Abstract
Our previous studies have confirmed that α2δ1 has the potential to function as a cancer stem cell marker, and CACNA2D1 is the coding gene of α2δ1. But it is unclear how microRNAs regulate the expression of the CACNA2D1 gene in laryngeal cancer cells. We detected the expressions of α2δ1 protein, microRNA-107, and CACNA2D1 in 40 pairs of laryngeal cancer tissues and adjacent normal tissues. Laryngeal squamous cell carcinoma cells, TU212 and TU686, were cultured and transfected in the blank control group, the agomiR negative control group, the agomiR-107 group, the antagomiR negative control group, or the antagomiR-107 group, and the dual-luciferase reporter assay was employed to assess the regulatory effect of microRNA-107 on CACNA2D1. Then, the effects of microRNA-107 on the biological function of laryngeal squamous cell carcinoma cells were detected by qRT-PCR, Western blot, MTT, cell migration/invasion assay, and cell colony-formation assay. Our data suggested that the protein level of α2δ1, encoded by CACNA2D1, in laryngeal carcinoma tissues was higher than that in adjacent normal tissues, while the expression of microRNA-107 was significantly decreased in laryngeal carcinoma tissues. The dual-luciferase reporter gene assay confirmed that microRNA-107 bound to the 3′-UTR two positions (202-209, 902-908) of CACNA2D1 mRNA. Moreover, the expression of CACNA2D1 and α2δ1 protein were significantly decreased in TU212 and TU686 cells transfected with microRNA-107 expression vectors (P < 0.05), and proliferation, clone formation, migration, and invasion of these cells were also reduced. Furthermore, after knocking down microRNA-107, exactly opposite results were obtained. Overexpression of microRNA-107 can inhibit the proliferation and invasion of laryngeal carcinoma cells in vitro.
Keywords: CACNA2D1, laryngeal squamous cell carcinoma, microRNA-107
Introduction
The incidence of head and neck squamous cell carcinoma (HNSCC) ranks sixth among systemic malignancies in China, and laryngeal squamous cell carcinoma (LSCC) ranks second among HNSCCs [1]. The main treatments for LSCC include surgery, adjuvant radiotherapy, and chemotherapy. Although the survival rate of patients has improved, it has not been significantly improved [2], and the quality of life of patients remains poor due to complications from surgery and adverse reactions of chemoradiotherapy. Recurrence and lymph node metastasis are regarded as the leading causes of death. Previous studies have shown that calcium channels are involved in the progression of various malignant tumors, such as invasion, metastasis, proliferation, and angiogenesis [3,4]. Our previous studies have demonstrated that voltage-gated calcium channel protein α2δ1 is related to the development of laryngeal cancer [5]. However, the underlying mechanism of α2δ1 expression and its role in progression of laryngeal cancer remain unclear.
MicroRNAs (miRNAs), approximately 18–22 nucleotides in length, are an endogenous family of non-coding small RNA molecules, which degrade the mRNA or inhibit post-transcriptional translation of the mRNA through complete or incomplete binding to the target gene mRNA [6]. The role of miRNAs in cancer has been extensively studied, and dysregulated miRNAs are frequently detected in cancer tissues and cells, which act as oncogenes or tumor suppressors [7]. Recent studies have shown that microRNA-107 (miR-107) is abnormally expressed in many solid tumors such as lung cancer [8], esophageal cancer [9], hepatocellular carcinoma [10], and cervical cancer [11]), and it is associated with tumor cell proliferation, invasion, apoptosis, and tumorigenic ability. Here, we hypothesized that miR-107 has a potential binding site for the α2δ1 encoding gene CACNA2D1 using bioinformatic analysis. However, it is unclear whether this binding affects the biological characteristics of laryngeal cancer cells.
In this study, we found that both miR-107 and CACNA2D1 were abnormally expressed in LSCC tissues, and their expression levels were negatively correlated. We predicted the potential binding sites of miR-107 and CACNA2D1 through online databases (Targetscan, PicTar, miRanda, and miRWalk), and the dual-luciferase reporter gene assay confirmed that CACNA2D1 is a target gene of miR-107. The expression levels of CACNA2D1 were decreased by miR-107. We subsequently observed that miR-107 inhibited the proliferation, migration, invasion, and clonality of LSCC cells. Therefore, the aforementioned data suggested that CACNA2D1 is a target gene, and miR-107 can inhibit the proliferation and invasion of LSCC cells through suppressing CACNA2D1 expression.
Materials and methods
Study subjects and patient tissue samples
This study included 40 patients (all male) who underwent surgery at Beijing Friendship Hospital, and it was approved by the institutional ethical committee of Beijing Friendship Hospital, Capital Medical University (# 2017-P2-187-01). All patients who signed the informed consent form and underwent surgery for the first time did not receive any adjuvant therapy such as radiotherapy or chemotherapy. All the specimens were pathologically confirmed. A tumor tissue and an adjacent normal tissue were collected from each patient. Adjacent normal tissue was obtained 2 cm away from the edge of the tumor and was confirmed by pathological examination as normal mucosa. The specimen obtained during surgery was immediately placed in liquid nitrogen and refrigerated in a −80°C refrigerator until it was used. Clinical pathology data were collected from hospital clinical records.
Cell culture
Human LSCC cell lines, TU212 (highly malignant) and TU686 (less malignant), were obtained from Shanghai Cell Bank, Chinese Academy of Sciences. Both TU212 and TU686 cell lines were routinely cultured in Dulbecco’s modified Eagle medium (DMEM, #11965118; Gibco, New York, USA), supplemented with 10% fetal bovine serum (FBS, #16000; Gibco), 100 µ/ml penicillin, and 100 mg/ml streptomycin (#15140-122; Gibco) in a humidified atmosphere containing 5% CO2 at 37°C.
Cell transient transfection
TU212 and TU686 LSCC cells were cultured in a six-well plate to ~70% density and collected by digestion and centrifugation (5 × 105 cells/well). Then agomiR-107 and antagomiR-107 (0.2 nM; GenePharma Co. Ltd, Shanghai, China) were transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, California, USA) and Opti-MEM medium (Invitrogen), respectively, according to the manufacturers’ instructions, and a negative control (NC) (GenePharma Co. Ltd.) was used for both reactions.
Immunofluorescence staining
Frozen tissues were sectioned using a cryostat and fixed with methanol for 30 seconds. After blocking with 5% nonfat milk in PBS, we added goat serum and blocked the tissues at room temperature for 30 minutes. Then, slices were incubated with the CACNA2D1 mAb (dilution ratio 1:100) (#MA3-921l; Thermo Fisher Scientific, Rockford, Illinois, USA) at 4°C overnight, and the NC group was added to PBS. This was followed by incubation with Cy3-labeled goat anti-mouse IgG (#BA1031; Wuhan Boster Biological Technology Ltd., Wuhan, China) for 1 hour at 37°C. The slices were rinsed four times with PBST for 3 minutes each time. Nuclei were stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI; #C1002; Biyuntian Biotechnology Co., Ltd., Shanghai, China) at 5 μg/ml. The slides were sealed with Fluoromount-G (#0100-01; Southern Biotech, Birmingham, Alabama, USA). Slices were examined with an Olympus BX53 confocal microscope (Olympus, Tokyo, Japan).
Quantitative real-time PCR
Total RNA was extracted using Trizol (#15596026; Invitrogen) from frozen tissues (100 mg) or LSCC cell lines. RNA concentration and purity were determined by the optical density ratio (OD260/OD280) of 1.8–2.0. The cDNA template was synthesized after reverse transcription of the RNA, and then, it was subjected to quantitative real-time PCR (qRT-PCR) using an ABI7500 real-time PCR instrument (ABI Company, New York, USA). The reaction system (20 μl in total) included the following: SYBR Green Master Mix (10 μl; Vazyme Biotech Co., Ltd, Nanjing, China), forward primer (0.4 μl; 10 μM), reverse primer (0.4 μl; 10 μM), cDNA (4 μl; dilution ratio 1:10), 50× ROX Reference Dye 2 (0.4 μl; Vazyme Biotech Co., Ltd.), and distilled deionized H2O (4.8 μl). The reaction conditions were as follows: 2 minutes at 50°C, pre-denaturation for 10 minutes at 95°C, 40 cycles of denaturation for 30 seconds at 95°C, annealing for 30 seconds at 60°C, and extension for 30 seconds at 72°C. Using U6 small nuclear RNA (Tianyihuiyuan, Co., Ltd., Beijing, China) and GAPDH (Tianyihuiyuan, Co., Ltd) selected as the internal reference, respectively, the relative expression of the target gene was calculated by the 2−ΔΔ Ct method. The threshold cycle (Ct) was obtained when the fluorescence intensity reached a certain threshold. All tests were repeated three times. The primers used were as follows:
Hsa-miR-107:
Forward:5′- TGCGCAGCAGCATTGTACAGGGC -3′
Reverse:5′- CCAGTGCAGGGTCCGAGGTATT -3′
U6:
Forward: 5′- CGCTTCGGCAGCACATATAC -3′
Reverse: 5′- AAATATGGAACGCTTCACGA -3′
Homo CACNA2D1:
Forward: 5′- GCATTGGAAGCGGAGAAAGT -3′
Reverse: 5′- GGAATATGGACTGCTGCGTG -3′
Homo GAPDH:
Forward: 5′- TCAAGAAGGTGGTGAAGCAGG -3′
Reverse: 5′- TCAAAGGTGGAGGAGTGGGT -3′
Western blot
Total proteins from tissues and cells transfected with specific plasmids were extracted with the radioimmunoprecipitation buffer (#P0013B; Biyuntian Biotechnology Co., Ltd). The protein concentrations were determined using the BCA protein assay kit (#P0012; Biyuntian Biotechnology Co., Ltd) based on the manufacturer’s description. Protein samples (40 μg/well) were separated by 12% SDS−polyacrylamide gel electrophoresis under denaturing conditions and transferred to a polyvinylidene difluoride (PVDF) membrane (#IPVH00010; Millipore Inc., Massachusetts, USA) using the wet transfer method (200 mA, 90 minutes). The PVDF membrane was soaked in TBST (blocking solution) containing 5% skim milk, blocked at room temperature for 2 hours, and then incubated with the CACNA2D1 mAb (1:500, #MA3-921; Thermo Fisher Scientific) and the Rabbit anti-GAPDH (1:1000, #AB-P-R 001; Hangzhou Goodhere Biotechnology Co., Ltd., Hangzhou, China) antibody at 4°C overnight. The PVDF membrane was washed five to six times with TBST, for 5 minutes every time. The blots were then washed and incubated for 2 hours with the HRP-labeled goat anti-mouse secondary antibody (1:50000, # BA1051; Wuhan Boster Biological Technology Ltd.). After washing with TBST (5–6 times, 5 minutes every time), the Electro-Chemi-Luminescence substrate solution (#NCI5079; Thermo Fisher Scientific) was mixed with the stable peroxidase solution in a 1:1 ratio, and it was added to the PVDF membrane. Immunoreactive bands were detected using the UVP image analysis system (BD Biosciences, New Jersey, USA). BandScan 5.0 was used to estimate the expression level of the target protein.
Cell proliferation assay
After transfection, the cells were divided into the following five groups: the blank control group, the agomiR NC group, the agomiR-107 group, the antagomiR NC group, and the antagomiR-107 group. The transfected cells were inoculated into 96-well plates at 5 × 103 cells, and each group had three replicate wells. The cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C for 48 hours. Furthermore, 10 μl of MTT (3-(4, 5-dimethylthiazol-2yl)-2, 5-diphenyl tetrazolium bromide, #M5655; Sigma, Missouri, USA) was added to each well, the cells were incubated at 37°C for 4 hours, the medium was aspirated, and the cells were incubated with 150 μl of DMSO (#HZB1133; Sigma) for 10 minutes. Absorbance was measured using a microplate reader at 490 nm.
Transwell assay
After 24 hours of transfection, TU212 and TU686 cells were digested with 0.25% trypsin (#15050065; Gibco). After centrifugation at 1200 rpm for 3 minutes, cell suspension was prepared and the number of cells was counted using the cell counting plate. Then 4 × 104 cells were seeded into the Transwell upper chamber (#353097; BD Biosciences). The upper chamber was added with serum-free medium DMEM (#1670291; Gibco), and the lower chamber was added with culture medium containing 10% FBS. Cells were cultured in an incubator for 48 hours, and the chamber was carefully rinsed with PBS. The cells were fixed with 70% ice ethanol solution for 1 hour, stained with 0.5% crystal violet stain, placed at room temperature for 20 minutes, and the unmigrated cells on the upper chamber side were wiped clean with a sterile cotton ball. After observation and photography under the Olympus IX51 microscope (Olympus), each chamber was randomly selected for six fields of view for counting.
A Matrigel (#354234; Corning, New York, USA) was melted at 4°C, and it was diluted to a final concentration of 1 mg/ml with serum-free medium. Then 800 μl of 10% FBS DMEM medium was added to a 24-well plate, and 100 μl of Matrigel was added to the upper chamber of the transwell and incubated at 37°C for 4–5 hours to form a gel. After Matrigel was gelatinized, a 4 × 104 cell suspension was inserted into the transwell upper chamber and cultured at 37°C in a 5% CO2 incubator for 48 hours. Subsequent steps were similar to those in the cell migration experiments. The number of cells passing through the matrigel is considered to be the ability of cell invasion.
Colony-formation assay
The transfected cells were digested with 0.25% trypsin (#15050065; Gibco) and pipetted into individual cells. The cell suspension was diluted, and each group was seeded at a cell density of 250 cells/well in six-well plates with three replicate wells per group. The cells were incubated for 2 weeks in a humidified atmosphere containing 5% CO2 at 37°C. When a macroscopic clone was noted, the supernatant was discarded, rinsed twice with PBS, and pre-cooled with 70% ethanol for 20 minutes. Crystal violet staining was performed for 30 minutes. The cells were photographed with a microscope (Olympus IX51), and the number of colonies was counted.
Vector construction and dual-luciferase reporter assay
Using the online databases such as TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), miRDB (mirdb.org/miRDB), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) to predict the potential upstream miRNAs of CACNA2D1, it was found that miR-107 may possess two potential binding sites in the conserved region of the 3′-UTR of CACNA2D1 (202-209, 902-908). To construct the luciferase reporter vectors, 3′-UTR fragments covering the putative hsa-miR-107 binding sites on the CACNA2D1 of interest were amplified from genomic DNA. The PCR products were restricted with XhoI/Not I (XhoI #1094A, Not I #1166A; TaKaRa, Kusatsu, Japan) enzymes. The primer sequence was as follows: Forward: 5′- GGTTCTTTTCCAACGCTATT -3′. Reverse: 5′- GACTCATTTAGATCCTCACAC -3′. The amplified fragments were inserted into pYr-MirTarget (Yingrun Biotechnologies Inc., Changsha, China) within the XhoI/Not I sites. All constructs were verified by DNA sequencing. Luciferase reporter experiments were performed in the TU686 LSCC cell line. Cells were seeded at 2 × 105 cells/well in 12-well plates one day prior to transfection, and then pYr-MirTarget-homo CACNA2D1 3′-UTR(202-209, 2 μg) and pYr-MirTarget-homo CACNA2D1 3′-UTR (902-908, 2 μg) were cotransfected with agomiR-107 (0.1 nM), agomiR NC (0.1 nM), antagomiR-107 (0.1 nM), or antagomiR NC (0.1 nM). The relative luciferase activity was measured after 48 hours of transfection by using the dual-luciferase assay kit (#RG027; Biyuntian Biotechnology Co., Ltd) according to the manufacturer’s instructions. Using firefly luciferase as an internal reference, the ratio of relative light units of renilla luciferase and firefly luciferase was used to compare the activation level of the target reporter gene between different samples. Mutated vectors were also constructed simultaneously. Two mutated vectors, which were concerned with two seed regions in the CACNA2D1 3′-UTR, were constructed. The first vector was pYr-MirTarget-homo CACNA2D1 3′-UTR 202-209mut, and the second vector was pYr-MirTarget-homo CACNA2D1 3′-UTR 902-908mut. The vectors with wild-type 3′-UTR (100 ng) or mutated 3′-UTR (100 ng) were cotransfected with agomiR-107, agomir NC, antagomiR-107, or antagomir NC into TU686 cells in a 96-well plate. Transfection and luciferase assay procedures were similar to those described above. All transfection experiments were performed in triplicate.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, California, USA). All quantitative data are expressed as mean ± SD. Student’s t-test was performed when only two groups were compared. Two-way analysis of variance was used to analyze the results of the luciferase assays. Variance analysis was used to analyze the association of miR-107 and CACNA2D1 expression levels with clinicopathological factors. P < 0.05 was considered to be statistically significant.
Results
Differential expression of microRNA-107 and CACNA2D1 in patients with laryngeal squamous cell carcinoma and cell lines
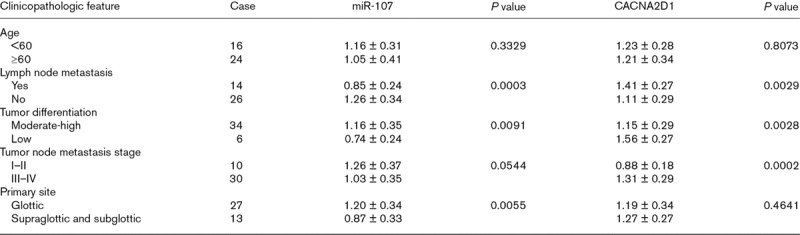
Immunofluorescence and Western blot (WB) results indicated that α2δ1 was highly expressed in LSCC tissues compared with adjacent normal tissues (P < 0.05; Fig. 1a and b). qRT-PCR results indicated that compared with adjacent normal tissues, the expression of miR-107 was significantly decreased in LSCC tissues (P < 0.05; Fig. 1c and d). MiR-107 expression was associated with lymph node metastasis, tumor differentiation, and primary site in patients with LSCC (both P < 0.05), but it was not associated with the age and Tumor Node Metastasis (TNM) staging of patients with LSCC (both P > 0.05) (Table 1). At the same time, the expression level of CACNA2D1 was closely related to LSCC lymph node metastasis, tumor differentiation, and TNM stage (P < 0.05), but it was not related to the age of LSCC patients and the primary site of the tumor (both P > 0.05) (Table 1).
Fig. 1.
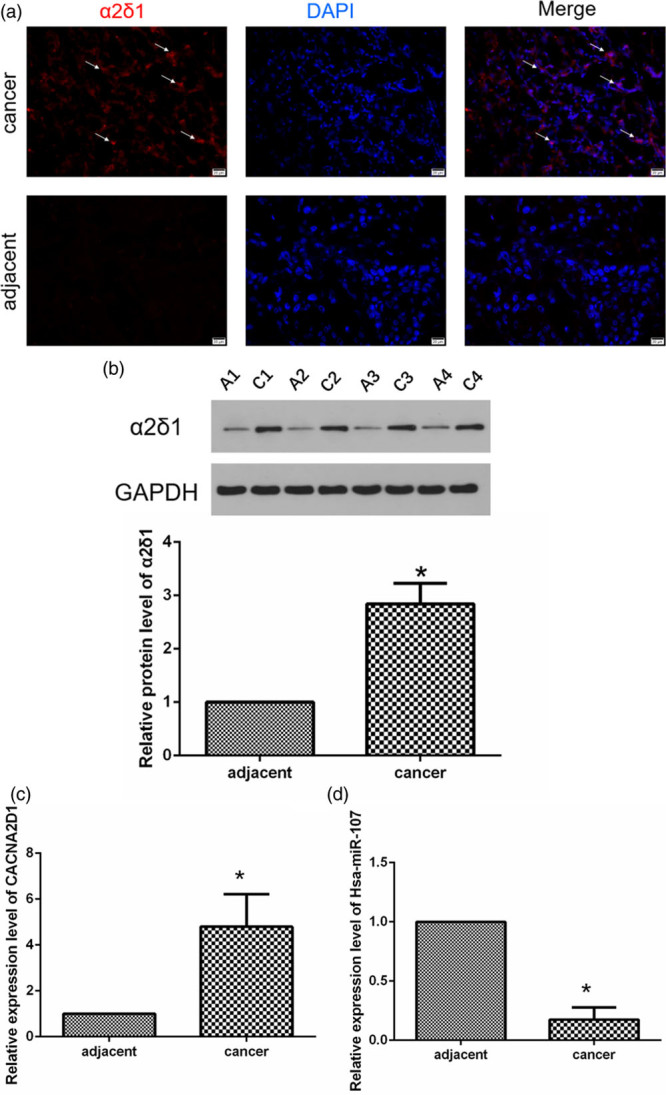
CACNA2D1 and miR-107 are inversely correlated in tissues. (a) This is an immunofluorescence photograph of a LSCC patient (T4N2M0, moderately differentiated, ×400). The arrow shows the typical α2δ1 expression (red). (b) The images of α2δ1 protein expression detected by WB in adjacent normal tissues and LSCC tissues are as follows: A1 represents the adjacent tissue from patient No.1; C1 represents the LSCC tissue from patient No.1; A2 represents the adjacent tissue from patient No.2; and C2 represents the LSCC tissue from patient No. 2, and so on. *P < 0.05. (c and d) qPCR detection of relative expressions of CACNA2D1 and miR-107 in adjacent normal tissues and LSCC tissues (*P < 0.05). It was easily observed that they are inversely related to normal and LSCC tissues. LSCC, laryngeal squamous cell carcinoma; miR-107, microRNA-107; WB, Western blot.
Table 1.
Associations of microRNA-107 and CACNA2D1 expressions with clinicopathologic features of laryngeal squamous cell carcinoma patients
Differential expression of microRNA-107 and CACNA2D1 in laryngeal squamous cell carcinoma cell lines
In the cell experiments, the expression level of miR-107 in TU212 and TU686 cells was significantly changed after transfection (Fig. 2a and b). Compared with the control group, the expression levels of CACNA2D1 mRNA and protein (α2δ1) in the agomiR-107 group were significantly reduced (Fig. 2c–f), while CACNA2D1 mRNA and α2δ1 protein expressions were significantly increased in the antagomiR-107 group (Fig. 2c–f). These results suggest a significant inverse correlation between miR-107 and CACNA2D1.
Fig. 2.
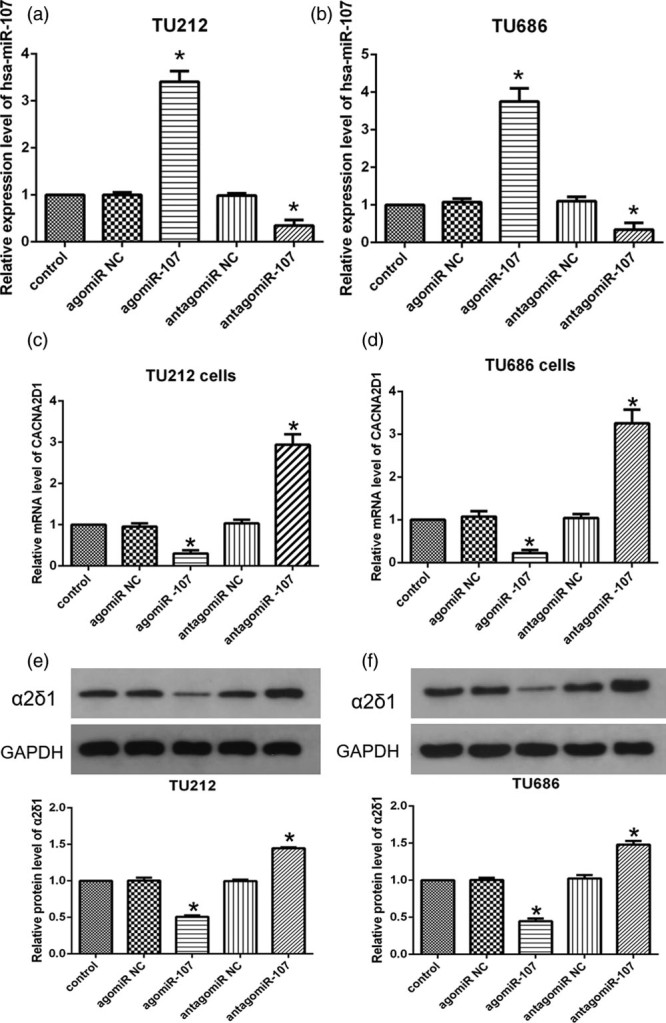
CACNA2D1 and miR-107 are inversely correlated in LSCC cell lines. (a and b) Transfection efficiency among the following five groups: TU212 and TU686 cells transfected with blank control, agomiR NC, agomiR-107, antagomiR NC, or antagomiR-107, and the expression of miR-107 was significantly increased or decreased, respectively (compared with the control group *P < 0.05). (c and d) qPCR analysis of relative expression level of CACNA2D1 mRNA after transfection of TU212 and TU686 cells (compared with the control group *P < 0.05). (e and f) Western blot analysis of relative expression of α2δ1 protein in transfected TU212 and TU686 cells (compared with the control group, *P < 0.05). LSCC, laryngeal squamous cell carcinoma; miR-107, microRNA-107; NC, negative control.
MicroRNA-107 directly targets CACNA2D1
We detected an inverse correlation between the expression of hsa-miR-107 and CACNA2D1 in LSCC tissues and adjacent normal tissues by qRT-PCR and WB. Generally, miRNAs inhibit post-transcriptional translation by binding to the 3′-UTR of the target gene mRNA. By searching for upstream microRNAs of the CACNA2D1 gene in TargetScan, miRDB, PicTar, and miRWalk databases, we predicted that miR-107 might target CACNA2D1. TargetScanHuman 7.1 suggested that there are two putative miR-107 binding sites in the 3′-UTR conserved regions of CACNA2D1 (site A: 202-209; site B: 902-908), and the reporter gene expression vectors containing the CACNA2D1 wild type or mutant 3′-UTR were cloned downstream of the luciferase gene (Fig. 3a and c). The dual-luciferase reporter assay was performed to verify whether CACNA2D1 is a direct target gene for hsa-miR-107. At site A (Fig. 3b), agomiR-107 blocked the luciferase activity by 55.6% (P < 0.05). At site B (Fig. 3d), agomiR-107 blocked the luciferase activity by 80.6% (P < 0.05). This indicates that the inhibitory effect of agomiR-107 at point A is weaker than that at point B. In the meantime, antagomiR-107 was able to enhance the luciferase activity at both binding sites (both P < 0.05). This result indicates that miR-107 functions by directly targeting CACNA2D1.
Fig. 3.
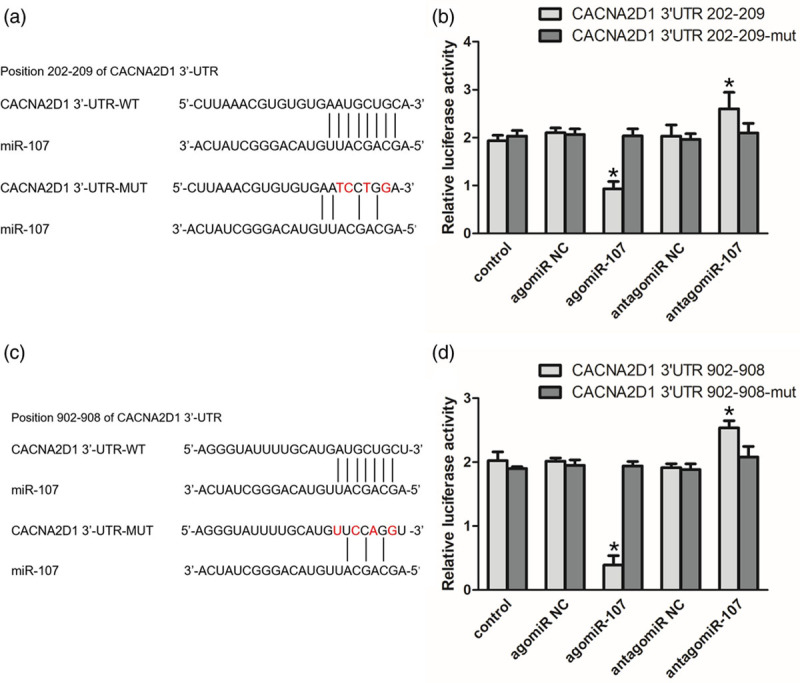
CACNA2D1 is a direct downstream target of miR-107. (a) Wild type and mutant of the 3′-UTR of CACNA2D1 mRNA and miR-107 binding site (site A: 202-209). (b) Luciferase activity controlled by the 3′-UTR of CACNA2D1 was inhibited by ectopic expression of miR-107 at site A. TU686 cells were co-transfected with wild-type or mutant CACNA2D1 3′-UTR and agomiR-107, antagomiR-107 and the corresponding negative controls. Renilla and firefly luciferase activities were measured using a dual-luciferase reporter assay system. The ratio of renilla luciferase activity to firefly luciferase activity was normalized, and the data are expressed as mean ± SEM (compared with the control group, *P < 0.05). (c) Wild type and mutant of the 3′-UTR of CACNA2D1 mRNA and miR-107 binding site (site B: 902-908). (d) Luciferase activity controlled by the 3′-UTR of CACNA2D1 was inhibited by ectopic expression of miR-107 at site B. TU686 cells were co-transfected with wild-type or mutant CACNA2D1 3′-UTR and agomiR-107, antagomiR-107, and the corresponding negative controls. The dual-luciferase reporter gene system indicates that miR-107 can bind to the site B of CACNA2D1 3′-UTR and inhibit the luciferase activity (compared with the control group, *P < 0.05). miR-107, microRNA-107
MicroRNA-107 inhibits proliferation of laryngeal squamous cell carcinoma cells
To determine the role of miR-107 in LSCC cell proliferation, we performed MTT assays in TU212 and TU686 cells, and transfected agomiR-107, antagomiR-107, and corresponding NC into these two cell lines. MTT assay indicated that the proliferation rate of the two cells cultured after 48 hours was significantly impacted. Compared with the control group, the proliferation rate of TU212 cells in the agomiR-107 group was decreased to 30.1% (P < 0.05), while the proliferation rate of TU212 cells in the antagomiR-107 group was increased to 130.3% (P < 0.05; Fig. 4a). Similarly, in TU686 cells, the proliferation rate in the agomiR-107 group was decreased to 38.4% (P < 0.05), and the proliferation rate in the antagomiR-107 group was increased to 138.9% (P < 0.05; Fig. 4b). These results suggest that miR-107 significantly inhibits the proliferation of LSCC cells.
Fig. 4.
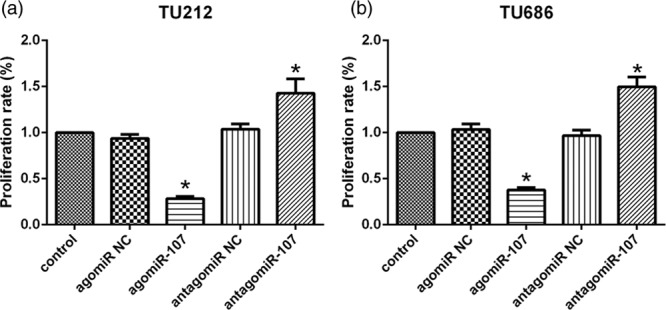
miR-107 inhibits the proliferation rate of LSCC cells. (a and b) After 48 hours, MTT assay showed that the proliferation rates of TU212 and TU686 cells transfected with agomiR-107, antagomiR-107, and corresponding negative controls were significantly reduced or increased compared with those in the control (*P < 0.05). LSCC, laryngeal squamous cell carcinoma; miR-107, microRNA-107.
MicroRNA-107 inhibits laryngeal squamous cell carcinoma cell migration and invasion
We further evaluated the effect of miR-107 on the migration and invasion abilities of TU212 and TU686 cells using the Transwell assay. After transfection of the two LSCC cells in the Transwell chamber for 48 hours, the migration ability of the TU212 and TU686 cells in the agomiR-107 group was significantly weaker (P < 0.05), while the migration ability of the TU212 and TU686 cells in the antagomiR-107 group was significantly enhanced (P < 0.05; Fig. 5a and b). Similar to the results of the migration experiment, the invasive ability of the two LSCC cells in the agomiR-107 group was significantly weaker than that in the control group (P < 0.05), and the invasive ability of the two LSCC cells in the antagomiR-107 group was significantly improved (P < 0.05; Fig. 5c and d). These results suggest that miR-107 can significantly inhibit the migration and invasion of LSCC cells.
Fig. 5.
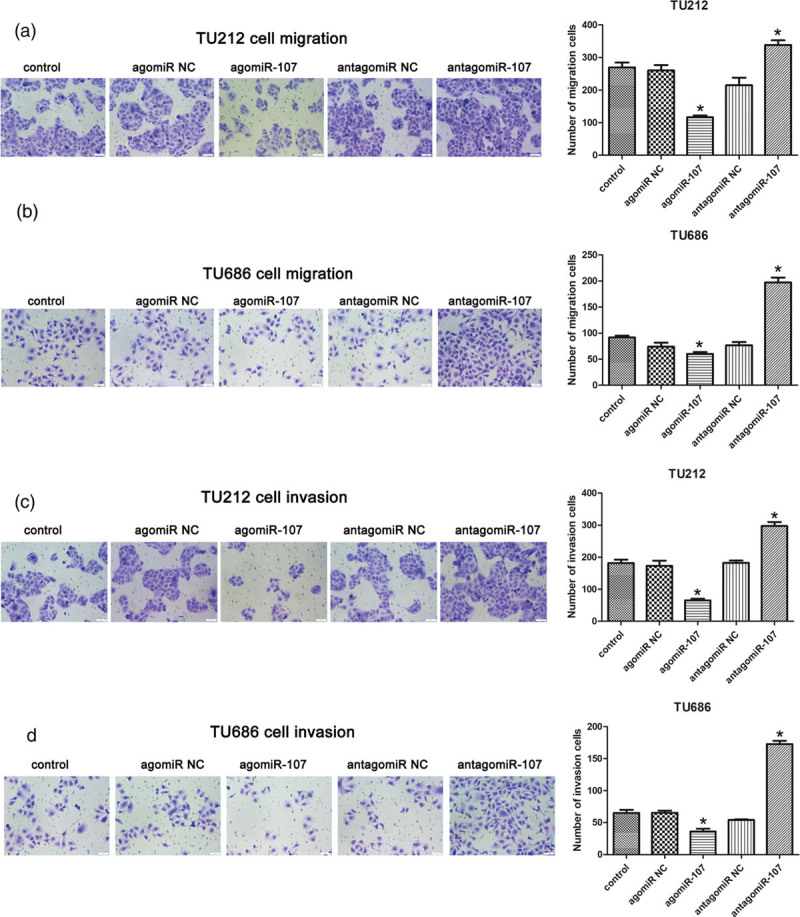
The migration and invasion abilities of LSCC cells were compared among the five groups. (a and b) After transfection, the migration ability of TU212 and TU686 cells among the five groups detected by Transwell assay, and the comparison of the number of polycarbonate membrane cells among the five groups. Compared to the control group, *P < 0.05. (c and d) The invasive ability of TU212 and TU686 cells in the five groups detected by Transwell assay, and the comparison of the number of cells in each group of Matrigel in the five groups. Compared to the control group, *P < 0.05. LSCC, laryngeal squamous cell carcinoma.
MicroRNA-107 inhibits laryngeal squamous cell carcinoma colony formation
Finally, we performed the colony-formation assay for the transfected TU212 and TU686 cells. The results suggested that the number of clones in the two LSCC cell lines in the agomiR-107 group was significantly lower than that in the control group (P < 0.05; Fig. 6a and b), and the number of clones formed in the antagomiR-107 group was higher than that in the control group (P < 0.05; Fig. 6a and b). The histogram visually displays these differences. This means that miR-107 is able to inhibit clonal formation of LSCC cells.
Fig. 6.
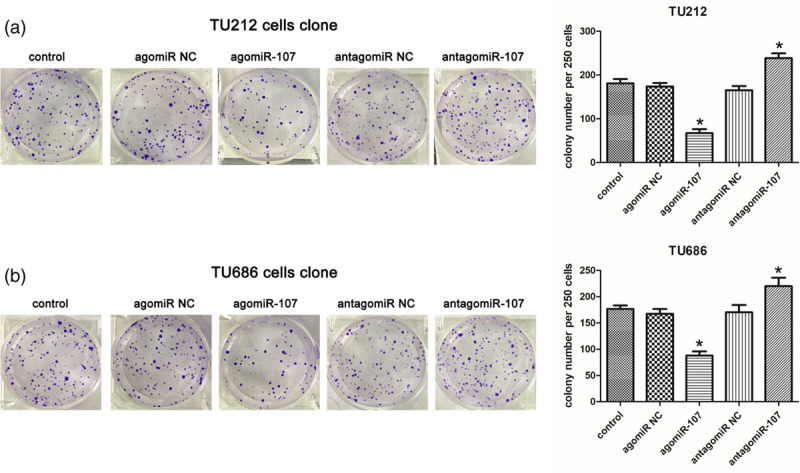
miR-107 inhibits clonal formation of LSCC cells. (a and b) Photographs of TU212 and TU686 cell colony formation and histograms of cell clone numbers. TU212/TU686 cells (250/24 mm plate) were inoculated on the medium for 2 weeks, stained with crystal violet, and the number of colonies was counted. Compared to the control group, *P < 0.05. LSCC, laryngeal squamous cell carcinoma; miR-107, microRNA-107.
Discussion
Being a common malignant tumor of the head and neck region, LSCC poses a serious threat to human health. Despite advances in surgery and chemoradiotherapy, the overall survival rate of patients with LSCC has not significantly improved in recent years [12]. Therefore, it is particularly important to study and identify the molecular mechanisms involved in the pathogenesis of LSCC. The ‘cancer stem cell’ theory provides a new research perspective on the incidence, progress, prognosis, and intervention of malignant tumors. W ZHAO et al. confirmed that α2δ1 plays the role of a marker for liver cancer and lung cancer tumor initiation cells [13,14]. Based on this research, our team initially confirmed that the voltage-gated calcium channel subunit α2δ1 has the potential to function as a marker of LSCC stem cells, and the α2δ1 subunit promotes the progression of LSCC. We then studied how endogenous genes regulate α2δ1 and its coding gene CACNA2D1.
α2δ1 is a T-type calcium channel subunit. It is well known that Ca2+ concentration is essential for cell proliferation, apoptosis, and regulation of the cell cycle [15]. This effect is mainly achieved by calcium oscillation. Calcium oscillation, as a major form of calcium signaling, can promote the expression of specific genes, which is related to the magnitude and duration of calcium transients. It has been suggested that α2δ1 may be involved in this ‘amplitude-encoding’ signal [13], thereby maintaining the stem cell characteristics of LSCC. High expression of T-type calcium channels has been found in malignant tumors, such as esophageal cancer, glioma, breast cancer, and prostate cancer [3]. This is consistent with our findings in the LSCC tissue, suggesting that α2δ1 may be involved in the key steps in the pathogenesis and progression of LSCC.
Various tumor studies have shown that miRNAs can regulate cancer stem cells and affect the characteristics of cancer cells [16–18]. MiR-107 is located on chromosome 10 in the human body, which corresponds to intron 5 of the pantothenate kinase gene, and it was discovered in 2003. MiR-107 plays a role in a variety of diseases, including hypoxia [19], neurodegenerative diseases [20], lipid metabolism [21], cell cycle arrest [22,23], angiogenesis [24], and tumor suppression and occurrence [25]. MiR-107 has been reported to be abnormally expressed in many solid malignancies. In most malignant tumors, including glioma [26], liver cancer [27], esophageal cancer [28], cervical cancer [11], and HNSCC [29], miR-107 expression is significantly decreased in the cancer tissues, while overexpression of miR-107 inhibits the proliferation of tumor cells, and it exists as a tumor suppressor gene. However, there are exceptions. In the study of miR-107 in gastric cancer [30,31] and pancreatic cancer [32,33], different research teams have obtained different results. miR-107 targets different genes, thereby exerting anti-cancer or cancer-promoting effects. We used online tools, such as Targetscan, PicTar, and miRanda, to predict the potential binding sites for CACNA2D1 and miR-107, and CACNA2D1 may be a target for miR-107. However, it is unclear whether miR-107 affects the biological function of laryngeal cancer cells by targeting CACNA2D1.
In this study, we analyzed the expressions of miR-107 and CACNA2D1 in 40 LSCC patients and adjacent normal tissues. We observed low expression of miR-107 in human LSCC tissues, while the CACNA2D1 mRNA and protein were highly expressed. We also analyzed the correlation between miR-107/CACNA2D1 and clinicopathological parameters, and we found that the lower miR-107 level was related to lymph node metastasis, tumor cell differentiation, and primary site. Higher levels of CACNA2D1 were associated with lymph node metastasis, tumor differentiation, and TNM stage. Therefore, these experimental results suggest that miR-107 is inversely related to the level of CACNA2D1, and miR-107 may have an effect in inhibiting the progression of LSCC. It is suggested that miR-107 may function as a tumor suppressor gene in LSCC.
We used TU212 and TU686 LSCC cells to explore the relationship between miR-107 and CACNA2D1. Knockdown of miR-107 was closely related to a significant surge in CACNA2D1 expression levels. In addition, dual-luciferase reporter assays revealed that miR-107 binds to CACNA2D1 3′-UTR and two sites, negatively affecting its stability and ultimately reducing CACNA2D1 levels. These results confirmed that CACNA2D1 is a direct target of miR-107. In the biological function study, we found that overexpression of miR-107 not only reduces the expression level of the CACNA2D1 gene, but it also curbs the expression level of CACNA2D1 protein (α2δ1). After knocking down miR-107, the expression levels of the CACNA2D1 gene and α2δ1 were increased. In addition, we demonstrated that overexpression of miR-107 inhibited the proliferation, migration, and invasion of TU212/TU686 cells and reduced the LSCC colony-forming ability. In contrast, knockdown of miR-107 promoted proliferation, migration, invasion, and colony formation of LSCC cells.
These results revealed that abnormal miR-107 and CACNA2D1 expression is involved in the progression of LSCC: overexpression of miR-107 results in downregulation of CACNA2D1 and significant inhibition of the malignant biological properties of LSCC cells. In addition, we also found that low expression of miR-107 and/or high expression of CACNA2D1 are associated with poor prognosis in patients with LSCC. Therefore, miR-107 and CACNA2D1 may have the potential to be a prognostic and therapeutic target for LSCC.
Certainly, our research is not profound enough. We plan to further explore the effect of miR-107 on the biological function of sorted α2δ1+/α2δ1− cells by flow cytometry. In addition, all our experiments were performed in vitro, and we need to perform tumorigenicity assays in nude mice to further elucidate the role of miR-107 and CACNA2D1 in the progression of LSCC and the possibility of their use as targets for future clinical applications.
Conclusion
Our results suggest that miR-107 can inhibit LSCC cell proliferation, invasion, and colony formation, and this inhibition is achieved by targeted regulation of CACNA2D1, CACNA2D1 is a direct downstream target of miR-107, and miR-107 is a tumor suppressor of LSCC. Future challenges include identifying other targets for miR-107 to further identify its function and to improve its applicability in LSCC therapy.
Acknowledgements
This research was supported by the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding (#XLMX201701), and The Research Fund of Chengdu Medical College (#CYZ17-33).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Liu Y, Zhao Q, Ding G, Zhu Y, Li W, Chen W. Incidence and mortality of laryngeal cancer in China, 2008-2012. Chin J Cancer Res. 2018; 30:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlito A, Thompson LD, Cardesa A, Gnepp DR, Devaney KO, Rodrigo JP, et al. The importance of histological types for treatment and prognosis in laryngeal cancer. Eur Arch Otorhinolaryngol. 2013; 270:401–403 [DOI] [PubMed] [Google Scholar]
- 3.Prevarskaya N, Skryma R, Shuba Y. Calcium in tumour metastasis: new roles for known actors. Nat Rev Cancer. 2011; 11:609–618 [DOI] [PubMed] [Google Scholar]
- 4.Zhu P, Fan Z. Cancer stem cells and tumorigenesis. Biophys Rep. 2018; 4:178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Li Y, Zhao W, Zhang A, Lu C, Wang Z, Liu L. Α2δ1 may be a potential marker for cancer stem cell in laryngeal squamous cell carcinoma. Cancer Biomark. 2019; 24:97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan SH, Wang LH. Regulation of cancer metastasis by micrornas. J Biomed Sci. 2015; 22:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P, Liu H, Wang Z, He F, Wang H, Shi Z, et al. Micrornas in laryngeal cancer: implications for diagnosis, prognosis and therapy. Am J Transl Res. 2016; 8:1935–1944 [PMC free article] [PubMed] [Google Scholar]
- 8.Qu L, Li L, Zheng X, Fu H, Tang C, Qin H, et al. Circulating plasma micrornas as potential markers to identify EGFR mutation status and to monitor epidermal growth factor receptor-tyrosine kinase inhibitor treatment in patients with advanced non-small cell lung cancer. Oncotarget. 2017; 8:45807–45824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma P, Saini N, Sharma R. Mir-107 functions as a tumor suppressor in human esophageal squamous cell carcinoma and targets cdc42. Oncol Rep. 2017; 37:3116–3127 [DOI] [PubMed] [Google Scholar]
- 10.Su SG, Yang M, Zhang MF, Peng QZ, Li MY, Liu LP, Bao SY. Mir-107-mediated decrease of HMGCS2 indicates poor outcomes and promotes cell migration in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017; 91:53–59 [DOI] [PubMed] [Google Scholar]
- 11.Dong P, Xiong Y, Hanley SJB, Yue J, Watari H. Musashi-2, a novel oncoprotein promoting cervical cancer cell growth and invasion, is negatively regulated by p53-induced mir-143 and mir-107 activation. J Exp Clin Cancer Res. 2017; 36:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issa MR, Samuels SE, Bellile E, Shalabi FL, Eisbruch A, Wolf G. Tumor volumes and prognosis in laryngeal cancer. Cancers (Basel). 2015; 7:2236–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W, Wang L, Han H, Jin K, Lin N, Guo T, et al. 1B50-1, a mab raised against recurrent tumor cells, targets liver tumor-initiating cells by binding to the calcium channel α2δ1 subunit. Cancer Cell. 2013; 23:541–556 [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Wang S, Zhao W, Duan J, Wang Z, Chen H, et al. Mechanistic exploration of cancer stem cell marker voltage-dependent calcium channel α2δ1 subunit-mediated chemotherapy resistance in small-cell lung cancer. Clin Cancer Res. 2018; 24:2148–2158 [DOI] [PubMed] [Google Scholar]
- 15.Taylor JT, Zeng XB, Pottle JE, Lee K, Wang AR, Yi SG, et al. Calcium signaling and T-type calcium channels in cancer cell cycling. World J Gastroenterol. 2008; 14:4984–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dziegielewska B, Gray LS, Dziegielewski J. T-type calcium channels blockers as new tools in cancer therapies. Pflugers Arch. 2014; 466:801–810 [DOI] [PubMed] [Google Scholar]
- 17.Lou W, Liu J, Gao Y, Zhong G, Ding B, Xu L, Fan W. Microrna regulation of liver cancer stem cells. Am J Cancer Res. 2018; 8:1126–1141 [PMC free article] [PubMed] [Google Scholar]
- 18.Xu YM, Liao XY, Chen XW, Li DZ, Sun JG, Liao RX. Regulation of mirnas affects radiobiological response of lung cancer stem cells. Biomed Res Int. 2015; 2015:851841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayremlou N, Mozdarani H, Mowla SJ, Delavari A. Increased levels of serum and tissue mir-107 in human gastric cancer: correlation with tumor hypoxia. Cancer Biomark. 2015; 15:851–860 [DOI] [PubMed] [Google Scholar]
- 20.Moncini S, Lunghi M, Valmadre A, Grasso M, Del Vescovo V, Riva P, et al. The mir-15/107 family of microrna genes regulates CDK5R1/p35 with implications for Alzheimer’s disease pathogenesis. Mol Neurobiol. 2017; 54:4329–4342 [DOI] [PubMed] [Google Scholar]
- 21.Daimiel-Ruiz L, Klett-Mingo M, Konstantinidou V, Micó V, Aranda JF, García B, et al. Dietary lipids modulate the expression of mir-107, a mirna that regulates the circadian system. Mol Nutr Food Res. 2015; 59:1865–1878 [DOI] [PubMed] [Google Scholar]
- 22.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. Mir-107 and mir-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. Plos One. 2009; 4:e6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng L, Xie Y, Zhang H, Wu Y. Mir-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012; 29:856–863 [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Mao L, Gao Y, Baral S, Zhou Y, Hu B. Microrna-107 contributes to post-stroke angiogenesis by targeting dicer-1. Sci Rep. 2015; 5:13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci U S A. 2012; 109:14470–14475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Chen XR, Chen FF, Liu Y, Li P, Zhang R, et al. Microrna-107 inhibits U87 glioma stem cells growth and invasion. Cell Mol Neurobiol. 2013; 33:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Chen F, Zhao M, Yang Z, Zhang S, Ye L, et al. Mir-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mrna 3’UTR. Biochem Biophys Res Commun. 2016; 480:455–460 [DOI] [PubMed] [Google Scholar]
- 28.Sharma P, Saraya A, Gupta P, Sharma R. Decreased levels of circulating and tissue mir-107 in human esophageal cancer. Biomarkers. 2013; 18:322–330 [DOI] [PubMed] [Google Scholar]
- 29.Datta J, Islam M, Dutta S, Roy S, Pan Q, Teknos TN. Suberoylanilide hydroxamic acid inhibits growth of head and neck cancer cell lines by reactivation of tumor suppressor micrornas. Oral Oncol. 2016; 56:32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song YQ, Ma XH, Ma GL, Lin B, Liu C, Deng QJ, Lv WP. Microrna-107 promotes proliferation of gastric cancer cells by targeting cyclin dependent kinase 8. Diagn Pathol. 2014; 9:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng F, Yang Z, Huang F, Yin L, Yan G, Gong G. Microrna-107 inhibits gastric cancer cell proliferation and metastasis by targeting PI3K/AKT pathway. Microb Pathog. 2018; 121:110–114 [DOI] [PubMed] [Google Scholar]
- 32.Imamura T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Ohashi T, et al. Depleted tumor suppressor mir-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017; 7:5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong J, Wang D, Wei A, Lu H, Tan C, Li A, et al. Deregulated expression of mir-107 inhibits metastasis of PDAC through inhibition PI3K/akt signaling via caveolin-1 and PTEN. Exp Cell Res. 2017; 361:316–323 [DOI] [PubMed] [Google Scholar]


