ABSTRACT
We aimed to identify potential mechanism associated with acute kidney injury (AKI) after kidney transplantation. The dataset GSE53771, which contained 18 zero-hour (ZERO group) and 18 selected post-transplant (POST group) biopsy samples from 18 kidney allografts (8 AKI and 10 controls) was downloaded from GEO database. Differentially expressed miRNAs (DEMIs) were screened using limma package, and bidirectional hierarchical clustering of the DEMIs was performed using the pheatmap package. Target genes of DEMIs were predicted by miRWalk 2.0, miRNA-target genes networks were presented using Cytoscape, protein–protein interaction (PPI) networks were constructed by STRING (version:10.0) database, and competing endogenous RNAs (ceRNA) regulating network were constructed using Cytoscape. In ZERO and POST groups, a total of 4 and 24 differentially expressed miRNAs were obtained in AKI samples compared with control, respectively. Specifically, 71 lncRNAs were obtained to interact with five miRNAs (hsa-miR-215-5p, hsa-miR-192-5p, hsa-miR-422a, hsa-miR-212-3p and hsa-miR-122-5p). Histone chaperone ASF1A (ASF1A) and bromodomain and WD repeat-containing protein 1(BRWD1) were targeted by hsa-miR-212-3p in PPI network. In ceRNA network, lncRNA XIST could interact with four miRNAs (hsa-miR-212-3p, hsa-miR-122-5p, hsa-miR-215-5p, and hsa-miR-192-5p). LncRNA XIST might serve as a ceRNA to sponge hsa-miR-212-3p to regulate the development of AKI via altering the expression of ASF1A/BRWD1. Furthermore, lncRNA XIST could also interact with hsa-miR-122-5p to modulate the expression of PFKFB2 in thyroid hormone signaling pathway and AMPK signaling pathway. LncRNA XIST can serve as a ceRNA to sponge hsa-miR-212-3p and hsa-miR-122-5p to regulate AKI progression via modulating the expression of ASF1A, BRWD1, and PFKFB2.
KEYWORDS: Acute kidney injury, kidney transplantation, differentially expressed microRNAs, target genes, competing endogenous RNAs
Graphical Abstract
Introduction
Acute kidney injury (AKI) is a global public health concern associated with high mortality, morbidity, and healthcare costs [1]. In clinic, the two generally accepted definitions for AKI are the risk, injury, failure, end-stage renal disease (RIFLE) criteria [2] and acute kidney injury network (AKIN) criteria [3]. They mainly depend on the elevation of serum creatinine (SCr) levels from an established baseline level and/or a decrease in urine output [4]. AKI can be caused by several diseases, including sepsis, heart failure, rhabdomyolysis, liver disease, and ischemia, etc. [5]. The incidence of AKI varies from 5% in hospitalized patients to 30–50% of patients in intensive care units [6]. Furthermore, in developed countries, this disease is happened more common in elderly patients, while pediatric and young patients are predominant in developing countries [7,8]. Kidney transplantation is the most effective method to improve the quality of life for patients suffering from end-stage renal disease while post-transplant AKI is a prevalent complication associated with various potential inciting causes [9]. Although multiple efforts have been made in kidney transplantation, the incidence of post-transplant AKI is still 20–28% [10]. In addition, for renal allograft recipients, it is difficult to establish a baseline level due to the frequent and rapid fluctuations in graft function in the months after transplant, so there is no official diagnostic criterion for post-transplant AKI [11]. Therefore, it is needed to investigate the biomarkers and molecular mechanism in kidney transplant AKI.
Korbe´ly et al. firstly found that the expression of lipocalin 2 and kidney injury molecule 1 were significantly increased in tubulointerstitium in zero-hour biopsies of developing delayed graft function, and suggested that they could be early biomarkers for delayed graft function [12]. In addition, microRNAs (miRNAs), a class of non-coding small RNA-regulating genes at a posttranscriptional level, are reported to mediate various processes in the development and progression of disease [13]. For example, Domenico et al. revealed that the expression of miR-142-3p was significantly upregulated in peripheral blood and urine for kidney transplant recipients with acute tubular necrosis, and could be biomarkers [14]. It had been reported that renal miR-182-5p was significantly increased in post-transplant AKI patients, while the kidney function and morphology were observed to be improved by inhibiting miR-182-5p [15]. Although many studies have indicated miRNAs play critical roles in the progression of post-transplant AKI, the molecular mechanisms remain unclear.
Wilflingseder et al. [16] generated the dataset GSE53773 (composed of the subseries of GSE53769 and GSE53771) to assess the whole-genome mRNA and miRNA profiles of post-transplant AKI, and they found a molecular AKI signature holding 20 mRNAs and two miRNAs (miR-182-5p and miR-21-3p). While in our study, only the subseries GSE53771 was analyzed by a series of bioinformatics tools to investigate the molecular mechanism of post-transplant AKI. The differentially expressed miRNAs were screened, followed by the predation of targeted mRNA and the construction of miRNA-target genes networks. Then, the functional enrichment analysis and protein–protein interaction (PPI) network were performed for the targeted genes. Finally, the lncRNA was searched followed by construction of competitive endogenous RNA (ceRNA) network. This study was expected to provide potential regulatory mechanism underlying the development of post transplant AKI.
Materials and methods
Affymetrix microarray data
GSE53771 dataset deposited by Korbély et al. was downloaded from National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/)[21]. A total of 36 samples were included in this dataset, including 18 zero-hour (ZERO group) and 18 selected post-transplant (Tx) biopsy samples (POST group) from 18 renal allograft recipients. From the 18 renal allograft recipients, 8 had histological lesions of acute tubular necrosis in the post-Tx biopsies (AKI) and 10 allografts had primary function and protocol biopsies without pathology (Control). Samples included in this dataset were sequenced by the platform of GPL16384 ([miRNA-3] Affymetrix Multispecies miRNA-3 Array).
Data preprocessing
The original CEL data were transformed using oligo package (Version 1.34.2) [18] in R (Version 3.5.2), and preprocessed using the robust multiarray average (RMA) algorithm. Based on the information of miRNA_ID and probe name provided by the platform GPL16384, the probes were mapped into the obtained miRNA symbols. If different probes mapped into one miRNA symbol, the mean of different probes was calculated and regarded as the final expression value of this miRNA.
Screening of differentially expressed miRNAs
The limma package (Version 3.26.9) [19] was used to identify the differentially expressed miRNAs via calculating differentially expressed P value and fold change. Multiple-test correction was performed for P value using Benjamini and Hochberg’s method. The adj. P. value < 0.05 and |log2FC| > 0.585 were regarded as the thresholds of differentially expressed miRNAs.
Bidirectional hierarchical clustering of differentially expressed miRNAs
The pheatmap offers more control over dimensions and appearance, and allows to aggregate the rows using k means clustering [20]. The pheatmap package (Version 1.0.8) was used for the bidirectional hierarchical clustering, and the expression values were presented using heatmaps.
Construction of miRNA-target genes networks
Following the aforementioned investigations, overlapped differentially expressed miRNAs from the ZERO and POST groups were isolated, and their potential target genes were predicted by miRWalk 2.0 [21,22]. In addition, miRWalk [22], miRDB [23], miRMap [24], RNA22 [25], miRanda [26], and Targetscan [27] database were added to help predicting supposed target genes. The targeted genes verified by experiments were downloaded from validated target module, unions of predicted target genes and targeted genes verified by experiments were obtained, and differentially expressed miRNA-target genes pairs were screened. The network was presented using Cytoscape (Version 3.2.1) [28]. Gene ontology (GO) [29] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [30] pathway enrichment analyzes for target genes of miRNAs were performed using clusterProfiler [31] package (Version 2.4.3). BH adjusted P.adjust value<0.05 was regarded as threshold value.
Construction of PPI network
STRING (version:10.0, http://www.string-db.org/) database [32] was used to the construction of PPI network for target genes-encoded proteins. Required Confidence (combined score) > 0.4 was set as threshold value, and tsv format files were downloaded.
The network was constructed using Cytoscape. miRNA nodes of every gene were added to PPI network to analyze regulatory mechanism of miRNAs.
Construction of ceRNA regulating network
Overlapped miRNAs targeted lncRNAs were searched using starBase Version 2.0 [33], and the interacting relationships were visualized using Cytoscape. Following this, lncRNA-miRNA network and miRNA-target genes network were combined to obtain lncRNA-miRNA-target genes network as ceRNA network [34,35].
Results
Screening of differentially expressed miRNAs and bidirectional hierarchical clustering
After preprocessing, 1329 miRNAs were obtained. A total of 4 differentially expressed miRNAs (3 up-regulated and 1 down-regulated) were obtained in ZERO group, and 24 differentially expressed miRNAs (19 up-regulated and 5 down-regulated) were obtained in POST group. The heatmap is shown in Figure 1, which showed that AKI group and control group could be obviously distinguished by differentially expressed miRNAs
Figure 1.
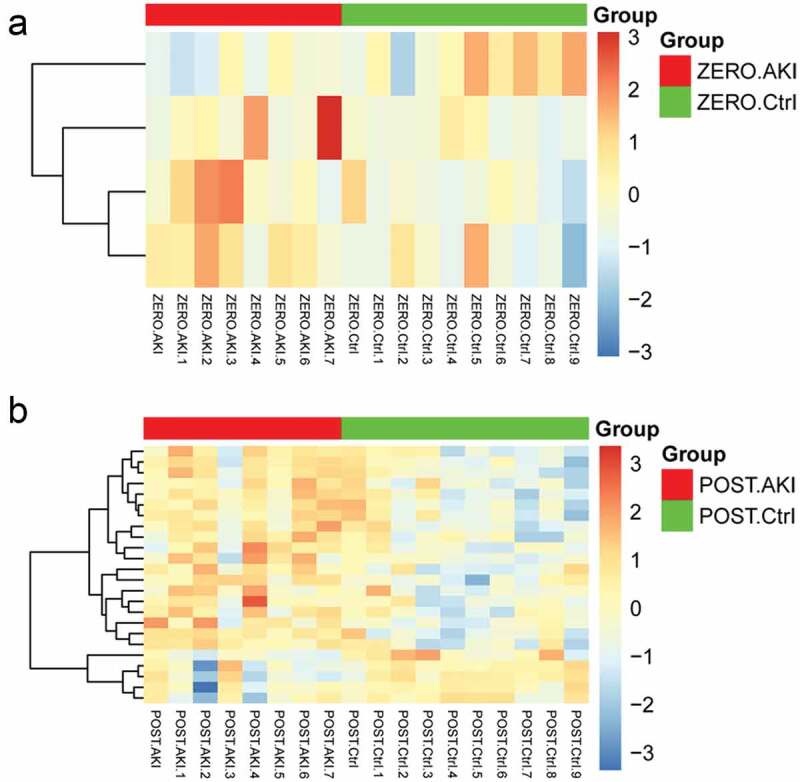
The heatmap for differentially expressed miRNAs. A: ZERO group; B: POST group. Top color represents grouping, red: disease group, green: control group; blue in the heatmap: down-regulated expression, orange: up-regulated expression.
Prediction of target genes of miRNAs and construction of the network
A total of 60 target genes regulated by up-regulated miRNAs and 40 target genes regulated by down-regulated miRNAs were obtained. A total of 13 miRNAs associated with above-mentioned 100 target genes were screened in the union of differentially expressed miRNAs of two groups. The network based on these 13 miRNAs and 100 target genes were constructed (Figure 2). Up-regulated miRNAs included miR-665, miR-3714, miR-122-5p, miR-1913, miR-762, and miR-212-3p, and down-regulated miRNAs included miR-215-5p, miR-192-5p, miR-3065-5p, miR-422a, miR-192-3p, miR-3065-3p, and miR-1184. The results of GO and KEGG pathway enrichment analyses for the miRNAs that the number of target genes ≥5 are presented in Figure 3. Target genes of hsa-miR-762 were significantly enriched in hippo signaling pathway-multiple species, target genes of hsa-miR-122-5p were significantly enriched in thyroid hormone signaling pathway and AMPK signaling pathway, target genes of hsa-miR-192-3p were significantly enriched in primary bile acid biosynthesis, target genes of hsa-miR-192-5p were significantly enriched in ubiquitin-mediated proteolysis, and target genes of hsa-miR-212-3p were enriched in tight junction.
Figure 2.
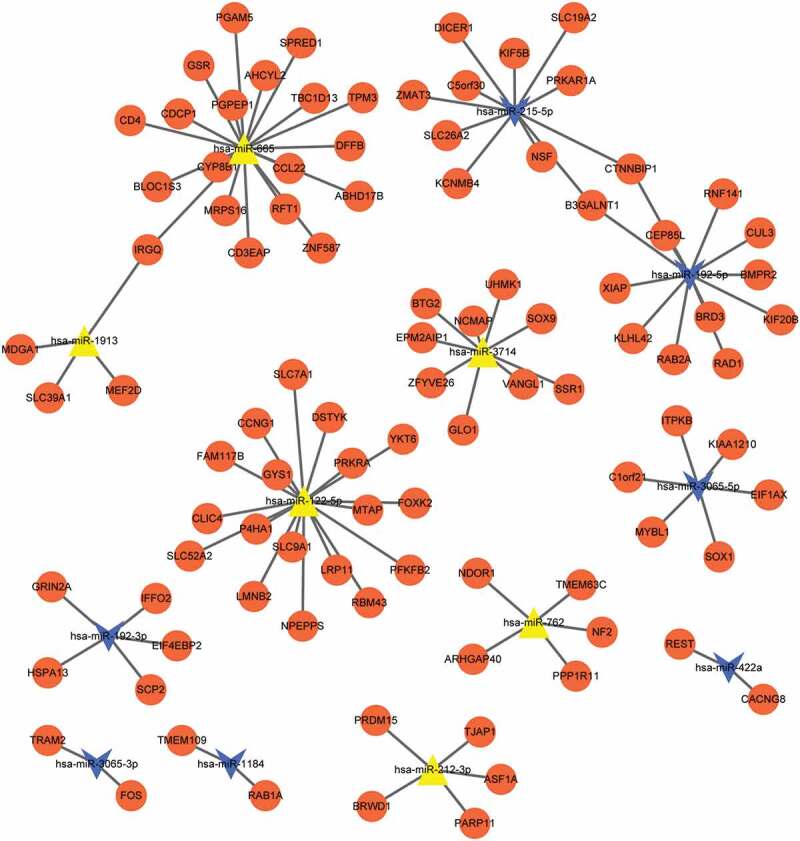
The network based on these 13 miRNAs and 100 target genes. Orange circle nodes: target genes; blue triangle: up-regulated miRNAs; Yellow arrow: down-regulated miRNAs.
Figure 3.
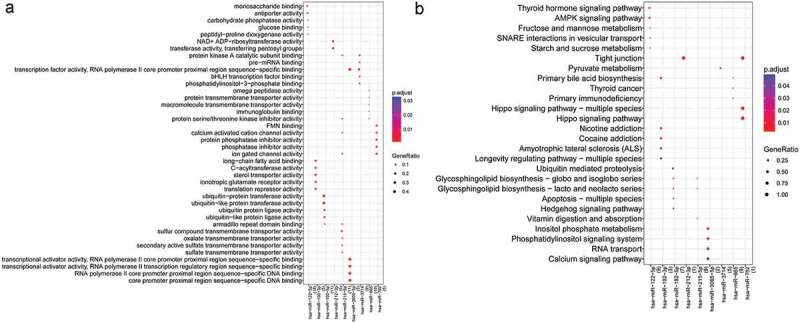
The result of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses for the miRNAs that the number of target genes≥5. A: GO, lateral axis: miRNAs, numbers in parentheses: the number of genes; vertical axis: the name of GO terms; the size of round dot: GeneRatio, bigger dot represents more miRNAs enriched in this term, smaller p value represents higher significance; B: KEGG, lateral axis: miRNAs, numbers in parentheses: the number of genes; vertical axis: the name of KEGG pathway; the size of round dot: GeneRatio, bigger dot represents more miRNAs enriched in this term, smaller p value represents higher significance.
Construction of PPI network
Proteins and their functional interactions form the backbone of the cellular machinery, and the connectivity networks are conducive to fully understand the biological phenomena [36]. STRING is an online tool to explore the known and predicted protein–protein interactions, including direct (physical) and indirect (functional) associations [36]. Therefore, based on the interactions retrieved from STRING, PPI network was constructed. PPI network showed that 34 nodes and 31 protein pairs were included in the network (Figure 4). In this network, CUL3, XIAP, BMPR2, RAD1, KIF20B, RAB2A, and BRD3 were targeted by hsa-miR-192-5p, but there is no interaction between the seven genes. PFKFB2, PRKRA, CCNG1, YKT6, SLC7A1, and CLIC4 were targeted by miR-122-5p, in which there exist interactions between CCNG1 and SLC7A1. BMPR2-CLIC4 interactions pair was found in the PPI network. ASF1A and BRWD1 were targeted by hsa-miR-212-3p.
Figure 4.
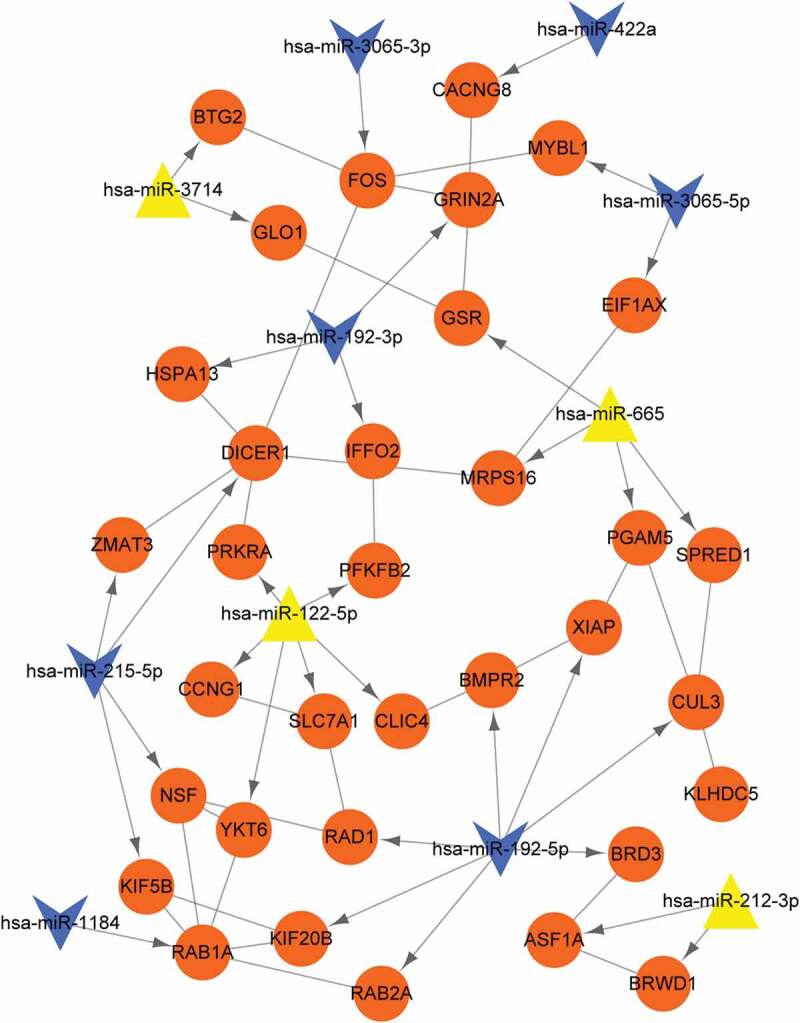
Protein–protein interaction (PPI) networks. Orange circle nodes: target genes; yellow triangle: up-regulated miRNAs; blue arrow: down-regulated miRNAs; line without arrow: PPI relations; line with arrow: miRNA-gene regulating relations.
CeRNA regulating network
To further reveal important lncRNAs targeted by miRNAs, down-regulated hsa-miR-215-5p, hsa-miR-192-5p, and hsa-miR-422a, and up-regulated hsa-miR-212-3p and hsa-miR-122-5p were chosen to match lncRNA. After searching, totally 71 matched lncRNAs and 83 regulatory relationships were obtained, and integrated with miRNA-target gene network to construct ceRNA regulating network (Figure 5). As shown in this network, LncRNA XIST matched with four miRNAs (hsa-miR-212-3p, hsa-miR-122-5p, hsa-miR-215-5p, and hsa-miR-192-5p), lncRNA RP11-216F19.2 matched with three miRNAs (hsa-miR-212-3p, hsa-miR-215-5p, and hsa-miR-192-5p), and lncRNA CTA-204B4.6 also matched with three miRNAs (hsa-miR-212-3p, hsa-miR-122-5p, and hsa-miR-422a). LncRNA XIST―hsa-miR-212-3p―ASF1A/BRWD1 may be one important ceRNA regulating relation.
Figure 5.
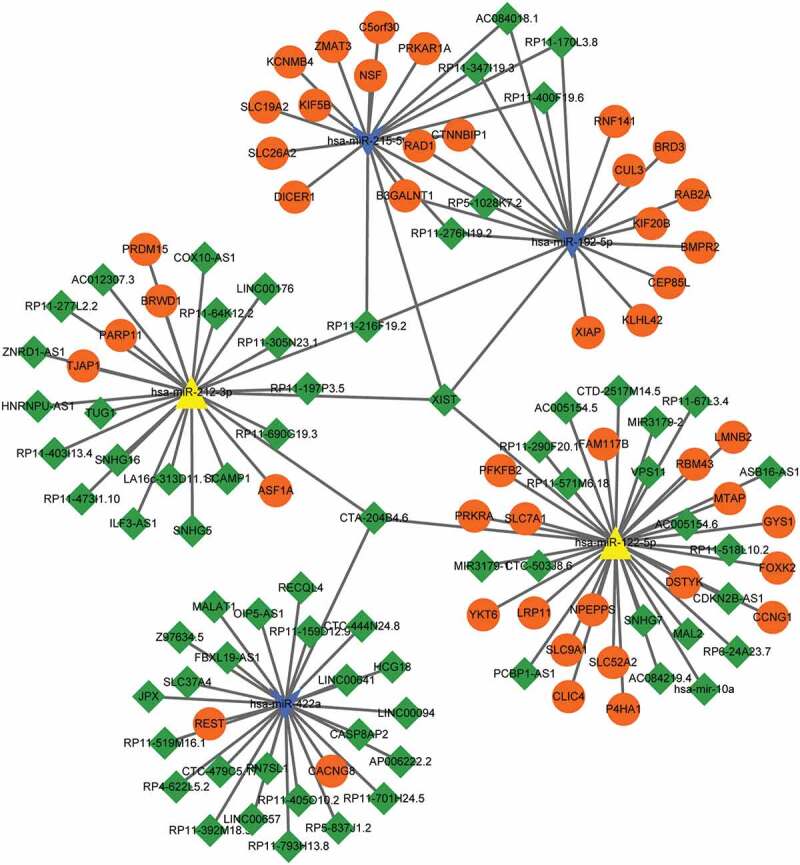
Competing endogenous RNAs (ceRNA) regulating networks. Orange circle nodes: target genes; green diamond: lncRNA; blue arrow: down-regulated miRNAs; yellow triangle: up-regulated miRNAs.
Discussion
In the present study, total of 13 differentially expressed miRNAs and 100 miRNA targeted genes were identified in both ZERO group and POST group. According to starBase database, 71 lncRNAs were identified to be targeted by five miRNAs (hsa-miR-215-5p, hsa-miR-192-5p, hsa-miR-422a, hsa-miR-212-3p and hsa-miR-122-5p, and lncRNA XIST could targeted by four miRNAs, including hsa-miR-212-3p, hsa-miR-122-5p, hsa-miR-215-5p, and hsa-miR-192-5p.). Then, a ceRNA network was constructed based on the miRNA-target gene and miRNA-lncRNA regulatory relationships. Specifically, lncRNA XIST might serve as a ceRNA to sponge hsa-miR-212-3p regulating the progression of AKI via ASF1A/BRWD1. Moreover, XIST also could sponge hsa-miR-122-5p to regulate the expression of PFKFB2 AMPK signaling pathway and fructose and mannose metabolism in AKI.
Previous study revealed that lncRNA XIST was significantly downregulated in several human cancers, suggesting that lncRNA XIST plays critical role in the development of tumors [37]. Tantai et al. documented that lncRNA XIST and HIF1A-AS1 in serum could serve as a predictive biomarker for non-small cell lung cancer [38]. Down-regulation of lncRNA XIST could inhibit neuro-inflammation by reducing expression of inflammatory cytokines in chronic constriction injury rats [39]. It is reported that lncRNA XIST contributes to neuronal apoptosis in rat spinal cord injury [40]. Cheng et al. have revealed that silenced lncRNA XIST significantly increased cell apoptosis compared with the control cells [41]. Besides, lncRNA XIST was significantly upregulated in urine samples of membranous nephropathy patients, indicating that lncRNA XIST might serve important role in the pathogenesis of kidney diseases [42]. However, the roles of lncRNA XIST in AKI were still rarely reported. In the present study, lncRNA XIST was identified to be targeted by four miRNAs (hsa-miR-212-3p hsa-miR-122-5p, hsa-miR-215-5p, and hsa-miR-192-5p), indicating that miRNAs could might involve in the progression of AKI via targeting lncRNA XIST. In addition, inflammation plays a key role in AKI [43], and some studies suggest the closely relations of apoptosis and AKI [44–46]. Considering of these evidence, we deduced that lncRNA XIST could interact with hsa-miR-212-3p and hsa-miR-122-5p to regulate the inflammatory and apoptosis in AKI.
CeRNA mechanism is an important regulatory manner in metabolism of cell activities as well as diseases. Zhang et al. [47] demonstrated that lncRNA XIST could act as a ceRNA to regulate TGF-β1 by sponging miR-185 in gastric cancer. Chen et al. have documented that lncRNA regulates the progression of gastric cancer via sponging miR-101 to modulate the expression of EZH2 [48]. In the present study, lncRNA XIST was identified to act as a ceRNA to regulate the expression of ASF1A and BRWD1 via sponging miR-212-3p. Inflammation plays a key role in AKI [43], and a previous study have showed that miR-212-3p downregulated LPS-induced inflammatory response via targeting HMGB1 [49]. Ramalinga et al. have revealed that miR-212 suppress starvation induced autophagy via attenuating the expression of SIRT1 in prostate cancer [50]. In addition, miR-212 could induce neuronal apoptosis in Alzheimer’s disease via downregulating the expression of FOXO2a death axis [51]. Considering of these finding, hsa-miR-212-3p might play important roles in inflammatory associated disease, including in kidney. ASF1A, histone chaperone ASF1A, functions in DNA replication and repair [52]. Decreasing DNA damage, inflammation, and oxidative stress can attenuate cisplatin-induced acute renal injury [53], and DNA repair plays roles in ischemic AKI [54]. BRWD1 encodes a member of the WD repeat protein family, members of which are involved in apoptosis, cell cycle progression, and gene regulation [55]. Multiple studies have suggested the inflammation, apoptosis and cell arrest are key mechanisms involved in AKI [44–46,56,57]. Taken together, we inferred that lncRNA XIST could regulate the inflammation and apoptosis of AKI via sponging hsa-miR-212-3p to modulate the expression of ASF1A and BRWD1.
Hsa-miR-122-5p was another miRNA-targeted lncRNA XIST. Hsa-miR-212-5p is an abundant liver-specific microRNA and plays critical roles in essential metabolic, anti-inflammatory, anti-apoptosis [58], and anti-tumorigenic function of liver [59]. Lian et al. have revealed that miR-122 promotes the proliferation and migration of renal carcinoma cells via PI3K/Akt signaling pathway significantly enriched in thyroid hormone signaling pathway and AMPK signaling pathway [60]. Besides, another study has demonstrated that miR-122 might play a key role in the recurrence of kidney carcinoma after nephrectomy [61]. These findings indicated that hsa-miR-122-5p might play critical role in kidney-associated disease, but the underlying mechanism of hsa-miR-122-5p remains unclear. In the current study, lncRNA XIST could sponge miR-122-5p to modulate the expression of PFKFB2. Functional enrichment analysis revealed that PFKFB2 was significantly enriched in thyroid hormone signaling pathway, AMPK signaling pathway, and fructose and mannose metabolism pathway. Muller et al. have demonstrated that PFKFB2 is associated with diabetic nephropathy, insulin secretion, and adiposity in American Indians [62]. Mishra et al. indicated that the thyroid hormone signaling could lead to induction of apoptosis in human breast cancer cells [63]. Modulation of AMPK signaling pathway can induce apoptosis in some cancer cells [64,65]. Taken together, lncRNA XIST might regulate the pathogenesis of AKI via sponging hsa-miR-122-5p to modulate the expression of PFKFB2 in thyroid hormone signaling pathway and AMPK signaling pathway.
In conclusion, lncRNA XIST can serve as a ceRNA to sponge hsa-miR-212-3p to regulate inflammatory and apoptosis in the progression of AKI via modulating the expression of ASF1A and BRWD1. Furthermore, lncRNA XIST could also sponge miR-122-5p to regulate thyroid hormone signaling pathway and AMPK signaling pathway via modulating the expression of PFKFB2 in AKI. The results may provide clues for the diagnosis and detection of this disease. However, there are limitations for the present study, such as without experimental verification, thus further studies are needed to verify the results of the present study.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Zuk A, Bonventre JV.. Acute kidney injury. Annu Rev Med. 2016;67:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rinaldo B. Defining, quantifying, and classifying acute renal failure. Crit Care Clin. 2005;21:223–237. [DOI] [PubMed] [Google Scholar]
- [3].Mehta RL, Kellum JA, Shah SV, et al. Acute kidney injury network: report of an initiative to improve outcomes in acute kidney injury. Crit Care. 2007;11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Soler YA, Mariely NP, Mónica P, et al. Pediatric risk, injury, failure, loss, end-stage renal disease score identifies acute kidney injury and predicts mortality in critically ill children: a prospective study. Pediatr Crit Care Med. 2013;14:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Uchino S, Kellum JA, Bellomo R, et al. Acute renal failure in critically ill patients: a multinational, multicenter study. Jama. 2005;294:813–818. [DOI] [PubMed] [Google Scholar]
- [6].Devarajan P. Emerging biomarkers of acute kidney injury. Contrib Nephrol. 2007;156:203–212. [DOI] [PubMed] [Google Scholar]
- [7].Xue JL, Daniels F, Star RA, et al. Incidence and mortality of acute renal failure in Medicare beneficiaries, 1992 to 2001. J Am Soc Nephrol. 2006;17:1135–1142. [DOI] [PubMed] [Google Scholar]
- [8].Kohli HS, Bhat A, Jairam A, et al. Predictors of mortality in acute renal failure in a developing country: a prospective study. Ren Fail. 2007;29:463–469. [DOI] [PubMed] [Google Scholar]
- [9].Zhang L, Yang YY, Fu P. The development and problems of continuous renal replacement therapy. Chin J Pract Internal Med. 2011;31(4):301–304. [Google Scholar]
- [10].Mittal T, Kohli HS. Post renal transplant acute kidney injury. Indian J Transplant. 2014;8:S33–S6. [Google Scholar]
- [11].Cooper JE, Wiseman AC. Acute kidney injury in kidney transplantation. Curr Opin Nephrol Hypertens. 2013;22:698–703. [DOI] [PubMed] [Google Scholar]
- [12].Korbély R, Wilflingseder J, Perco P, et al. Molecular biomarker candidates of acute kidney injury in zero-hour renal transplant needle biopsies. Transpl Int. 2011;24:143–149. [DOI] [PubMed] [Google Scholar]
- [13].Scian MJ, Maluf DG, Mas VR. MiRNAs in kidney transplantation: potential role as new biomarkers. Expert Rev Mol Diagn. 2013;13:93–104. [DOI] [PubMed] [Google Scholar]
- [14].Domenico TD, Joelsons G, Montenegro RM, et al. Upregulation of microRNA 142-3p in the peripheral blood and urinary cells of kidney transplant recipients with post-transplant graft dysfunction. Braz J Med Biol Res. 2017;50:e5533–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wilflingseder J, Jelencsics K, Bergmeister H, et al. miR-182-5p inhibition ameliorates ischemic acute kidney injury. Am J Pathol. 2017;187:70. [DOI] [PubMed] [Google Scholar]
- [16].Wilflingseder J, Sunzenauer J, Toronyi E, et al. Molecular pathogenesis of post-transplant acute kidney injury: assessment of whole-genome mRNA and MiRNA profiles. Plos One. 2014;9:e104164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Korbély R, Wilflingseder J, Perco P, et al. Molecular biomarker candidates of acute kidney injury in zero-hour renal transplant needle biopsies. Transplant Int. 2011;24:143–149. [DOI] [PubMed] [Google Scholar]
- [18].Parrish RS. Effect of normalization on significance testing for oligonucleotide microarrays. J Biopharm Stat. 2004;14:575–589. [DOI] [PubMed] [Google Scholar]
- [19].Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using Rand Bioconductor. New York: Springer; 2005; p. 397–420. [Google Scholar]
- [20].Kolde R pheatmap: Pretty Heatmaps. 2015.
- [21].Karin B, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. [DOI] [PubMed] [Google Scholar]
- [23].Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vejnar CE, Zdobnov EM. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40:11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rigoutsos I, Miranda K, Huynh T. rna22: a unified computational framework for discovering mirna precursors, localizing mature miRNAs, identifying 3’ UTR target-islands, and determining the targets of mature-miRNAs. United States: Ibm Corporation; 2007. Available from: https://domino.research.ibm.com/library/CyberDig.nsf/papers/EB2B39242822F96785256FAB006234A1/ [Google Scholar]
- [26].Turner DA Miranda: a non-strict functional language with polymorphic types. Proc of A Conference on Functional Programming Languages & Computer Architecture. New York: Springer-Verlag; 1985. [Google Scholar]
- [27].Igaz P, Racz K, Falus A, et al. MicroRNA target prediction: problems and possible solutions. Curr Bioinf. 2010;5:81–88. [Google Scholar]
- [28].Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. In: Hamacher M, Eisenacher M, Stephan C, editors. Data mining in proteomics. Methods in Molecular Biology (Methods and Protocols). Vol. 696. United States: Humana Press; 2011. p. 291–303. [DOI] [PubMed] [Google Scholar]
- [29].Sherlock G. Gene ontology: tool for the unification of biology. Can Inst Food Sci Technol J. 2009;22:415. [Google Scholar]
- [30].Ogata H, Goto S, Sato K, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li J-H, Liu S, Zhou H, et al. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013;42:D92–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [35].Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA–lncRNA Interactions. Methods Mol Biol. 2016;1402:271. [DOI] [PubMed] [Google Scholar]
- [36].Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weakley SM, Wang H, Yao Q, et al. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tantai J, Hu D, Yang Y, et al. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887. [PMC free article] [PubMed] [Google Scholar]
- [39].Jin H, Du XJ, Zhao Y, et al. XIST/miR‐544 axis induces neuropathic pain by activating STAT3 in a rat model. J Cell Physiol. 2018;233(8):5847–5855. [DOI] [PubMed] [Google Scholar]
- [40].Gu S, Xie R, Liu X, et al. Long coding RNA XIST contributes to neuronal apoptosis through the downregulation of AKT phosphorylation and is negatively regulated by miR-494 in rat spinal cord injury. Int J Mol Sci. 2017;18:732. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [41].Cheng Q, Xu X, Jiang H, et al. Knockdown of long non-coding RNA XIST suppresses nasopharyngeal carcinoma progression by activating miR-491-5p. J Cell Biochem. 2018;119(5):3936–3944. [DOI] [PubMed] [Google Scholar]
- [42].Huang YS, Hsieh HY, Shih HM, et al. Urinary Xist is a potential biomarker for membranous nephropathy. Biochem Biophys Res Commun. 2014;452:415–421. [DOI] [PubMed] [Google Scholar]
- [43].Fontana J, Vogt A, Hohenstein A, et al. Impact of steroids on the inflammatory response after ischemic acute kidney injury in rats. Indian J Nephrol. 2017;27:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Havasi A, Borkan SC. Apoptosis and acute kidney injury. Kidney Int. 2011;80:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bengatta S, Arnould C, Letavernier E, et al. MMP9 and SCF protect from apoptosis in acute kidney injury. J Am Soc Nephrol. 2009;20:787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sugiura H, Yoshida T, Mitobe M, et al. Klotho reduces apoptosis in experimental ischaemic acute kidney injury via HSP-70. Nephrol Dial Transplant. 2010;25:60–68. [DOI] [PubMed] [Google Scholar]
- [47].Zhang Q, Chen B, Liu P, et al. XIST promotes gastric cancer (GC) progression through TGF‐β1 via targeting miR‐185. J Cell Biochem. 2018;119(3):2787–2796. [DOI] [PubMed] [Google Scholar]
- [48].Chen D, Ju H, Lu Y, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen W, Ma X, Zhang P, et al. MiR-212-3p inhibits LPS-induced inflammatory response through targeting HMGB1 in murine macrophages. Exp Cell Res. 2017;350:318–326. [DOI] [PubMed] [Google Scholar]
- [50].Ramalinga M, Roy A, Srivastava A, et al. MicroRNA-212 negatively regulates starvation induced autophagy in prostate cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis and cellular senescence. Oncotarget. 2015;6:34446–34457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wong HK, Veremeyko T, Patel N, et al. De-repression of FOXO3a death axis by microRNA-132 and −212 causes neuronal apoptosis in Alzheimer’s disease. Hum Mol Genet. 2013;22:3077. [DOI] [PubMed] [Google Scholar]
- [52].Paul PK, Rabaglia ME, Wang CY, et al. Histone chaperone ASF1B promotes human β-cell proliferation via recruitment of histone H3.3. Cell Cycle. 2016;15:3191–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sahu BD, Kuncha M, Sindhura GJ, et al. Hesperidin attenuates cisplatin-induced acute renal injury by decreasing oxidative stress, inflammation and DNA damage. Phytomedicine. 2013;20:453–460. [DOI] [PubMed] [Google Scholar]
- [54].Pressly JD, Park F. DNA repair in ischemic acute kidney injury. Am J Physiol Renal Physiol. 2016;312:F551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ramos VC, Vidal-Taboada J, Bergoñon S, et al. Characterisation and expression analysis of the WDR9 gene, located in the Down critical region-2 of the human chromosome 21. Biochim Biophys Acta. 2002;1577:377. [DOI] [PubMed] [Google Scholar]
- [56].Price PM, Safirstein RL, Megyesi J. The cell cycle and acute kidney injury. Kidney Int. 2009;76:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Canaud G, Bonventre JV. Cell cycle arrest and the evolution of chronic kidney disease from acute kidney injury. Nephrol Dialysis Trans. 2014;30:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lin CJ, Gong HY, Tseng HC, et al. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. [DOI] [PubMed] [Google Scholar]
- [59].Hsu S, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Investig. 2012;122:2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lian JH, Wang WH, Wang JQ, et al. MicroRNA-122 promotes proliferation, invasion and migration of renal cell carcinoma cells through the PI3K/Akt signaling pathway. Asian Pac J Cancer Prev Apjcp. 2013;14:5017–5021. [DOI] [PubMed] [Google Scholar]
- [61].Wotschofsky Z, Busch J, Jung M, et al. Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin Chim Acta. 2013;416:5–10. [DOI] [PubMed] [Google Scholar]
- [62].Muller YL, Piaggi P, Hanson RL, et al. A cis-eQTL in PFKFB2 is associated with diabetic nephropathy, adiposity and insulin secretion in American Indians. Hum Mol Genet. 2015;24:2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mishra SK, Sar P, Rath B, et al. In human breast cancer cells thyroid hormone signaling overshadows estrogen signaling on SMP30 gene leading to induction of apoptosis. Aacr. 2012;72(2):4961. Supplement. DOI: 10.1158/1538-7445.AM2012-4961 [DOI] [Google Scholar]
- [64].Yi B, Liu D, He M, et al. Role of the ROS/AMPK signaling pathway in tetramethylpyrazine-induced apoptosis in gastric cancer cells. Oncol Lett. 2013;6:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yuan HD, Quan HY, Zhang Y, et al. 20(S)-Ginsenoside Rg3-induced apoptosis in HT-29 colon cancer cells is associated with AMPK signaling pathway. Mol Med Rep. 2010;3:825. [DOI] [PubMed] [Google Scholar]


