Summary
From the cellular perspective, organ growth is determined by production and growth of cells. Uncovering how these two processes are coordinated is essential for understanding organogenesis and regulation of organ growth.
We utilized phenotypic and genetic variation of 252 natural accessions of Arabidopsis thaliana to conduct genome‐wide association studies (GWAS) for identifying genes underlying root growth variation; using a T‐DNA line candidate approach, we identified one gene involved in root growth control and characterized its function using microscopy, root growth kinematics, G2/M phase cell count, ploidy levels and ribosome polysome profiles.
We identified a factor contributing to root growth control: Arabidopsis Adenylate Kinase 6 (AAK6). AAK6 is required for normal cell production and normal cell elongation, and its natural genetic variation is involved in determining root growth differences between Arabidopsis accessions. A lack of AAK6 reduces cell production in the aak6 root apex, but this is partially compensated for by longer mature root cells. Thereby, aak6 mutants exhibit compensatory cell enlargement, a phenomenon unexpected in roots. Moreover, aak6 plants accumulate 80S ribosomes while the polysome profile remains unchanged, consistent with a phenotype of perturbed ribosome biogenesis.
In conclusion, AAK6 impacts ribosome abundance, cell production and thereby root growth.
Keywords: Arabidopsis thaliana, cell elongation, cell proliferation, natural variation, ribosome biogenesis, root growth and development
Introduction
The growth of organisms depends on cell growth and cell division. These two processes need to be coordinated to maintain the characteristic sizes of cells (cell size homeostasis), organs and the whole organism. Two of the best‐characterized models of the coordination of cell division and growth are fission (Schizosaccharomyces pombe) and budding yeasts (Saccharomyces cerevisiae). There, cell size homeostasis is maintained by regulating cell division depending on growth. Blocking cell growth by nutrient deprivation leads to cell cycle arrest in G1 phase and cessation of cell division. Conversely, when cell division is blocked by arrest of cell cycle, at various stages, protein content continues increasing and cells keep growing until eventual cellular lysis (Mitchison & Creanor, 1971; Johnston et al., 1977). This led to the hypothesis that cell division depends on cell growth that is monitored by a certain parameter, which has to reach a threshold value before cell division is initiated. Parameters proposed to control cell division initiation include a cell's age (Trucco, 1970), size (Fantes & Nurse, 1977; Johnston et al., 1977) or biosynthetic capacity (the ability of a cell to synthesize organic molecules) (Unger & Hartwell, 1976; Brooks, 1977). The hypothesis of size‐dependent cell division is supported by the identification of the geometric size sensor Pom1 regulating cell division in fission yeast (Martin & Berthelot‐Grosjean, 2009; Moseley et al., 2009). In budding yeast, methionyl‐tRNA concentration has been suggested as the decisive signal reporting cell's capacity to synthesize proteins (Unger & Hartwell, 1976). It is not surprising that protein synthesis is viewed as a key measure of cell growth (Polymenis & Aramayo, 2015) – proteins are needed for all cellular functions and represent the most abundant macromolecules in cellular dry mass. Interestingly, a significant fraction of eukaryotic proteome – 10–20% in yeast (Liebermeister et al., 2014) and 7–16% in Arabidopsis accessions (Ishihara et al., 2017) – is dedicated to assembly of ribosomes that in turn synthesize more proteins. It is the ribosome abundance and energy that are implicated to be the growth‐limiting factors in higher eukaryotes (slow‐growing respiring cells), in contrast to limitation of growth by gene transcription and/or translation in rapidly growing microorganisms (Kafri et al., 2016).
However, there is a fine balance between the benefits of ribosomes and their costs. Specifically, the production and use of ribosomes are the most costly expenses of growth (Russell & Cook, 1995; Rolfe & Brown, 1997; Warner, 1999). The multi‐step, energy‐demanding ribosome biogenesis includes synthesis of pre‐ribosomal RNAs in the nucleolus, synthesis of ribosomal proteins in the cytoplasm, pre‐ribosome assembly in the nucleolus, processing of pre‐ribosomal RNAs and transport of ribosome subunits to their final workplace in the cytoplasm (excellently characterized in yeast; Woolford & Baserga, 2013).
Only a fraction of yeast ribosome biogenesis orthologues in plants have been functionally characterized (Barakat et al., 2001; Weis et al., 2015). At the same time, mutant phenotypes of genes involved, or with a suggested role, in ribosome biogenesis (toz, swa1, nuc‐l1/paralell1, ebp1) (Shi et al., 2005; Horváth et al., 2006; Griffith et al., 2007; Kojima et al., 2007; Petricka & Nelson, 2007) phenocopy ribosomal structural protein defects. The developmental impact of proteins involved structurally or functionally in ribosome biogenesis overlaps. The respective mutant phenotypes generally occur in tissues requiring high protein synthesis and/or transcript of a critical gene involved in promoting proliferation and development (reviewed by Byrne, 2009).
Mutations in a few Arabidopsis orthologues of yeast ribosomal assembly or structural proteins have been associated with a phenomenon termed compensatory cell enlargement (CCE) (Fujikura et al., 2009). CCE is an excessive postmitotic cellular growth that follows a decrease of cell production that is thereby compensated for at the organ level. CCE occurs in plants and animals as well (Hemerly et al., 1995; Neufeld et al., 1998; De Veylder et al., 2001; Ferjani et al., 2007; Roeder et al., 2010; Worley et al., 2013). In plants, CCE has been described in organs of determinate growth (e.g. leaves (Ferjani et al., 2007), petals (Randall et al., 2015) and sepals (Roeder et al., 2010)). However, for organs of indeterminate growth – such as roots – it has been thought not to occur (Ferjani et al., 2007).
Shoots and roots grow throughout the entire lifetime of higher plants. This growth is enabled by cell divisions and postmitotic cellular expansion specifically at the shoot and root apices. In the root apex, cell divisions occur in the meristematic zone directly adjacent to the quiescent centre and the stem cell niche, while the postmitotic growth starts when cells exit the meristematic zone. The postmitotic increase in size of root cells is accompanied by endoreplication (Hayashi et al., 2013). According to the current understanding, the coordination of cell division and cell growth in the root apex is flexibly coupled (Yang et al., 2017). Moreover, phytohormone crosstalk (e.g. auxins‐cytokinins, auxins‐brassinosteroids and gibberellins) determines the root apex zonation noncell‐autonomously (Ubeda‐Tomás et al., 2009; Moubayidin et al., 2010, Ubeda‐Tomás et al., 2012, Chaiwanon & Wang, 2015). Much of the work on growth regulation in plants has focused on environmental stresses (Guan et al., 2013; Wang et al., 2014), hormones (Ishida et al., 2009; Moubayidin et al., 2010) and cell cycle genes (De Schutter et al., 2007; Inagaki & Umeda, 2011). Despite indications of a role for ribosome biogenesis in plant development and growth, its involvement in the coordination of cell production and cell expansion in the root development remained unknown.
Here we report on the identification of a factor contributing to the control of root growth – the Arabidopsis Adenylate Kinase 6 (AAK6) that affects cellular growth by impacting the assembly of ribosomes. We show that 80S ribosome complexes overaccumulate in mutants of this gene (aak6), orthologue of yeast ribosome assembly factor Fap7. Cell production is lower in aak6 plants, resulting in fewer cells in aak6 roots. This reduced cell production is partially compensated for by additional postmitotic cellular growth. Nevertheless, aak6 roots are overall shorter and slower‐growing. Cyclin expression and ploidy levels in the aak6 mutant suggest that AAK6 activity is required for normal cell cycle progression during cell proliferation and endoreplication. Moreover, association of natural allelic variation of AAK6 with measured root growth differences suggest a contribution of ribosome assembly to root organ growth among natural Arabidopsis accessions. This work thereby demonstrates a link between ribosome assembly and growth rate modulation.
Materials and Methods
Plant material
We used seeds of 252 Arabidopsis thaliana accessions (Supporting Information Table S1; Notes S1) in the genome‐wide association study (GWAS).
Available T‐DNA insertion lines within 4000 bp upstream or downstream (Table S2) for two of the top five associated single nucleotide polymorphisms (SNPs) (Fig. S1d) were ordered from Nottingham Arabidopsis Stock Centre, for candidate gene prioritization (Notes S2).
Seed size variance was controlled by two sieving steps using Retsch sieves (60.131.000250 and 60.131.000280, Retsch GmbH, Haan, Germany), for all genotypes. Seeds < 250 µm and > 280 µm in size were excluded.
Growth conditions
Seeds were surface‐sterilized, plated, stratified and cultured as described in Slovak et al. (2014). In particular, after stratification, seeds were transferred to the growth chamber and allotted 48 h for germination. At this time point, on the second day after stratification (DAS2), the vast majority of the seedlings germinated.
Growth quantification
Plates with growing seedlings were moved to the image acquisition room once a day, from DAS3 (corresponding to the first day after germination) for 5 d until DAS7, scanned and, after imaging, returned immediately to the growth chamber. The root traits were quantified by high‐throughput plugin BRAT including manual quality control step, as described in Slovak et al. (2014).
Genome‐wide association studies
We evaluated 78 GWASs executed on 14 root morphology and two growth rate traits evaluated for five time points of a 5‐d‐long growth assay, four end points respectively for the ‘rate' traits. The GWASs are considered significant when they pass the 5% Benjamini–Hochberg–Yekutieli false discovery rate (FDR) threshold. Arithmetic means (n > 5) of 252 accessions were utilized for these GWASs. The computations were performed on a Gregor Mendel Institute computer cluster using algorithms described in Seren et al. (2012). We took into account SNPs with minor allele counts ≥ 12.
Effects of seed size restriction on GWASs
We compared counts of significant genome‐wide associations between the current size‐controlled dataset and our previous study that did not control seed size (Slovak et al., 2014). Both datasets were downsized to 160 accessions (representing the overlap of both studies). We compared cumulative counts from across 78 GWASs executed on 14 root morphology and two growth rate traits evaluated for five time points of a 5‐d‐long growth assay, four end points respectively for the ‘rate' traits.
Plasmid construction and plant transformation
The following plasmids were constructed by In‐Fusion cloning (Clontech Laboratories Inc., Mountain View, CA, USA) into the modified pGreen0229 that has p35S:PM‐mCherry reporter gene for visual selection of positive transformants (Hellens et al., 2000; Emami et al., 2013; Satbhai et al., 2017), kindly provided by our colleague Dr Santosh Satbhai. The following plasmids were verified by Sanger sequencing and the respective T–DNAs transferred by Agrobacterium‐mediated transformation by floral dipping (Weigel & Glazebrook, 2006). We used a Zeiss Discovery.V8 stereo‐microscope to visually select transgenic seeds, and finally obtained homozygous transgenic seeds in the third generation. The primers are listed in Table S3.
35S:AAK6 overexpressor line
The AAK6 coding sequence, amplified from Columbia‐0 (Col‐0) genomic DNA (primer pair P18, P19; Table S3), was inserted in the modified pGreen0229 under the control of the 35S promoter and the translational enhancer (TL‐TEV) (primer pair P16, P17; Table S3) to form an overexpression vector.
The 35S:AAK6 overexpression was validated (see Methods S1; Fig. S2a).
Complementation of aak6 by AAK6 natural alleles
The DNA fragment, amplified from genomic DNA of Bå4‐1, Col‐0 and Tha‐1 accessions, respectively (corresponding to the 1823 bp region in the reference genome of Col‐0 – covering 788 bp upstream to 241 bp downstream from CDS), was inserted into the modified pGreen0229 (primer pair P20, P21; Table S3).
We generated 50 independent insertion lines for each of the three natural AAK6 alleles, checked the segregation ratio of the mCherry marker in the T2 generation and chose seven independent lines, with segregation ratio closest to 3 : 1, for further propagation. All inserted AAK6 alleles complemented the dwarf phenotype of the original aak6 background. In the third generation, we obtained seven independent, homozygous insertion lines for each of the three AAK6 alleles (from Bå4‐1, Col‐0 and Tha‐1 accessions).
pAAK6:AAK6‐Clover reporter line
The expression vector was constructed using Multisite Three‐fragment Gateway cloning (Thermo Fisher Scientific, Vienna, Austria). The native promotor (788 bp upstream of the translation start site) and the AAK6 coding sequence without the stop codon were cloned into Gateway entry vectors pDONR P4‐P1r and pDONR221, respectively (primer pairs P22 and P23, and P24 and P25; Table S3). The Clover reporter transgene in pENTR P2r‐P3 was kindly provided by Dr Tomokazu Kawashima. These three entry vector clones and the destination vector pK7m34GW were then recombined in the Multisite Gateway LR+ recombination reaction to create the pAAK6:AAK6‐Clover expression vector. This plasmid was verified by Sanger sequencing and the respective T‐DNA transferred into the Col‐0 background using Agrobacterium transformation (Weigel & Glazebrook, 2006). Positive transformants were selected on kanamycin 1 × MS medium agar plates, plants were propagated on soil and second‐generation seedlings were used for analysis of the spatial pattern of AAK6‐Clover protein expression in vivo.
Root anatomy measurements
Static measurements of cell lengths and root zones by confocal microscopy
Wild‐type Col‐0 and aak6 mutant (SALK_015289) seeds were surface‐sterilized, stratified and cultured on 1× MS, 1% sucrose, 0.8% agar plates and seedlings were grown until DAS5. Seedlings were incubated for 10 s with 15 μg ml–1 propidium iodide, washed, transferred to standard microscopy slides, and mounted in MonoQ water (VBCF, Vienna, Austria) under coverslip. An LSM 700 Axio Observer.Z1 confocal microscope (Zeiss) with a motorized stage was used for confocal image acquisition. Using zen2010 software (Zeiss), ×20 dry objective (Zeiss Plan‐Apochromat ×20/0.8) we acquired 12 z‐stacks of > 20 planes spaced at 2.4 µm (400 × 400 μm) for each root, with a resolution of 0.78 μm per pixel. Propidium iodide was excited at 488 nm with 10% laser power and detected at 576–700 nm. Twelve stacks were stitched for each root using the fiji stitching plugin (Preibisch et al., 2009) post‐acquisition.
We measured root cortex cell lengths using the manual segmentation tool in fiji starting with the first cell after the endodermis/cortex initial and continuing cell‐by‐cell past the maturation zone boundary. The first cell being twice as long as the previous one, with a longer subsequent cell (to skip dividing cells) defined the rapid elongation zone start. The meristem zone length was measured from the root quiescent centre to the rapid‐elongation‐zone boundary using the zone determination as described earlier. The number of cells in the meristematic zone was determined by counting the cells after the endodermis/cortex initial until the first cell of the rapid elongation zone. The mature zone start is defined as a position after which cell elongation stops and is detected in the longitudinal dimension after the rapid elongation zone where a cell has approximately the same length as the following one. The mature cell length was calculated as the average of first five cells in the mature zone.
We evaluated these measurements for both right and left cortex cell files in the root middle plane and subsequently calculated the average for each seedling. We analysed 10 seedlings for wild‐type Col‐0 and aak6 genotypes each, in blinded analyses.
Kinematic analyses of root zones, cell production and postmitotic growth
Wild‐type Col‐0 and aak6 mutant (SALK_015289) seeds were surface‐sterilized (with 70% ethanol for 1 min and 5% bleach for 5 min and rinsed five times with MilliQ water) and plated on 1 × MS, 0.1% sucrose, 0.8% agar plates. Plates were stored for 3 d at 4°C, after which they were placed vertically in a growth chamber under constant conditions (16 h : 8 h, light : dark, 20°C, 74.5 μmol PAR m− 2 s−1).
Kinematic analysis of cell division and elongation was performed on DAS8 (5–6 d after germination for the majority of plants). To obtain velocity measurements, black‐toner particles (Hewlett‐Packard, Belgium BVBA, Diegem, Belgium) were applied on the roots and their displacement measured after 1 h. Images of the particle‐sprinkled roots at start and end time points were acquired directly from the plates using a binocular microscope SMZ1000 (Nikon) at ×8 magnification equipped with a digital camera (AxioCam ICc 1; Zeiss), connected to a PC running axiovision v.4.8.2 software (Zeiss). Roots were then whole‐mounted on microscope slides in 1 × MS liquid medium, 0.1% sucrose and cortical cells visualized by an Axio Scope.A1 microscope (Zeiss) using differential interference contrast (DIC) optics at ×20 magnification. Images were again captured with an AxioCam ICm 1 camera (Zeiss) controlled by axiovision v.4.8.2 software (Zeiss).
Cell division and elongation parameters were determined by kinematic calculations as described previously (Beemster & Baskin, 1998) with minor modifications. In particular, the cell production was calculated by dividing the final velocity by the mature cell length. The cortical cell lengths were measured in imagej (Schneider et al., 2012). A ruler image was used for calibration. The cell flux (h−1) was calculated by dividing the velocity (µm h−1) by the cell length (µm) at the respective discrete distance points. The meristematic zone length was determined as the distance between the quiescent centre and the first point where the flux became ≥ 95% of the average of the remaining flux data points in the growth zone (i.e. when the flux curve reaches the plateau). The elongation zone length was determined as the distance between the meristematic zone boundary and the first point where the velocity became ≥ 95% of the average of the remaining velocity data points in the growth zone (i.e. when the fitted velocity curve reaches the plateau).
Cell proliferation analysis
To count root meristematic zone cells in the G2/M phase, we introgressed the pCycB1;1:CycB1;1DB‐GFP reporter generated by P. Doerner (Ubeda‐Tomás et al., 2009) into Col‐0 and aak6 –/– backgrounds, respectively. We phenotyped the F3 generation homozygous for CycB1;1DB‐GFP (+/+) in Col‐0 and aak6 –/– homozygous backgrounds, respectively, on DAS5.
Before imaging, seedlings were stained 10 s by propidium iodide (15 μg ml–1), washed in MonoQ water and mounted on microscope slides in MonoQ water (VBCF, Vienna, Austria) under a coverslip. Z‐stacks were acquired using a confocal microscope LSM 700 Axio Observer Z1 (Zeiss) with PlanApochromat ×20/0.8 objective, controlled by zen2010 software (Zeiss). Z‐stacks covered the range of planes from the root surface past the root middle plane with a 2.4 µm interval. In XY dimension, the stack comprised longitudinally the root meristematic and part of the rapid elongation zone within a single tile (400 μm2). A 488 nm laser, at 10% power, was used for green fluorescent protein (GFP) and propidium iodide excitation. Emitted light was recorded in a single track separated into two channels (shortpass 555 nm; longpass 560 nm; dichroic beam splitter (DBS) 572 nm).
Using the z‐stack confocal images described earlier, we first established the position of the root middle plain and manually registered and counted CycB1;1DB‐GFP‐positive cells in the root meristematic zone in all cell files from the surface to the root middle plain (disregarding columella and lateral root cap) using the fiji roi manager tool. We scored 10 seedlings of wild‐type Col‐0 (CycB1;1DB‐GFP +/+) and aak6 −/− (CycB1;1DB‐GFP +/+) genotypes each, in a blinded analysis.
Flow‐cytometric ploidy level assessment
Five‐millimetre‐long root tips were harvested from c. 100 seedlings, on DAS5, and placed in 250 ml of cold nuclei Extraction Buffer (Sysmex Partec GmbH, Görlitz, Germany) in a Petri dish on ice. Root tips were chopped with a new razor blade exchanged between every sample. The nuclei suspension was incubated for 2 h in 1 ml of CyStain PI Absolute P (Sysmex Partec GmbH) staining buffer in the dark on ice. The stained homogenate was filtered through a 30 μm CellTrics filters (Sysmex Partec GmbH). Stained particles were excited with 561 nm (110 mW) and 488 nm (100 mW) lasers and emission was detected for propidium iodide‐stained particles (610/20 nm) and forward scattering (488/10 nm). Fluorescence intensity was recorded for 10 000 particles for each sample. facs diva software (BD Lifesciences, Franklin Lakes, NJ, USA) was used for gating the nuclear populations. We analysed six pooled biological samples each for the Col‐0 wild‐type and the aak6 mutant.
Ribosome/polysome profiles
Plants were grown on 1 × MS agar plates for 3 wk after germination. Fresh polysome extraction buffer was prepared as follows: 0.2 M Tris‐HCl (pH 9.8), 0.2 M KCl, 0.025 M EGTA, 0.035 M MgCl2, 1% Detergent mix (1% (w/v) Brij35, 1% Triton X‐100, 1% (v/v) Igepal CA 630, 1% Tween 20), 1% sodium deoxycholate, 1% polyoxyethylene 10 tridecyl ether, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 50 μg ml–1 cycloheximide, 50 μg ml–1 chloramphenicol and 0.5 mg ml–1 heparin. Whole seedlings were harvested and manually grounded in liquid nitrogen. A unit of weight (1 g) of tissue powder was incubated for 10 min with 2 units of volume (ml) of polysome extraction buffer, on ice. Samples were centrifuged at 16 000 g for 15 min at 4°C, in an Eppendorf microcentrifuge. Supernatant was poured through Miracloth (Merck, Darmstadt, Germany) into a new chilled tube. Centrifugation at 16 000 g for 15 min was repeated at 4°C. 12 ml of 5–45% (w/v) linear sucrose gradients were prepared in 0.04 M Tris, 0.02 M KCl and 0.01 M MgCl2 salt solution (with 50 μg ml–1 cycloheximide, 50 μg ml–1 chloramphenicol) in thick‐walled polycarbonate tubes using Gradient Master 107 (Biocomp Instruments, Fredericton, Canada). Clarified ribosome/polysome extracts (500 μl) were layered on top of sucrose gradients. Tubes with samples and sucrose gradients were centrifuged at 40 000 g for 2.5 h, at 4°C, in an SW40 rotor, Optima L‐70 ultracentrifuge (Beckman Coulter, Brea, CA, USA). UV 254 nm absorbance was measured using an ÄKTAFPLC machine (GE Healthcare Life Sciences, Vienna, Austria). We analysed three biological pooled replicates each originating from 144 plants for the wild‐type Col‐0 and aak6 mutant genotype.
Results
A seed size‐controlled root trait dataset identifies an unknown root growth factor
Previous studies on root growth and gravitropism suggested that early root traits are impacted by seed size and other maternal effects (Elwell et al., 2011). Additionally, we observed a correlation between average seed size and average root growth traits (published previously for 163 accessions) (e.g. for total lengthDAS7, r = 0.43; Fig. S1a; Table S4; Methods S2). To minimize maternal and seed size effects of 252 accessions, we used sieves to restrict seed sizes to 250–280 μm. We then quantified 16 root growth traits over 5 d, from DAS3 to DAS7, using the BRAT phenotyping (Slovak et al., 2014). While these new seed size‐controlled early root growth traits correlate with the traits from the dataset not controlling seed size (Slovak et al., 2014) (e.g. root lengthDAS7, Pearson's r = 0.75; Table S4), restricting seed size variance led to a decrease in variance within accessions (median accession's SDDAS7 = 2.6 mm → median accession's SDDAS7 = 1.8 mm) as well as between accessions (e.g. for primary root lengthDAS7, SDDAS7 = 2.9 mm → SDDAS7 = 2.3 mm) (Table S4). Despite the variance reduction in early root growth traits, the remarkable natural variation of root traits was still preserved (e.g. c. three‐fold difference in primary root growth on DAS7 between accessions: from 1.6 to 4.6 mm d–1; Fig. S1b). Upon restriction of seed size variance, broad‐sense heritability increased for most traits (e.g. primary root lengthDAS7, H = 0.49 → H = 0.50; average root growth rate: H = 0.45 → H = 0.50; Table S4). Using our seed size‐controlled phenotype dataset, we then conducted GWASs using the EMMA(X) method that corrects for population structure (Kang et al., 2008, 2010; Seren et al., 2012). We focused on dissecting the molecular basis of the quantitative root growth, despite having conducted 77 more GWASs for additional root growth traits (Table S5, Spreadsheet 1). Although no genome‐wide association crossed the 5% FDR significance threshold for the root growth rate on DAS7, we identified multiple distinct genome‐wide association peaks (Fig. S1c).
We focused on two of the top five associated genomic regions (Fig. S1d). Because the top SNP, identified by GWAS, frequently is not the causal SNP, but rather only marks the genomic region in which causal nucleotide polymorphisms reside, we screened the available T‐DNA lines targeting genes within 8 kb regions comprising the top associated marker SNPs (Table S2). One of these lines (SALK_015289) developed significantly shorter roots (P < 0.001; Fig. 1). The data therefore suggested that this line is defective in a gene required for root growth. SALK_015289 has a T‐DNA insertion in the 5′ untranslated region of Arabidopsis Adenylate Kinase 6 (AAK6; AT5G60340) leading to severe downregulation of AAK6 expression level (Fig. S2a). Importantly, using the Col‐0 genomic region (1823 bp, 788 bp upstream, 241 bp downstream) we could complement the known aak6 dwarf mutant phenotype (Feng et al., 2012) (Fig. S3) and partially complement the shorter root and slower growth (Fig. S4) excluding a background mutation causing these phenotypes. Consistent with a requirement in normal root growth, pAAK6:AAK6‐Clover reporter construct is expressed in nuclei of all cell types in primary and lateral root meristematic zones (Fig. 2). Furthermore, AAK6 shows general predominant expression in actively dividing tissues in Arabidopsis (TraVa RNAseq expression atlas; Klepikova et al., 2016). Nonetheless, overexpression of AAK6 under 35S promoter did not lead to longer roots or their faster growth (Fig. S2b). In conclusion, the data show that the AAK6 gene is required for normal root growth but its overexpression is not sufficient for faster root growth.
Figure 1.
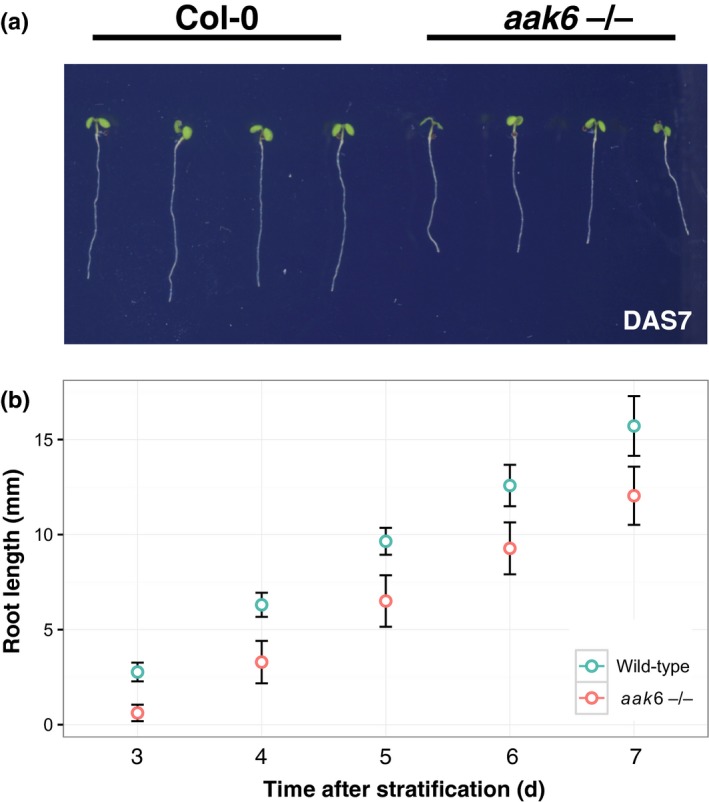
Primary root growth of Arabidopsis thaliana 1 wk after stratification. (a) Representative seedlings of wild‐type and aak6 genotype, on day 7 after stratification (DAS7). (b) Primary root length on DAS3–DAS7; t‐test, P < 0.001 on each of the 5 d, means ± SD.
Figure 2.
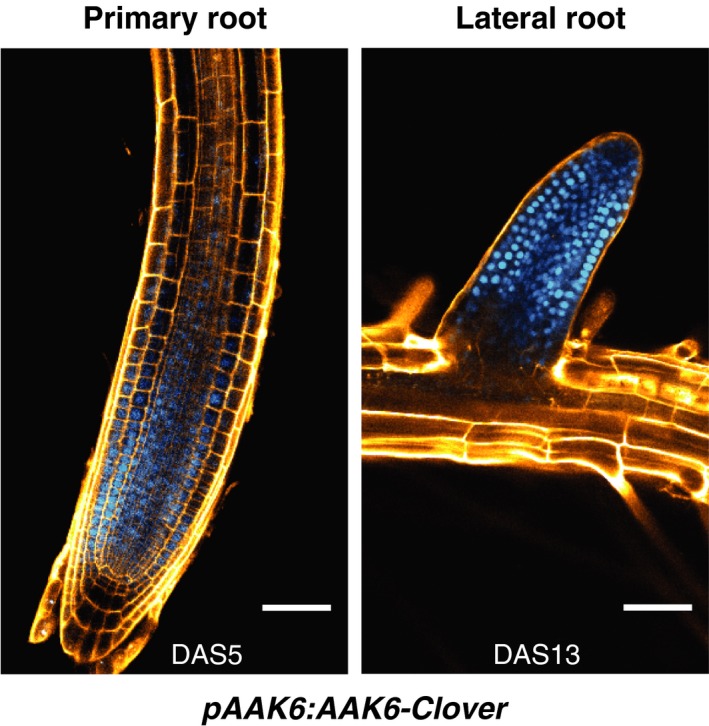
Expression of pAAK6:AAK6‐Clover translational reporter. AAK6‐Clover translational fusion protein (shown in blue) has nuclear localization in files of all cell types in primary and lateral root meristems of Arabidopsis thaliana (on 5th and 13th day after stratification, DAS5 and DAS13). Cell walls are stained by propidium iodide (shown in orange). Bars, 50 μm.
AAK6 natural allelic variation is involved in root growth rate determination
The GWAS, followed by the mutant phenotype, suggested that AAK6 natural genetic variation matters in determining the different root growth rates observed in our panel of accessions. However, to directly test this, we used a transgenic complementation approach. We classified the accessions into haplotypes and tested whether transforming the slow‐growing aak6 mutant with a genomic fragment – including AAK6 promotor and coding region – from one of the slower‐growing haplotypes (Bå4‐1) would result in plants displaying slower root growth than plants transformed with an allele from the faster‐growing haplotypes, such as Col‐0 or Tha‐1. We generated 50 independent complementation lines for each of the three alleles, and chose seven for each allele – with the mCherry selection marker segregation ratio closest to 3 : 1 in the T2 generation. We then phenotyped the T3 generation homozygous seedlings. Complementation of the loss‐of‐function aak6 mutant with the allele from the slower‐growing haplotype resulted in significantly slower root growth of seedlings than observed in plants complemented with alleles from the faster‐growing haplotypes (Tukey honestly significant difference, P adj = 0.02 and P adj = 0.09 for the Col‐0 and Tha‐1 genomic fragment, respectively; jointly tested allele effect via ANOVA, P = 0.02; Fig. 3). As there are four nonsynonymous amino acid substitutions in the coding region of AAK6 genomic fragments used for aak6 complementation (Table S6), these are the prime candidates for causing the phenotypes. In summary, we show that allelic variation of the tested AAK6 genomic fragments determines root growth differences and therefore conclude that natural genetic variation in the AAK6 gene is involved in determining the root growth differences between natural accessions.
Figure 3.
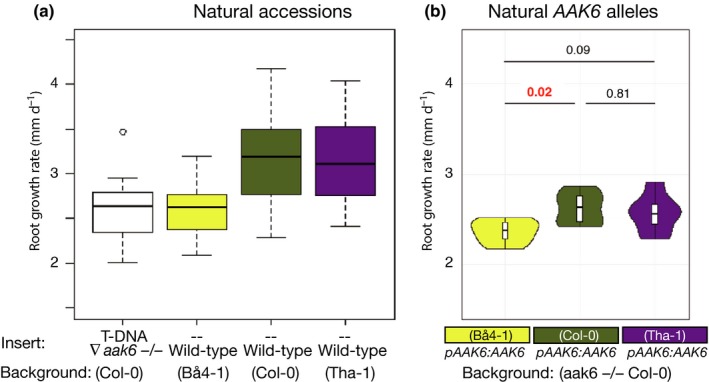
Complementation of Arabidopsis thaliana aak6 mutant with natural AAK6 alleles. (a) Root growth rate of natural accessions and aak6 mutant on day 7 after stratification (n > 15). Whiskers represent the last value at a distance smaller than 1.5 × the interquartile range (IQR). (b) Root growth rate of transgenic plants carrying natural AAK6 DNA fragment in aak6 −/− background on day 7 after stratification. Shown are medians with interquartile range (black line in white box) and density distribution in a violin plot trimmed to the range of the data (n = 7 independent insertion lines for each of the three constructs); Tukey honestly significant difference adjusted P‐values are indicated above the violin plots.
AAK6 is required for normal cell production and normal cell elongation
To understand the role of AAK6 in root development, we examined plants homozygous for aak6. These have shorter roots and decreased root growth rate. Therefore, cell proliferation and/or cell elongation could be altered as these are the two main processes that determine the root growth rate. We assessed this by two independent sets of experiments (confocal microscopy and kinematic analysis).
First, we investigated cell proliferation. Using confocal microscopy, we observed significantly fewer cells (P = 0.03) in the meristematic zone of aak6 roots (Figs 4a,c, S5a,b), although the meristematic zone length was unaffected (P = 0.97). Kinematic analyses of root apical growth revealed that the aak6 root meristematic zone produces fewer cells compared with wild‐type plants (P = 0.01; Fig. 4d). Consistent with the confocal microscopy‐based observations, we independently observed a lower number of meristematic cells in the aak6 mutant using DIC microscopy (P = 0.04). Furthermore, the length of the meristematic zone again did not differ between the aak6 and the wild‐type (Fig. S5c,d). The average cell cycle duration, determined by kinematic analysis, is similar in the aak6 mutant and the wild‐type (Fig. 4e). We therefore conclude that the lower number of the proliferating cells in the aak6 meristematic zone is the predominant reason for the significantly lower cell production in the aak6 roots.
Figure 4.
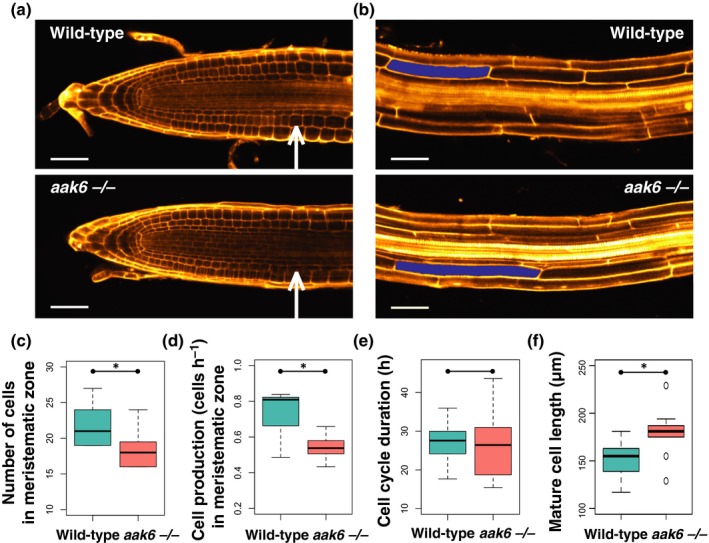
Effect of lack of AAK6 on root anatomy, cell production and elongation in Arabidopsis thaliana. (a) Wild‐type and aak6 −/− root meristematic zone. Cell walls are stained by propidium iodide (shown in orange). The white arrows indicate the first cortex cell in the zone of rapid elongation marking the end of the meristematic zone on day 5 after stratification (DAS5). Bars, 50 μm. (b) Image section from wild‐type and aak6 −/− root maturation zone on DAS5. Representative cortical cells are highlighted in blue. Cell walls are stained by propidium iodide (shown in orange). Bars, 50 μm. (c) Meristematic zone cell numbers in wild‐type and aak6 −/− roots evaluated in the cortex cell file on confocal microscopy images on DAS5. Whiskers represent the last value at a distance smaller than 1.5 × the interquartile range (IQR) (t‐test: *, P < 0.05). (d) Meristematic zone cell production (cells h–1), determined by kinematic analysis, on DAS8; whiskers < 1.5 × IQR (t‐test: *, P < 0.05). (e) Cell cycle duration (h), determined by kinematic analysis, on DAS8; whiskers < 1.5 × IQR (P = 0.88). (f) Wild‐type and aak6 −/− mature cortical cell length in the root, evaluated on confocal microscopy images, on DAS5; whiskers < 1.5 × IQR (t‐test: *, P < 0.05).
Next, we investigated the involvement of cell elongation in the decreased growth of the aak6 roots. We measured cell lengths along the longitudinal axis of the root (Fig. S5). While the length of cells leaving the meristematic zone was not significantly different between aak6 and the wild‐type, the final mature cell length of the aak6 mutants was, contrary to our expectations, 19% longer in the slower‐growing roots of the aak6 mutant (cortex cell tier, confocal microscopy, P < 0.01 (Fig. 4b,f); kinematic analyses, P‐value < 0.01 (Fig. S5d)). The aak6 mature cortical cells are generally affected – the whole mature cell size distribution is shifted towards longer cells (Fig. 4f). This could be the result of faster local cell elongation rates and/or the longer time a cell spends transiting the elongation zone (average duration of cell elongation). However, the profile of local relative cell elongation rates was not significantly different between aak6 mutant and wild‐type roots (kinematic analysis of the average local cell elongation rate, P = 0.93; Fig. S6a,b) and we observed only a statistically insignificant increase in the average time a cell spends in the elongation zone (9 h for wild‐type cells and 13 h for aak6 cells, P = 0.50; Fig. S6c). Given the exponential nature of the elongation process, a small increase in the duration of the postmitotic cell elongation has a strong effect on the final mature cell size and eventually leads to significantly larger mature cells.
Taken together, we observed a reduced cell production and an increased cell growth in the aak6 root. AAK6 activity is thus required for normal cell production and normal cell elongation. Moreover, given that the AAK6‐Clover translational fusion protein was expressed in nuclei of cells in the primary and lateral root meristematic zones (Fig. 2), we conclude that the AAK6 effect on the root cell expansion in the elongation zone is rather indirect. Last but not least, the aak6 mutation uncovered an unexpected case of a phenomenon termed the compensatory cell enlargement – in an organ of indeterminate growth – the Arabidopsis root.
AAK6 is required for normal cell proliferation while lack of AAK6 enhances endoreplication
The kinematic analysis of the cellular dynamics suggested that average cell cycle duration is similar in the aak6 mutant and the wild‐type. We therefore independently assessed if the duration of cell cycle phases is altered in aak6 roots. For this, we introgressed the G2/M phase reporter (pCycB1;1:CycB1;1DB‐GFP) into aak6 and wild‐type plants. Interestingly, the aak6 mutant showed significantly more CycB1;1‐GFP‐positive cells than did the wild‐type (P < 0.001; Fig. 5). The higher number of cells observed in the G2/M phase suggests an increase of G2/M phase duration. Longer G2/M despite the same cell cycle duration in turn suggests that G1/S is shorter.
Figure 5.
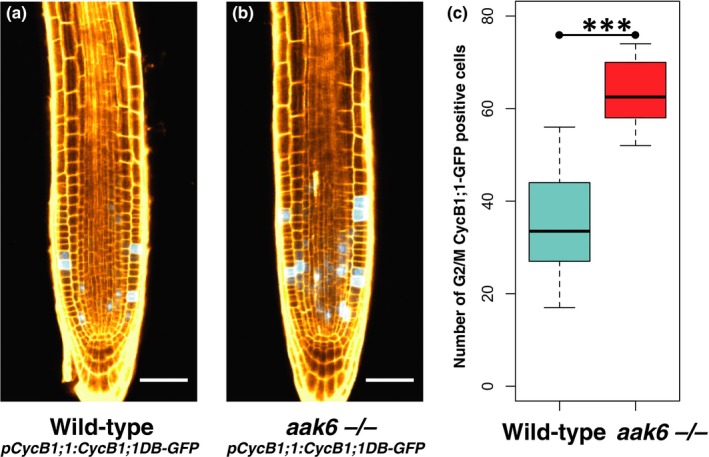
Arabidopsis thaliana aak6 −/− root meristematic zone contains more G2/M‐positive cells. (a, b) Representative confocal images showing the expression pattern of the pCycB1;1:CycB1;1DB‐GFP marker (shown in cyan), on day 5 after stratification in median longitudinal optical sections of wild‐type (a) and aak6 −/− root apices (b). Cell walls are stained by propidium iodide (shown in orange). Bars, 50 μm. (c) Box plot of G2/M CycB1;1‐GFP‐positive cells in wild‐type and aak6 −/− root apices; whiskers represent minimum and maximum values (t‐test: ***, P < 0.001).
As the endocycle only contains the G1/S transition, accelerated G1/S would lead to faster endocycling. We therefore set out to assess the extent of cell cycling and endocycling in the root tip cells. The nuclear ploidy levels were measured by flow‐cytometric analysis of propidium iodide‐stained nuclei isolated from 5‐mm‐long root tips. Interestingly, aak6 roots have significantly lower proportions of 2C and 4C nuclei and a significantly increased proportion of 16C nuclei (t‐test, P < 0.001; Fig. 6; Table S7). Therefore these data do indeed demonstrate increased endoreplication in the aak6 mutant root (for further discussion, see Notes S3).
Figure 6.
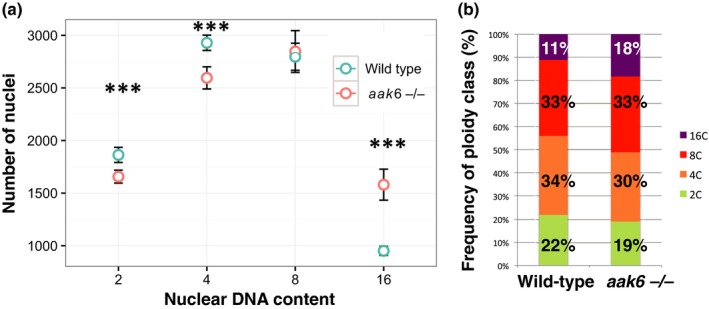
Nuclear ploidy levels in Arabidopsis thaliana wild‐type and aak6 root apices. For each of the two genotypes, we collected six pooled 5 mm root tip samples on day 5 after stratification. 10 000 Arabidopsis nuclei were analysed from each sample. (a) Flow‐cytometry profile (means ± SD) (t‐test: ***, P < 0.001). (b) Frequency of nuclear ploidy classes.
Overall, AAK6 is required for normal cell production in the root meristematic zone and normal progression through the cell cycle. A lack of AAK6 significantly lowers cell production and alters cell cycle progression, leading to increased endoreplication and CCE.
A lack of AAK6 leads to accumulation of 80S ribosomes
While previous work has demonstrated an in vitro adenylate kinase activity of AAK6, its functional role remained unclear (Feng et al., 2012). Interestingly, yeast (Fap7) and human (hCINAP) homologues of AAK6 are implicated in small ribosomal subunit maturation (Strunk et al., 2012; Bai et al., 2016). To test the hypothesis of AAK6 involvement in ribosome biogenesis, we evaluated ribosome‐polysome profiles from wild‐type and aak6 mutant plants. The respective cell lysates were fractionated by centrifugation in 5–45% sucrose gradient to examine the relative abundance of ribosomal subunits, ribosomes and polysomes. Compared with the wild‐type, aak6 seedlings contained increased amounts of 80S ribosomes (Figs 7, S7). These data suggest that upon AAK6 deficiency, the ribosome biogenesis becomes stalled at the 80S maturation step. Overall, our results show that an absence of AAK6 results in accumulation of 80S ribosomes in Arabidopsis, thereby linking ribosome biogenesis, cell production and postmitotic cellular growth in determining root growth.
Figure 7.
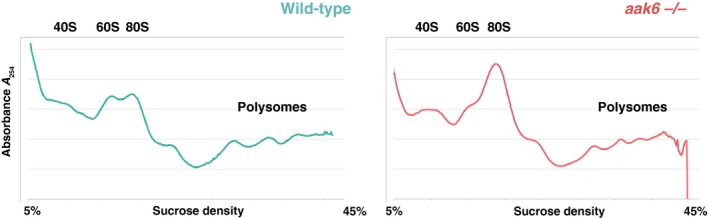
Ribosome/polysome profiles. Spectrophotometric profiles of UV absorbance (254 nm) of sugar gradients. Ribosome/polysome extracts were obtained from 3‐wk‐old whole seedlings of Arabidopsis thaliana. Ribosomal particles were fractionated through a 5–45% (w/v) sucrose gradient by 2.5‐h‐long ultracentrifugation.
Discussion
Despite the essentiality of ribosomes for growth and development, it was not known how modulating ribosome abundance impacts cell production and cell expansion in the root development. This study reports on the identification of AAK6 as a factor of quantitative root growth control, using phenotypic and genetic variation of natural populations of Arabidopsis thaliana. Our results demonstrate that AAK6 is required for normal cell production in the root meristematic zone and is consistent with repression of excessive cell expansion in the elongation zone. We have thereby characterized a case of a CCE in an organ of indeterminate growth (the root). Furthermore, the orthologue of AAK6 encodes the well‐characterized yeast ribosome assembly factor Fap7 (Loc'h et al., 2014; Peña et al., 2016). Our data suggest that the molecular function of AK6 has been conserved between yeast and plants, Arabidopsis AK6 is involved in the assembly of ribosomes, and its natural alleles confer differential root growth.
Controlling seed size leads to the identification of higher number of genome‐wide significant associations
High variance within samples reduces the statistical power of hypothesis testing and obscures the detection of growth differences between samples when they truly are present. As such, the variance of seed sizes impacts early root growth traits (this study; Elwell et al. 2011) and complicated identification of quantitative root growth factors. To reduce the variance of root growth as a result of variance in seed sizes, we used seeds of relatively homogeneous class (250–280 μm). Controlling the variance of seed sizes led to the identification of 138 genome‐wide significant associations across all the 78 studied traits cumulatively (14 morphology traits and two growth rate traits, each analysed during a 5‐d‐long growth assay; see Table S5, Spreadsheet 1). When we downsize the dataset from 252 to 160 accessions, present in our previous early‐root‐growth study (comparability described in the Materials and Methods section), then controlling the variance of seed sizes leads to the identification of 76 genome‐wide significant associations (Table S5, Spreadsheet 2) in contrast to 35 associations (Slovak et al., 2014); these counts refer to the genome‐wide significant associations across all the 78 studied traits cumulatively. Furthermore, this is also indicated by the GWAS association that led to the identification of AAK6 – while the most significantly associated AAK6 marker SNP displayed a P‐value of 0.1889936 in a previous dataset that did not control seed size (Slovak et al., 2014), the current seed size‐controlled dataset led to an association of this AAK6 SNP with a P‐value of 0.0000223 (downsized seed size‐controlled dataset, P = 0.0019019). In conclusion, controlling seed size can be beneficial to decrease the variance in early root growth traits that was present as a result of maternal and seed size effects. The limitation of controlling seed size before a growth assay is that we will probably miss out on the identification of genetic loci controlling seed size and the studied root growth trait simultaneously. Overall, we find that this practice can decrease the variance of early root growth traits within (and between) accessions, can increase estimated heritability of the studied traits (Table S4), and can lead to the identification of considerably higher numbers of genome‐wide significant associations.
Perturbation of ribosome biogenesis can lead to compensatory cell enlargement
We observed decreased cell production in the aak6 root meristematic zone that is partially compensated for by an increased postmitotic cellular growth in the elongation zone and results in significantly longer mature root cells. Such compensation of the decrease in cell number by excessive cell enlargement was previously characterized in plant organs of determinate growth – the leaves (Tsukaya, 2003; Fujikura et al., 2009). Here, we have shown unexpected evidence of CCE in the Arabidopsis root, an organ that continues to grow throughout the plant's lifetime. Furthermore, our findings are consistent with observed postmitotic cell enlargement being a consequence of a loss‐of‐function mutation of a gene which we show impacts ribosome abundance. While a previous study linked CCE in plants with ribosome‐related processes (Fujikura et al., 2009), it relied on the annotation of yeast orthologues of Arabidopsis oligocellula (oli) genes. In this study we provide experimental evidence of altered plant ribosome abundance and CCE in the aak6 mutant. Our evidence for the induction of CCE in a mutant of the ribosomal assembly factor gene AAK6 is in agreement with previous evidence of CCE in oli2 oli5 and oli2 oli7 double mutants (Fujikura et al., 2009) (genes orthologous to yeast ribosomal assembly and structural proteins encoding genes Nop2, RPL5A and RPL5B). This strongly suggests that perturbation of ribosome biogenesis can lead to CCE and that efficiency of ribosome biogenesis plays a role in cell production and determination of sizes of mature cells and plant organs. Given the large scale of the eukaryotic ribosome assembly machinery (reviewed by Peña et al., 2017; Weis et al., 2015), we can therefore hypothesize that additional genes involved in ribosome biogenesis will be identified, that mutations in these genes are likely to reduce cell production and that this in turn will lead to CCE in the respective mutant roots.
The role of AAK6 in ribosome maturation and its link to root growth
While a previous study reported an in vitro protein interaction of Arabidopsis AK6 with the AtRPS14 ribosomal protein (Feng et al., 2012), the role of AK6 in plants remained largely ill‐defined. Well‐studied homologous proteins of AAK6 (Fap7/aFap7) provide potential clues for a discussion of its possible mechanisms of action. AK6 interacts with the small ribosomal subunit (SSU) protein Rps14 in humans, yeast and archaea (Granemann et al., 2005; Zhang et al., 2012; Hellmich et al., 2013). In yeast and archaea, its binding to uS11/RPS14 occludes the uS11/RPS14 RNA‐binding interface, suggesting that Fap7/aFap7 stabilizes uS11/RPS14 before being incorporated into pre‐SSU during ribosome biogenesis (Hellmich et al., 2013; Loc'h et al., 2014). This interaction is proposed to facilitate conformational change of the maturing SSU (Loc'h et al., 2014). Recent genetic, biochemical and structural data indicate that Fap7 contributes to induction of a rotated state during pre‐SSU subunit maturation, thereby facilitating the release of the essential assembly factor Dim1 from pre‐SSU subunits late in their maturation (Ghalei et al., 2017; Rai et al., 2019). Lastly, Fap7, together with other assembly factors and ribosomal structural proteins, mediates the Nob1 executed SSU rRNA maturation (Granneman et al., 2005; Strunk et al., 2012), indicating that these proteins play a role in correct positioning of rRNA cleavage site with respect to the Nob1 nuclease active site. The final steps of ribosome biogenesis in yeast involve the joining of a pre‐SSU with a large ribosomal subunit to form an 80S complex in which final SSU maturation steps take place (Lebaron et al., 2012; Strunk et al., 2012). Ribosomal subunits are then dissociated before they enter the translational pool. Several defects in SSU subunit maturation as a result of assembly factor depletion (e.g. Fap7 and Rio1) and incomplete assembly cause overaccumulation of 80S ribosomes (Strunk et al., 2012; Ferreira‐Cerca et al., 2014; Loc'h et al., 2014). Importantly, in agreement with these previous findings, we have shown that plants lacking AAK6 overaccumulate 80S ribosomes relative to polysome levels (Figs 7, S7) and this is consistent with AAK6 role in ribosome maturation. It therefore seems reasonable to assume that mechanisms of AK6 action described in yeast might be relevant for Arabidopsis. Nevertheless, further experiments will be required to test whether any of the roles of yeast AAK6 homologue (Fap7) (i.e. facilitation of a conformational change during ribosome assembly/maturation) are conserved in plants.
We propose that the observed root growth defects of aak6 plants are a result of costs of overproduction of 80S ribosomes and that this 80S overaccumulation depletes organismal resources (nucleotides, rRNA, amino acids, ribosomal proteins as well as energy and molecular machinery used for their production) that cannot be utilized and turned into cell production. Consistently, an inverse relationship between ribosome abundance and biomass of Arabidopsis accessions was shown previously (Ishihara et al., 2017). A higher protein turnover and the connected costs were suggested as the likely cause.
It remains to be established whether the mRNA‐free 80S ribosome quality control checkpoint that precedes translation, characterized in yeasts, exists in plants as well, but the aak6‐driven overaccumulation of 80S ribosomes suggests this possibility. Moreover, our results suggest that AK6 function in the final steps of ribosome assembly is conserved between yeast and Arabidopsis.
Natural variation of AAK6 coding sequence probably impacts the efficiency of ribosome assembly
How, then, does the AAK6 function relate to effects of its natural allelic variants? The data presented here show that natural AAK6 alleles modulate root growth of complemented aak6 plants. While we did not directly investigate the molecular mechanism responsible for these allelic effects, we have gained some hints as to likely mechanisms. There are several polymorphisms in the noncoding sequences in the genomic fragments of AAK6; however, we did not find any correlation with steady‐state levels of AAK6 mRNA and growth rate among several tested accessions. Moreover, the ectopic overexpression of AAK6 does not significantly impact root growth in Arabidopsis (Fig. S2b). This is consistent with the wild‐type‐like phenotype of the Fap7 overexpression line in yeast (Juhnke et al., 2000). We therefore propose that the need for AAK6 abundance is saturated in Arabidopsis accessions and that it is rather the natural genetic variation in AAK6 coding region that affects the AAK6 function and leads to root growth differences. Although the four nonsynonymous amino acid substitutions in the coding region of AAK6 genomic fragments used for aak6 complementation are the prime candidates for the functional basis for observed variation, we cannot exclude differential post‐transcriptional regulation of natural AAK6 alleles either. We hypothesize that the natural variation modulates the efficiency of AAK6‐facilitated ribosome assembly, which impacts cell production and finally root growth.
Author contributions
RS and WB conceived the study; RS, MR and JB developed the methodology and carried out the formal analysis; RS, CS, MR and JB carried out the investigation; CS and KJ were responsible for resources and data curation; CG, RS, JB, GTSB and WB developed and operated the software; GTSB and WB supervised the research; and RS and WB wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Natural phenotypic variation and GWAS of root growth rate 7th day after stratification.
Fig. S2 Relative AAK6 expression levels and root growth in wild‐type, SALK_015289 (aak6 ‐/‐) and AAK6 overexpression line (35S:AAK6).
Fig. S3 Complementation of aak6 ‐/‐ by pAAK6:AAK6‐Clover, in the shoots.
Fig. S4 Complementation of aak6 ‐/‐ by pAAK6:AAK6, in the root.
Fig. S5 Root anatomy parameters of aak6 ‐/‐ compared with the wild‐type.
Fig. S6 Kinematic analysis of root growth.
Fig. S7 Replicates of ribosome/polysome profiles.
Methods S1 Relative expression fold change of the AAK6 gene.
Methods S2 Seed size measurements.
Notes S1 Precautions taken in propagation of natural accessions.
Notes S2 Precautions taken with SALK T‐DNA lines.
Notes S3 Increase in endopolyploidy levels accompanies an increase in mature cell size in the aak6 mutant.
Table S1 252 Arabidopsis thaliana accessions used in this study.
Table S2 Phenotypes of T‐DNA insertion mutant lines (t‐test, P‐values).
Table S3 Primers used in this study.
Table S4 Impact of seed size on root growth traits.
Table S5 Genome‐wide significant GWAS associations (FDR 5%) aggregated for 16 root growth traits and time‐course experiment, 3rd – 7th day after stratification (DAS).
Table S6 Alignment of AAK6 coding sequences (amino acids) of Bå4‐1, Col‐0 and Tha‐1.
Table S7 Proportions of nuclear ploidy classes in the root apices.
Acknowledgements
We are grateful to Dr Peter Doerner for the pCycB1;1:CycB1;1DB‐GFP marker line. We thank Dr Daniele Filliault for helpful discussions and for her support with the haplotype analyses. We would like to acknowledge Ms Sevgi Oden for assistance with the kinematics image analysis. The authors are grateful to Prof. Isabella Moll and Dr Fernando A. Rabanal for fruitful discussions and valuable feedback regarding the ribosome/polysome profiles. We are grateful for the support by the BioOptics FACS Facility and the Plant Sciences Facility at Vienna BioCenter Core Facilities (VBCF). RS is a recipient of an EMBO Long‐Term Fellowship.
References
- Bai D, Zhang J, Li T, Hang R, Liu Y, Tian Y, Huang D, Qu L, Cao X, Ji J, Zheng X. 2016. The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth. Nature Communications 7: 12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Baskin TI. 1998. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana . Plant Physiology 116: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Szick‐Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey‐Serres J. 2001. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiology 127: 398–415. [PMC free article] [PubMed] [Google Scholar]
- Brooks RF. 1977. Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell 12: 311–317. [DOI] [PubMed] [Google Scholar]
- Byrne ME. 2009. A role for the ribosome in development. Trends in Plant Science 14: 512–519. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY. 2015. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Current Biology 25: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Joubès J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inzé D, De Veylder L. 2007. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D. 2001. Functional analysis of cyclin‐dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell AL, Gronwall DS, Miller ND, Spalding EP, Brooks TLD. 2011. Separating parental environment from seed size effects on next generation growth and development in Arabidopsis. Plant, Cell & Environment 34: 291–301. [DOI] [PubMed] [Google Scholar]
- Emami S, Mc Yee, Dinneny JR. 2013. A robust family of Golden Gate Agrobacterium vectors for plant synthetic biology. Frontiers in Plant Science 4: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P, Nurse P. 1977. Control of cell size at division in fission yeast by a growth‐modulated size control over nuclear division. Experimental Cell Research 107: 377–386. [DOI] [PubMed] [Google Scholar]
- Feng X, Yang R, Zheng X, Zhang F. 2012. Identification of a novel nuclear‐localized Adenylate Kinase 6 from Arabidopsis thaliana as an essential stem growth factor. Plant Physiology and Biochemistry 61: 180–186. [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. 2007. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiology 144: 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira‐Cerca S, Kiburu I, Thomson E, LaRonde N, Hurt E. 2014. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre‐40S biogenesis factors in 80S‐like ribosomes. Nucleic Acids Research 42: 8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H. 2009. Coordination of cell proliferation and cell expansion mediated by ribosome‐related processes in the leaves of Arabidopsis thaliana . Plant Journal 59: 499–508. [DOI] [PubMed] [Google Scholar]
- Ghalei H, Trepreau J, Collins JC, Bhaskaran H, Strunk BS, Karbstein K. 2017. The ATPase Fap7 tests the ability to carry out translocation‐like conformational changes and releases Dim1 during 40S ribosome maturation. Molecular Cell 6: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Nandineni MR, Baserga SJ. 2005. The putative NTPase Fap7 mediates cytoplasmic 20S pre‐rRNA processing through a direct interaction with Rps14. Molecular and Cellular Biology 25: 10352–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith ME, Mayer U, Capron A, Ngo QA, Surendrarao A, McClinton R, Jürgens G, Sundaresan V. 2007. The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis. Plant Cell 19: 2246–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Wu J, Yue X, Zhang Y, Zhu J. 2013. A nuclear calcium‐sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLoS Genetics 9: e1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Hasegawa J, Matsunaga S. 2013. The boundary of the meristematic and elongation zones in roots: endoreduplication precedes rapid cell expansion. Scientific Reports 3: 2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Molecular Biology 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hellmich UA, Weis BL, Lioutikov A, Wurm JP, Kaiser M, Christ NA, Hantke K, Kötter P, Entian KD, Schleiff E et al. 2013. Essential ribosome assembly factor Fap7 regulates a hierarchy of RNA–protein interactions during small ribosomal subunit biogenesis. Proceedings of the National Academy of Sciences, USA 110: 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, Engler JdA, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. 1995. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO Journal 14: 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RGF, Bachem CWB, Bögre L. 2006. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO Journal 25: 4909–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Umeda M. 2011. Cell‐cycle control and plant development. International Review of Cell and Molecular Biology 291: 227–261. [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. 2009. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71. [DOI] [PubMed] [Google Scholar]
- Ishihara H, Moraes TA, Pyl ET, Schulze WX, Obata T, Scheffel A, Fernie AR, Sulpice R, Stitt M. 2017. Growth rate correlates negatively with protein turnover in Arabidopsis accessions. The Plant Journal 91: 416–429. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae . Experimental Cell Research 105: 79–98. [DOI] [PubMed] [Google Scholar]
- Juhnke H, Charizanis C, Latifi F, Krems B, Entian KD. 2000. The essential protein fap7 is involved in the oxidative stress response of Saccharomyces cerevisiae . Molecular Microbiology 35: 936–948. [DOI] [PubMed] [Google Scholar]
- Kafri M, Metzl‐Raz E, Jona G, Barkai N. 2016. The cost of protein production. Cell Reports 14: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. 2010. Variance component model to account for sample structure in genome‐wide association studies. Nature Genetics 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Na Zaitlen, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. 2016. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA‐seq profiling. The Plant Journal 88: 1058–1070. [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, Ki E, Sato S, Kato T, Tabata S, Saez‐Vasquez J, Echeverria M, Nakagawa T, Ishiguro S, Nakamura K. 2007. Sugar‐inducible expression of the nucleolin‐1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. The Plant Journal 49: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Bottcher B, Granneman S, Watkins NJ, Tollervey D. 2012. Proofreading of pre‐40S ribosome maturation by a translation initiation factor and 60S subunits. Nature Structural and Molecular Biology 19: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R. 2014. Visual account of protein investment in cellular functions. Proceedings of the National Academy of Sciences, USA 111: 8488–8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc'h J, Blaud M, Réty S, Lebaron S, Deschamps P, Bareille J, Jombart J, Robert‐Paganin J, Delbos L, Chardon F, Zhang E, Charenton C, Tollervey D, Leulliot N. 2014. RNA mimicry by the Fap7 adenylate kinase in ribosome biogenesis. PLoS Biology 12: e1001860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Berthelot‐Grosjean M. 2009. Polar gradients of the DYRK‐family kinase Pom1 couple cell length with the cell cycle. Nature 459: 852–856. [DOI] [PubMed] [Google Scholar]
- Mitchison JM, Creanor J. 1971. Induction synchrony in the fission yeast Schizosaccharomyces pombe . Experimental Cell Research 67: 368–374. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. 2009. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459: 857–860. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Peña C, Hurt E, Panse VG. 2017. Eukaryotic ribosome assembly, transport and quality control. Nature Structural & Molecular Biology 24: 689–699. [DOI] [PubMed] [Google Scholar]
- Peña C, Schütz S, Fischer U, Chang Y, Panse VG. 2016. Prefabrication of a ribosomal protein subcomplex essential for eukaryotic ribosome formation. eLife 5: e21755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Nelson TM. 2007. Arabidopsis nucleolin affects plant development and patterning. Plant Physiology 144: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M, Aramayo R. 2015. Translate to divide: control of the cell cycle by protein synthesis. Microbial Cell 2: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai J, Parker MD, Ghalei H, Johnson MC, Karbstein K, Stroupe ME. 2019. Subunit joining exposes nascent pre‐40S rRNA for processing and quality control. bioRxiv. 10.1101/617910. [DOI] [Google Scholar]
- Randall RS, Sornay E, Dewitte W, Murray JAH. 2015. AINTEGUMENTA and the D‐type cyclin CYCD3;1 independently contribute to petal size control in Arabidopsis: evidence for organ size compensation being an emergent rather than a determined property. Journal of Experimental Botany 66: 3991–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Chickarmane V, Cunha A, Obara B, Meyerowitz EM. 2010. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana . PLoS Biology 8: e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. 1997. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiology Reviews 77: 731–758. [DOI] [PubMed] [Google Scholar]
- Russell JB, Cook GM. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiology Reviews 59: 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satbhai SB, Setzer C, Freynschlag F, Slovak R, Kerdaffrec E, Busch W. 2017. Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency. Nature Communication 8: 15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü, Vilhjálmsson BJ, Horton MW, Meng D, Forai P, Huang YS, Long Q, Segura V, Nordborg M. 2012. GWAPP: a web application for genome‐wide association mapping in Arabidopsis. Plant Cell 24: 4793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. 2005. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovak R, Göschl Christian SuX, Shimotani K, Shiina T, Busch W. 2014. A scalable open‐source pipeline for large‐scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. 2012. A translation‐like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco E. 1970. On the average cellular volume in synchronized cell populations. Bulletin of Mathematical Biophysics 32: 459–473. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2003. Organ shape and size: a lesson from studies of leaf morphogenesis. Current Opinion in Plant Biology 6: 57–62. [DOI] [PubMed] [Google Scholar]
- Ubeda‐Tomás S, Beemster GTS, Bennett MJ. 2012. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends in Plant Science 17: 326–331. [DOI] [PubMed] [Google Scholar]
- Ubeda‐Tomás S, Federici F, Casimiro I, Beemster GTS, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ. 2009. Gibberellin signaling in the endodermis controls arabidopsis root meristem size. Current Biology 19: 1194–1199. [DOI] [PubMed] [Google Scholar]
- Unger MW, Hartwell LH. 1976. Control of cell division in Saccharomyces cerevisiae by methionyl‐tRNA. Proceedings of the National Academy of Sciences, USA 73: 1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WH, Chen J, Liu TW, Chen J, Han AD, Simon M, Dong XJ, He JX, Zheng HL. 2014. Regulation of the calcium‐sensing receptor in both stomatal movement and photosynthetic electron transport is crucial for water use efficiency and drought tolerance in Arabidopsis. Journal of Experimental Botany 65: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends in Biochemical Science 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. 2006. In planta transformation of Arabidopsis. CSH Protocols 2006: pdb.prot4668. [DOI] [PubMed] [Google Scholar]
- Weis BL, Kovacevic J, Missbach S, Schleiff E. 2015. Plant‐specific features of ribosome biogenesis. Trends in Plant Science 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae . Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley MI, Setiawan L, Hariharan IK. 2013. TIE‐ DYE: a combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development 140: 3275–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Dong G, Palaniappan K, Mi G, Baskin TI. 2017. Temperature‐compensated cell production rate and elongation zone length in the root of Arabidopsis thaliana . Plant, Cell & Environment 40: 264–276. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bai D, Ma X, Guan J, Zheng X. 2012. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene 33: 246–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Natural phenotypic variation and GWAS of root growth rate 7th day after stratification.
Fig. S2 Relative AAK6 expression levels and root growth in wild‐type, SALK_015289 (aak6 ‐/‐) and AAK6 overexpression line (35S:AAK6).
Fig. S3 Complementation of aak6 ‐/‐ by pAAK6:AAK6‐Clover, in the shoots.
Fig. S4 Complementation of aak6 ‐/‐ by pAAK6:AAK6, in the root.
Fig. S5 Root anatomy parameters of aak6 ‐/‐ compared with the wild‐type.
Fig. S6 Kinematic analysis of root growth.
Fig. S7 Replicates of ribosome/polysome profiles.
Methods S1 Relative expression fold change of the AAK6 gene.
Methods S2 Seed size measurements.
Notes S1 Precautions taken in propagation of natural accessions.
Notes S2 Precautions taken with SALK T‐DNA lines.
Notes S3 Increase in endopolyploidy levels accompanies an increase in mature cell size in the aak6 mutant.
Table S1 252 Arabidopsis thaliana accessions used in this study.
Table S2 Phenotypes of T‐DNA insertion mutant lines (t‐test, P‐values).
Table S3 Primers used in this study.
Table S4 Impact of seed size on root growth traits.
Table S5 Genome‐wide significant GWAS associations (FDR 5%) aggregated for 16 root growth traits and time‐course experiment, 3rd – 7th day after stratification (DAS).
Table S6 Alignment of AAK6 coding sequences (amino acids) of Bå4‐1, Col‐0 and Tha‐1.
Table S7 Proportions of nuclear ploidy classes in the root apices.


