Abstract
Background
Glioblastoma is one of the most common malignant brain tumors. Conventional clinical treatment of glioblastoma is not sufficient, and the molecular mechanism underlying the initiation and development of this disease remains unclear. Our study aimed to explore the expression and function of miR-873a-5p in glioblastoma and related molecular mechanism.
Methods
We analyzed the most dysregulated microRNAs from the Gene Expression Omnibus (GEO) database and examined the expression of miR-873-5p in 20 glioblastoma tissues compared with ten normal brain tissues collected in the Zhejiang Tongde Hospital. We then overexpressed or inhibited miR-873-5p expression in U87 glioblastoma cell lines and analyzed the phenotype using the cell counting kit-8 assay, wound healing assay, and apoptosis. In addition, we predicted upstream and downstream genes of miR-873-5p in glioblastoma using bioinformatics analysis and tested our hypothesis in U87 cells using the luciferase reporter gene assay and Western blotting assay. The differences between two groups were analyzed by Student's t test. The Kruskal-Wallis test was used for the comparison of multiple groups. A P < 0.05 was considered to be significant.
Results
The miR-873-5p was downregulated in glioblastoma tissues compared with that in normal brain tissues (normal vs. tumor, 0.762 ± 0.231 vs. 0.378 ± 0.114, t = 4.540, P < 0.01). Overexpression of miR-873-5p inhibited cell growth (t = 6.095, P < 0.01) and migration (t = 3.142, P < 0.01) and promoted cell apoptosis (t = 4.861, P < 0.01), while inhibition of miR-873-5p had the opposite effect. Mechanistically, the long non-coding RNA HOTAIRM1 was found to act as a sponge of miR-873-5p to activate ZEB2 expression in U87 cells.
Conclusions
We uncovered a novel HOTAIRM1/miR-873-5p/ZEB2 axis in glioblastoma cells, providing new insight into glioblastoma progression and a theoretical basis for the treatment of glioblastoma.
Keywords: MicroRNA, Long non-coding RNA, ZEB2, Glioblastoma
Introduction
Glioma is a type of tumor that occurs in the human central nervous system; 80% of primary malignant brain tumors are gliomas. Based on histological features together with molecular biomarkers, gliomas can be divided into four grades according to the World Health Organization classification of tumors.[1,2] Grade IV glioma, also known as glioblastoma, accounts for 60% of all high-grade gliomas.[3] Glioblastoma is the most aggressive form of glioma and is associated with poor prognosis and the highest mortality of all types of gliomas.[4] Current clinical therapy for glioblastoma involves surgical resection followed by radiotherapy together with chemotherapy (usually temozolomide treatment).[5,6] However, current treatment is insufficient due to the high recurrence of glioblastoma, its drug resistance, and the adverse complications of treatment. Thus, future research is needed to uncover the molecular mechanism underlying the initiation and development of glioblastoma and to test novel treatments for this disease.[7]
Protein-coding genes comprise only around 2% of the whole human genome. Over the past several decades, research has shown that non-coding genes are multiplying functional in various biological processes. MicroRNAs (miRNAs) are a subclass of small non-coding RNAs with a length of 18 to 25 nucleotides. MiRNAs participate in gene silencing and are thought to affect more than 90% of protein-coding gene loci in the human genome.[8,9] Generally, mature miRNAs inhibit gene expression through binding to the complementary site of the 3′-untranslated region (UTR) of target mRNAs; this leads to inhibition of mRNA translation and degradation of the indicated mRNAs.[10] In 2005, Ciafrè et al[11] first detected miRNA expression levels in glioblastoma and Chan et al[12] were the first to report that miRNA-21 is remarkably overexpressed in glioblastoma and inhibits the cell apoptosis pathway by repressing key molecules related to apoptosis. Since then, numerous studies have shown that miRNAs play vital roles in glioblastoma initiation and development. In addition to miR-21, many other miRNAs have been found to be upregulated in glioblastoma tissues compared to normal human brain tissues, including miR-10b, the miR17-92 cluster, and miR-93.[13] Nevertheless, downregulated miRNAs in glioblastoma are also critical gene regulators and have potential therapeutic significance for glioblastoma treatment.[14,15]
Competing endogenous RNAs are an important mechanism related to miRNA functions. RNA transcripts, including mRNAs, pseudogene transcripts, long non-coding RNAs (lncRNAs), and circular RNAs, competitively bind to certain miRNA recognition/response elements forming scaffolds; these scaffolds block the binding abilities of other transcripts to miRNAs, resulting in gene modulation.[16] In this process, lncRNAs can act as miRNA decoys, inhibiting the binding of miRNAs to coding mRNAs, thereby regulating protein expression.[17] The ZEB2 protein was found to mediate glioma cell proliferation, metastasis, and apoptosis pathways,[18] and is regulated by miRNAs in glioma.[19]
This study aimed to investigate the role of miR-873-5p in glioblastoma progression and the potential molecular mechanism.
Methods
Ethical approval
All the experimental procedures were approved by the Ethics Committee of Zhejiang Tongde Hospital (No. ZJTDLL-2017-039). Patients were aware of the studies and gave signatures to the informed consent.
Clinical samples and histology
A total of 30 human tissue samples including ten normal brain samples and 20 glioblastoma samples were collected from the Zhejiang Tongde Hospital. Normal brain tissues were obtained from normal adjacent tissues away from tumor tissues of glioblastoma patients. Samples were frozen immediately into liquid nitrogen for the following RNA analysis or fixed into formaldehyde solution for hematoxylin-eosin staining (H&E). H&E staining assay was performed following the described protocol.[20]
Cell lines and cell culture
The normal brain cells human astrocytes (HA) and different glioblastoma cell lines (U87, LN-229, U-251, and A172) were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Science (Shanghai, China). Cells were cultured in the Dulbecco modified Eagle medium (Thermo Fisher Scientific, San Jose, CA, USA) supplemented with 10% fetal bovine serum (Biological Industries, Beit Haemek, Israel) and 100× penicillin/streptomycin (Thermo Fisher Scientific). Incubators containing 5% CO2 and provided with a humid atmosphere were used to maintain the cell lines at 37°C.
Cloning and cell transfection
pcDNA vectors which expressed ZEB2 or HOTAIRM1 and HOTAIRM1 shRNA vector were constructed by the Sangon Biotech (Shanghai, China). Wild-type (WT) and binding sites deleted fragments of ZEB2 3′UTR or HOTAIRM1 were cloned into luciferase reporter vector pmirGLO by BGI (Shenzhen, China). The mimics of miR-873-5p, anti-miR-873-5p, and mock were purchased from GenePharm (Shanghai, China). The sequences of mimics were shown as followed: miR-873-5p mimic, 5′-GCAGGAACUUGUGAGUCUCCU-3′; anti-miR-873-5p, 5′-AGGAGACTCTGTGTTCCTGC-3′; Mock: 5′-UUCUCCGAACGUGUCACGUUU-3′. The vectors and indicated miRNA mimics were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated from tissues and cell lines using TRIzol reagent (Invitrogen). RNA samples were reversed into complementary DNA (cDNA) using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Synthesized cDNA was used to perform qRT-PCR assay using FastStart Universal SYBR Green Master (Roche Molecular Systems, Inc., Pleasanton, CA, USA) on an Applied Biosystems 7500 real-time PCR system (Applied Biosystems). Expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 transcripts were detected as endogenous controls for mRNA or miRNA expression analysis. Primers used for qRT-PCR were: miR-873-5p forward: 5′-GCAGGAACTTGTGAG-3′, reverse: 5′- GTGCAGGGTCCGAGGT-3′. miR-129-5p forward: 5′-ACCCAGTGCGATTTGTCA-3′, reverse: 5′-ACTGTACTGGAAGATGGACC-3′. miR-433 forward: 5′-TACGGTGAGCCTGTCA-3′, reverse: 5′-ACTGTACTGGAAGATGGACC-3′. miR-128 forward: 5′-CGGGGCCGTAGCACTGT-3′, reverse: 5′-ACTGTACTGGAAGATGGACC-3′. miR-491-5p forward: 5′-AGTGGGGAACCCTTCC-3′, reverse: 5′-ACTGTACTGGAAGATGGACC-3′. U6 forward: 5′-GGTCGGGCAGGAAAGAGGGC-3′, reverse: 5′-CTAATCTTCTCTGTATCGTTCC-3′. ZEB2 forward: 5′-AACAAGCCAATCCCAGGAG-3′, reverse: 5′-ACCGTCATCCTCAGCAATATG-3′. HOTAIRM1 forward: 5′-CCATCAACAGCTGGGAGATT-3′, reverse: 5′-TACAGAAACCCCCAACTCCA-3′. Actin forward: 5′-TCATGTTTGAGACCTTCAA-3′, reverse: 5′-GTCTTTGCGGATGTCCACG-3′.
Western blotting assay
Transfected cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) on ice for 15 min and centrifuged at 13,000 × g for 10 min at 4°C. Protein levels were measured by Enhanced bicinchoninic acid (BCA) Protein Assay Kit (Beyotime) and calculated evenly to load onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGEs) for the following blotting assays. Proteins were then transferred onto polyvinylidene difluoride (PVDF) membranes (Roche) using a semi-dry transfer cell (Bio-Rad, Hercules, CA, USA). After blocking with 5% skim milk, membranes were incubated with corresponding primary antibodies overnight at 4°C. Primary antibodies used in our study are obtained from Abcam (Cambridge, MA, USA).
Cell proliferation assay
Cell proliferation rate was detected by the cell counting kit-8 (Boster Biological Technology, Wuhan, China). Transfected cells were plated onto 96-well plates at a density of 3000 cells per well with six replicates. Cell amounts were measured every 24 h by a Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA) for a total of 3 days.
In vitro wound-healing assay
Cells were seeded onto six-well plates and cultured in the incubator overnight until becoming confluent. 200-μL pipette tips were then used to scratch on the cell monolayers. After the 24-h incubation, images of annealing wounds were photographed by an inverted microscope.
Flow cytometry and cell apoptosis detection
Cell apoptosis was examined by the fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, USA) following the manufacturer's instructions. Briefly, 1 × 106 cells were collected and re-suspended in 100 μL binding buffer. Five microlitres of FITC-Annexin V stain and 5 μL of PI stain were added into each tube. The mixtures were incubated in the dark for 15 min and added 400 μL binding buffer, respectively. Cell apoptosis was then evaluated by flow cytometry within 1 h.
Dual-luciferase activity assay
Luciferase reporter vectors of WT or mutant fragments described formerly were used to assess luciferase activity in cell lines. Distinct pmirGLO vectors were co-transfected with appropriate miRNA mimics into cells using Lipofectamine 2000 (Invitrogen). After 48-h incubation, Firefly luciferase activity representing expression of target transcripts and Renilla luciferase activity considered as control of transfection efficiency was examined by the Dual-Luciferase Reporter Assay (Promega, Madison, WI, USA) referring to manufacturer's instructions.
RNA immunoprecipitation (RIP)
RIP assay was performed utilizing Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Sigma-Aldrich, St. Louis, MO, USA) according to manufacturer's instructions. Ago2 antibody was used to precipitate HOTAIRM1 and miR-873-5p transcripts in cell lysates. Collected RNAs were then reversely transcribed into cDNAs. qRT-PCR assay was used to detect RNA expression levels as described in previous methods.
Statistical analysis
All experiments were performed three times independently. The Kolmogorov-Smirnov test was used to examine whether the data were normally distributed and quantitative data are represented as the mean ± standard deviation. GraphPad Prism 8.0.1 (GraphPad Software, La Jolla, CA) was used to compare and evaluate data among groups. The differences between two groups were analyzed by Student's t test. The Kruskal-Wallis test was used for the comparison of multiple groups. A P < 0.05 was considered to be significant.
Results
MiR-873-5p down-regulated in glioma
To explore the biological function of miRNAs in glioblastoma, we analyzed the GSE103228 data from the Gene Expression Omnibus (GEO) database. We found that five miRNAs were down-regulated in glioblastoma tissues; among these, miR-873-5p was the most significantly down-regulated miRNA in glioblastoma compared to normal brain tissues (P < 0.01) [Figure 1A]. Then, we analyzed miR-873-5p mRNA levels in collected clinical samples [Figure 1B]. qRT-PCR indicated that miR-873-5p was down-regulated in glioblastoma tissues compared with that in normal brain tissues (normal vs. tumor, 0.762 ± 0.231 vs. 0.378 ± 0.114, t = 4.540, P < 0.01) [Figure 1C]. Moreover, we examined miR-873-5p levels in normal HA and different glioblastoma cell lines (U87, LN-229, U-251, and A172) and got similar results (P < 0.01 in all glioblastoma cell lines) [Figure 1D]. MiR-873-5p expression in U87 was the lowest of all glioblastoma cell lines; thus, we used U87 in the following experiments.
Figure 1.
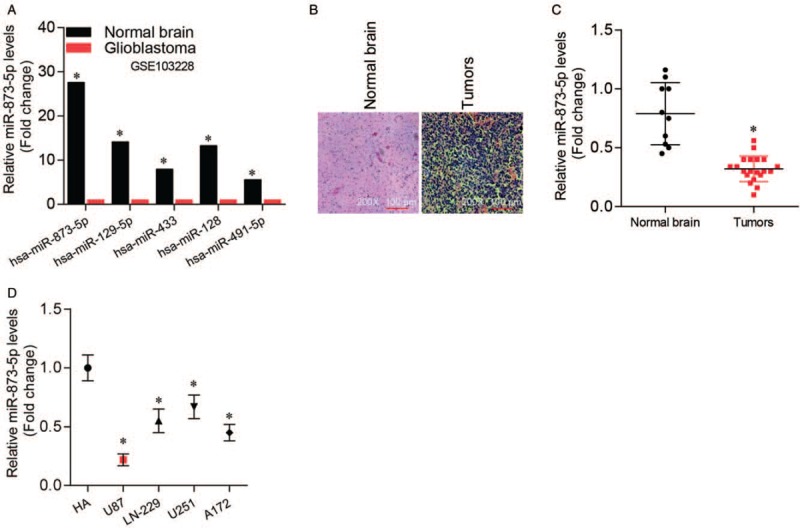
MiR-873-5p was down-regulated in glioma. (A) Expression of the five most down-regulated miRNAs in glioblastoma tissues compared with normal brain tissues in GSE103228 from GEO database. ∗P < 0.01 compared with normal brain tissues. (B) Representative H&E staining images of collected normal brain and glioblastoma tissues (original magnification ×200). (C) qRT-PCR analysis of miR-873-5p expression levels in collected normal brain and glioblastoma tissues. ∗P < 0.01 compared with normal brain tissues. (D) qRT-PCR analysis of miR-873-5p expression levels in normal brain cells HA and different glioblastoma cell lines (U87, LN-229, U-251, and A172). ∗P < 0.01 compared with HA control. Data were shown as mean ± standard deviation of three independent experiments. GEO: Gene expression omnibus, HA: Human astrocytes; H&E: Hematoxylin and eosin, qRT-PCR: Quantitative real-time polymerase chain reaction.
miR-873-5p suppressing cell growth and migration and promotes cell apoptosis in glioblastoma
To investigate the function of miR-873-5p in glioblastoma progression, we transfected U87 cells with miR-873-5p mimics or inhibitor. We performed qRT-PCR to analyze the expression of miR-873-5p and confirmed that miR-873-5p expression was up-regulated in U87 cells. (P < 0.01) [Figure 2A]. We found that miR-873-5p overexpression repressed cell growth, while miR-873-5p silencing accelerated cell growth (P < 0.01) [Figure 2B]. A wound-healing assay revealed a similar effect of miR-873-5p on cell migration (P < 0.01) [Figure 2C and 2D]. In addition, flow cytometry results suggested a similar function of miR-873-5p on cell apoptosis (P < 0.01) [Figure 2E and 2F].
Figure 2.
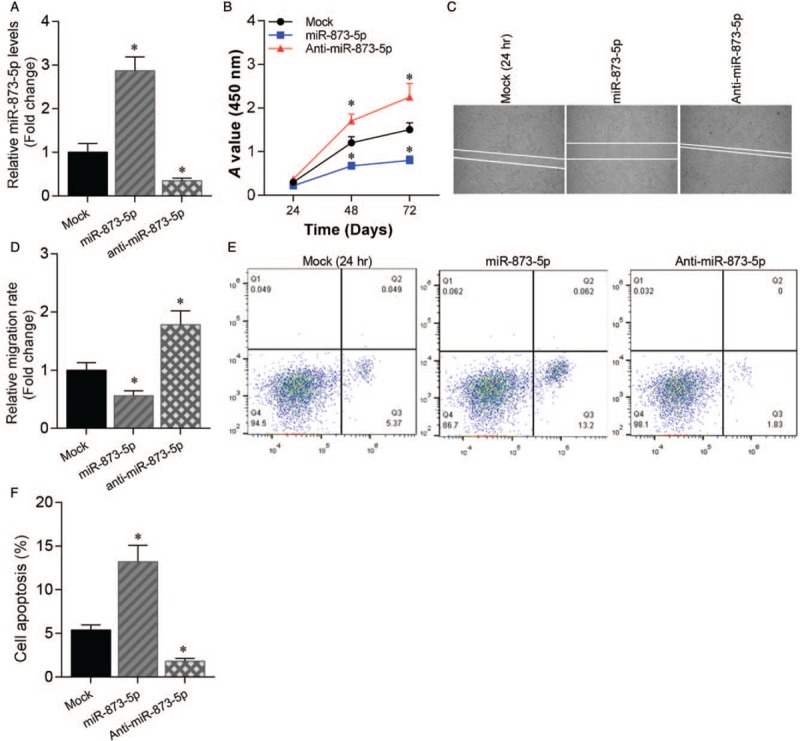
MiR-873-5p suppressed cell growth and migration and promoted cell apoptosis in glioblastoma. (A) qRT-PCR analysis of miR-873-5p expression levels in U87 cells transfected with miR-873-5p mimics or inhibitor compared with mock control. ∗P < 0.01 compared with mock control. (B) Cell growth of U87 cells transfected with miR-873-5p mimics or inhibitor was assessed by cell counting kit-8 assay. ∗P < 0.01 compared with mock control. (C) Cell migration of U87 cells transfected with miR-873-5p mimics or inhibitor was assessed by wound healing assay. (D) Statistics data of cell migration rate. ∗P < 0.01 compared with mock control. (E) Cell apoptosis of U87 cells transfected with miR-873-5p mimics or inhibitor was assessed by cell flow cytometry. (F) Statistics data of cell apoptosis. ∗P < 0.01 compared with mock control. Data were shown as mean ± standard deviation of three independent experiments. qRT-PCR: Quantitative real-time polymerase chain reaction.
ZEB2 as a target of miR-873-5p in glioblastoma
Next, we noticed that the overexpression or silencing of miR-873-5p suppressed or activated ZEB2 expression in the glioblastoma cells (P < 0.01) [Figure 3A]. Luciferase reporter gene assay results indicated that miR-873-5p mimics inhibited luciferase activities of vectors containing the WT ZEB2 3′UTR (Mock vs. miR-873-5p, 1.000 ± 0.113 vs. 0.378 ± 0.033, t = 7.241, P < 0.01) but not the ZEB2 deletion vector (Mock vs. miR-873-5p, 1.000 ± 0.121 vs. 0.968 ± 0.105, t = 0.451, P > 0.05) [Figure 3B]. The findings indicate that ZEB2 is a target gene of miR-873-5p in glioblastoma. We then tested the possibility that enforced expression of ZEB2 would compensate for miR-873-5p overexpression. As expected, proliferation caused by miR-873-5p overexpressed in U87 cells was restored by ectopic expression of ZEB2 [Figure 3C]. Similar results were observed for cell migration [Figure 3D and 3E] and cell apoptosis [Figure 3F and 3G]. These effects were accompanied by increased expression of Cyclin A1, Cyclin D1, and Bcl-2, and decreased expression of cleaved Caspase-3 [Figure 3H and 3I].
Figure 3.
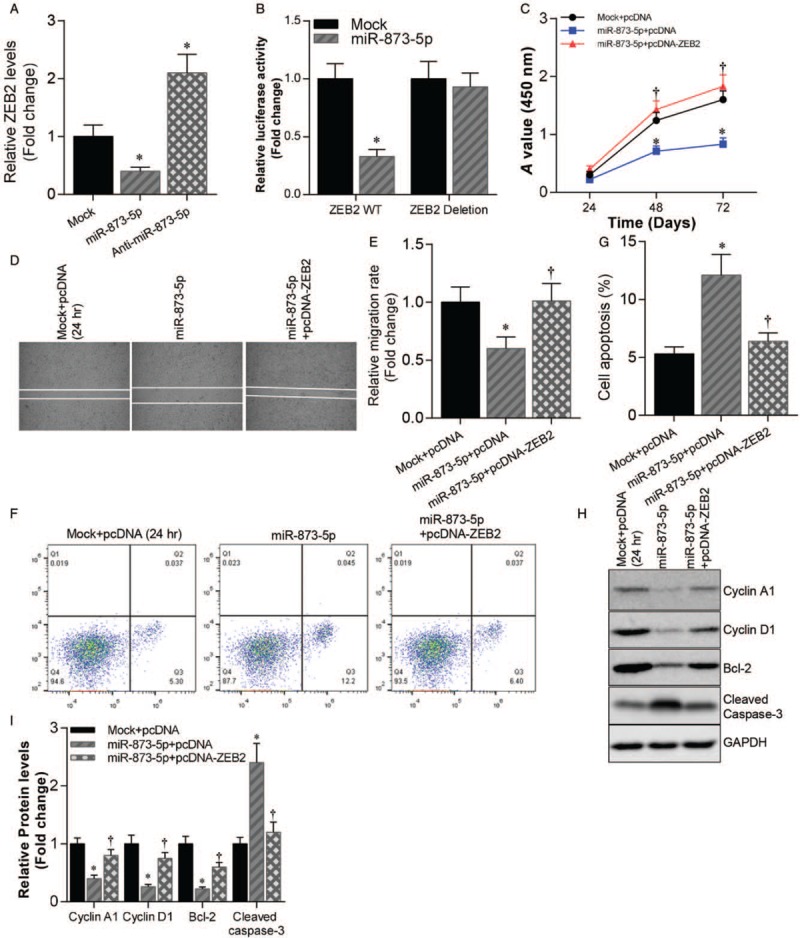
ZEB2 was a target of miR-873-5p in glioblastoma. (A) qRT-PCR analysis of ZEB2 mRNA levels in U87 cells transfected with miR-873-5p mimics or inhibitor compared with mock control. ∗P < 0.01 compared with mock control. (B) Luciferase reporter gene assay was used to analyze luciferase activity of WT/deleted ZEB2 3′UTR co-transfected with miR-873-5p mimics or mock control in U87 cells. ∗P < 0.01 compared with mock control. (C) Cell growth of U87 cells transfected with miR-873-5p mimics with or without pcDNA-ZEB2 and mock control U87 cells. ∗P < 0.01 compared with mock + pcDNA group. †P < 0.01 compared with miR-873-5p + pcDNA group. (D) Cell migration of U87 cells transfected with miR-873-5p mimics with or without pcDNA-ZEB2 and mock control U87 cells was assessed by wound healing assay. (E) Statistics data of cell migration rate. ∗P < 0.01 compared with mock + pcDNA group. †P < 0.01 compared with miR-873-5p + pcDNA group. (F) Cell apoptosis of U87 cells transfected with miR-873-5p mimics with or without pcDNA-ZEB2 and mock control U87 cells was assessed by cell flow cytometry. (G) Statistics data of cell apoptosis. ∗P < 0.01 compared with mock + pcDNA group. †P < 0.01 compared with miR-873-5p + pcDNA group. (H) Western blotting analysis of Cyclin A1, Cyclin D1, Bcl-2, and Cleaved Caspase-3 protein levels in U87 cells transfected with miR-873-5p mimics with or without pcDNA-ZEB2 and mock control U87 cells. GAPDH was used as loading control. (I) Statistics data of Cyclin A1, Cyclin D1, Bcl-2, and Cleaved Caspase-3 protein levels in H. ∗P < 0.01 compared with mock + pcDNA group. †P < 0.01 compared with miR-873-5p + pcDNA group. Data were shown as mean ± standard deviation of three independent experiments. qRT-PCR: Quantitative real-time polymerase chain reaction; UTR: Untranslated region.
HOTAIRM1 promotes cell growth and inhibits apoptosis in glioblastoma
We further explored the mechanism of miR-873-5p in glioblastoma progression and found that miR-873-5p expression was inhibited in lncRNA HOTAIRM1 overexpressed U87 cells (pcDNA vs. HOTAIRM1, 1.000 ± 0.091 vs. 0.618 ± 0.065, t = 9.231, P < 0.01) [Figure 4A]. Meanwhile, ZEB2 mRNA (pcDNA vs. HOTAIRM1, 1.000 ± 0.052 vs. 2.368 ± 0.155, t = 8.236, P < 0.01) and protein levels were increased in HOTAIRM1 overexpressed U87 cells [Figure 4B–D]. Importantly, Kaplan-Meier survival analysis showed that glioblastoma patients with high HOTAIRM1 expression had shorter overall survival (P = 0.023) [Figure 4E]. In addition, HOTAIRM1 knockdown inhibited cell growth and promoted apoptosis of U87 cells, and this effect was counteracted by miR-873-5p inhibitor transfection [Figure 4F–I].
Figure 4.
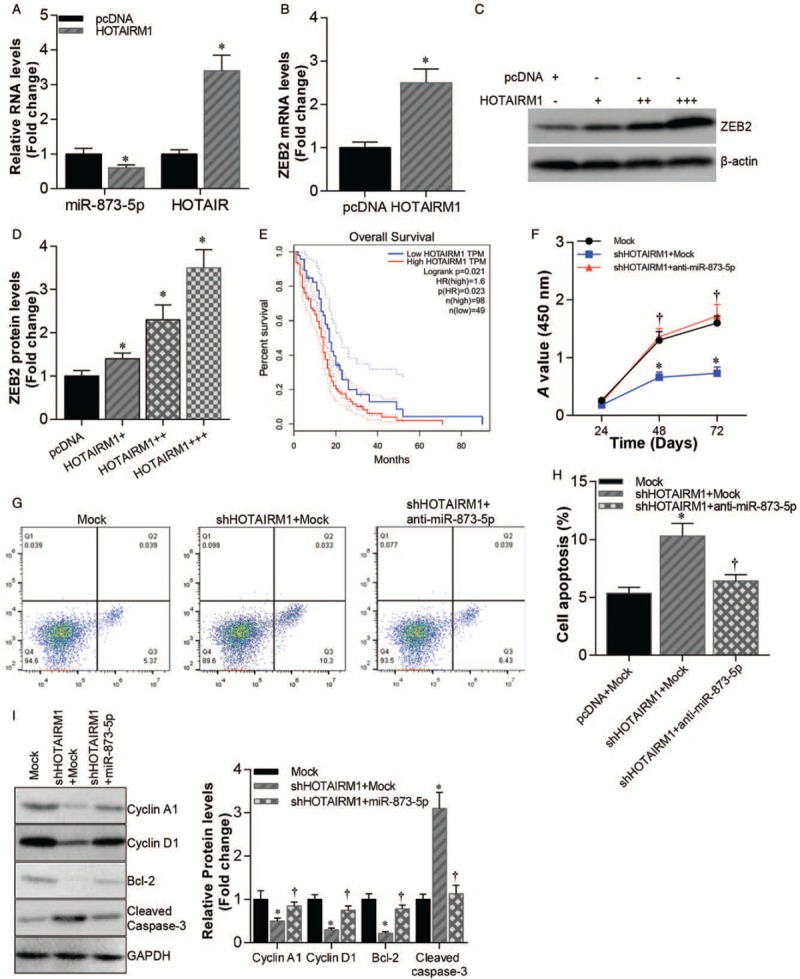
HOTAIRM1 promoted cell growth and inhibited apoptosis in glioblastoma. (A) qRT-PCR analysis of miR-873-5p and HOTAIRM1 expression levels in HOTAIRM1 overexpressed U87 cells compared pcDNA control. ∗P < 0.01 compared with pcDNA control. (B) qRT-PCR analysis of ZEB2 mRNA levels in HOTAIRM1 overexpressed U87 cells compared pcDNA control. ∗P < 0.01 compared with pcDNA control. (C) Western blotting analysis of ZEB2 protein levels in U87 cells transfected with pcDNA-HOTAIRM1 (0.5, 1, and 2 μg, respectively) compared with pcDNA vector control. (D) Statistics data of ZEB2 protein levels in C. ∗P < 0.01 compared with pcDNA control. (E) Kaplan-Meier plots of overall survival of glioblastoma patients stratified by HOTAIRM1 expression. (F) Cell growth of HOTAIRM1 knockdown U87 cells transfected with or without miR-873-5p mimics and mock U87 cells. ∗P < 0.01 compared with pcDNA + mock group. †P < 0.01 compared with shHOTAIRM1 + mock group. (G) Cell apoptosis of HOTAIRM1 knockdown U87 cells transfected with or without miR-873-5p and mock U87 cells. (H) Statistics data of cell apoptosis. ∗P < 0.01 compared with mock group. †P < 0.01 compared with shHOTAIRM1 + mock group. (I) Western blotting analysis of Cyclin A1, Cyclin D1, Bcl-2, and Cleaved Caspase-3 protein levels in HOTAIRM1 knockdown U87 cells transfected with or without miR-873-5p mimics and mock U87 cells (right). Statistics data of Cyclin A1, Cyclin D1, Bcl-2, and Cleaved Caspase-3 protein levels (left). ∗P < 0.01 compared with mock group. †P < 0.01 compared with shHOTAIRM1 + mock group. Data were shown as mean ± SD of three independent experiments. qRT-PCR: Quantitative real-time polymerase chain reaction.
HOTAIRM1 activates ZEB2 expression through sponging miR-873-5p
To further confirm the regulatory relationship among HOTAIRM1, miR-873-5p, and ZEB2, we analyzed the binding sites of miR-873-5p on the HTAIRM1 gene [Figure 5A]. Then, we used a luciferase reporter gene assay to test our prediction. The result showed that transfection with miR-873-5p mimics suppressed the luciferase activity of the WT HOTAIRM1 vector (Mock vs. miR-873-5p, 1.000 ± 0.098 vs. 0.753 ± 0.044, t = 7.521, P < 0.01), while miR-873-5p inhibitor increased the luciferase activity of the WT HOTAIRM1 vector (Mock vs. Anti-miR-873-5p, 1.000 ± 0.098 vs. 1.823 ± 0.136, t = 8.210, P < 0.01) [Figure 5B]. RNA immunoprecipitation assay showed that Ago2 interacted with HOTAIRM1 (Ago2 vs. IgG, 1.000 ± 1.062 vs. 8.632 ± 00.975, t = 9.241, P < 0.01) and miR-873-5p (Ago2 vs. IgG, 1.000 ± 0.083 vs. 29.962 ± 3.625, t = 12.241, P < 0.01) [Figure 5C]; this suggests that HOTAIRM1 may act as a sponge of miR-873-5p in glioblastoma cells. Then, we analyzed the regulatory effects of HOTAIRM1 and miR-873-5p on ZEB2 expression. Luciferase reporter gene assay showed that transfection of miR-873-5p mimics inhibited luciferase activity of the vector containing the 3′UTR of ZEB2 (Mock vs. miR-873-5p, 1.000 ± 0.116 vs. 0.506 ± 0.044, t = 8.239, P < 0.01), while HOTAIRM1 transfection diminished this effect [Figure 5D]. Moreover, miR-873-5p mimics inhibited ZEB2 expression in U87 cells and HOTAIRM1 overexpression attenuated this regulatory effect [Figure 5E]. Similarly, miR-873-5p mimics significantly impaired the activation effect of HOTAIRM1 on ZEB2 expression [Figure 5F].
Figure 5.
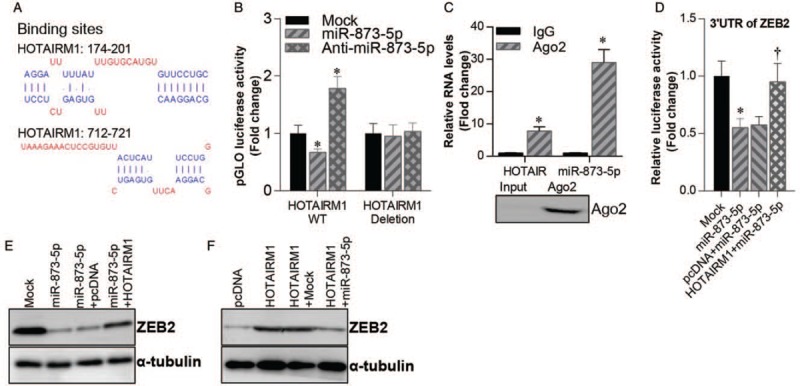
HOTAIRM1 activated ZEB2 expression through sponging miR-873-5p. (A) Predicted binding sites of miR-873-5p on HOTAIRM1 mRNA. (B) Luciferase activity of WT/deleted HOTAIRM1 vector co-transfected with miR-873-5p mimics or miR-873-5p inhibitor or mock control. ∗P < 0.01 compared with mock control. (C) qRT-PCR analysis of AGO2 immunoprecipitated HOTAIRM1 and miR-873-5p levels compared with IgG control (top). Western blotting analysis of immunoprecipitated AGO2 protein (bottom). ∗P < 0.01 compared with IgG control. (D) Luciferase activity of ZEB2 3′UTR vector in mock, miR-873-5p, pcDNA + miR-873-5p and HOTAIRM1 + miR-873-5p U87 cells. ∗P < 0.01 compared with mock control. †P < 0.01 compared with pcDNA + miR-873-5p U87 cells. (E) Western blotting analysis of ZEB2 protein levels in mock, miR-873-5p, pcDNA + miR-873-5p and HOTAIRM1 + miR-873-5p U87 cells. (F) Western blotting analysis of ZEB2 protein levels in pcDNA, HOTAIRM1, HOTAIRM1 + mock, and HOTAIRM1 + miR-873-5p U87 cells. Tubulin was used as a loading control. Data were shown as mean ± standard deviation of three independent experiments. qRT-PCR: Quantitative real-time polymerase chain reaction; UTR: Untranslated region; WT: Wild type.
Discussion
Glioblastoma is one of the most common and aggressive tumors that originates in the brain or spinal cord. Most patients with glioblastoma die within 2 years of diagnosis due to limited therapeutic approaches.[3] Thus, it is important to study the pathogenesis of glioblastoma and develop new therapeutic strategies. In recent years, with the development of high-throughput sequencing technology, numerous non-coding RNAs have been found to play crucial roles in human diseases, including cancer.[21–23] In this study, we analyzed a GEO dataset of miRNA expression profiles in human glioblastoma and selected miR-873-5p for further study. We confirmed that miR-873-5p was significantly down-regulated in glioblastoma tissues and cell lines. A previous study reported that miR-873-5p was overexpressed in lung cancer tissues and promoted cancer progression.[24] Further, in colon cancer, miR-873-5p was found to inhibit cancer progression via repression of the tumor suppressor candidate 3 (TUSC3)/protein kinase B (AKT) pathway.[25] The function of miR-873-5p varies in different cancer types, and to date, there is no report of the role of miR-873-5p in glioblastoma. Therefore, we overexpressed or silenced miR-873-5p expression in U87 cells and found that cell growth, migration, and apoptosis were altered. Our findings indicate that miR-873-5p acts as a tumor suppressor in glioblastoma progression.
ZEB2 is a zinc finger-containing transcription factor that is essential during early embryonic development.[26] It has been demonstrated that ZEB2 functions as an oncogene and is correlated with poor prognosis in many cancers. ZEB2 has been found to be overexpressed at the invasion front of colorectal cancer and was found to promote tumor invasion.[27] ZEB2 has also been found to suppress the expression of E-cadherin, an epithelial marker, and promote epithelial-mesenchymal transition in gastric cancer.[28] Similarly, ZEB2 was reported to promote epithelial-mesenchymal transition and tumor progression in glioblastoma.[29] Here, we found that ZEB2 is a target of miR-873-5p, and miR-873-5p can directly bind to the 3′UTR of ZEB2 mRNA and inhibit ZEB2 expression in glioblastoma cells. Moreover, reintroduction of ectopic ZEB2 abrogated the effect of miR-873-5p in glioblastoma cells. These findings suggest that the function of miR-873-5p in glioblastoma cells depends on ZEB2.
The ceRNA regulatory axis is wildly used in molecular biology to describe the function of lncRNAs and miRNAs in gene expression regulation. Typically, lncRNAs interact with miRNAs leading to expression changes in the target gene of the miRNA. Our understanding of the ceRNA signaling pathway in gene regulation is incomplete; further research in this field is required. The lncRNA HOTAIRM1 is encoded in the human HOXA gene cluster and plays a vital role in myeloid cell development.[30,31] HOTAIRM1 is reported to act as a tumor suppressor and a potential biomarker in colorectal cancer.[32] Further, HOTAIRM1 was found to inhibit tumor progression through regulation of the miR-17-5p/PTEN axis in gastric cancer.[33] Here, we found that HOTAIRM1 expression was negatively correlated with overall survival of glioblastoma patients. Moreover, HOTAIRM1 interacted with miR-873-5p to suppress its expression and promoted ZEB2 expression in glioblastoma cells. Our findings demonstrate that HOTAIRM1 promotes cell growth and inhibits cell apoptosis in glioblastoma cells and activates ZEB2 expression via sponging of miR-873-5p.
In conclusion, taken together, the above findings suggest that miR-873-5p acts as a tumor suppressor and HOTAIRM1 acts as an oncogene in glioblastoma progression. We have also demonstrated, for the first time, a novel HOTAIRM1/miR-873-5p/ZEB2 regulatory axis involved in the development of glioblastoma. These findings provide a theoretical basis for the development of alternative strategies for the clinical treatment of glioblastoma.
Conflicts of interest
None.
Footnotes
How to cite this article: Lin YH, Guo L, Yan F, Dou ZQ, Yu Q, Chen G. Long non-coding RNA HOTAIRM1 promotes proliferation and inhibits apoptosis of glioma cells by regulating the miR-873-5p/ZEB2 axis. Chin Med J 2019;133:174–182. doi: 10.1097/CM9.0000000000000615
References
- 1.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas implications for classification and therapy. Nat Rev Clin Oncol 2017; 14:434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016; 131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Wu BW, Tang C, Wang Y, Li ZQ, Hu SK, Hua W, et al. High-grade thalamic gliomas: microsurgical treatment and prognosis analysis. J Clin Neurosci 2018; 49:56–61. doi: 10.1016/j.jocn.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Miyai M, Tomita H, Soeda A, Yano H, Iwama T, Hara A. Current trends in mouse models of glioblastoma. J Neurooncol 2017; 135:423–432. doi: 10.1007/s11060-017-2626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Deng SL, Li YQ, Zhao G. Imaging gliomas with nanoparticle-labeled stem cells. Chin Med J 2018; 131:721–730. doi: 10.4103/0366-6999.226900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozdemir-Kaynak E, Qutub AA, Yesil-Celiktas O. Advances in glioblastoma multiforme treatment: new models for nanoparticle therapy. Front Physiol 2018; 9:170.doi: 10.3389/fphys.2018.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci 2008; 13:2537–2547. doi: 10.2741/2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai H, Zhu XX, Li ZF, Zhu YP, Lang JH. MicroRNA dysregulation and steroid hormone receptor expression in uterine tissues of rats with endometriosis during the implantation window. Chin Med J 2018; 131:2193–2204. doi: 10.4103/0366-6999.240808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahir BK, Ozer H, Engelhard HH, Lakka SS. MicroRNAs in glioblastoma pathogenesis and therapy: a comprehensive review. Crit Rev Oncol Hematol 2017; 120:22–33. doi: 10.1016/j.critrevonc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 2005; 334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65:6029–6033. doi: 10.1158/0008-5472.Can-05-0137. [DOI] [PubMed] [Google Scholar]
- 13.Moller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol 2013; 47:131–144. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008; 6:14.doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Li WQ, Yang YJ, Lu YC, He C, Hu GH, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res 2009; 1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 16.Li MJ, Zhang J, Liang Q, Xuan CH, Wu JX, Jiang P, et al. Exploring genetic associations with ceRNA regulation in the human genome. Nucleic Acids Res 2017; 45:5653–5665. doi: 10.1093/nar/gkx331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conte F, Fiscon G, Chiara M, Colombo T, Farina L, Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. Plos One 2017; 12:e0171661.doi: 10.1371/journal.pone.0171661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi ST, Song Y, Peng YP, Wang H, Long H, Yu XL, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. Plos One 2012; 7:e38842.doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Yuan J, Yuan XR, Zhao J, Zhang ZP, Weng L, et al. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int J Oncol 2016; 48:541–550. doi: 10.3892/ijo.2015.3267. [DOI] [PubMed] [Google Scholar]
- 20.Wu HH, Jovonovich SM, Randolph M, Post KM, Sen JD, Curless K, et al. Utilization of cell-transfer technique for molecular testing on hematoxylin-eosin-stained sections: a viable option for small biopsies that lack tumor tissues in paraffin block. Arch Pathol Lab Med 2016; 140:1383–1389. doi: 10.5858/arpa.2015-0454-OA. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 22.Zhi H, Li YS, Wang L. Profiling DNA methylation patterns of non-coding RNAs (ncRNAs) in human disease. Adv Exp Med Biol 2018; 1094:49–64. doi: 10.1007/978-981-13-0719-5_6. [DOI] [PubMed] [Google Scholar]
- 23.Xiao Y, Xu J, Yin W. Aberrant epigenetic modifications of non-coding RNAs in human disease. Adv Exp Med Biol 2018; 1094:65–75. doi: 10.1007/978-981-13-0719-5_7. [DOI] [PubMed] [Google Scholar]
- 24.Luo JD, Zhu H, Jiang H, Cui YY, Wang MJ, Ni XY, et al. The effects of aberrant expression of LncRNA DGCR5/miR-873-5p/TUSC3 in lung cancer cell progression. Cancer Med 2018; 7:3331–3341. doi: 10.1002/cam4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, Zhang X, Qi M, Zhang Y, Ding F. miR-873-5p inhibits the progression of colon cancer via repression of TUSC3/AKT signaling. J Gastroenterol Hepatol 2019; [Epub ahead of print] doi: 10.1111/jgh.14697. [DOI] [PubMed] [Google Scholar]
- 26.Goossens S, Janzen V, Bartunkova S, Yokomizo T, Drogat B, Crisan M, et al. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood 2011; 117:5620–5630. doi: 10.1182/blood-2010-08-300236. [DOI] [PubMed] [Google Scholar]
- 27.Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Cancer Res 2011; 17:7654–7663. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 28.Dai YH, Tang YP, Zhu HY, Lv L, Chu Y, Zhou YQ, et al. ZEB2 promotes the metastasis of gastric cancer and modulates epithelial mesenchymal transition of gastric cancer cells. Dig Dis Sci 2012; 57:1253–1260. doi: 10.1007/s10620-012-2042-6. [DOI] [PubMed] [Google Scholar]
- 29.Feng S, Cai XM, Li YY, Jian XG, Zhang LX, Li B. Tripartite motif-containing 14 (TRIM14) promotes epithelial-mesenchymal transition via ZEB2 in glioblastoma cells. J Exp Clin Cancer Res 2019; 38:57.doi: 10.1186/s13046-019-1070-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 2017; 24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XQ, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol 2014; 11:777–787. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan LD, Kong JL, Tang JL, Wu YH, Xu EP, Lai MD, et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med 2016; 20:2036–2044. doi: 10.1111/jcmm.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu RQ, Zhao G, Yang YL, Jiang ZY, Cai JL, Zhang ZJ, et al. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/PTEN axis in gastric cancer. J Cell Biochem 2019; 120:4952–4965. doi: 10.1002/jcb.27770. [DOI] [PubMed] [Google Scholar]


