Abstract
Background
The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor, which is critically involved in the pathogenesis of a variety of skin diseases. The aim of this study was to detect AhR and its downstream regulators including cytochrome P450 (CYP1A1), AhR nuclear translocation (ARNT), and aryl hydrocarbon receptor repressor (AhRR) in serum, peripheral blood mononuclear cells (PBMCs), and skin lesions in patients with atopic dermatitis (AD).
Methods
Twenty-nine AD patients defined according to the criteria of Hanifin and Rajka and Chinese criteria of AD were included. Subjects without allergic and chronic diseases were recruited as controls. Patients and controls were selected from the dermatology outpatient clinic of Peking University People's Hospital from August 1 to December 31 in 2018. Enzyme-linked immunosorbent assay was performed to detect serum AhR level. The mRNA of AhR, AhRR, ARNT, and CYP1A1 in PBMCs were measured by real-time quantitative polymerase chain reaction. AhR expression in skin lesions was measured by immunohistochemistry.
Results
AhR was significantly higher expressed in serum (41.26 ± 4.52 vs. 33.73 ± 2.49 pmol/L, t = 6.507, P < 0.001) and skin lesions (0.191 ± 0.041 vs. 0.087 ± 0.017, t = 10.036, P < 0.001) of AD patients compared with those of controls. The mRNA levels of AhR (1.572 ± 0.392 vs. 1.000 ± 0.173, t = 6.819, P < 0.001), AhRR (2.402 ± 1.716 vs. 1.000 ± 0.788, t = 3.722, P < 0.001), CYP1A1 (2.258 ± 1.598 vs. 1.000 ± 0.796, t = 3.400, P = 0.002) in PBMCs of AD patients were higher compared with those of controls. The difference in mRNA levels of ARNT was not statistically significant between the patients and controls (1.383 ± 0.842 vs. 1.000 ± 0.586, t = 1.653, P = 0.105). AhR mRNA levels in PBMCs positively correlated with eczema area and severity index score and serum interleukin-6 levels.
Conclusion
AhR and its downstream regulators were highly expressed in serum, PBMCs, and skin of AD patients, which might contribute to the pathogenesis of AD.
Keywords: Aryl hydrocarbon receptor, Cytochrome P450, Atopic dermatitis, Peripheral blood mononuclear
Atopic dermatitis (AD) is an inflammatory skin disease characterized by chronic recurrent dermatitis with severe pruritus. Abnormal immunity and impaired skin barrier function played important roles in the pathogenesis of AD.[1] Aryl hydrocarbon receptor (AhR) is a transcription factor expressed in all cells types. It also emerged as a key transcription factor controlling many physiological processes including cell proliferation, apoptosis, differentiation, adhesion, and migration.[2] It responds to exogenous and endogenous chemicals by inducing or repressing genes such as cytochrome P4501A1 (CYP1A1) with toxic or protective effects in a wide range of species and tissues.[3] Some studies indicated that AhR expression appears to be aberrantly induced in some skin diseases, such as psoriasis and vitiligo.[4] However, little was known about AhR expression in AD.
Methods
Ethical approval
The study was approved by the Ethnic Committee of Peking University People's Hospital (No. 2018PHA033). The written informed consents were obtained from each subject.
Subjects
Patients and controls were selected from the dermatology outpatient clinic of the Peking University People's Hospital from August 1 to December 31 in 2018. A total of 29 adult patients with AD were recruited. All the patients satisfied the criteria of Hanifin and Rajka[5] and the Chinese criteria of AD.[6] Exclusion criteria included treatment with systemic glucocorticoids or other immunosuppressive agents as well as local treatment with a glucocorticoid within the previous 6 weeks. All the AD patients received dermatologic examination and assessment of the severity. The severity of AD was determined by the eczema area and severity index (EASI) score. Subjects without allergic and chronic diseases were recruited as controls.
Measurement of AhR and its downstream regulators’ mRNA in peripheral blood mononuclear cells (PBMCs)
Five milliliters of venous blood was obtained from all patients and controls under total aseptic technique in blood collection tubes containing dipotassium ethylenediamine tetra-acetic acid for serum and PBMCs separation. PBMCs were separated from peripheral blood by Ficoll Hypaque density gradient centrifugation.
Total RNA was extracted from PBMCs using RNA blood mini kit (Qiagen, Germany) according to the manufacturer's instructions. The concentration and purity of RNA were measured at 260 and 280 nm using NanoDrop2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The integrality of RNA was observed by agarose gel electrophoresis. RNA was then reverse-transcribed with the Revert Aid First Strand complementary DNA (cDNA) Synthesis Kit (Reverse Transcription Kit, Tiangen, China). The reverse transcription mixture was incubated for 15 min at 42°C followed by 5 min at 95°C. The mRNA levels of AhR, CYP1A1, AhR repressor (AhRR), and AhR nuclear translocation (ARNT) were tested by real-time quantitative polymerase chain reaction (RQ-PCR). RQ-PCR was performed using the SYBR Green Dye method which was carried out using cDNAs supplemented with SYBR Green supermix (Bio-rad, Hercules, CA, USA). The PCR amplification used gene-specific primers for β-actin (forward, 5′-TGGCACCCAGCACAATGAA-3′; reverse, 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′), AhR (forward, 5′-ATACCGAAGACCGAGCTGAAT-3′; reverse, 5′-CCAGCAGACACCTTAGACGACT-3′), CYP1A1 (forward, 5′-GTCATCTGTGCCATTTGCTTTG-3′; reverse, 5′-CAACCACCTCCCCGAAATTATT-3′), AhRR (forward, 5′-AGGTTTGGTTGGCAGGACT-3′; reverse, 5′-CAACCACCTCCCCGAAATTATT-3′) and ARNT (forward, 5′-TCTGGAAACTCTGGACCTGGAA-3′; reverse, 5′-GGCAAACCGCTCCTTATCGT-3′). The PCR protocol consisted of a cycle at 95°C for 60 s followed by 40 cycles consisting of 15 s at 95°C, 15 s at 60°C, and 15 s at 72°C. The average Ct was calculated for the target genes and internal control (β-actin). ΔCt (Cttarget – Ctβ-actin) values were determined. The expression levels of target genes in AD patients was determined relative to controls as 2−ΔΔCt, where ΔΔCt = ΔCt (patient or control) – ΔCt (average for controls). The results were converted into relative expression.
AhR and cytokines measurement
Patients’ sera were stocked at –80°C to measure cytokine levels and serum AhR. AhR, interleukin (IL)-1β, IL-6, tumor necrosis factor (TNF)-α, IL-4, and IL-22 levels were analyzed by enzyme-linked immunosorbent assay (eBioscience, Germany) according to the manufacturer's instructions. Spontaneous release of the above cytokines was undetectable in both controls and AD patients.
Immunohistochemistry
Punch biopsies of 5 mm were taken from the skin lesions of AD patients. Control biopsies were obtained from normal skin area of pigmented nevus surgeries excision. The biopsies were formalin-fixed and tissue slides were subjected to hematoxylin and eosin staining. Immunohistochemical stainings were performed by using formalin-fixed, paraffin-embedded specimens with mouse anti-human monoclonal anti-AhR (Abcam, Cambridge, MA, USA). Samples were incubated by overnight exposure in dilution 1:500. The secondary antibody was biotinylated rabbit anti-mouse IgG (Servicebio, China). An image analysis technique was employed for quantitative scoring of the stained slides. A color deconvolution technique was applied to separate the color mixture of hematoxylin. Image pro plus was used to obtain absorbance (A) of the AhR antibody.
Statistical analysis
All the data were input using Epidata 3.1 software (The EpiData Association, http://www.epidata.dk/, Denmark) and analyzed by SPSS 22.0 (SPSS Inc., Chicago, IL, USA). Qualitative data were described using number and percent. Statistical significance was determined using an unpaired student t test to compare the expression of AhR and its downstream regulators in serum, PBMCs and skin lesions from patients with those of controls. Pearson correlation index was applied to describe the correlation between AhR, serum cytokines levels and EASI score. P < 0.05 was considered statistically significant.
Results
Subjects
The mean age of AD patients was 48.7 ± 18.3 years including 11 women and 18 men. The mean age of controls for blood samples was 39.4 ± 14.4 years including ten women and seven men. Twenty-one biopsies were obtained from 17 patients with pigmented nevus with a mean age of 40.9 ± 10.6 years including eight women and nine men. No significant difference was found in demographical features between patients and controls.
Serum AhR levels and AhR mRNA expression in PBMCs
The expression levels of AhR and its downstream regulators in serum and PBMCs of AD patients and controls are shown in Figure 1. The mean serum AhR levels in AD patients were significantly higher than those of controls (41.26 ± 4.52 vs. 33.73 ± 2.49 pmol/L, t = 6.507, P < 0.001). The mRNA levels of AhR (1.572 ± 0.392 vs. 1.000 ± 0.173, t = 6.819, P < 0.001), AhRR (2.402 ± 1.716 vs. 1.000 ± 0.788, t = 3.722, P < 0.001), and CYP1A1 (2.258 ± 1.598 vs. 1.000 ± 0.796, t = 3.400, P = 0.002) in PBMCs of AD patients were significantly higher compared with those of controls. The difference in mRNA levels of ARNT was not statistically significant between the patients and controls (1.383 ± 0.842 vs. 1.000 ± 0.586, t = 1.653, P = 0.105). AhR mRNA level in PBMCs was associated with disease severity of AD [Figure 2A] (r = 0.448, P = 0.019).
Figure 1.
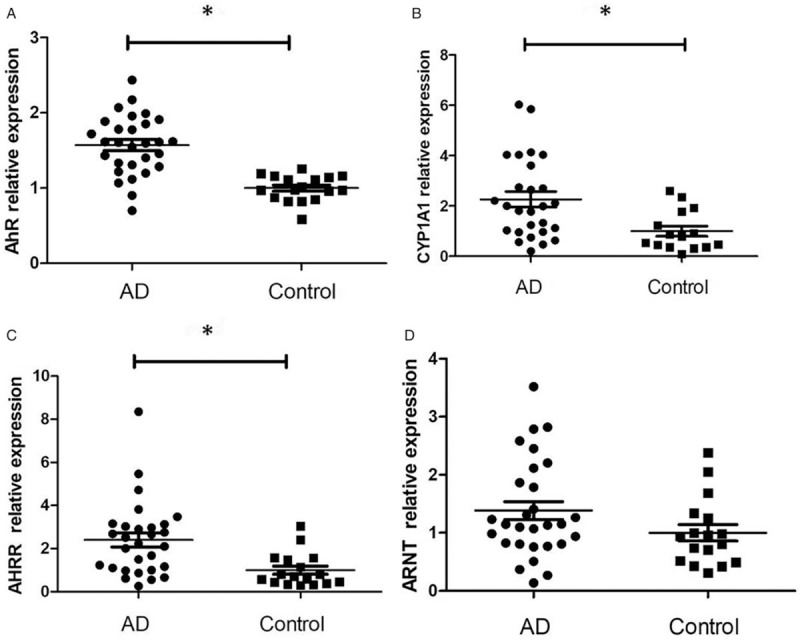
AhR (A), CYP1A1 (B), AhRR (C), and ARNT (D) mRNA expression in PBMCs of 29 patients with atopic dermatitis and 17 controls. The mRNA was detected by real-time quantitative polymerase chain reaction and expressed as 2−ΔΔCt. ∗P < 0.01. The results were converted into relative expression. AD: Atopic dermatitis; AhR: Aryl hydrocarbon receptor; CYP1A1: Cytochrome P450; ARNT: AhR nuclear translocation, AhRR: Aryl hydrocarbon receptor repressor.
Figure 2.
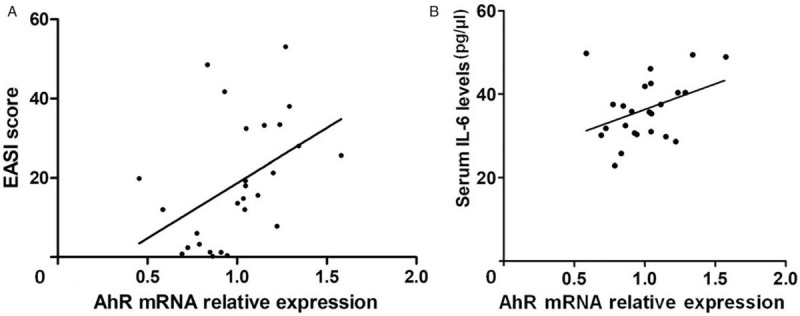
Correlation of AhR mRNA expression with EASI score (A, r = 0.448, P = 0.019) and serum IL-6 levels (B, r = 0.377, P = 0.046) in 29 AD patients. AhR: Aryl hydrocarbon receptor; EASI: Eczema area and severity index; IL: Interleukin.
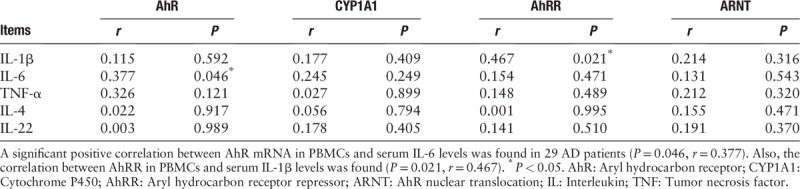
Relationship between mRNA expression of AhR and its downstream regulators in PBMCs and cytokines in sera of AD patients
The relationship between mRNA expression of AhR, CYP1A1, AhRR, ARNT in PBMCs and serum cytokines are shown in Table 1. A significant positive correlation was found between AhR mRNA in PBMCs and serum IL-6 levels in AD patients [Figure 2B] (r = 0.377, P = 0.046). Also, the expression of AhRR in PBMCs significantly correlated with serum IL-1β levels (P = 0.021, r = 0.467).
Table 1.
Correlations between mRNA expression of AhR, CYP1A1, AhRR, ARNT and IL-1β, IL-6, TNF-α, IL-4, IL-22 in sera.
Expression of AhR in skin lesions of AD
In normal skin, AhR was mainly expressed at the basal layer of epidermis [Figure 3A]. In AD patients; however, AhR was expressed mainly in the epidermis especially in granular layers [Figure 3B and 3C]. The AhR expression in epidermis of AD patients was significantly higher than that of controls (t = 10.036, P < 0.001). AhR also expressed in endothelial cells of blood vessels and infiltrating inflammatory cells in AD which consist of mainly lymphocytes [Figure 3B and 3C].
Figure 3.
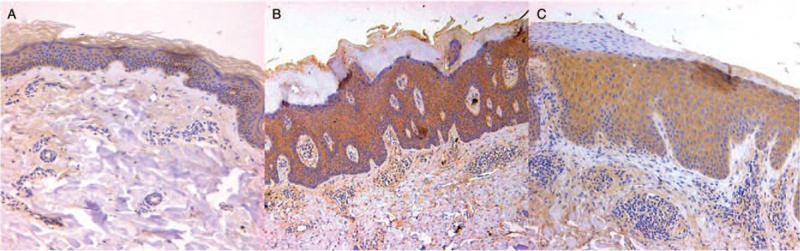
The immunohistochemical staining results of AhR antibodies in normal skin area of pigmented nevus surgeries excision and the skin lesions of AD patients (immunoperioxidase for AhR, A and B, original magnification ×40; C, original maginification ×200). (A) Expression of AhR in normal skin was mainly distributed at the basal layer of epidermis. (B and C) AhR mainly expressed in endothelial cells of blood vessels and infiltrating inflammatory cells of AD patients in dermis. The mean absorbance of epidermis in AD group was 0.191 ± 0.041 compared with 0.087 ± 0.017 in controls (t = 10.036, P < 0.001). No statistical significance was found in mean absorbance of dermis. AD: Atopic dermatitis; AhR: Aryl hydrocarbon receptor.
Discussion
AhR (also known as dioxin-receptor) is a proteic ligand-activated transcription factor expressed in many cell types. It is a member of the helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) family of transcription factors known for mediating the toxic effects of environmental contaminants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and a range of other xenobiotic substances.[7] In addition to the regulation of xenobiotic metabolism, several alternative functions have been characterized such as cell cycle regulation, cell migration, hematopoiesis, vascular development, and lymphocyte differentiation.[8]
Canonical and non-canonical signaling pathways activated by AhR have been identified and canonical pathways were more common.[9] When inactive, AhR is trapped in a cytosolic multiprotein complex (interactions with several chaperone proteins, including heat shock proteins 90, aryl hydrocarbon receptor interacting protein, and putative 23,000 Da protein). Various ligands such as dioxine, flavonoids, tryptophan photoproducts can activate AhR to translocate into the nucleus, where it is released from the complex and dimerizes with a partner molecule to form a transcription factor.[10] The first AhR dimerization partner discovered was the ARNT. The binding of ligand/AhR/ARNT complex to specific DNA recognition site, namely xenobiotic-responsive element (XRE), induces the transcription of genes encoding xenobiotic metabolism enzymes such as CYP1A1 and CYP1B1.[11] CYP1A1 was the most well-known downstream regulator of AhR, the function of CYP1A1 activation deal mostly with hydrocarbon detoxication.[12] Another key target gene activated in the AhR genomic pathway is the AhRR. It allows the binding of corepressors which are involved in a negative regulatory loop for AhR. AhRR, thus, is able to modulate the transcription of AhR-dependent genes. The expression of AhRR is regulated by XRE gene and enhanced upon the AhR ligand activation.[13]
It has been reported that AhR signaling is involved in the pathogenesis of inflammatory skin disease such as AD, psoriasis, and vitiligo. Recently, studies have demonstrated that the activation of AhR could be beneficial in inflammatory skin diseases.[14] Therefore, AhR is increasingly considered an attractive therapeutic target. For example, coal tar, an AhR agonist, has been proven to restore filaggrin expression and counteract Th2 cytokine-mediated downregulation of skin barrier proteins in topical use.[15] Benvitimod is another natural AhR agonist which can alleviate skin inflammation in both mice and human.[16] However, the functions of AhR are still unclear and contradictory. The altered balance of the receptor is poorly understood. In healthy skin, AhR signaling pathways driven by endogenous ligands can regulate keratinocyte differentiation and skin barrier function, which can significantly reduce skin inflammation.[17] On the other hand, in xenobiotic AhR ligand-exposed skin, canonical signaling may be dominant and lead to a set of adverse effects characterized by impairment of epidermal barrier, release of cytokines, oxidative stress, and cancer promotion. Kim et al[18] reported that the expression of AhR mRNA was increased in skin lesions of patients with psoriasis. Besides that, AhR and ARNT were found to colocalize in the nuclei of keratinocytes at the lower epidermis in psoriatic lesions, which suggested activation of the AhR pathway in psoriasis. In this study, we found increased levels of AhR, AhRR, CYP1A1, and ARNT in PBMCs, sera, and lesional skin of AD patients and AhR mRNA expression in PBMCs positively correlated with disease severity. This study provided new evidences that AhR signal pathway was involved in pathogenesis of AD.
AhR has been recently recognized as modulating expression of various cytokines and chemokines including IL-1β, IL-6, IL-10, IL-22, and TNF-α. Some of these cytokines were regulated by the canonical AhR signaling cascade, involving activation of XREs.[19] The cytokines expression was regulated by direct AhR-mediated gene transcription or, alternatively, through indirect AhR-related regulation of the cell types secreting cytokines/chemokines.[20] However, AhR can also act as an anti-inflammatory factor in some physiological or pathological situations. In our research, we found positive correlation between the mRNA levels of AhR and serum IL-6 in AD patients. Recent studies have found similar results. Kim et al[18] reported that AhR endogenous ligands such as TCDD can suppress IL-6 secretion in a dose-dependent manner in normal human epidermal keratinocytes. However, IL-6 regulation in response to AhR ligands appears to be complex and it depends on cell types and cellular inflammatory environment. We also found that the mRNA levels of AhRR positively correlated with IL-1β. However, the explicit association between AhRR and IL-1β remains unknown. Further studies are still needed.
Our results added new insight into the pathophysiology of AD. Increased expression of AhR and its downstream regulators in AD and its association with disease severity suggest that AhR signaling pathways play an important role in pathogenesis of AD.
Acknowledgements
The authors thank all the investigators from our department and also thank Dr. Qi Zhang from the laboratory of Peking University People's Hospital for her assistance in the design of the experimental procedure.
Conflicts of interest
None.
Footnotes
How to cite this article: Hu YQ, Liu P, Mu ZL, Zhang JZ. Aryl hydrocarbon receptor expression in serum, peripheral blood mononuclear cells, and skin lesions of patients with atopic dermatitis and its correlation with disease severity. Chin Med J 2019;133:148–153. doi: 10.1097/CM9.0000000000000591
References
- 1.Wollenberg A, Oranje A, Deleuran M, Simon D, Szalai Z, Kunz B, et al. ETFAD/EADV eczema task force 2015 position paper on diagnosis and treatment of atopic dermatitis in adult and paediatric patients. J Eur Acad Dermatol Venereol 2016; 30:729–747. doi: 10.1111/jdv.13599. [DOI] [PubMed] [Google Scholar]
- 2.Furue M, Tsuji G, Mitoma C, Nakahara T, Chiba T, Morino-Koga S, et al. Gene regulation of filaggrin and other skin barrier proteins via aryl hydrocarbon receptor. J Dermatol Sci 2015; 80:83–88. doi: 10.1016/j.jdermsci.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Napolitano M, Patruno C. Aryl hydrocarbon receptor (AhR) a possible target for the treatment of skin disease. Med Hypotheses 2018; 116:96–100. doi: 10.1016/j.mehy.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Di Meglio P, Duarte JH, Ahlfors H, Owens ND, Li Y, Villanova F, et al. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity 2014; 40:989–1001. doi: 10.1016/j.immuni.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanifin JM. Diagnostic criteria for atopic dermatitis: consider the context. Arch Dermatol 1999; 135:1551. [PubMed] [Google Scholar]
- 6.Liu P, Zhao Y, Mu ZL, Lu QJ, Zhang L, Yao X, et al. Clinical features of adult/adolescent atopic dermatitis and Chinese criteria for atopic dermatitis. Chin Med J 2016; 12:757–762. doi: 10.4103/0366-6999.178960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okey AB. Special contribution – an aryl hydrocarbon receptor odyssey to the shores of toxicology: the Deichmann lecture, international congress of toxicology-XI. Toxicol Sci 2007; 98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- 8.Fardel O. Cytokines as molecular targets for aryl hydrocarbon receptor ligands: implications for toxicity and xenobiotic detoxification. Expert Opin Drug Met 2013; 9:141–152. doi: 10.1517/17425255.2013.738194. [DOI] [PubMed] [Google Scholar]
- 9.Larigot L, Juricek L, Dairou J, Coumoul X. AhR signaling pathways and regulatory functions. Biochim Open 2018; 7:1–9. doi: 10.1016/j.biopen.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roman AC, Carvajal-Gonzalez JM, Merino JM, Mulero-Navarro S, Fernandez-Salguero PM. The aryl hydrocarbon receptor in the crossroad of signalling networks with therapeutic value. Pharmacol Ther 2018; 185:50–63. doi: 10.1016/j.pharmthera.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Costa C, Catania S, De Pasquale R, Stancanelli R, Scribano GM, Melchini A. Exposure of human skin to benzo[a]pyrene: role of CYP1A1 and aryl hydrocarbon receptor in oxidative stress generation. Toxicology 2010; 271:83–86. doi: 10.1016/j.tox.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Nebert DW, Puga A, Vasiliou V. Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann N Y Acad Sci 1993; 685:624–640. [DOI] [PubMed] [Google Scholar]
- 13.Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology 2009; 127:299–311. doi: 10.1111/j.1365-2567.2009.03054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuji G, Hashimoto-Hachiya A, Kiyomatsu-Oda M, Takemura M, Ohno F, Ito T, et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis 2017; 8:e2931.doi: 10.1038/cddis.2017.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest 2013; 123:917–927. doi: 10.1172/JCI65642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SH, Jayawickreme C, Rickard DJ, Nicodeme E, Bui T, Simmons C, et al. Tapinarof is a natural AhR agonist that resolves skin inflammation in mice and humans. J Invest Dermatol 2017; 137:2110–2119. doi: 10.1016/j.jid.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Haarmann-Stemmann T, Esser C, Krutmann J. The Janus-faced role of aryl hydrocarbon receptor signaling in the skin: consequences for prevention and treatment of skin disorders. J Invest Dermatol 2015; 135:2572–2576. doi: 10.1038/jid.2015.285. [DOI] [PubMed] [Google Scholar]
- 18.Kim HO, Kim JH, Chung BY, Choi MG, Park CW. Increased expression of the aryl hydrocarbon receptor in patients with chronic inflammatory skin diseases. Exp Dermatol 2014; 23:278–281. doi: 10.1111/exd.12350. [DOI] [PubMed] [Google Scholar]
- 19.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 2014; 32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 20.Veldhoen M, Duarte JH. The aryl hydrocarbon receptor: fine-tuning the immune-response. Curr Opin Immunol 2010; 22:747–752. doi: 10.1016/j.coi.2010.09.001. [DOI] [PubMed] [Google Scholar]


