (Vermes, Haanen, & Reutelingsperger, 2000). The apoptotic rates of GMECs were higher in the miR‐574‐5p mimics group and lower in the miR‐574‐5p inhibitors group (Figures 3a and 2A). As several crucial genes including Bcl‐2 (Cory & Adams, 2002) and Bax (Chipuk et al., 2004) are reported to be related to apoptosis, we performed western blot analysis and showed significant increased proapoptotic Bax and decreased antiapoptotic Bcl‐2 protein expression in cells with miR‐574‐5p mimics (Figure 3b). Afterward, we investigated whether miR‐574‐5p functioned in cell apoptosis depends on MAP3K9 partly. Here we illustrated the elevated count of apoptotic cells after cells treated with si‐MAP3K9 (Figures 3a and S2B), and inducing MAP3K9 restrained the apoptotic rates of GMECs, oppositely (Figures 3a and S2C). Cocultured cells were then subjected and MAP3K9 partially counteracted the influence of miR‐574‐5p on cell apoptosis (Figure 3a and S2C). Western blot showed that loss of MAP3K9 elevated the protein level of Bax accompanied by a suppressed expression of Bcl‐2, whereas active MAP3K9 showed the adverse results and alleviated the impact of miR‐574‐5p (Figure 3b). It suggested that miR‐574‐5p promoted apoptosis of GMECs, at least in part, depended on its mediation of MAP3K9.
Figure 3.
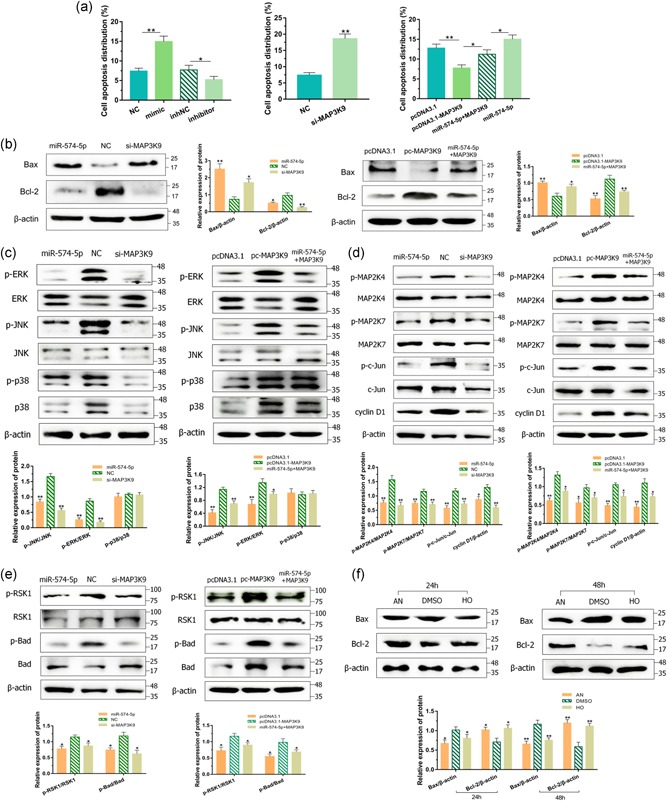
miR‐574‐5p regulated apoptosis and mitogen‐activated protein kinase pathways via MAP3K9 in goat mammary epithelial cells (GMECs). (a) The apoptosis of GMECs transfected with miR‐574‐5p mimics, inhibitors, si‐MAP3K9 or pcDNA3.1‐MAP3K9 expression vectors or cotransfected MAP3K9 with miR‐574‐5p for 24 hr. The histogram showed the apoptotic cell percentage detected by flow cytometry method, and error bars denote mean ± SD. After 48 hr transfection, (b) western blot analysis was used to detect the expression of Bcl‐2 and Bax in GMECs. (c, d) Cytosolic proteins were analyzed by western blot for extracellular‐signal‐regulated kinase, p38, c‐Jun N‐terminal kinase, MAP2K4, MAP2K7, c‐Jun, RSK1, Bad, and cyclin D1. (e) GMECs were treated with 5 μM anisomycin (AN) and honokiol (HO) in 24 and 48 hr. The expression of Bcl‐2 and Bax was analyzed by western blot. β‐Actin was detected as a loading control. *p < .05; **p < .01
3.7. miR‐574‐5p inhibited MAPK signaling pathways via MAP3K9 in GMECs
MAPKs, including ERK, JNK, and p38 MAPK, are pivotal signaling molecules in cell signaling processes and MAPK pathways participate in the regulation of cell apoptosis and cell proliferation (Bisson et al., 2008; Cargnello & Roux, 2011). To confirm whether the apoptosis and proliferation of GMECs is mediated by miR‐574‐5p through MAPK pathways, phosphorylation of JNK, ERK, and p38 MAPKs were evaluated by western blot. Induced miR‐574‐5p or deficiency of MAP3K9 decreased ERK1/2 and JNK activation, but without remarkable suppressive impact on the activation of p38 MAPK (Figure 3c). Conversely, the elevation of MAP3K9 enhanced the increase in phosphorylation of ERK1/2 and JNK (Figure 3c). Then we examined the protein levels of up or downstream targets of ERK/JNK pathways. Induced miR‐574‐5p and blockage of MAP3K9 caused downregulation of cyclin D1 and phosphorylation of MAP2K4/7, c‐Jun, RSK1, Bad, whereas contrary results were exhibited after MAP3K9 overexpressed (Figure 3d,e). As expected, the effects of miR‐574‐5p on these key proteins' motivation were partially mitigated by MAP3K9. Afterward, we treated GMECs with anisomycin and honokiol in 24 and 48 hr, inducing the expression of JNK and ERK to identify the functions to Bcl‐2. We measured that the expression of Bcl‐2 was promoted by JNK and ERK, accompanying with blockage of Bax (Figure 3f). These findings evinced that miR‐574‐5p was capable of suppressing the activities of MAPK signaling pathways via MAP3K9 and then facilitating cell apoptosis and repressing cell proliferation in GMECs.
3.8. miR‐574‐5p inhibited the milk synthesis via MAP3K9 in GMECs
To estimate whether the secretion of β‐casein and triglycerides is dominated by miR‐574‐5p and MAP3K9, the content of β‐casein in the cell‐free supernatants and triglycerides were identified using detection kits. GMECS with enhanced miR‐574‐5p and depressed MAP3K9 exhibited notably fewer production of β‐casein and triglycerides compared to the NC group (Figure 4a,b) oppositely, upon MAP3K9 overexpressed, the production was induced and enhanced MAP3K9 triggered the assuasive impacts of miR‐574‐5p (Figure 4a,b) indicating that miR‐574‐5p suppressed the secretion of β‐casein and triglycerides via MAP3K9 in GMECs.
Figure 4.
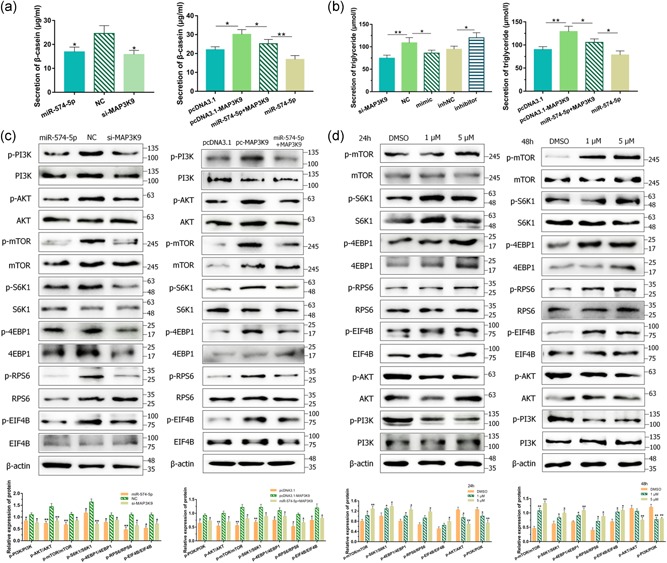
miR‐574‐5p regulated milk synthesis via mitogen‐activated protein kinase kinase kinase 9 (MAP3K9) in goat mammary epithelial cells (GMECs). GMECs were posttransfected with miR‐574‐5p mimics, si‐MAP3K9, pcDNA3.1‐MAP3K9 expression vectors or cotransfected MAP3K9 with miR‐574‐5p for 48 hr. (a) The secretion of β‐casein in the cell‐free supernatants in GMECs was measured by enzyme‐linked immunosorbent assay kit. (b) The secretion of triglycerides in GMECs was measured by the detection kit. (c) Cytosolic proteins and related phosphorylation levels were analyzed by western blot for protein kinase B (AKT), phosphoinositide 3 kinase (PI3K), mammalian target of rapamycin (mTOR), S6K1, 4EBP1, RPS6, and EIF4B. (d) GMECs were treated with 1 and 5 μM anisomycin in 24 and 48 hr. The expression of AKT, PI3K, mTOR, S6K1, 4EBP1, RPS6, and EIF4B was analyzed by western blot. β‐Actin was detected as a loading control. *p < .05; **p < .01
Previous studies indicate that phosphoinositide 3 kinase (PI3K)/AKT–mTOR signaling pathway regulates milk synthesis (Samant & Sylvester, 2006; L. Wang et al., 2014); thus, we speculated whether miR‐574‐5p inhibited milk synthesis via its negative effect on MAP3K9 in GMECs. The protein involved in that signaling pathway expression levels of PI3K, mTOR, AKT, S6K1, and 4EBP1 were analyzed by western blot. Groups transfected with miR‐574‐5p mimics and si‐MAP3K9 decreased the protein levels of p‐PI3K, p‐mTOR, p‐AKT, p‐S6K1, p‐4EBP1, p‐RPS6, and p‐EIF4B (Figure 4c). MAP3K9 overexpression clearly increased these phosphorylation levels compared with the control group, at the same time, rescued the negative functions of miR‐574‐5p (Figure 4c). The consequence provided further evidence that miR‐574‐5p inhibited the expression of milk protein synthesis‐associated proteins via downregulating MAP3K9 in GMECs.
It is reported that MAPK/JNK promotes mTOR activities (Basu, Rajakaruna, Reyes, Van Bockstaele, & Menko, 2014), accordingly, we speculated whether JNK could regulate mTOR in GMECs. We induced the expression of JNK using pharmacological anisomycin in different concentrations and times to detect the conceivable roles. Inducement of JNK gave rise to positive phosphorylation of mTOR, S6K1, 4EBP1, RPS6, and EIF4B which are members of the downstream targets of mTOR (Figure 4d). Intriguingly, blockade of activation in AKT and PI3K was observed under this condition (Figure 4d). These data supported a role for MAPK/JNK motivation of the mTOR‐S6K1/4EBP1 pathway in GMECs.
3.9. miR‐574‐5p inhibited milk synthesis via AKT3 in GMECs
To further understand the regulatory mechanism of milk synthesis by miR‐574‐5p, we selected the target genes of miR‐574‐5p again. From these predicted targets, AKT3, as a pivotal regulator of milk synthesis, engaged our attention. To produce reporter structures for the luciferase assay, approximately 204 bp fragment incorporating with predicted miRNA target sites in the 3′‐UTR of AKT3 was inlet into the psiCHECK‐2 vector. The reporter assay exhibited that miR‐574‐5p mimics reduced the luciferase activities of the psC‐AKT3‐3′‐UTR‐wt reporter and the inhibitors showed the promoted‐activities, while luciferase activities of the mutant reporter vector did not change (Figure 5a). Consistently, miR‐574‐5p mimics or inhibitors suppressed or elevated not only in mRNA level but also in the protein level of AKT3 (Figure 5b), revealing that AKT3 was a direct target of miR‐574‐5p in GMECs. Then we detected the content of β‐casein and triglycerides after cells treated with variant expressions of AKT3. The diagrams exhibited lessened or incremental contents after cells transfected with si‐AKT3 or pcDNA3.1‐AKT3 vectors (Figure 5c). AKT3 treatment alleviated miR‐574‐5p induced inhibitory impacts on the production of β‐casein and triglycerides (Figure 5c). As AKT3 is a pivotal hinge in PI3K/AKT–mTOR pathway, we then detected the total and phosphorylated protein levels of pathway pivotal members after miR‐574‐5p and AKT3 elevated. Figure 5d showed that AKT3 promoted p‐mTOR, p‐S6K1, p‐4EBP1, p‐RPS6, and p‐EIF4B significantly, and these phosphorylated expressions were recovered when coexpression of miR‐574‐5p with AKT3. On the contrary, knocked down AKT3 using siRNA exhibited depressed protein levels (Figure 5d). We hence suggested that miR‐574‐5p could attenuate milk synthesis via AKT3 in GMECs.
Figure 5.
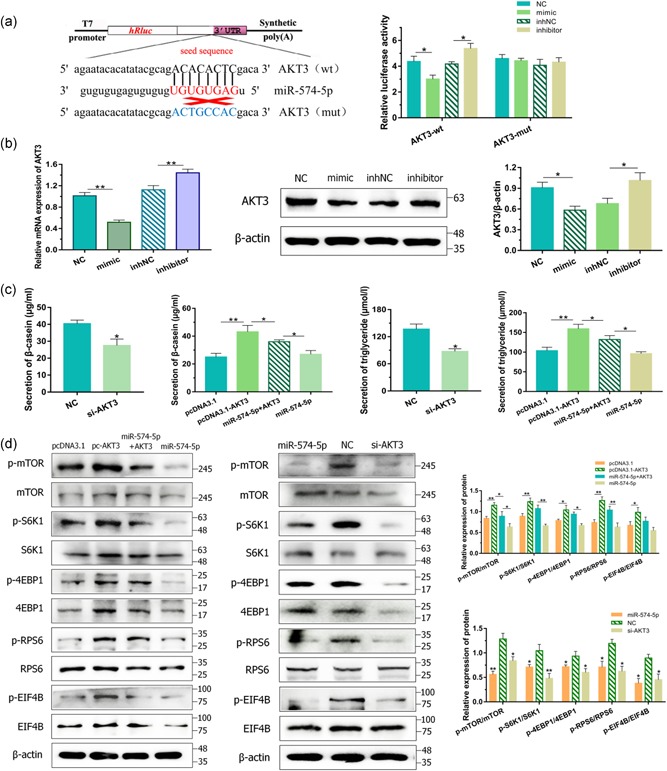
miR‐574‐5p regulated milk synthesis via AKT3 in goat mammary epithelial cells (GMECs). (a) The target site for miR‐574‐5p in the AKT3 3′‐untranslated region (3′‐UTR) and the construction of the luciferase expression vector (Luc) fused with the AKT3 3′‐UTR. After 24 hr, the luciferase activities were measured. (B) Messenger RNA and protein expressions of AKT3 in GMECs transfected with miR‐574‐5p mimics or inhibitors for 24 and 48 hr. Expression was quantified by quantitative real‐time polymerase chain reaction and western blot, β‐actin was used as an internal control. (c) GMECs were posttransfected with miR‐574‐5p mimics, si‐AKT3 or pcDNA3.1‐AKT3 expression vectors or cotransfected AKT3 with miR‐574‐5p for 48 hr. The secretion of β‐casein in the cell‐free supernatants and triglycerides in GMECs was measured by enzyme‐linked immunosorbent assay kit and detection kit. (d) Cytosolic proteins and related phosphorylation levels were analyzed by western blot for mammalian target of rapamycin, S6K1, 4EBP1, RPS6, and EIF4B. β‐Actin was detected as a loading control. *p < .05; **p < .01
3.10. circ‐016910 acted as a sponge for miR‐574‐5p
Incremental clues uncover that circRNAs undertake the effect of miRNAs sponges then further regulate downstream gene expression (Memczak et al., 2013). After we used the bioinformatics prediction approach to find the target circRNAs based on the constructed circRNA libraries, we predicted the target circRNAs of miR‐574‐5p to make an underlying inquiry of its molecular mechanisms. We found that circ‐016910 contained predicted complementary sequences from the constructed circRNA libraries in our laboratory (Figure 6a and Table S8). circ‐016910 fragment was constructed and then we performed a luciferase assay by cotransfection of miR‐574‐5p mimics or inhibitors with wild or mutant luciferase reporters into GMECs, separately. Notably, depletion of luciferase activities was examined compared mimics group with the NC group and the inhNC group with inhibitors group (Figure 6b). Transfection of miR‐574‐5p had no remarkable impact on luciferase activities when the binding sites were mutated (Figure 6b). In addition, miR‐574‐5p expression was improved and the mRNA and protein levels of miR‐574‐5p targeted genes: MAP3K9 and AKT3, were restrained along with the interference of circ‐016910 (Figure 6c,d). Advancing circ‐016910 exhibited the inverse results and the coexpression of miR‐574‐5p with circ‐016910 partly neutralize that effect (Figure 6c,d). Hence, the direct correlation of miR‐574‐5p and circ‐016910 was affirmed.
Figure 6.
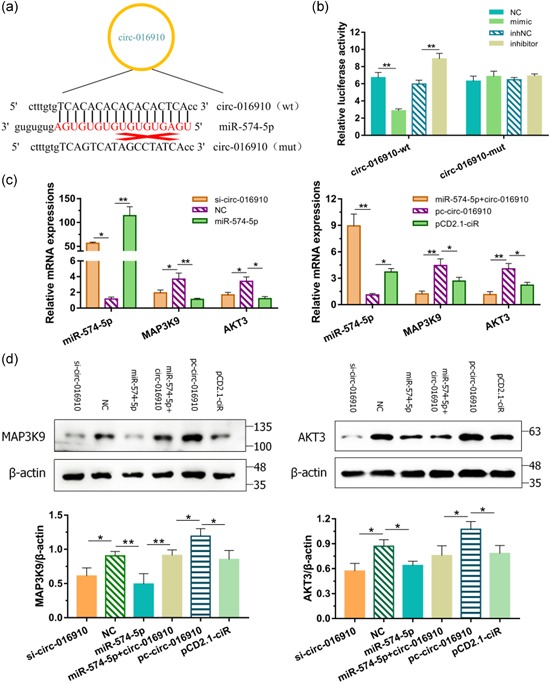
circ‐016910 acted as a sponge for miR‐574‐5p. (a) We performed a luciferase vector assay to confirm the direct binding between circ‐016910 and miR‐574‐5p in terms of their complementary sequences. (b) After cotransfecting wt‐ or mut‐ plasmids with miRNA mimics in goat mammary epithelial cells (GMECs) for 24 hr, luciferase activities were measured. After transfected si‐circ‐016910, miR‐574‐5p mimics, pc‐circ‐016910 or cotransfected circ‐016910 with miR‐574‐5p, (c) messenger RNA expressions, and (d) cytosolic proteins expressions of miR‐574‐5p, MAP3K9, and AKT3 in GMECs were quantified by quantitative real‐time polymerase chain reaction for 24 hr and western blot for 48 hr, respectively. U6 and β‐actin were used as an internal control. *p < .05; **p < .01
3.11. circ‐016910 promoted the proliferation and inhibited the apoptosis of GMECs via miR‐574‐5p
As we clarified that circ‐016910 significantly restrained miR‐574‐5p in GMECs, we speculated that circ‐016910 could regulate the survival capabilities of GMECs in accordance with proliferation and apoptosis. Owing to this hypothesis, we transfected circ‐016910 siRNA and circ‐016910 overexpression vector into GMECs. CCK‐8 and EdU assays were accomplished to verify the proliferation of GMECs, and the apoptosis of GMECs was analyzed by FCM assay and western blot. circ‐016910 blocking suppressed the cell viability and proliferation apparently, while the elevation of circ‐016910 upregulated these abilities (Figure 7a). In addition, we illustrated the improved or depressed apoptotic rates of GMECs by down‐ or upregulation of circ‐016910 (Figures 7b and S3). Western blot found out that circ‐016910 silencing repressed antiapoptotic Bcl‐2 expression cooperated with induced expression of proapoptotic Bax, whereas induced circ‐016910 exerted the contrary results (Figure 7c). Neutral impacts were observed after circ‐016910 and miR‐574‐5p coexpressed, providing evidence that circ‐016910 mediated the proliferation and apoptosis of GMECs via miR‐574‐5p.
Figure 7.
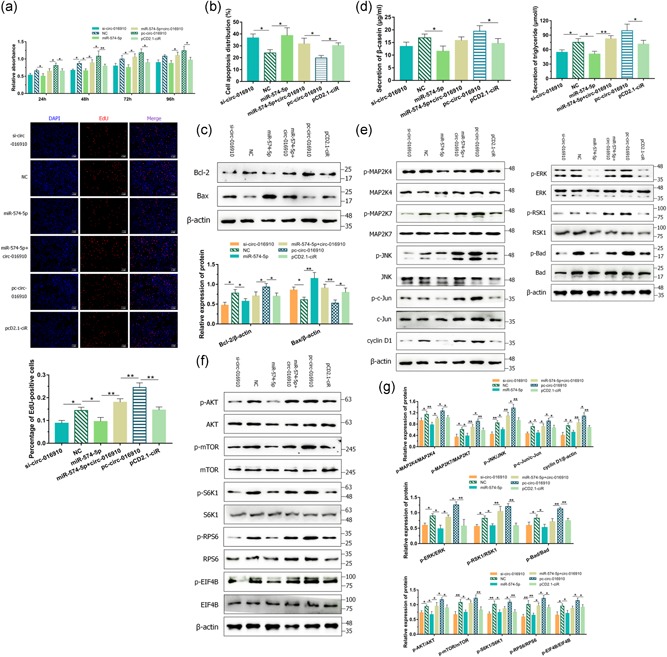
The regulation of circ‐016910 via miR‐574‐5p in goat mammary epithelial cells (GMECs). (a) Cell viability was determined using the cell counting kit‐8 assay after GMECs were posttransfected for 24, 48, 72, and 96 hr. After 24 hr transfection, GMECs in the S phase were stained with EdU in red, while cell nuclei were dyed with DAPI in blue. (b) The apoptosis of GMECs was measured 24 hr posttransfection. The histogram showed the apoptotic cell percentage detected by flow cytometry method, and error bars denote mean ± standard deviation. (c) Western blot analysis was used to detect the expression of Bcl‐2 and Bax in GMECs for 48 hr. (d) The secretion of β‐casein in the cell‐free supernatants and triglycerides in GMECs was measured by enzyme‐linked immunosorbent assay kit and detection kit. (e) After 48 hr treatment, cytosolic proteins and related phosphorylation levels were analyzed by western blot for c‐Jun N‐terminal kinase, MAP2K4/7, c‐Jun, cyclin D1, and extracellular‐signal‐regulated kinase, RSK1, Bad, and (f) protein kinase B, mammalian target of rapamycin, S6K1, RPS6, and EIF4B. (g) Histograms represented the analysis of the data. β‐Actin was detected as a loading control. *p < .05; **p < .01
3.12. circ‐016910 promoted the secretion of β‐casein and triglycerides via miR‐574‐5p in GMECs
To further inquire into the function of circ‐016910 in GMECs, we analyzed the secretion of β‐casein and triglycerides using detection kits. Successful circ‐016910 knockdown or overexpression decreased or increased these secretions and we found that miR‐574‐5p advancement partial abrogated that biological effects of circ‐016910 (Figure 7d). Thus circ‐016910 might facilitate the secretion of β‐casein and triglycerides due to its mediation of miR‐574‐5p.
3.13. circ‐016910 promoted MAPK and mTOR signaling pathways via miR‐574‐5p in GMECs
Then we wonder whether circ‐016910 could impact MAPK pathways, thus the protein expressions of these key pathway members were detected. Transfection of si‐circ‐016910 caused a marked depletion of phosphorylation of JNK, ERK, MAP2K4/7, c‐Jun, RSK1, and Bad and cyclin D1 protein level, which was adequately reversed by pretreatment with pc‐circ‐016910 (Figures 7e and 7g). The protein involved in milk synthesis signaling pathway and respective phosphorylated expressions were analyzed by western blot. Compared with NC group, using circ‐016910 siRNA raised against activation of AKT, mTOR, S6K1, RPS6, and EIF4B (Figure 7f,g), but without noteworthy differences in expression of PI3K and 4EBP1 (data not shown). Meanwhile, GMECs transfected with pc‐circ‐016910 vectors gave rise to phosphorylation of AKT, mTOR, S6K1, RPS6, and EIF4B and miR‐574‐5p blanked that impacts (Figure 7f,g). These data implicated that circ‐016910 upregulated JNK/ERK MAPK and AKT–mTOR pathways by performing its “miR‐574‐5p sponge” abilities.
4. DISCUSSION
Recently, extensive studies have been devoted to milk components and mammary gland development regulated by miRNAs (Bionaz & Loor, 2007; C. Wang & Li, 2007), but limited numbers of genes directly or indirectly regulated by miRNAs are reported in GMECs. Thus, the detection and identification of relevant genes are preconditioned for studying their functions. In previous studies, after a comparative analysis of our sequence data with published mammary gland transcriptome data during colostrum and common milk stages using high‐throughput sequence, miR‐574‐5p has been screened because it significantly influences mammary gland development and lactation performance (Hou et al., 2017). Since miRNAs can trigger inhibition of protein translation, regulation of miR‐574‐5p during lactation indicates that it may play important roles in GMECs. At present, the discovery and identification of DEGs regulated by miR‐574‐5p directly or indirectly rely on high‐throughput sequencing technologies. An overall of 332 genes was distinguishingly expressed between MC and NC libraries, with 74 genes upregulated including ISG15, ISG20, RAB38, GAPDHS, and LIPH and 258 genes downregulated including DGKI, DGKH, MAP3K9, MAP3K2, SVEP1, RGS9BP, and IGFBP5. Ribosomal, spliceosome, axon guidance, amebiasis, and mucin‐type O‐glycan biosynthesis were among the significantly enriched pathways. Therefore, miR‐574‐5p can play roles in the regulation of lactation or mammary gland development by these differential expression genes and pathways.
Previous studies about miR‐574‐5p have elaborated on its antioncogenic and antimetastatic activities, whereas, the effect of miR‐574‐5p in healthy mammary glands remains relatively uncharacterized. We illustrated that overexpression of miR‐574‐5p enhanced cell apoptosis and decreased cell proliferation in GMECs, and inhibition of miR‐574‐5p showed the opposite results. Meanwhile, expression levels of β‐casein and triglycerides significantly decreased in the MC group compared with the negative control. It was known that the viability, proliferation and apoptosis of MECs are associated with milk production, so an increased number of MECs, enhanced cell proliferation, and decreased apoptosis will contribute to lactation (Boutinaud, Guinardflamenta, & Jammes, 2004). Hence, we further explored the molecular mechanism of that effect of miR‐574‐5p in GMECs.
Our study found that miR‐574‐5p targeted seed sequences of one of the DEGs, MAP3K9 and then suppressed its mRNA and protein expressions. MAPKs are crucial regulators of evolutionarily conservative signaling networks that take a predominant part in a variety of cell physiology, mammary development, and milk synthesis. As a group of serine‐threonine kinases, MAPKs are classified into three primary MAPK subfamilies, including JNK, p38 MAPK, and ERK (Cargnello & Roux, 2011; Slattery et al., 2012). All the known MAP3K9 is an imperative component and can activate the three MAPKs signal transduction pathways (Bisson et al., 2008; Durkin et al., 2004). Therefore, we explored whether miR‐574‐5p functions in GMECs via its depletion of MAP3K9 by MAPK signaling pathways. In our study, upon transfection of GMECs with si‐MAP3K9 and pcDNA3.1‐MAP3K9 plasmids, we found that MAP3K9 depressed cell apoptosis and enhanced cell proliferation in GMECs, and coexpression of miR‐574‐5p with MAP3K9 demonstrated an adiaphorous effect. MAP3K9 elevation or inhibition significantly decreased or increased phosphorylation of ERK1/2 and JNK, but not of p38, indicating that MAP3K9 could active MAPK pathways via JNK and ERK. MAP3K9 is known to motivate JNK in vivo by phosphorylating and irritating the JNK kinase SEK1 (MAP2K4 and MAP2K7) directly and then motivate the c‐Jun amino‐terminal kinase pathway (Lawler, Fleming, Goedert, & Cohen, 1998; Xu, Maroney, Dobrzanski, Kukekov, & Greene, 2001). Relying on c‐Jun, JNK activity improves AP‐1 complex synthesis and transcription of genes incorporating AP‐1‐binding sites, including genes that regulate cell growth, such as cyclin D1 (Sabapathy et al., 2004). Cyclin D1 is found to control G1 to the S phase of the cell cycle and is a vital mediator of cell growth and proliferation (Guttridge, Albanese, Reuther, Pestell, & Jr, 1999). So then MAP2K4, MAP2K7, c‐Jun phosphorylation levels, and cyclin D1 protein level were analyzed and our data showed that MAP3K9 enhanced their expressions, revealing that miR‐574‐5p repressed cell proliferation via MAP3K9 through JNK‐c‐Jun pathway in GMECs. Previous studies clarify that RSK is a kinase downstream of ERK then contributes to the phosphorylation of Bad, which restrains its proapoptotic ability by facilitating its dissociation from Bcl‐xL (Shimamura, Ballif, Richards, & Blenis, 2000). We demonstrated that MAP3K9 gave rise to the phosphorylation of Bad in GMECs, suggesting a mechanism of cell apoptosis owing to the ERK‐Bad pathway. JNK1 is best known to forthrightly motivate Bcl‐2 at Ser70 in vitro and colocalize with Bcl‐2 in the mitochondria (Deng et al., 2001; Wei, Pattingre, Sinha, Bassik, & Levine, 2008), meanwhile, studies identify ERK1/2 as SRKs that can function in concert with PKC to phosphorylate Bcl‐2 (Deng, Ruvolo, Carr, & May, 2000). Bax activation is thought to be mediated, in part, by sequestration of Bcl‐2 proteins (Zong, Lindsten, Ross, Macgregor, & Thompson, 2001). Our data supported the observation that miR‐574‐5p promoted proapoptotic Bax and inhibited antiapoptotic Bcl‐2 expressions, whereas MAP3K9 showed the reverse results. To evaluate whether JNK and ERK function Bcl‐2 in GMECs, the expressions of JNK and ERK were induced and we measured that Bcl‐2 was activated by JNK and ERK, accompanying with downregulation of Bax. Together we draw a conclusion that miR‐574‐5p motivated cell apoptosis via MAP3K9 through JNK/ERK pathways in GMECs.
While our work focused on the mechanism of miR‐574‐5p inhibition on milk synthesis, detection kit analysis evinced that MAP3K9 facilitated the secretion of β‐casein and triglycerides in GMECs, and partially alleviated the negative effect of miR‐574‐5p. There are many transcription factors and signal pathways connected with lactation, studies over the past decade have highlighted the impact for mTOR in modulating mammary development and milk synthesis. AKT is a key regulatory factor in cell signal transduction pathways, and the PI3K/AKT pathway is imperative for the production of milk components such as caseins and triglycerides (Samant & Sylvester, 2006). The motivation of AKTs occurs in answer to signaling by PI3K and PI3K then stimulates the generation of the second messenger PIP3 (Hutchinson, Jin, Cardiff, Woodgett, & Muller, 2001). AKTs, in turn, bring about activation of the mTOR, whose downstream effectors, S6K1 and 4EBP1 (Dann, Selvaraj, & Thomas, 2007; L. Wang, Rhodes, & Lawrence, 2006). Recent work has clarified that activation of S6K1 results in the enrichment of protein synthetic pathways and 4EBP1 improves translation initiation via the binding of EIF4E to EIF4G, revealing that S6K1 and 4EBP1 regulate many fields of cell physiology, consist of milk production (Bian et al., 2010). Meanwhile, RPS6 and EIF4B are extensively studied targets for S6K1, responsible for the initiation of translation (Roux et al., 2007). On the basis of these reports, we measured changes in the protein expression levels of AKT, mTOR, PI3K, S6K1, 4EBP1, RPS6, and EIF4B. The results showed that phosphorylation levels of these proteins were decreased in MC and si‐MAP3K9 groups, while enhanced after MAP3K9 overexpressed. At the same time, miR‐574‐5p targeted AKT3 directly and then inhibited the AKT–mTOR pathway, causing suppressed production of β‐casein and triglycerides. The regulatory associated protein of mTOR, complex 1 (RPTOR) is found to affect the formation of the mTORC1 complex and JNK1 can point RPTOR29 with osmotic stress, (Foster et al., 2010; Kwak et al., 2012). To evaluate whether JNK functions mTOR in GMECs, JNK was induced by anisomycin and that resulted in the inactivation of mTOR, S6K1, 4EBP1, RPS6, and EIF4B, which implicated that MAP3K9 stimulated JNK and then improved mTOR‐S6K1/4EBP1 pathway. Interestingly, the phosphorylation levels of AKT and PI3K were restrained, which showed a feedback loop in GMECs. These observations further supported our hypothesis that miR‐574‐5p repressed milk synthesis via repressing MAP3K9 and AKT3 motivation of PI3K/AKT–mTOR pathway in GMECs.
Recently, the circRNAs–miRNAs–mRNA networks are brought to the forefront. circRNAs incorporating introns mainly store up in the nucleus and regulate gene transcription, whereas exonic circRNAs serve on protein or miRNAs sponges to restrain their activities in the cytoplasm. In terms of the function of circRNAs as “miRNA sponges,” we specialize in miR‐574‐5p upstream mediators. We selected circ‐016910 for further research via the bioinformatics prediction approach in terms of the constructed circRNA libraries. We found the negative interaction between miR‐574‐5p and circ‐016910 and positive interaction between circ‐016910 and MAP3K9 and AKT3 in GMECs, account for circ‐016910 acting as a sponge for miR‐574‐5p. Significantly, circ‐016910 not only had a proliferative and antiapoptotic effect on GMECs but also induced the secretion of β‐casein and triglycerides via its blockage of miR‐574‐5p. To provide the potential mechanisms of circ‐016910 in biological processes, we further detected the momentous members’ protein levels of MAPK and AKT–mTOR pathways. Conform to our anticipation, circ‐016910 activated JNK‐c‐Jun and ERK‐Bad pathways. AKT–mTOR‐S6K1 signaling was also elevated, however, PI3K and 4EBP1 showed no distinctiveness, which means resided in another regulatory mechanism probably.
In conclusion, 74 upregulated and 258 downregulated genes were detected in the MC group by transcriptome sequencing. GO terms and KEGG pathway analysis indicated that differential expression of genes can take part in the regulation of lactation or mammary gland development. circ‐016910 acts as a sponge for miR‐574‐5p and miR‐574‐5p not only inactivates the MAPK pathways, results in suppressing cell proliferation and enhancing cell apoptosis, but also represses PI3K/AKT–mTOR pathway and the expression of β‐casein and triglycerides by downregulation of MAP3K9, showing a circRNA–miRNA–mRNA network (Figure 8). Further comprehending of molecular mechanisms based on these specialties will offer available information for optimizing milk quality in dairy goats.
Figure 8.
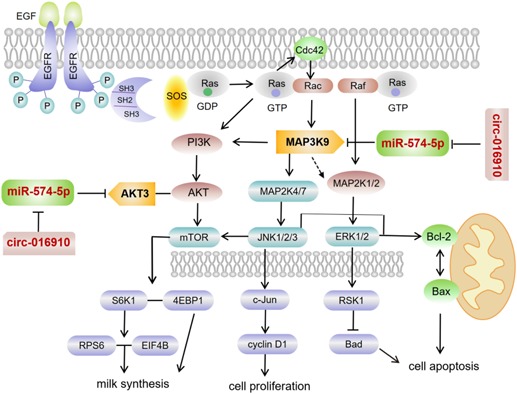
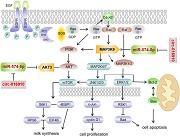
A proposed model linking the miR‐574‐5p locus to the biology of goat mammary epithelial cells (GMECs). miR‐574‐5p promotes cell apoptosis, reduces cell proliferation and milk synthesis in GMECs. miR‐574‐5p targets mitogen‐activated protein kinase kinase kinase 9 (MAP3K9) and AKT3 directly and MAP3K9 stimulates different signal pathways. MAP3K9 actives JNK‐c‐Jun‐cyclin D1 pathway resulting in enhanced cell proliferation, JNK/ERK‐Bcl‐2/Bax pathway and ERK‐RSK1‐Bad pathway accompanied by weakening cell apoptosis. phosphoinositide 3 kinase/protein kinase B–mammalian target of rapamycin pathway is also stimulated by MAP3K9, further accelerates milk synthesis. Meanwhile, circ‐016910 acts as a sponge for miR‐574‐5p and shows a circRNA–miRNA–mRNA network to undertake biological effects
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31601925), China Postdoctoral Science Foundation (2016T90954 and 2014M552498), Shaanxi Science and Technology Innovation Project Plan (2017ZDXM‐NY‐081 and 2016KTZDNY02–04), and Natural Science Foundation of Shaanxi Province (2018JM3006).
Liu Y, Hou J, Zhang M, et al. circ‐016910 sponges miR‐574‐5p to regulate cell physiology and milk synthesis via MAPK and PI3K/AKT–mTOR pathways in GMECs. J Cell Physiol. 2020;235:4198–4216. 10.1002/jcp.29370
Contributor Information
Binyun Cao, Email: caobinyun@126.com.
Xiaopeng An, Email: anxiaopengdky@163.com.
REFERENCES
- Ambros, V. (2003). MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell, 113(6), 673–676. [DOI] [PubMed] [Google Scholar]
- Basu, S. , Rajakaruna, S. , Reyes, B. , Van Bockstaele, E. , & Menko, A. S. (2014). Suppression of MAPK/JNK‐MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during the terminal differentiation of lens fiber cells. Autophagy, 10(7), 1193–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Annals of Statistics, 29(4), 1165–1188. [Google Scholar]
- Bian, C. X. , Shi, Z. Q. , Meng, Q. , Jiang, Y. , Liu, L. Z. , & Jiang, B. H. (2010). P70S6K 1 regulation of angiogenesis through VEGF and HIF‐1alpha expression. Biochemical & Biophysical Research Communications, 398(3), 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bionaz, M. , & Loor, J. J. (2007). Identification of reference genes for quantitative real‐time PCR in the bovine mammary gland during the lactation cycle. Physiological Genomics, 29(3), 312–319. [DOI] [PubMed] [Google Scholar]
- Bisson, N. , Tremblay, M. , Robinson, F. , Kaplan, D. R. , Trusko, S. P. , & Moss, T. (2008). Mice lacking both mixed‐lineage kinase genes Mlk1 and Mlk2 retain a wild type phenotype. Cell Cycle, 7(7), 909–916. [DOI] [PubMed] [Google Scholar]
- Boutinaud, M. , Guinardflamenta, J. , & Jammes, H. (2004). The number and activity of mammary epithelial cells, determining factors for milk production. Reproduction, Nutrition, Development, 44(5), 499–508. [PubMed] [Google Scholar]
- Cai, P. , Yang, T. , Jiang, X. , Zheng, M. , Xu, G. , & Xia, J. (2017). Role of miR‐15a in intervertebral disc degeneration through targeting MAP3K9. Biomedicine & Pharmacotherapy, 87, 568–574. [DOI] [PubMed] [Google Scholar]
- Camon, E. , Magrane, M. , Barrell, D. , Lee, V. , Dimmer, E. , Maslen, J. , … Apweiler, R. (2004). The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Research, 32, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello, M. , & Roux, P. P. (2011). Activation and function of the MAPKs and their substrates, the MAPK‐activated protein kinases. Microbiology & Molecular Biology Reviews Mmbr, 75(1), 50–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Luo, J. , Sun, S. , Cao, D. , Shi, H. , & Loor, J. J. (2017). miR‐148a and miR‐17‐5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biology, 14(3), 326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk, J. E. , Kuwana, T. , Bouchier‐Hayes, L. , Droin, N. M. , Newmeyer, D. D. , Schuler, M. , & Green, D. R. (2004). Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science, 303(5660), 1010–1014. [DOI] [PubMed] [Google Scholar]
- Cory, S. , & Adams, J. M. (2002). The Bcl2 family: Regulators of the cellular life‐or‐death switch. Nature Reviews Cancer, 2(9), 647–656. [DOI] [PubMed] [Google Scholar]
- Cui, Z. , Tang, J. , Chen, J. , & Wang, Z. (2014). Hsa‐miR‐574‐5p negatively regulates MACC‐1 expression to suppress colorectal cancer liver metastasis. Cancer Cell International, 14(1), 47–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, H. , Han, G. , Yan, Y. , Zhang, F. , Liu, Z. , Li, X. , … Zhang, Z. (2013). Transcript assembly and quantification by RNA‐Seq reveals differentially expressed genes between soft‐endocarp and hard‐endocarp hawthorns. PLoS one, 8(9), e72910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann, S. G. , Selvaraj, A. , & Thomas, G. (2007). mTOR Complex1–S6K1 signaling: At the crossroads of obesity, diabetes, and cancer. Trends in Molecular Medicine, 13(6), 252–259. [DOI] [PubMed] [Google Scholar]
- Deng, X. , Ruvolo, P. , Carr, B. , & May, W. S., Jr (2000). Survival function of ERK1/2 as IL‐3‐activated, staurosporine‐resistant Bcl2 kinases. Proceedings of the National Academy of Sciences of the United States of America, 97(4), 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Xiao, L. , Lang, W. , Gao, F. , Ruvolo, P. , & May, M. W., Jr (2001). Novel role for JNK as a stress‐activated Bcl2 kinase. Journal of Biological Chemistry, 276(26), 23681–23688. [DOI] [PubMed] [Google Scholar]
- Durkin, J. T. , Holskin, B. P. , Kopec, K. K. , Reed, M. S. , Spais, C. M. , Steffy, B. M. , … Meyer, S. L. (2004). Phosphoregulation of mixed‐lineage kinase 1 activity by multiple phosphorylations in the activation loop. Biochemistry, 43(51), 16348–16355. [DOI] [PubMed] [Google Scholar]
- Eulalio, A. , Huntzinger, E. , & Izaurralde, E. (2008). Getting to the root of miRNA‐mediated gene silencing. Cell, 132(1), 9–14. [DOI] [PubMed] [Google Scholar]
- Foster, K. G. , Acostajaquez, H. A. , Romeo, Y. , Ekim, B. , Soliman, G. A. , Carriere, A. , … Fingar, D. C. (2010). Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. Journal of Biological Chemistry, 285(1), 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M. G. , Haas, B. J. , Yassour, M. , Levin, J. Z. , Thompson, D. A. , Amit, I. , … Regev, A. (2011). Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology, 29(7), 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J. U. , Agarwal, V. , Guo, H. , & Bartel, D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biology, 15(7), 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge, D. C. , Albanese, C. , Reuther, J. Y. , Pestell, R. G. , & Baldwin, B. A., Jr (1999). NF‐κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Molecular & Cellular Biology, 19(8), 5785–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, H. , & Liu, X. (2013). Characterization of transcriptional complexity during longissimus muscle development in bovines using high‐throughput sequencing. PLoS one, 8(6), e64356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, J. , An, X. , Song, Y. , Cao, B. , Yang, H. , Zhang, Z. , … Li, Y. (2017). Detection and comparison of microRNAs in the caprine mammary gland tissues of colostrum and common milk stages. BMC Genetics, 18(1), 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, J. , Jin, J. , Cardiff, R. D. , Woodgett, J. R. , & Muller, W. J. (2001). Activation of Akt (protein kinase B) in mammary epithelium provides a critical cell survival signal required for tumor progression. Molecular & Cellular Biology, 21(6), 2203–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck, W. R. , & Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nature Biotechnology, 32(5), 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, S. , Ye, G. , Zhang, J. , Wang, L. , Wang, Z. , Zhang, T. , … Yang, J. Y. (2013). miR‐574‐5p negatively regulates Qki6/7 to impact β‐catenin/Wnt signaling and the development of colorectal cancer. Gut, 62(5), 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , & Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research, 27(1), 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Goto, S. , Sato, Y. , Kawashima, K. , Furumichi, M. , & Tanabe, M. (2014). Data, information, knowledge, and principle: Back to metabolism in KEGG. Nucleic Acids Research, 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, D. , Choi, S. , Jeong, H. , Jang, J. H. , Lee, Y. , Jeon, H. , … Ryu, S. H. (2012). Osmotic stress regulates the mammalian target of rapamycin (mTOR) complex 1 via c‐Jun N‐terminal kinase (JNK)‐mediated Raptor protein phosphorylation. Journal of Biological Chemistry, 287(22), 18398–18407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler, S. , Fleming, Y. , Goedert, M. , & Cohen, P. (1998). Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Current Biology, 8(25), 1387–1390. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Li, X. , Guo, Z. , Xu, F. , Xia, J. , Liu, Z. , & Ren, T. (2012). MicroRNA‐574‐5p was pivotal for TLR9 signaling enhanced tumor progression via down‐regulating checkpoint suppressor 1 in human lung cancer. PLoS one, 7(11), e48278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. M. , Wang, C. M. , Li, Q. Z. , & Gao, X. J. (2012). MiR‐15a decreases bovine mammary epithelial cell viability and lactation and regulates growth hormone receptor expression. Molecules, 17(10), 12037–12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, M. (2009). MicroRNA: A new entrance to the broad paradigm of systems molecular medicine. Physiological Genomics, 38(2), 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPiccolo, J. , Blumenthal, G. M. , Bernstein, W. B. , & Dennis, P. A. (2008). Targeting the PI3K/Akt/mTOR pathway: Effective combinations and clinical considerations. Drug Resistance Updates, 11(1), 32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L. M. , Li, Q. Z. , Huang, J. G. , & Gao, X. J. (2013). Proteomic and functional analyses reveal MAPK1 regulates milk protein synthesis. Molecules, 18(1), 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, E. C. , Qureshi, A. , Dasa, V. , Freitas, M. A. , Gimble, J. M. , & Davis, T. A. (2016). MicroRNA regulation of stem cell differentiation and diseases of the bone and adipose tissue: Perspectives on miRNA biogenesis and cellular transcriptome. Biochimie, 124, 98–111. [DOI] [PubMed] [Google Scholar]
- Memczak, S. , Jens, M. , Elefsinioti, A. , Torti, F. , Krueger, J. , Rybak, A. , … Rajewsky, N. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. [DOI] [PubMed] [Google Scholar]
- Nie, F. , Liu, T. , Zhong, L. , Yang, X. , Liu, Y. , Xia, H. , … Chen, T. (2016). MicroRNA‐148b enhances proliferation and apoptosis in human renal cancer cells via directly targeting MAP3K9. Molecular Medicine Reports, 13(1), 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, P. P. , Shahbazian, D. , Vu, H. , Holz, M. K. , Cohen, M. S. , Taunton, J. , … Blenis, J. (2007). RAS/ERK signaling promotes site‐specific ribosomal protein S6 phosphorylation via RSK and stimulates cap‐dependent translation. Journal of Biological Chemistry, 282(19), 14056–14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabapathy, K. , Hochedlinger, K. , Nam, S. Y. , Bauer, A. , Karin, M. , & Wagner, E. F. (2004). Distinct roles for JNK1 and JNK2 in regulating JNK activity and c‐Jun‐dependent cell proliferation. Molecular Cell, 15(5), 713–725. [DOI] [PubMed] [Google Scholar]
- Salzman, J. , Gawad, C. , Wang, P. L. , Lacayo, N. , & Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS one, 7(2), e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant, G. V. , & Sylvester, P. W. (2006). Gamma‐tocotrienol inhibits ErbB3‐dependent PI3K/Akt mitogenic signaling in neoplastic mammary epithelial cells. Cell Proliferation, 39(6), 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiah, S. G. , Hsiao, J. R. , Chang, W. M. , Chen, Y. W. , Jin, Y. T. , Wong, T. Y. , … Chang, J. Y. (2014). Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt‐7b. Cancer Research, 74(24), 7560–7572. [DOI] [PubMed] [Google Scholar]
- Shimamura, A. , Ballif, B. A. , Richards, S. A. , & Blenis, J. (2000). Rsk1 mediates a MEK‐MAP kinase cell survival signal. Current Biology, 10(3), 127–135. [DOI] [PubMed] [Google Scholar]
- Shukla, G. C. , Singh, J. , & Barik, S. (2011). MicroRNAs: Processing, maturation, target recognition, and regulatory functions. Molecular & Cellular Pharmacology, 3(3), 83–92. [PMC free article] [PubMed] [Google Scholar]
- Slattery, M. L. , Lundgreen, A. , & Wolff, R. K. (2012). MAP kinase genes and colon and rectal cancer. Carcinogenesis, 33(12), 2398–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G. K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology, 3(3), Article3. [DOI] [PubMed] [Google Scholar]
- Stahl, J. M. , Sharma, A. , Cheung, M. , Zimmerman, M. , Cheng, J. Q. , Bosenberg, M. W. , … Robertson, G. P. (2004). Deregulated Akt3 activity promotes the development of malignant melanoma. Cancer Research, 64(19), 7002–7010. [DOI] [PubMed] [Google Scholar]
- Stark, M. S. , Woods, S. L. , Gartside, M. G. , Bonazzi, V. F. , Dutton‐Regester, K. , Aoude, L. G. , … Hayward, N. K. (2011). Frequent somatic MAP3K5 and MAP3K9 mutations in metastatic melanoma identified by exome sequencing. Nature Genetics, 44(2), 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona, S. , Furió‐Tarí, P. , Turrà, D. , Pietro, A. D. , Nueda, M. J. , Ferrer, A. , & Conesa, A. (2015). Data quality aware analysis of differential expression in RNA‐seq with NOISeq R/Bioc package. Nucleic Acids Research, 43(21), e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. , & Salzberg, S. L. (2009). TopHat: Discovering splice junctions with RNA‐Seq. Bioinformatics, 25(9), 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie, S. , Ashburner, M. , Falls, K. , Leyland, P. , McQuilton, P. , Marygold, S. , … Zhang, H. (2009). FlyBase: Enhancing Drosophila Gene Ontology annotations. Nucleic Acids Research, 37, 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes, I. , Haanen, C. , & Reutelingsperger, C. (2000). Flow cytometry of apoptotic cell death. Journal of Immunological Methods, 243(1), 167–190. [DOI] [PubMed] [Google Scholar]
- Wang, C. , & Li, Q. (2007). Identification of differentially expressed microRNAs during the development of the Chinese murine mammary gland. Journal of Genetics & Genomics, 34(11), 966–973. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Lin, Y. , Bian, Y. , Liu, L. , Shao, L. , Lin, L. , … Li, Q. (2014). Leucyl‐tRNA synthetase regulates lactation and cell proliferation via mTOR signaling in dairy cow mammary epithelial cells. International Journal of Molecular Sciences, 15(4), 5952–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Rhodes, C. J. , & Lawrence, J. C., Jr (2006). Activation of the mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. Journal of Biological Chemistry, 281(34), 24293–24303. [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Pattingre, S. , Sinha, S. , Bassik, M. , & Levine, B. (2008). JNK1‐mediated phosphorylation of Bcl‐2 regulates starvation‐induced autophagy. Molecular Cell, 30(6), 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Maroney, A. C. , Dobrzanski, P. , Kukekov, N. V. , & Greene, L. A. (2001). The MLK family mediates c‐Jun N‐terminal kinase activation in neuronal apoptosis. Molecular & Cellular Biology, 21(14), 4713–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F. , Lv, J. , Gan, H. , Li, Y. , Wang, R. , Zhang, H. , … Chen, Y. (2015). MiRNA profile of osteosarcoma with CD117 and stro‐1 expression: miR‐1247 functions as an onco‐miRNA by targeting MAP3K9. International Journal of Clinical & Experimental Pathology, 8(2), 1451–1458. [PMC free article] [PubMed] [Google Scholar]
- Zhou, R. , Zhou, X. , Yin, Z. , Guo, J. , Hu, T. , Jiang, S. , … Wu, G. (2016). MicroRNA‐574‐5p promotes metastasis of non‐small cell lung cancer by targeting PTPRU. Scientific Reports, 6, 35714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, W. X. , Lindsten, T. , Ross, A. J. , MacGregor, G. R. , & Thompson, C. B. (2001). BH3‐only proteins that bind pro‐survival Bcl‐2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes & Development, 15(12), 1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information


