Abstract
Liver X receptors limit cellular lipid uptake by stimulating the transcription of Inducible Degrader of the LDL Receptor (IDOL), an E3 ubiquitin ligase that targets lipoprotein receptors for degradation. The function of IDOL in systemic metabolism is incompletely understood. Here we show that loss of IDOL in mice protects against the development of diet-induced obesity and metabolic dysfunction by altering food intake and thermogenesis. Unexpectedly, analysis of tissue-specific knockout mice revealed that IDOL affects energy balance, not through its actions in peripheral metabolic tissues (liver, adipose, endothelium, intestine, skeletal muscle), but by controlling lipoprotein receptor abundance in neurons. Single-cell RNA sequencing of the hypothalamus demonstrated that IDOL deletion altered gene expression linked to control of metabolism. Finally, we identify VLDLR rather than LDLR as the primary mediator of IDOL effects on energy balance. These studies identify a role for the neuronal IDOL-VLDLR pathway in metabolic homeostasis and diet-induced obesity.
Introduction
Lipoprotein receptors are key determinants of cardiovascular disease due to their pivotal roles in regulating blood cholesterol levels. Dissecting the function and regulation of members of the LDL receptor (LDLR) superfamily has advanced our understanding of fundamental processes such as receptor-mediated endocytosis1, neuronal development2, and lipid-responsive transcription3. Cells maintain optimal cholesterol levels in part by regulating the uptake of cholesterol from circulating lipoproteins via the LDL receptor. Sterol regulatory element binding proteins (SREBPs) are transcription factors activated by low cholesterol levels that stimulate the expression of genes that drive cholesterol synthesis and uptake, including LDLR. Conversely, when cells accumulate excess cholesterol, activation of the Liver X Receptors (LXRs) promotes the expression of genes that restore homeostasis by increasing cholesterol efflux and transport4,5. Both the LXR and SREBP pathways also exert negative feedback on each other6,7.
One LXR-mediated feedback mechanism is to limit the influx of lipoprotein cholesterol into cells via the Inducible Degrader of the LDL Receptor (IDOL), an E3 ubiquitin ligase that targets members of the LDLR family for degradation8,9. Previous work elucidated the molecular mechanisms underlying IDOL-dependent degradation of its lipoprotein receptor targets9–13, and addressed its role in the species-specific regulation of hepatic cholesterol metabolism14–18. IDOL and its targets are expressed in a number of metabolically-active tissues other than liver, but the physiological role of IDOL in systemic lipid metabolism remains poorly understood. Furthermore, the lipoprotein receptor(s) through which IDOL regulates metabolism are unknown.
Here we report that mice globally deficient in IDOL expression are protected against diet-induced obesity and metabolic dysfunction. Unexpectedly, loss of IDOL in individual metabolic tissues, including liver, skeletal muscle, adipose and intestine, failed to recapitulate the phenotype of the whole-body knockouts, prompting us to search for an alternative mechanism. Ultimately, we traced this metabolic phenotype to loss of IDOL in the central nervous system, and to the consequent dysregulation of VLDLR protein levels. Deletion of IDOL from mouse neurons phenocopies the whole-body IDOL knockout mice. Our data identify the IDOL-VLDLR axis in neurons as a regulatory pathway impacting systemic energy balance.
Results
Global IDOL-knockout mice are protected from diet-induced metabolic dysfunction
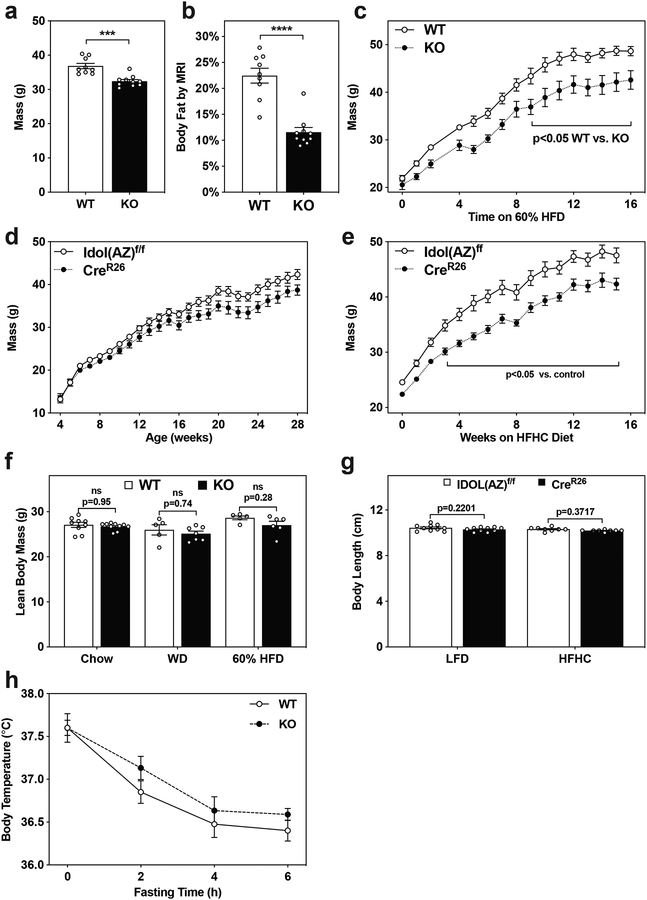
Previous efforts to characterize the physiological role of IDOL have focused primarily on its ability to regulate cholesterol metabolism. Unexpectedly, in the course of these studies we noted that 18-month old chow-fed IDOL-KO mice (developed at UCLA) demonstrated that deletion of IDOL protected against age-induced obesity, even when the mice were fed standard chow diet (Extended Data Fig. 1A, 1B). We then proceeded to challenge mice with obesogenic diets. Group-housed IDOL-deficient knockout mice fed a western diet (WD) enriched in fat and cholesterol for 15 weeks were 22% lighter than WT controls (Fig. 1A). Body composition analysis by MRI revealed that IDOL-KO mice were leaner, averaging 11% body fat compared to 32% in the WT mice (Fig. 1B). This was accompanied by reduced hepatic lipid accumulation and smaller adipocytes in the inguinal white adipose tissue depot (Fig. 1C). As the gene encoding IDOL is an LXR target, we postulated that the differential response to diet may require dietary cholesterol to stimulate LXR activity; however, the IDOL-KO mice were also leaner when fed a high-fat diet (HFD) that contained comparatively little cholesterol (Extended Data Fig. 1C).
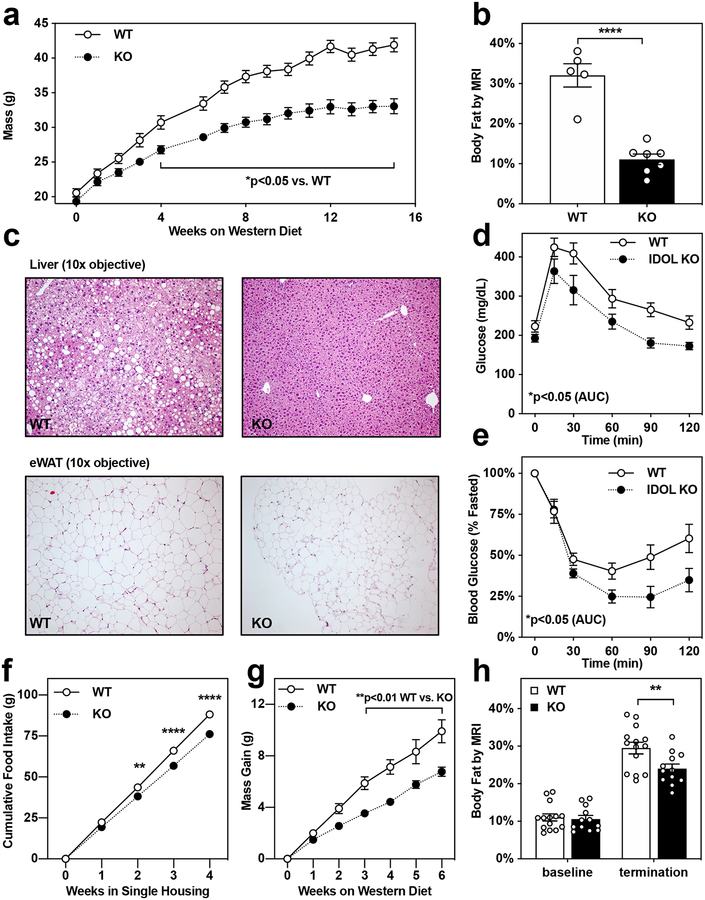
Figure 1: Global IDOL knockout mice are protected from diet-induced metabolic dysfunction.
(a) Growth curve for wild-type (WT) vs. Idol knockout (KO) mice fed western diet from 5–6 weeks of age. The mean masses are shown +/− the standard error of the mean (SEM); n=12 WT, n=9 KO mice *p<0.05 by repeat measures two-way ANOVA. (b) Adiposity reported as body fat percentage +/− the SEM measured by MRI after 15 weeks of western diet feeding; n=5 WT, n=7 KO mice ****p<0.0001 by 2-tailed t-test. (c) H&E-stained sections of liver and epididymal white adipose tissue (eWAT) depots from mice maintained on western diet for 15 weeks; these images are representative of tissues analyzed from both the UCLA-produced and AstraZeneca-produced lines. (d) Intraperitoneal glucose tolerance test (1 mg/kg) administered after 9 weeks of western diet feeding. The mean blood glucose levels are shown +/− the SEM; n=10 WT, n=9 KO mice *p<0.05 by 2-tailed t-test of the area under the curve (AUC) (e) Intraperitoneal insulin tolerance test (1 U/kg) administered after 14 weeks of western diet feeding. The mean blood glucose levels are shown +/− SEM; n=7 WT, n=6 KO mice *p<0.05 by 2-tailed t-test of the area under the curve (AUC). (f-h) Single-housed IDOL KO mice consume less food than WT littermates, protecting them from diet-induced adipose expansion; n=14 WT, n=12 KO mice **p<0.01, ****p<0.0001 WT vs. KO by repeat measures ANOVA. (f) The mean food consumed per mouse is labeled +/− SEM. (g) The mean mass gained per mouse after being placed in single housing +/− SEM. (h) Adiposity reported as body fat percentage +/− the SEM measured by MRI. The precise n-number, p-value, and details of the statistical testing are provided in the source data file.
We also analyzed a second, independently-derived IDOL knockout mouse line generated by crossing Idol-floxed mice developed at AstraZeneca, denoted IDOL(AZ), with a Rosa26-Cre transgenic line, denoted CreR26 (Supplemental Fig. 1). When maintained on low-fat low-cholesterol diet (LFD), these CreR26/IDOL(AZ) mice also showed a lean phenotype after 20-weeks of age (Extended Data Fig. 1D). The phenotype was exaggerated when the mice were challenged with a high-fat high-cholesterol diet (HFHC; Extended Data Fig. 1E).
We found no evidence of a generalized developmental growth defect in either strain of IDOL-knockout mice. There were no differences in lean body mass (Extended Data Fig. 1F), nasal to anal length (Extended Data Fig. 1G), or to body temperature attributable to genotype (Extended Data Fig. 1H). The reduced adiposity in the WD-fed IDOL-KO mice was associated with improved glucose clearance (Fig. 1D; nine weeks of diet) and insulin tolerance (Fig. 1E; fourteen weeks of diet). Although we did not detect differences in food intake in short term studies, analysis of mice housed singly over the course of weeks (WT, n=14; IDOL KO, n=12) revealed reduced food intake in the genetic absence of IDOL (Fig 1F), associated with reduced diet-induced adipose expansion (Fig 1G, 1H).
Deletion of IDOL from peripheral metabolic tissues does not protect from obesity
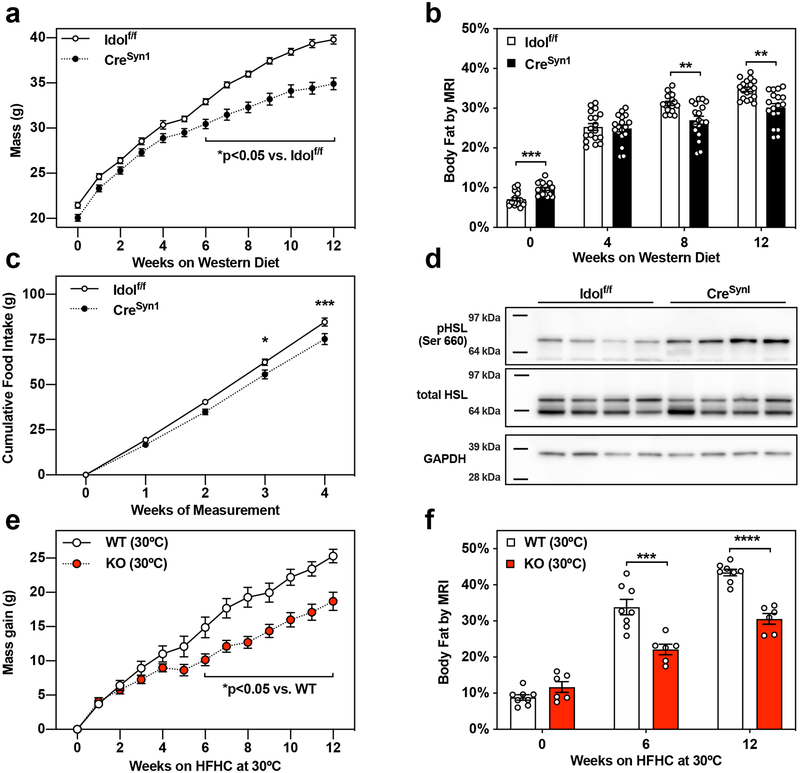
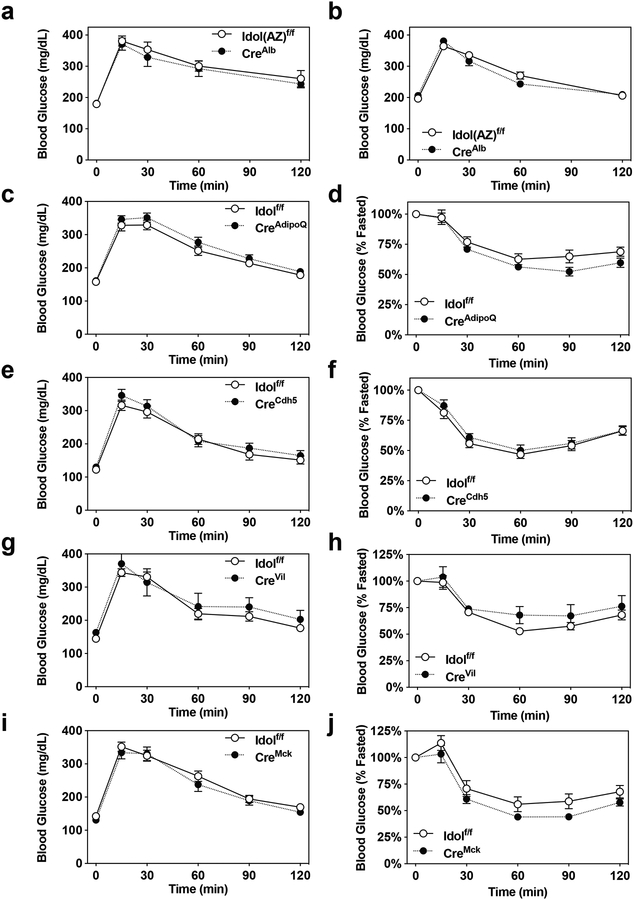
We hypothesized that the metabolic phenotype of IDOL-deficient mice resulted from primary actions of the IDOL pathway in one or more metabolic tissues. To test this idea, we crossed Idol-floxed mice to a range of tissue-specific Cre transgenic lines to generate tissue-selective IDOL knockouts. We first analyzed the contribution of the liver to the IDOL-KO phenotype using Alb-Cre-IDOL(AZ)-floxed mice (Idol(AZ)f/f; CreAlb) generated at AstraZeneca. Hepatic deletion of IDOL had no effect on the obesity of mice challenged with the obesogenic HFHC diet (Fig. 2A, 2F), nor on glucose metabolism (Extended Data Fig 2A, 2B), suggesting that the liver is not the primary driver of the global IDOL-KO phenotype.
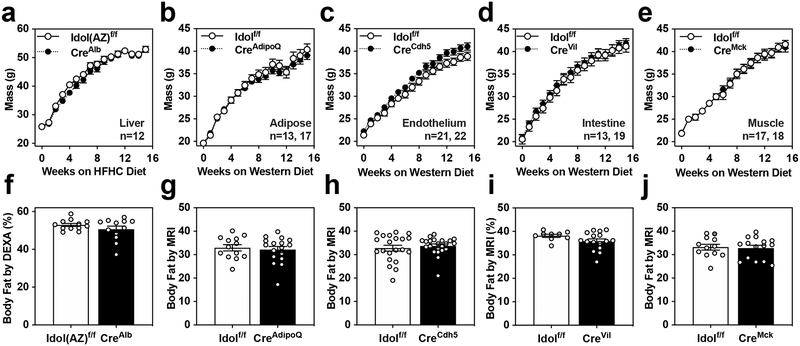
Figure 2: Deletion of IDOL from peripheral tissues does not protect against diet-induced metabolic dysfunction.
(a-e) Growth curves showing the mean mass +/− SEM for male mice from each conditional knockout mouse strain tested. (f-j) Adiposity reported as body fat percentage +/− SEM measured by MRI or DEXA after 15 weeks consuming the indicated test diet. (a, f) Liver-specific IDOL KO mice (n=12 Idol(AZ)f/f, n=12 Idol(AZ)f/f CreAlb) fed a high-fat, high-cholesterol (HFHC) diet for 15 weeks beginning when mice were eight weeks old. Adiposity measured by DEXA scan. (b,g) Adipose-specific IDOL KO mice (n=13 Idolf/f, n=17 Idolf/fCreAdipoQ) fed a western diet for 15 weeks starting when the mice were six weeks old. Adiposity measured by MRI. (c,h) Endothelium-specific IDOL KO mice (n=21 Idolf/f, n=22 Idolf/f CreCdh5) fed a western diet for 15 weeks starting when the mice were six weeks old. Adiposity measured by MRI. (d,i) Intestine-specific IDOL KO mice (n=13 Idolf/f, n=19 Idolf/f CreVil) fed a western diet for 15 weeks starting when the mice were six weeks old. Adiposity measured by MRI. (e,j) Muscle-specific IDOL KO mice (n=11 Idolf/f, n=18 Idolf/f CreMck) fed a western diet for 15 weeks starting when the mice were six weeks old. Adiposity measured by MRI. There were no statistically significant differences between groups in any panel of this figure. The precise n-number, p-value, and details of the statistical testing are listed in the source data file.
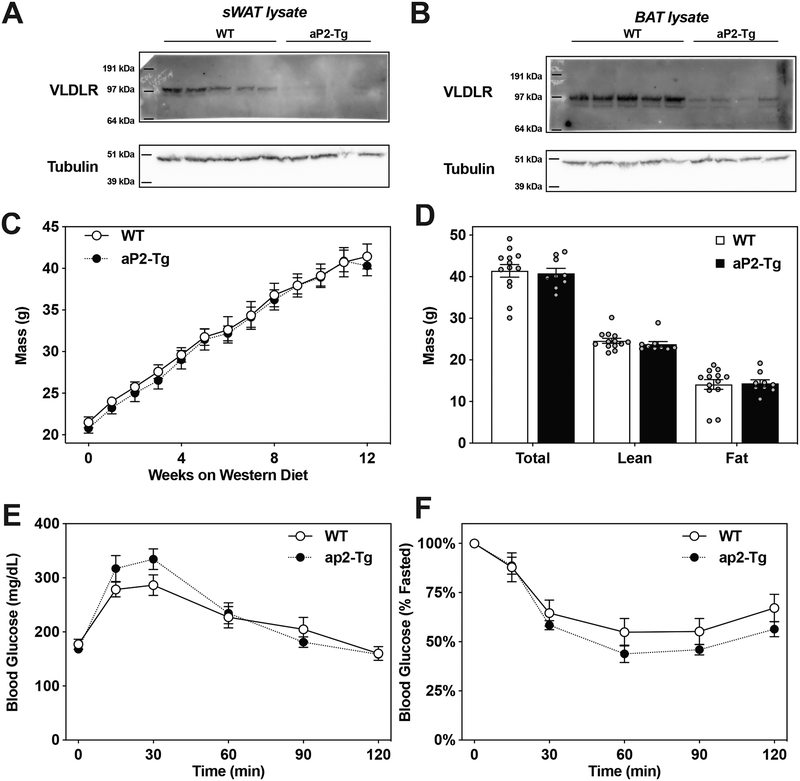
Given the reduced adiposity in IDOL-KO mice, we next considered that a direct effect of the IDOL pathway on adipose tissue might explain the phenotype. We crossed AdipoQ-Cre transgenics19 to Idolf/f mice to generate mice lacking IDOL expression in both white and brown fat (Idolf/f; CreAdipoQ). We challenged the mice with 15 weeks of western-diet feeding but saw no effect on mass or adiposity (Fig. 2B, 2G) or on glucose or insulin tolerance (Extended Data Fig. 2C, 2D). In a complimentary approach, we developed a transgenic mouse expressing a dominant active form of human IDOL under the control of Fabp4 promoter (aP2-Tg). This line showed nearly complete ablation of VLDLR protein levels in both white and brown adipose depots, confirming the activation of the IDOL pathway (Extended Data Fig. 3A, 3B). Despite this change in VLDLR levels, we observed no differences in mass between the aP2-IDOL Tg mice and their littermate controls fed WD for twelve weeks (Extended Data Fig. 3C). Accordingly, neither adiposity nor responses to glucose and insulin challenges were affected by the transgene (Extended Data Fig. 3D–3F). Collectively the data collected from both gain- and loss-of-function mice lead us to conclude that adipose-intrinsic actions of IDOL cannot explain the phenotype of global IDOL-knockout mice.
We next considered the possibility that IDOL-dependent changes in LDLR or VLDLR protein levels may be acting in the vasculature to alter lipid delivery and metabolism in adipocytes secondarily. This would be consistent with studies showing that VLDLR affects lipoprotein lipase activity20–23. We crossed Idolf/f mice with a line expressing Cre from the Cadherin 5 promoter (commonly referred to as VE Cadherin-Cre)24 to generate an endothelial-specific IDOL knockout line (Idolf/f; CreCdh5). Loss of IDOL activity in the endothelium had no effect on the initiation or progression of obesity after challenge with WD for 15 weeks (Fig. 2C, 2H). There was also no difference in glucose or insulin tolerance between groups (Extended Data Fig. 2E, 2F).
An alternative possibility was that altered caloric intake from the diet could be causing the change in weight gain. Idol is highly expressed in the intestine8, and it is plausible that altering lipoprotein receptor expression could affect dietary lipid absorption or efflux. We crossed the Idol-floxed mice with Villin-Cre transgenics to generate intestine-specific knockout mice (Idolf/f; CreVil). However, loss of IDOL activity in the intestine did not affect mass gain or adiposity (Fig. 2D, 2I) and did not affect glucose or insulin tolerance (Extended Data Fig. 2G, 2H). Prompted by the high level of expression of Vldlr in muscle, we also generate muscle-specific IDOL-knockouts (Idolf/f; CreMck) by crossing the IDOL floxed mice to MCK-Cre transgenics25. Again, the tissue-selective knockout failed to recapitulate the effects of whole-body IDOL loss (Fig. 2E, 2J and Extended Data Fig. 2I, 2J).
IDOL regulates systemic energy balance through the VLDL receptor
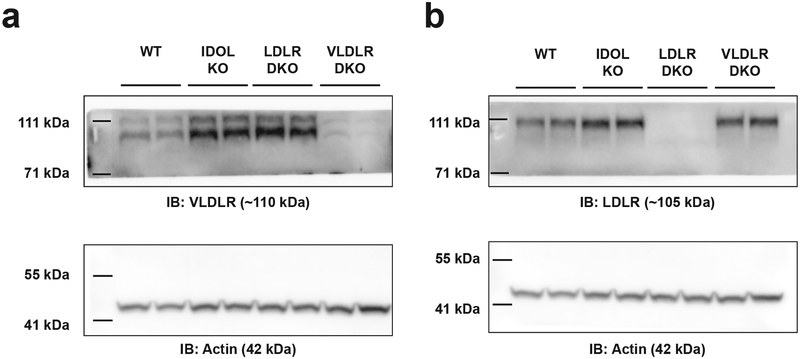
IDOL controls the abundance of three proteins (LDLR, VLDLR, and ApoER2) via ubiquitination, which targets the proteins for lysosomal degradation9,13. We therefore hypothesized that the metabolic phenotype of the global IDOL knockout was due to changes in levels of one or more of these lipoprotein receptors. To test this idea, we developed two double-knockout (DKO) mouse lines in which IDOL and either LDLR or VLDLR were deleted simultaneously. We did not pursue an ApoER2 compound mutant line as global ApoER2 knockout mice are sterile26, have severe neurological defects at baseline2, and floxed-Apoer2 mice are not available. We challenged cohorts of single and double IDOL-knockout mice with HFHC diet. Consistent with the result of prior cohorts, the mass (Fig. 3A and 3B) and adiposity (Fig. 3E and 3F) of global IDOL-KO mice diverged rapidly from the WT group, reaching statistical significance after two to four weeks on diet (p<0.05 by repeat measures two-way ANOVA). Throughout the study, the IDOL KO and IDOL-LDLR DKO groups were indistinguishable with respect to body mass and adiposity (Fig. 3A, 3E). LDLR is the only IDOL target expressed in the liver, further supporting the conclusion that the liver is not the primary driver of the global IDOL-KO phenotype.
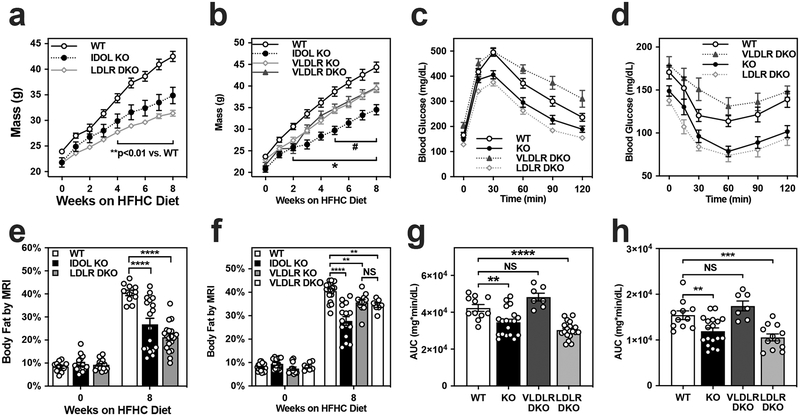
Figure 3: IDOL regulates systemic energy balance through the VLDL receptor.
(a) Growth curves showing the mean mass and the standard error of the mean (SEM) for male mice placed on a high fat high cholesterol (HFHC) diet from 6–8 weeks of age; n=11 wild type (WT), n=18 Idol−/− (KO), n=18 Idol−/−Ldlr−/− (LDLR DKO) mice. (b) Growth curves showing the mean mass +/− SEM for male wild-type (WT) n=20, Idol−/− (IDOL KO) n=15, Vldlr−/− (VLDLR KO) n=12, and Idol−/−Vldlr−/− (VLDLR DKO) n=7 mice fed a HFHC diet from 6–8 weeks of age. (c) Intraperitoneal glucose tolerance test (1 g/kg) administered after six weeks on the HFHC diet. The values shown are the mean blood glucose levels +/− SEM. (d) Intraperitoneal insulin tolerance test (1U/kg) administered after ten weeks on the HFHC diet. The values shown are the mean blood glucose levels +/− SEM. (e) Adiposity reported as body fat percentage +/− SEM measured by MRI for the mice reported in panel (a) at baseline and after 8 weeks on the HFHC diet. (f) Adiposity reported as body fat percentage +/− SEM measured by MRI for the mice reported in panel (b) at baseline and after 8 weeks on the HFHC diet. (g) The area under the curve (AUC) was calculated for the glucose tolerance test for each mouse and the mean values +/− SEM and individual values are plotted. (h) The area under the curve (AUC) was calculated for the insulin tolerance test for each mouse and the mean values +/− SEM and individual values are plotted. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. The precise n-number, p-value, and details of the statistical testing are provided in the source data file.
Whole body deletion of VLDLR has previously been shown to be protective against diet-induced obesity22,27,28. To further understand the contributions of IDOL to body weight we compared the responses of WT, VLDLR-KO, IDOL-KO, and IDOL-VLDLR DKO mice to HFHC diet feeding (Fig. 3B, 3F). The VLDLR single knockout mice and IDOL-VLDLR DKO showed comparable protection against adipose expansion upon high-fat diet feeding, confirming previous reports that deletion of VLDLR is protective against diet-induced obesity. They further show that deletion of IDOL, which leads to an over-abundance of VLDLR protein (Extended Data Fig. 4A, 4B), has an even more pronounced protective effect on WT mice, but has no additional effect on the background of VLDLR deficiency. These data strongly suggest that altered abundance of the VLDLR receptor and not the LDLR is an important contributor to systemic energy balance.
We assessed metabolic fitness in the compound mutant mice by performing a glucose tolerance test after six weeks on HFHC diet (Fig. 3C) and an insulin tolerance test after ten weeks on diet (Fig. 3D). Both the IDOL-KO and IDOL-LDLR DKO groups had improved glucose clearance and enhanced response to a bolus of insulin relative to the WT group; however, the IDOL-VLDLR DKO group was indistinguishable from the WT group. These data were analyzed by comparing the AUCs using a one-way ANOVA (Fig. 3G, 3H). Collectively, these data provide strong evidence for the hypothesis that regulation of VLDLR protein levels is important for the maintenance of metabolic homeostasis.
Acute knockdown of IDOL in the central nervous system increases energy expenditure
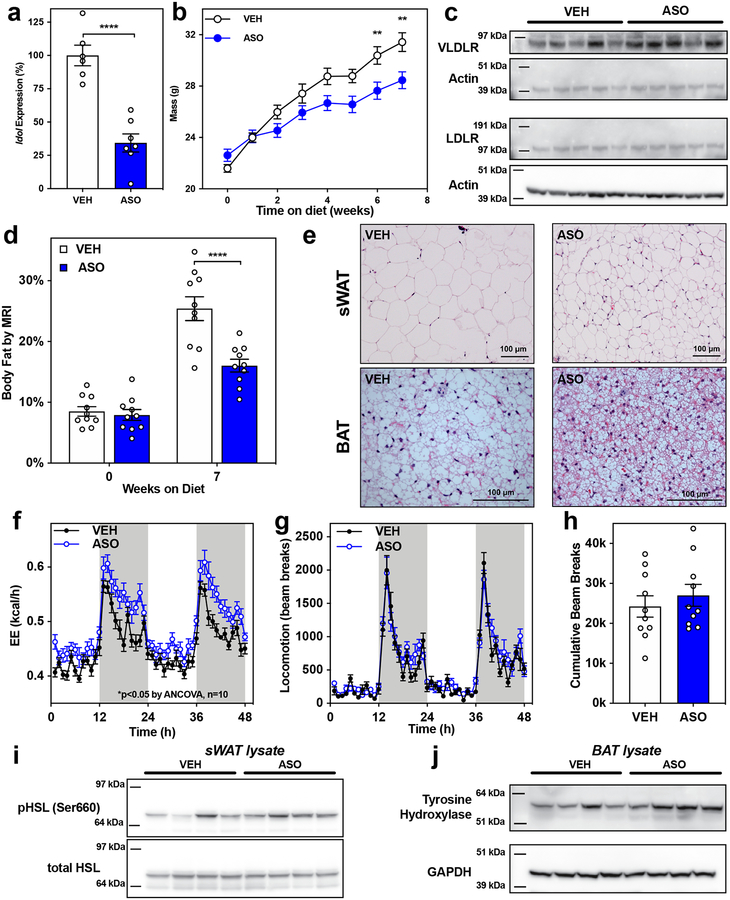
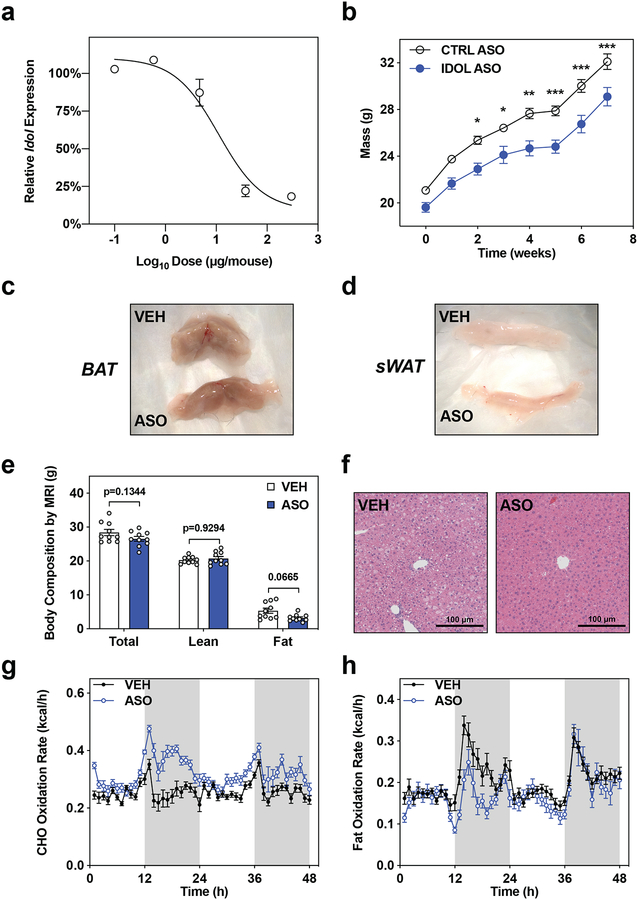
VLDLR is most highly expressed in the brain, muscle, fat, and the endothelium. Having ruled out the peripheral metabolic tissues as the source of the IDOL-VLDLR effect on energy balance, we considered the CNS as a potential site of action. Prior studies showed that the IDOL pathway in the CNS affects the function of both microglia and neurons29,30; however, the possibility that central actions of IDOL affect metabolism has not been addressed. To test whether IDOL was acting centrally, we developed a CNS-optimized antisense oligonucleotide (ASO) targeting Idol. Dose-response studies determined that 40 μg given by intracerebroventricular injection was the minimum dose required for maximal suppression of Idol expression in whole brain homogenates collected two weeks after the injection (Extended Data Fig. 5A). Using this optimized protocol, we knocked down Idol expression in 5-week old C57Bl/6J mice and initiated a western diet-induced obesity study one-week post-injection. At the end of the seven-week western diet study, Idol expression was suppressed by 65% in the hypothalamus of ASO-injected mice relative to vehicle-treated control mice (Fig. 4A). As expected, Idol knockdown in the CNS was associated with increased VLDLR protein in the hypothalamus (Fig. 4C). The mice treated with the IDOL ASO weighed less than their control counterparts after six weeks on diet (Fig. 4B; p<0.01 by repeat measures two-way ANOVA). These findings were confirmed in a second cohort of mice using a CNS-optimized negative control ASO (Extended Data Fig. 5B). Consistent with the whole-body knockout, the Idol ASO-treated mice were leaner by MRI analysis (Fig. 4D) and had smaller fat pads upon dissection (Extended Data Fig. 5C, 5D).
Figure 4: Acute knockdown of IDOL in the central nervous system reduces adiposity by increasing energy expenditure.
(a) Intracerebroventricular injection of a CNS-optimized antisense oligonucleotide (ASO) targeting Idol reduced Idol expression by 65% in the hypothalamus measured at study termination. The mean Idol expression +/− the standard error of the mean (SEM) is shown, p<0.0001 by two-tailed t-test n=8. (b) Growth curve showing the mean mass +/− SEM for male mice placed on western diet one week after ICV injection of Idol-targeting ASO or vehicle control (VEH); **p<0.01 by repeat measures ANOVA, n=10. (c) Increased VLDLR protein levels in the hypothalamus of IDOL KO mice 8 weeks after the ASO injection, the blots are representative of two independent analyses. (d) Adiposity reported as body fat percentage +/− SEM measured by MRI; n=10, ****p<0.0001 by repeat measures ANOVA (e) Hematoxylin and eosin stained subcutaneous white adipose tissue (sWAT) and interscapular brown adipose tissue (BAT) sections from mice after seven weeks of western diet feeding. (20x objective lens for sWAT and 40x objective lens for BAT, these images are representative of five independent biological replicates) (f) Energy expenditure measured by indirect calorimetry in mice after 2–3 weeks on western diet showing the mean value per hour and +/− SEM; n=10 *p<0.05 VEH vs. ASO by ANCOVA using either total body mass or lean body mass as covariates. (g) Locomotor activity as measured when a mouse crossed multiple IR beams during the calorimetry experiment; mean values +/− SEM are shown for n=10 mice. (h) No difference in the overall locomotion of mice as quantified by the cumulative number of beam breaks in a 48-hour period +/− SEM; p=0.4772 by two-tailed t-test; n=10. (i) Western blot analysis of the phosphorylation of hormone sensitive lipase (HSL) in subcutaneous white adipose tissue. This blot is representative of two independent analyses (j) Western blot analysis of tyrosine hydroxylase (TH) abundance in the brown adipose tissue. This blot is representative of two independent analyses. The precise n-number, p-value, and details of the statistical testing are provided in the source data file.
Although IDOL ASO-treated mice phenocopied the whole-body IDOL KO mice, we did not observe changes to food intake. We hypothesized that the body weight difference may involve changes in energy expenditure. We performed indirect calorimetry (Oxy-Max CLAMS, Columbus Instruments) on the cohorts after 2–3 weeks on diet (before their body mass and adiposity diverged; Extended Data Fig. 5E). The ASO-treated mice exhibited increased energy expenditure (Fig. 4F), determined with two different data analysis methods: 1. the NIDDK Mouse Metabolic Phenotyping Centers energy expenditure analysis page (http://www.mmpc.org/shared/regression.aspx) and 2. the BWH-Harvard web application for indirect calorimetry analysis (https://calrapp.org)31. The treatment effect was significant (p<0.05 by ANCOVA) when either the overall mean total body mass or the mean lean body mass of each treatment group was used as the covariate. The increased energy expenditure could not be explained by changes in locomotor activity (Fig. 4G, 4H).
Livers of the IDOL ASO-treated mice had less lipid deposition, similar to the global IDOL KO mice (Extended Data Fig. 5F). The Idol-knockdown mice also had smaller white adipose tissue depots containing smaller adipocytes, but we did not find an obvious increase in multi-locular cells resembling beige adipocytes (Fig. 4E). Histological analysis of brown adipose tissue revealed smaller cells with more abundant lipid droplets (Fig. 4E), consistent with increased energy expenditure. Increased BAT activity is associated with alterations in glucose catabolism32 while acute pharmacological stimulation of BAT can reduce circulating lipids by driving fatty acid oxidation33. The ASO-treated mice appeared to have increased carbohydrate metabolism, but we did not observe a consistent change in fat oxidation (Extended Data Fig. 5G, 5H). ASO treatment increased phosphorylation of hormone sensitive lipase in the subcutaneous white adipose depot (Fig. 4I), a canonical downstream target of adrenergic receptor mediated PKA stimulation32; and we noted increased abundance of tyrosine hydroxylase in the BAT (Fig. 4J), consistent with increased catecholamine production by sympathetic nerves. These observations linked acute knockdown of Idol in the central nervous system to systemic changes in energy expenditure; however, they did not provide insight into the brain cell type(s) responsible for the phenotype.
IDOL deletion alters hypothalamic gene expression linked to energy balance
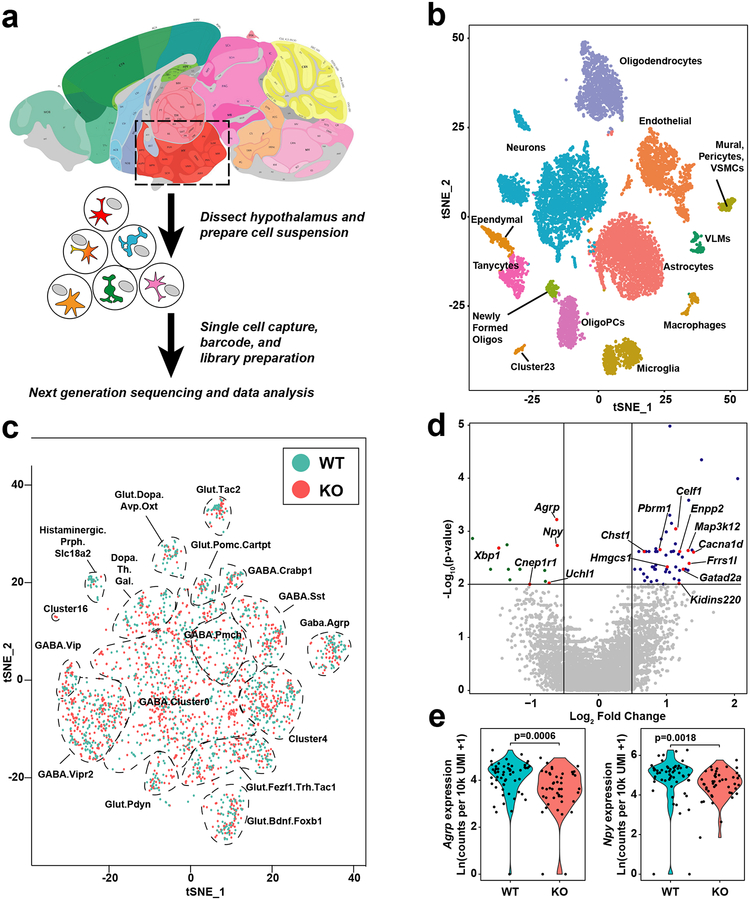
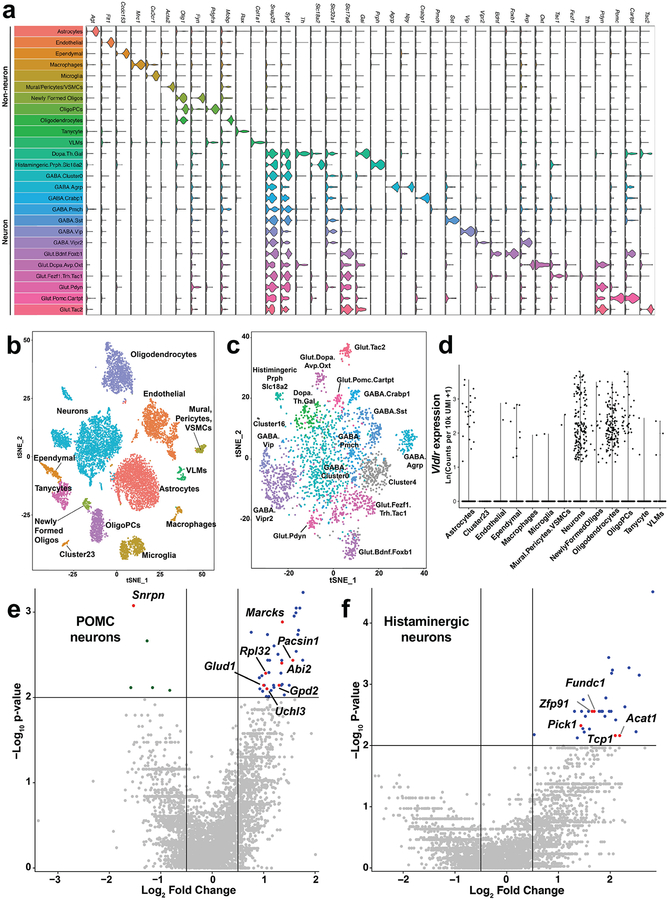
The increased energy expenditure seen when CNS expression of Idol was knocked down suggests that IDOL regulates one or more energy homeostasis circuits in the brain. We employed Drop-seq, a single cell RNA-sequencing technique, to obtain an overview of the changes in the transcriptional landscape of individual cell populations in the hypothalamus provoked by IDOL deletion (Fig. 5A). We placed wild-type and IDOL-KO mice on HFHC for two weeks and then analyzed hypothalamic gene expression. Clustering algorithms successfully identified twenty-six populations of cells in the hypothalamus, including eleven non-neuronal populations and fifteen distinct populations of neurons (Fig. 5B and Extended Data Fig. 6A–C). We analyzed the dataset for Vldlr gene expression and found two clusters with significant numbers of cells with high Vldlr expression: oligodendrocytes and neurons (Extended Data Fig 6D). When the genotype of the neurons was unmasked, the clustering analysis revealed several populations of neurons that had incompletely overlapping clusters of wild-type and IDOL knockout cells (Fig. 5C). Such separations indicate altered global transcriptional profiles in these populations. We interrogated two populations of cells that have been extensively studied for their contributions to energy homeostasis: AGRP and POMC neurons34,35. A third population of cells, histaminergic neurons expressing Slc18a2, was also profiled because of its apparent reduction in the IDOL knockout mice; however, our analysis of transcriptional changes did not reveal any reason for the difference in this population. It is likely that the observed changes in cell abundance are an artifact of it being a relatively under-represented cluster even in the wild-type mice.
Figure 5: The single cell transcriptional landscape of the hypothalamus is affected by deletion of IDOL.
(a) Simplified schematic of the single cell RNA-sequencing experimental design. 5-week old mice were placed on a high fat, high cholesterol diet for two weeks. Hypothalamic tissues from six mice (n=3/group) were dissected and dissociated into single-cell suspension. Single cells and barcoded beads were captured into droplets followed by cDNA synthesis, amplification, and library preparation. The library was sequenced with an Illumina HiSeq 4000 next-generation instrument using the Drop-seq custom read 1B primer. (b) Global gene expression relationships in the 11,453 single cells projected onto two dimensions using t-distributed Stochastic Neighbor Embedding (tSNE). The clusters were defined using shared nearest neighbor graph-based clustering. (c) tSNE plot demonstrating the effect of IDOL knockout on the clustering of single cells in fifteen different neuron clusters; n=11,453 cells. (d) Volcano plot of the differentially expressed genes in AGRP neurons; expression data calculated as counts per 10k unique molecular identities (UMI); the p-values were calculated using a two-tailed Wilcoxon rank sum tests from n=56 WT and n=45 Idol−/− neurons identified within the AGRP cluster. Labeled genes are linked to whole-body metabolic homeostasis – see Extended Data Table 1 for details). (e) Violin plots demonstrating that IDOL deletion reduced gene expression of Agrp and Npy in AGRP neurons. Individual data points indicate the magnitude of gene expression in a single cell. These are superimposed on a probability density plot for the distribution of the data; statistics were calculated using two-tailed Wilcoxon rank sum tests on non-Ln transformed data; n=56 WT and n=45 IDOL KO neurons identified within the AGRP cluster.
Analysis of transcript abundance in the AGRP neurons revealed that loss of IDOL altered the expression of a battery of genes linked to energy balance. Each of the labeled genes in each of the volcano plot of Fig. 5D are annotated in GWAS, OMIM or PubMed as being associated with energy homeostasis (Supplemental Data Table 2). The differentially expressed genes identified in AGRP neurons showed an enrichment of these genes (Fig. 5D). More modest differences were observed between genotypes in POMC and histaminergic neurons (Extended Data Fig. 6E, F). Interestingly, two key orexigenic genes expressed were downregulated in the AGRP neurons of IDOL-KO mice: the genes encoding agouti-related peptide and neuropeptide Y (Fig. 5E). AGRP neurons have also been associated with the negative regulation of energy expenditure36–38. Collectively, these data reveal that deletion of IDOL in the CNS altered the transcriptional regulation of several key neural circuits known to regulate energy balance.
IDOL deletion in neurons regulates energy balance
We followed up on the ASO and single cell RNA-sequencing results by creating pan-neuron conditional knockout mice by crossing the Idolf/f mice with SynI-Cre transgenics (Idolf/f; CreSynI). Similar to the global-KO mice, the CreSynIIdolf/f mice had reduced mass and adiposity when challenged with the western diet (Fig. 6A, 6B; n=18). Again, we conducted long-term food intake measurements to maximize our ability to detect small changes in food intake. As shown in Fig. 6C, the CreSynIIdolf/f mice consumed less food per week than their Idolf/f littermates. This finding shows that the changes in food intake in IDOL-deficient mice are the result of primary IDOL effects in neurons. We assessed energy expenditure by indirect calorimetry but did not detect any statistically significant differences between the two groups. Western blot analysis of subcutaneous white adipose tissue showed enhanced phosphorylation of HSL in CreSynIIdolf/f mice (Fig. 6D). The activation of HSL clearly indicates that deletion of IDOL from neurons affected peripheral energy homeostasis, however if changes in whole-body energy expenditure were present they were too subtle for the calorimeter to detect.
Figure 6: Conditional deletion of Idol from neurons drives the metabolic protection seen in global IDOL KO mice.
(a) Growth curve showing the mean mass +/− SEM and (b) Adiposity reported as body fat percentage +/− SEM measured by MRI for three cohorts of neuron-specific IDOL conditional knockout mice fed a western diet starting at 5–6 weeks old; n=18 per genotype *p<0.05, **p<0.01, ***p<0.001 ****p<0.0001 Idolf/f vs. Idolf/f CreSynI mice by repeat measures ANOVA. (c) Neuron-specific deletion of IDOL reduces food intake. The mean cumulative food consumption is shown +/− SEM; n=7 per genotype *p<0.05, ***p<0.001 Idolf/f vs. Idolf/f CreSynI mice by repeat measures ANOVA. (d) Increased phosphorylation of hormone sensitive lipase in the subcutaneous white adipose tissue of neuron-specific conditional IDOL KO mice. Each lane was loaded with a protein lysate from independent mice. This blot is representative of two independent repeats. (e,f) Global IDOL KO mice are protected from diet-induced mass gain and adiposity when group-housed at thermoneutrality; n=6 WT, n=8 IDOL KO mice (e) Growth curve showing the mean mass gained +/− SEM. (f) Adiposity reported as body fat percentage +/− SEM measured by MRI for adiposity. *p<0.05, ***p<0.001, ****p<0.0001 WT vs. KO by repeat-measures ANOVA. The precise n-number, p-value, and details of the statistical testing are provided in the source data file.
To distinguish the relative contributions of reduced food intake and increased energy expenditure to the IDOL-KO phenotype, we endeavored to minimize the energy expenditure effects by housing the mice at thermoneutrality (30 °C for 12 weeks). Relative to WT mice, global IDOL-KO mice gained less mass and adiposity on the obesogenic HFHC diet, suggesting that changes in food intake were sufficient to affect body weight in the absence of energy expenditure differences (Fig. 6E, 6F).
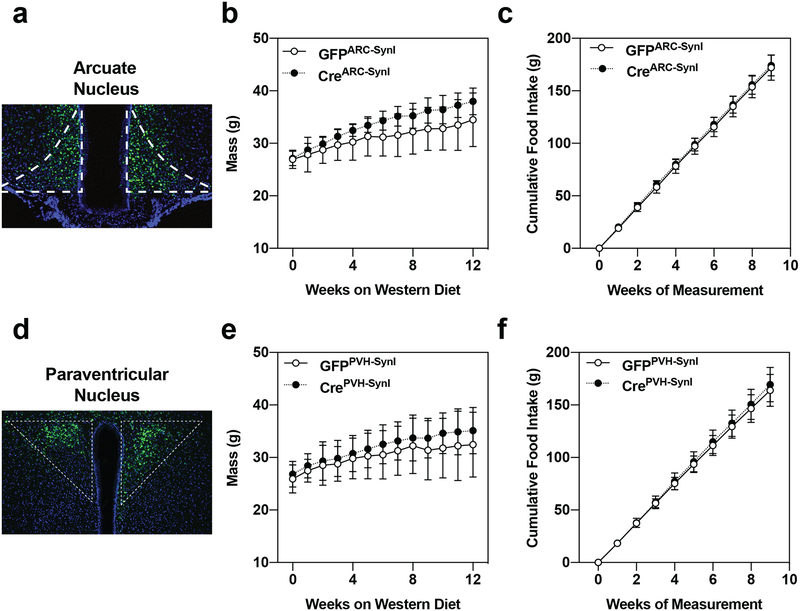
We endeavored to identify a specific hypothalamic locus driving the IDOL KO phenotype; however, viral-Cre-mediated deletion of IDOL in neurons in the arcuate nucleus (ARC), ventromedial hypothalamus (VMH), or the paraventricular nucleus of the hypothalamus (PVH) was not sufficient to recapitulate the CreSynIIdolf/f mouse phenotype (Extended Data Fig. 7). While the results of our viral experiments did not allow us to identify a precise neuron population responsible for the IDOL KO phenotype, our work with the SynI-Cre mice clearly demonstrated that deletion of IDOL from neurons protected mice against diet-induced obesity by reducing food intake while enhancing energy expenditure pathways. They further identify IDOL-mediated regulation of neuronal VLDL receptor as a previously unrecognized modulator of whole-body energy homeostasis.
Discussion
The ability of an organism to adapt to changing nutritional availability is fundamental to its survival. A plurality of mechanisms maintaining whole-body energy homeostasis have been selected for over the course of mammalian evolution. In this study, we identify the IDOL-VLDLR pathway in neurons as a previously unknown modulator of whole-body energy homeostasis. Two independent lines of global IDOL-KO mice (one derived at UCLA and one at AstraZeneca) exhibited a highly reproducible phenotype of resistance to diet-induced obesity. Despite the abundant expression of IDOL and its targets in adipose tissue, tissue-specific studies pointed to the CNS as the primary site of IDOL action in the regulation of energy balance. Loss of VLDLR but not LDLR expression on an IDOL-KO background abolished the protective effects of IDOL deficiency on the development of obesity.
Previous studies have established the molecular mechanisms by which IDOL regulates the abundance of its lipoprotein receptor targets. IDOL forms a homodimer which recognizes a specific conserved amino acid sequence in its membrane-bound targets for recognition and ubiquitin transfer. To date only three proteins have been identified that meet this stringent criterion: the LDL receptor and its two most closely-related family members, VLDL receptor and ApoER2. Although unexpected, our finding that VLDLR expression is required for the effects of IDOL on whole body metabolism is consistent with prior work demonstrating that VLDLR is important to metabolic homeostasis. One of the initial observations made by Herz and colleagues when they developed the VLDLR-KO mice was that the mice exhibited a reduction in mass and BMI relative to WT controls on a mixed background27. These findings were confirmed by other groups22,28 and we replicated this finding again here (Fig. 3). Prior mechanistic analyses have suggested that VLDLR in the periphery acts in the endothelium to enhance lipoprotein lipase-dependent hydrolysis of triglycerides20–23. It is important to point out, however, that the consequence of IDOL deficiency is increased, rather than decreased, VLDLR protein levels. Coupled with prior work, our results suggest that the appropriate balance of VLDLR protein levels in the CNS is important for the maintenance of whole-body energy homeostasis.
While this study was under review, Van Loon et al.39 described a similar phenotype of obesity resistance in the UCLA-generated IDOL knockout mouse line that we had previously provided them. These authors attributed the body weight differences in global IDOL-KO mice to IDOL activity in the liver, based on correlations between hepatic Idol expression and metabolic parameters in a BXD dataset. By contrast, our data do not support a causal role for hepatic IDOL in the phenotype. We tested the role of IDOL in the liver directly with hepatocyte-specific conditional IDOL-KO mice (Albumin-Cre; Fig. 2) and found that loss of liver IDOL did not confer protection against diet-induced metabolic dysfunction. Furthermore, the only lipoprotein receptor target of IDOL that is expressed by the liver is the LDL receptor, which we unequivocally demonstrated was not required for IDOL’s effects on the response to high-fat diet (Fig. 3A, 3B). Nevertheless, we conclude that the phenotype of the UCLA global IDOL KO mice is consistent between different vivarium conditions.
Although energy balance is maintained by multiple brain regions40,41, the role of the hypothalamus is particularly prominent34,42. Acting via the sympathetic nervous system32,43,44. the hypothalamus is a key effector of CNS influence on adipose tissue, including response to cold. It has also been implicated in the metabolic actions of Leptin36,45, FGF2146, PPARɣ agonists47,48, and thyroid hormone49,50. We observed increased VLDLR protein levels in hypothalamus of both the IDOL-KO mice and IDOL-ASO-treated mice, suggesting that this region likely contributes to IDOL’s metabolic effects. Furthermore, our single cell RNA-seq study demonstrated that deletion of IDOL altered the transcriptional landscape in several populations of neurons with well-established roles in the regulation of energy balance.
We investigated the potential for IDOL to regulate central control of metabolism using a combination of acute and chronic interventions, each of which provided insight into the physiological role of IDOL. The acute nature of the ASO studies allowed us to detect increased energy expenditure at a whole-organism level, linking IDOL in the CNS to peripheral energy homeostasis. We did not detect changes to food intake during a two week-long evaluation, perhaps due to differences between chronic deletion of IDOL compared with acute knockdown, or due to the duration of the food intake study.
Based on the Vldlr expression pattern seen in our single cell RNA-sequencing data, we focused our attention on IDOL in neurons. The complete loss of IDOL from neurons provided evidence that IDOL regulates food intake, consistent with the reduced expression of Agrp and Npy seen in our single cell RNA-sequencing data of whole-body IDOL knockout mice. The relative importance of altered food intake to the phenotype was confirmed by thermoneutrality studies showing that body weight differences persisted when the role of thermogenesis was minimized. Unfortunately, we have not yet been able to identify specific hypothalamic regions involved in the effects of IDOL on energy balance. We acknowledge that the viral-Cre approach used here suffers from the inherent weakness that the SynI promoter-driven Cre will be expressed in all infected neurons, which might might the role of specific neuronal populations51. Individual populations of neurons within a given hypothalamic nucleus may have opposing effects, as with POMC and AGRP neurons in the arcuate nucleus52.
Collectively, our studies identify the IDOL-VLDLR pathway as a previously unrecognized mode of regulating whole-body energy homeostasis. We expect that this observation will prompt further investigation into the specific cellular mechanism(s) by which cell-surface VLDLR levels regulate energy balance. The CNS contains a specific set of lipoproteins. Both ApoE and ApoJ (also called Clusterin)–two apolipoproteins found in cerebrospinal fluid–are known ligands for VLDLR26,53,54. Several non-apolipoprotein ligands have also been proposed for VLDLR, including Reelin2 and F-spondin55. While there is evidence that ApoJ can bind and modulate Leptin response in cell culture56, to date none of these ligands have conclusively been shown to regulate whole body energy homeostasis. In the periphery, VLDLR has a clear role facilitating lipolytic activity of LPL20,22, and thereby enhancing the production of free fatty acids. Free fatty acid sensing by the hypothalamus is a well-established modulator of neuronal activity57, affecting peripheral metabolism58–60, but the role of central VLDLR in this process has not been explored. Future studies will be required to define precisely how IDOL activity in the brain conveys a VLDLR-dependent signal to the periphery to regulate energy balance and which specific neuronal populations are involved.
Methods
Contact for Reagent and Resource Sharing:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Peter Tontonoz (PTontonoz@mednet.ucla.edu).
The antisense oligonucleotides were provided by IONIS Pharmaceuticals and are subject to an MTA between Ionis Pharmaceuticals and the University of California, Los Angeles.
Experimental Models:
The UCLA line of IDOL-KO mice was generated with a gene trap cassette inserted in intron 1, which could be released with FRT recombination to create conditional knockout mice in which exon 2 was “floxed”61. All UCLA mice were studied after ten backcrosses to C57Bl/6J.
The AstraZeneca line of IDOL KO mice was bred to a pure C57Bl/6N strain such that exon 2 was flanked by loxP sites. Whole body IDOL KO mice were derived from this strain by crossing the Idolf/f mice with Rosa26 Cre transgenic mice. This strain was also used to create hepatocyte-specific Idol−/− mice by crossing with Albumin Cre transgenic mice62.
The aP2-IDOL transgenic mice were generated at the UCLA core facility. We cloned a dominant active form of human IDOL (K293R, K309R, K310R, K320R) into a pBluescript vector containing the 5.4kb Fabp4 enhancer/promoter (A gift from Ronald Kahn) followed by bovine growth hormone polyA. The linearized construct was gel purified and microinjected into C57Bl/6J mice fertilized zygotes. Founders were identified using the following PCR primers (Fwd- GCCAGGAGGGCTCTGTACA; Rev-GAATAGAATGACACCTACTCAGACAAT). The mouse studies conducted at UCLA were reviewed and approved by the Chancellor’s Animal Research Committee. The mouse studied conducted by AstraZeneca were approved by Gothenburg Ethics Committee for Experimental Animals, license nr: 38–2011.
Mouse studies:
All studies performed with male mice in climate-controlled facilities maintained on a 12h light/dark cycle constant temperature of 22°C with group housed mice, unless otherwise noted. Pilot studies used both genders of mice; however, female mice are resistant to diet-induced obesity and required prolonged challenges with more severe diets to elicit adiposity changes. Consequently, male mice were used for all subsequent studies. LFD and HFHC studies with the AstraZeneca mouse lines: Dietary challenge was initiated in 8-week-old littermate mice. Body weights were recorded on a weekly basis. Body length (nose to base of the tail) was assessed at 4, and 20 weeks of age. Food intake in AZ mice was measured at 15-weeks of age by fasting mice for 12h during the dark period before single-housing them in cages (23 × 16cm) with pre-weighed aliquots of food for 48h. Body composition was assessed in 20-week-old animals by dual energy X-ray absorptiometry (DEXA, GE Lunar, Madison, USA) in isoflurane anaesthetized mice. Metabolic fitness was determined by oral glucose tolerance tests (2g/kg) performed at 22 weeks of age. The energy content of the feces was determined using a bomb calorimeter (C 5000, IKA Werke GmbH & Co. KG, Germany). Preliminary WD and HFD studies (Fig. 1 and Extended Data Fig. 1): we placed 5–6-week old littermate mice on test diets for 15 weeks and measured body mass weekly. We separated the mice to measure food intake at week 12 before returning them to group housing. Metabolic fitness was determined by administering an intraperitoneal glucose tolerance test (1g/kg) at week 9, and an intraperitoneal insulin tolerance test (1U/kg) at week 14. We measured body composition by MRI after 15 weeks on western diet (EchoMRI 3-in-1, Houston, TX, USA) Western diet-fed conditional knockout studies: we placed 5–6-week littermate mice on diet for 15 weeks and measured their body mass weekly. We measured adiposity by MRI at baseline, then every four weeks until study termination. Metabolic fitness was determined with an intraperitoneal glucose tolerance test (1g/kg) at 6-weeks and an intraperitoneal insulin tolerance test (1U/kg) at 10-weeks.
HFHC diet fed single and double knockout studies: we placed male 7–8-week-old mice on the HFHC diet and measured mass, adiposity, and metabolic fitness as described for the western diet conditional knockout studies above. Each cohort in the double knockout studies was arranged to maximize the number of littermate controls. Double knockout mice had single knockout littermate controls, while single knockouts had wild-type littermate controls).
Western diet ASO study: We performed a pilot dose-response study in five-week-old male C57Bl/6J mice to determine the minimum dose of ASO given by intracerebroventricular injection required for the maximal suppression of Idol expression in whole brain homogenates relative to vehicle control-treated mice (Extended Data Figure 5A). For the adiposity studies, five-week-old male C57Bl/6J mice ordered from The Jackson Laboratory were administered 40μg ASO or 10μL vehicle control by intracerebroventricular injection. The study was repeated using 40μg IDOL-targeting ASO or 40μg control ASO. The mice recovered for one week before we initiated the dietary challenge. We measured energy expenditure by indirect calorimetry after 2–3 weeks of western diet feeding (Columbus Instruments Oxymax CLAMS, Columbus, OH, USA). We measured adiposity by MRI at baseline, immediately prior to the calorimetry experiments, at 4 weeks on diet at 7 weeks on diet just prior to study termination. Mice were singly housed from the time they were moved to the metabolic chambers until the end of the study. We monitored food intake from week 4 to 7.
Food Intake studies: Littermate mice were singly-housed in standard housing and fed Western Diet ad libitum. The mass of diet administered and consumed was recorded weekly.
Thermoneutrality studies: Littermate mice were group housed in monitored climate-controlled facilities maintained on a 12h light/dark cycle constant temperature of 30°C.
Virogenetic studies: Ten-week-old littermate UCLA-derived Idolf/f male mice were anaesthetized with isofluorane and received analgesics (0.01mg/mL buprenorphine, 0.58mg/mL carprofen) pre- and post- surgery. The AAV-hSyn-GFP or AAV-hSyn-GFP-Cre (UNC Vector Core, titer ≥ 6×1012 vg/mL) was injected bilaterally into the following brain regions with the listed coordinates and volumes: Arcuate Nucleus (ARC): A-P: −1.57 mm from Bregma; lateral: ±0.2 mm from Bregma; D-V: 5.8 mm from the cortex, 125nL/side; Paraventricular nucleus of the Hypothalamus (PVH): A-P: −.33 mm from Bregma; lateral: ±0.25 mm from Bregma; D-V: 4.4 mm from the cortex, 250nL/side; Ventromedial Hypothalamus (VMH): A-P: −1.6 mm from Bregma; lateral: ±0.45 mm from Bregma; D-V: 5.4 mm from the cortex, 200nL/side. Mice were allowed to recover for 1 week before starting them on western diet for one week before we measured energy expenditure using the OxyMax CLAMS system. Upon completion of the energy expenditure study, the mice were singly-housed and food intake was measured as described above.
Study termination (all cohorts): mice were fasted for 6 hours beginning at 8:00am, then euthanized by isoflurane overdose, exsanguinated by cardiac puncture before cervical dislocation. Blood was collected in EDTA-coated tubes and centrifuged at 2000g for 15min to separate plasma. Tissues were collected from the mice and either snap frozen in liquid nitrogen or fixed in 4% paraformaldehyde.
Antisense oligonucleotide production:
Antisense oligonucleotides (ASOs) targeting IDOL (5’-TTCCTTTTTTCCACACGCCA-3’) were provided by Ionis Pharmaceuticals, Carlsbad CA. Complete ASO chemistry information is as follows. IDOL ASO (Tes Tes mCes mCeo Tes Tds Tds Tds Tds Tds mCds mCds Ads mCds Ads mCeo Ges mCes mCes Ae) and control ASO (mCes mCeo Teo Aeo Tes Ads Gds Gds Ads mCds Tds Ads Tds mCds mCds Aeo Geo Geo Aes Ae) where capital letters indicate base abbreviation, m=5-methylcytosine, e=2’-O-methoxyethylribose (MOE), k= (S)-2’,4’-constrained 2’-O-ethyl (cEt), d=deoxyribose, s=phosphorothioate, and o=phosphodiester63–65.
Gene expression:
Differences in gene expression were determined by real-time RT-qPCR (Diagenode, Denville, NJ, USA) from RNA isolated from frozen tissues with TRIzol (Life Technologies, Carlsbad, CA, USA) according the manufacturer’s instructions.
Western blot analysis:
Proteins were isolated from frozen tissue using a Dounce homogenizer on ice with RIPA buffer (Boston Bioproducts, Ashland, MA, USA) containing phosphatase and protease inhibitors (Roche, Mannheim, Germany). Equal masses of proteins were separated by electrophoresis through Bis-Tris gels before being transferred to PVDF membrane. The membranes were probed using the antibodies listed in the key resources table.
Histology:
Tissues were fixed in 4% paraformaldehyde and stored in ethanol before being mounted in paraffin. 10μm sections were cut and stained by hematoxylin and eosin by the UCLA Translational Pathology Core.
Single Cell RNA Sequencing
Single cell preparation:
Single cell suspensions were generated at a final concentration of 100 cells/μl in 0.01% BSA-PBS by digesting freshly dissected hypothalamus tissue with papain (Worthington, Lakewood, NJ, USA). Briefly, the hypothalamus was rapidly dissected then transferred into 4 mL ice-cold Hibernate A (HA, BrainBits LLC, Springfield, IL, USA) / B27 (Invitrogen, Carlsbad, CA, USA)/Glutamax Fisher Scientific, Hampton, NH, USA) (HABG) and incubated in a water bath at 30°C for 8 min66,67. The supernatant was discarded, and the remaining tissue was incubated with papain (12 mg in 6 ml HA-Ca) at 30 °C for 30 min. After incubation, the papain solution was removed from the tissue and washed with HABG three times. Using a siliconized 9-in Pasteur pipette with a fire-polished tip, the suspension was triturated approximately ten times in 45 sec. Next, the cell suspension was carefully applied to the top of the prepared OptiPrep density gradient (Sigma Aldrich, St. Louis, MO, USA). The gradient was then centrifuged at 800g for 15 min at 22 °C. We aspirated the top 6 ml containing cellular debris. To dilute the gradient material, we mixed the desired cell fractions with 5 ml HABG. The cell suspension containing the desired cell fractions was centrifuged for 3 min at 22°C at 200g, and the supernatant containing the debris was discarded. Finally, the cell pellet was loosened by flicking the tube and the cells were re-suspended in 1 ml 0.01% BSA (in PBS). This final cell suspension solution was passed through a 40-micron strainer (Fisher Scientific, Hampton, NH, USA) to discard debris, followed by cell counting.
Drop-seq single cell barcoding, library preparation and sequencing:
Drop-seq was performed using standard methodology68, with the following modifications: 1). The number of beads in a single PCR tube was increased to 4,000/tube. 2). The number of PCR cycles was increased to 4+11 cycles. 3). Multiple PCR tubes were pooled prior to size selection and purification with AMPure XP (Beckman Coulter, Brea, CA). The libraries were then checked on a TapeStation (Agilent, Santa Clara, CA, USA) for library quality, average size, and concentration estimation. The samples were then tagmented using the Nextera DNA Library Preparation kit (Illumina, San Diego, CA, USA) and multiplex indices were added. The Drop-seq library molar concentration was quantified by Qubit Fluorometric Quantitation (ThermoFisher, Canoga Park, CA, USA) and library fragment length was estimated using a TapeStation. Sequencing was performed on an Illumina HiSeq 4000 (Illumina, San Diego, CA, USA) instrument using the Drop-seq custom read 1B primer (IDT, Coralville, IA, USA). 100bp paired end reads were generated with an 8bp index read for multiplexing. Read 1 consists of the 12bp cell barcode, followed by the 8bp UMI. Read 2 contains the single cell transcripts.
Drop-seq data pre-processing and quality control:
The demultiplexed fastq files from the Drop-seq sequencing data were processed into a digital gene expression matrix using the dropSeqPipe (https://github.com/Hoohm/dropSeqPipe) SnakeMake wrapper for Drop-seq tools version 1.13. Briefly, read pairs with quality score less than 10 were discarded, transcript reads were trimmed to remove TSO contamination and polyA tails and then aligned to mm10 using STAR v2.5.0c with default settings. A digital gene expression (DGE) matrix for each sample was generated where each row is the read count of a gene and each column a unique single cell. The transcript counts of each cell were normalized by the total number of UMIs for that cell. These values were then multiplied by 10,000 and Ln transformed. Digital gene expression matrices from the six samples (3 WT and 3 KO samples) were combined to create a pooled digital gene expression matrix. Single cells were identified from background noise by using a threshold of at least 250 genes and 500 transcripts, resulting in a dataset of 6600 cells from IDOL knock out samples and 4800 cells from wild type samples.
Identification of cell clusters:
The Seurat R package (version 2.3.1; https://github.com/satijalab/seurat) was used to project all sequenced cells onto two dimensions using t-Distributed Stochastic Neighbor Embedding (tSNE) and shared nearest neighbor graph-based clustering69 was used to define clusters. To further refine the neuronal cell clusters, clusters expressing neuronal-specific markers were pooled and re-analyzed separately in a similar fashion, only considering this neuronal subset. Briefly, the most highly variable genes were identified using the mean and dispersion (variance/mean) which were used to scale and center the data. Principal Components Analysis (PCA) was performed on this normalized data and significant PCs were identified using the JackStraw permutation-based approach70. These significant PCs were used in tSNE to project the data onto two dimensions and the graph-based clustering to identify cell clusters.
Resolving cell identities of the cell clusters:
To resolve the identities of the cell clusters, known cell-type specific markers from previous studies were curated and checked for expression patterns within the cell clusters. A cluster showing high expression levels of a known marker gene specific for a particular cell type was considered to carry the identity of that cell type. Known markers for major hypothalamus cell types and neuronal subtypes were retrieved from Romanov71, Chen et al.72, and Campbell et al.73. These markers were sufficient to define all major cell types as well as neuronal subpopulations.
Identification of differentially expressed genes between WT and KO:
Within each identified cell type, single cells from WT and KO samples were compared for differential gene expression using a Wilcoxon Rank Sum Test. To be considered in the analysis, the gene had to be expressed in at least 25% of the single cells from one of the two groups within that cell type and there had to be at least a 0.25 log fold change in gene expression between the groups.
Data quantification and statistical analysis:
All data except the Energy Expenditure data were analyzed using GraphPad Prism v7 with a predetermined alpha value of 0.05. The values reported in the text and figures are the group means ± SEM where the n-number refers to the number of animals per group. The data were analyzed for the assumptions of parametric statistical methods (equal variance and normal distribution) and log-transformed if required. If the log-transformed data did not meet the assumptions of parametric statistical methods, the non-transformed data were analyzed using a non-parametric method, including the Mann Whitney test and the Wilcoxon rank sum test, as indicated in the appropriate figure legend. Parametric comparisons between two groups were done by t-test and of multiple groups with a single independent factor by one-way ANOVA. If a parameter was measured over time, then data were analyzed by repeat measures two-way ANOVA where time is one independent factor. The glucose- and insulin-tolerance tests were analyzed by calculating the area under the curve (AUC) for each mouse and then analyzing the AUC data set by t-test or one-way ANOVA, as appropriate.
The energy expenditure data generated by the indirect calorimetry experiment were analyzed using two methods: 1. the NIDDK Mouse Metabolic Phenotyping Centers using their energy expenditure analysis page (http://www.mmpc.org/shared/regression.aspx) and 2. the R programming language with CalR, a custom package for analysis of indirect calorimetry using analysis of covariance with a graphical user interface (https://calr.bwh.harvard.edu)31.
Data Availability:
The data that support the findings of this study are available from the corresponding author upon request, furthermore the Reporting Summary is available from the Nature Metabolism website. We deposited the single-cell RNA sequencing data in the NCBI Gene Expression Omnibus - accession number GSE119960.
Extended Data
Extended Data Figure 1. Metabolic phenotype of IDOL-deficient mice.
(a,b) 18-month-old IDOL KO mice are protected against age-induced adiposity. (a) Mean mass and standard error of the mean; n=9 WT, n=10 KO mice, ***p<0.001, ****p<0.0001 vs. WT by two-tailed t-test. (b) Mean body fat percentage +/− the standard error of the mean; n=9 WT, n=10 KO mice, ***p<0.001, ****p<0.0001 vs. WT 2-tailed t-test. (c) Growth curve for littermate male mice fed a 60% kcal high fat diet (HFD) starting when the mice were 6 weeks old. The mean values are shown +/− the standard error of the mean; n=5 WT and n=6 KO mice *p<0.05 by repeat measures ANOVA. (d) Growth curve for littermate male IDOL(AZ)f/f mice with or without whole-body Rosa26 Cre fed a low-fat diet from 8 weeks of age. The mean mass is shown +/− SEM; n=10 mice per group. (e) Growth curve for littermate male IDOL(AZ)f/f mice with or without whole-body Rosa26 Cre fed a high fat high cholesterol diet from 8-weeks of age; n=8 Idol(AZ)f/f mice n=9 CreR26+Idol(AZ)f/f mice; *p<0.05 by repeat measures ANOVA (f) Unchanged lean body mass in male IDOL knockout mice administered test diets despite adiposity changes measured by MRI. Mean lean mass is shown +/− SEM; n=9 WT and n=10 KO mice fed Chow, n=5 WT and n=7 mice fed western diet, n=5 WT and n=6 KO mice fed the 60% HFD, p-values calculated by two-way ANOVA with Sidak post hoc tests. (g) Body length measured from nose to anus at 20 weeks of age for male mice fed either a low-fat diet (LFD) or a high-fat high-cholesterol diet (HFHC) for 12 weeks. The mean values are shown +/− the standard error of the mean. Low-fat diet: n=10 mice per genotype. High Fat High Cholesterol diet: n=8 Idol(AZ)f/f mice and n=9 CreR26+Idol(AZ)f/f mice. P-values calculated by repeat measures ANOVA. (h) Mean body temperature +/− SEM measured rectally in response to fasting in mice fed a high-fat high-cholesterol diet; n=8 WT and n=9 IDOL KO mice. The precise n-number, p-value, and details of all statistical testing are provided in the source data file.
Extended Data Figure 2: Glucose and insulin tolerance tests conducted on Idolf/f and conditional knockouts with tissue-specific deletion of IDOL.
(a-b) Mean blood glucose levels +/− SEM for liver-specific conditional IDOL KO mice challenged with an oral glucose tolerance test (2g/kg) at 22 weeks of age fed either (a) the low-fat diet; n=12 mice per genotype or (b) the high-fat high-cholesterol diet for 16 weeks; n=11 mice per genotype. (c-j) Mean blood glucose levels +/− SEM for male mice fed a western diet challenged with an intraperitoneal glucose tolerance test (1 g/kg, shown on the left) after six weeks on diet and an intraperitoneal insulin tolerance test (1 U/kg, shown on the right) after ten weeks on diet. (c,d) Adipose-specific conditional IDOL KO; n=11 Idolf/f, n=9 CreAdipoQIdolf/f mice. (e,f) Endothelium-specific conditional IDOL KO; n=13 Idolf/f, n=10 CreCdh5Idolf/f mice. (g,h) Intestine-specific conditional IDOL KO; n=8 Idolf/f, n=4 CreVilIdolf/f mice. (i,j) Muscle-specific conditional IDOL KO; n=10 mice per genotype for the GTT and n=10 Idolf/f, n=9 CreMckIdolf/f mice for the ITT.
Extended Data Figure 3: Adipose-specific transgenic IDOL mice are not protected from diet-induced obesity.
(A) Ablation of VLDLR protein levels in the subcutaneous inguinal white adipose tissue of the aP2-IDOL transgenic mice. This blot is representative of many independent experiments conducted by two independent researchers. (B) Reduced VLDLR protein in the interscapular brown adipose tissue of the aP2-IDOL transgenic mice. This blot is representative of many independent experiments conducted by two independent researchers. (c) Growth curve for WT and aP2-IDOL transgenic mice fed a western diet from 5 weeks of age showing the mean mass +/− SEM; n=13 WT and n=9 aP2-IDOL transgenic mice (d) Mean body fat percentage measured by MRI +/− SEM for n=13 WT and n-9 aP2-IDOL transgenic mice after 12 weeks of western diet feeding. (e) Intraperitoneal glucose tolerance test (1g/kg) administered after six weeks of western diet feeding; n=13 WT and n-9 aP2-IDOL transgenic mice. (f) Intraperitoneal insulin tolerance test (1U/kg) administered after ten weeks of western diet feeding n=13 WT and n-9 aP2-IDOL transgenic mice.
Extended Data Figure 4: IDOL regulates systemic energy balance through the VLDL receptor.
100ug of RIPA isolate from the hypothalamus of individual wild-type (WT), Idol−/− (KO), Idol−/−Ldlr−/− (LDLR DKO), or Idol−/−Vldlr−/− (VLDLR DKO) mice was loaded per lane of a Tris-Acetate NuPAGE gel. The image is representative of many independent experiments. These samples were repeated twice to assess reproducibility. (a) Western blot analysis of VLDLR protein levels in the hypothalamus at study termination. (b) Western blot analysis of LDLR protein levels in the hypothalamus at study termination.
Extended Data Figure 5: Acute knockdown of Idol in the central nervous system increases energy expenditure.
(a) Optimization of the dose of ASO required to suppress Idol expression in whole-brain homogenates measured 8-weeks post-injection. The values represent the mean expression of Idol in a whole brain homogenate 8-weeks post-injection +/− SEM; n=4 mice per time point. (b) Growth curve for male mice placed on western diet one week after intracerebroventricular injection of Idol-targeting antisense oligonucleotide (IDOL ASO) or CNS-optimized control (CTRL ASO). The mean masses are shown +/− SEM; *p<0.05, **p<0.01, ***p<0.001 by repeat measures two-way ANOVA, n=10 mice treated with CTRL and n=5 mice treated with IDOL ASO. The (c) Macroscopic view of interscapular brown adipose tissue depots after seven weeks on western diet. These images are representative of the ten mice per group in the ASO study. (d) Macroscopic view of subcutaneous (inguinal) white adipose tissue depots after seven weeks on western diet. These samples are representative of the ten mice per group in the ASO study. (e) No statistically significant differences in body composition at the onset of CLAMS experiment. The mean total body mass, lean body mass, and fat body mass are shown +/− SEM; statistical significance determined by two-way ANOVA to account for multiple testing, n=10 mice per group. The precise n-number, p-value, and details of all statistical testing are provided in the source data file file. (f) Reduced lipid accumulation in the livers of ASO treated mice evident with hematoxylin and eosin staining of 5μm sections of liver (10x objective). These images are representative of three mice per treatment group that were analyzed for histology. (g) Calculation of carbohydrate metabolism in the n=10 mice per group. The mean energy expenditure derived from carbohydrate metabolism is shown +/− SEM for each time point. (h) Calculation of lipid oxidation in the n=10 mice per group. The mean energy expenditure derived from lipid metabolism is shown +/− SEM for each time point.
Extended Data Figure 6: Single cell RNA sequencing examination of the transcriptional landscape of the hypothalamus with Drop-seq.
Clustering analysis combined with expression profiling of a panel of marker genes allowed us to discriminate 26 unique clusters of cells in the hypothalamus. (a) Violin plots demonstrate the expression patterns of the 38 marker genes used to identify the cell clusters. Individual data points indicating the magnitude of gene expression in a single cell are superimposed on a probability density plot for the distribution of the data; the expression analysis is based on the data collected from n=11,453 single cells. (b) Global gene expression relationships in the 11,453 single cells isolated from the hypothalamic tissues of six mice projected onto two dimensions using t-distributed Stochastic Neighbor Embedding (tSNE). The clusters were defined using shared nearest neighbor graph-based clustering. (c) tSNE plot of the neuronal cells identified in the Drop-seq experiment (n=3369 single cells). (d) Violin plot demonstrating that Vldlr is only appreciably expressed in neuron and oligodendrocyte cell populations. Individual data points indicating the magnitude of gene expression in a single cell; the expression analysis is based on the unique molecular identities (UMI) data collected from n=11,453 single cells. (e-f) Volcano plots of the differentially expressed genes analyzed by two-sided Wilcoxon rank sum tests in (e) POMC+ (n=24 WT and n=26 Idol−/− cells) and (f) Histaminergic neurons (n=23 WT and n=11 Idol−/− neurons. Labeled genes are linked to whole body metabolic homeostasis – see Supplemental Data Table 2 for details.
Extended Data Figure 7: Neuron-specific virogenetic deletion of IDOL from individual hypothalamic nuclei is insufficient to protect against diet-induced obesity.
(a-c) Adeno-associated virus (AAV) expressing either GFP-Cre or GFP regulated by the Synapsin I (SynI) promoter were injected into the arcuate nucleus (ARC: panels a-c) or the paraventricular nucleus of the hypothalamus (PVH: panels d-f). (a-c) Deletion of IDOL from neurons in the ARC had no effect on body mass or food intake for mice fed a western diet for 12 weeks; n=8 mice injected with AAV-expressing GFP (GFPARC-SynI), n=8 mice injected with AAV expressing Cre-GFP (CreARC-SynI). (a) An image of the ARC showing GFP-positive cells to demonstrate successful infection of neurons; the image is representative of the sixteen mice injected in the ARC study. (b) Growth curve showing the mean mass of the mice from each treatment group +/− one standard deviation. (c) The mean cumulative mass of food consumed per mouse +/− one standard deviation. (d-f) Deletion of IDOL from neurons in the PVH had no effect on body mass or food intake for mice fed a western diet for 12 weeks; n=8 mice injected with AAV expressing GFP (GFPPVH-SynI), n=9 mice injected with AAV expressing Cre-GFP (CrePVH-SynI). (d) An image of the PVH showing GFP-positive cells to demonstrate successful infection of neurons; the image is representative of the seventeen mice injected in the PVH experiment. (e) Growth curve showing the mean mass of the mice from each treatment group +/− one standard deviation. (f) The mean cumulative mass of food consumed per mouse +/− one standard deviation.
Supplementary Material
Acknowledgments
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Peter Tontonoz (PTontonoz@mednet.ucla.edu).
Funding for this project was provided by grants to PT from the National Institutes of Health (HL066088, HL136618, DK063491). SDL was supported by a fellowship from the Canadian Institutes of Health Research and by a grant from the National Institutes of Health (P30 DK063491). CP was supported by a F32 fellowship from the National Institutes of Health (HL123236). ACC was supported by a fellowship from the National Heart Foundation of Australia (O 08M 3934) JG was supported by a K99/R00 pathway to independence award from the National Institutes of Health (AG054736). CH was supported by AHA grant 3BGIA17110079 and ADA grant 1–14-JF-33.
Footnotes
Competing Interests Statement
MB, HA, MP, AC, JJ, AA, FS, IM, PE, MA, PA, DL, HB, MB-Y are employees of AstraZeneca. TAC and RL are employees of Ionis Pharmaceuticals.
References
- 1.Goldstein JL & Brown MS A Century of Cholesterol and Coronaries: From Plaques to Genes to Statins. Cell 161, 161–172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trommsdorff M et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97, 689–701 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Goldstein JL, DeBose-Boyd RA & Brown MS Protein sensors for membrane sterols. Cell 124, 35–46 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Lee SD & Tontonoz P Liver X receptors at the intersection of lipid metabolism and atherogenesis. Atherosclerosis 242, 29–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong C & Tontonoz P Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug. Discov 13, 433–444 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Rayner KJ et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallam T et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zelcer N, Hong C, Boyadjian R & Tontonoz P LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong C et al. The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J. Biol. Chem 285, 19720–19726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scotti E et al. IDOL Stimulates Clathrin-Independent Endocytosis and Multivesicular Body-Mediated Lysosomal Degradation of the Low-Density Lipoprotein Receptor. Mol. Cell. Biol 33, 1503–1514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Xu M, Scotti E, Chen ZJ & Tontonoz P Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. J. Lipid Res. 54, 1410–1420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L et al. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 25, 1262–1274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calkin AC et al. FERM-dependent E3 ligase recognition is a conserved mechanism for targeted degradation of lipoprotein receptors. Proc. Natl. Acad. Sci. U.S.A. 108, 20107–20112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong C et al. The LXR-Idol Axis Differentially Regulates Plasma LDL Levels in Primates and Mice. Cell Metab. 20, 910–918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorrentino V et al. Identification of a loss-of-function inducible degrader of the low-density lipoprotein receptor variant in individuals with low circulating low-density lipoprotein. Eur. Heart J. 34, 1292–1297 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Weissglas-Volkov D et al. The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. J. Clin. Invest 121, 3062–3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calkin AC et al. Transgenic expression of dominant-active IDOL in liver causes diet-induced hypercholesterolemia and atherosclerosis in mice. Circ. Res 115, 442–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eguchi J et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13, 249–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagyu H et al. Very low density lipoprotein (VLDL) receptor-deficient mice have reduced lipoprotein lipase activity. Possible causes of hypertriglyceridemia and reduced body mass with VLDL receptor deficiency. J. Biol. Chem 277, 10037–10043 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Arcos I et al. Adipose-specific lipoprotein lipase deficiency more profoundly affects brown than white fat biology. J. Biol. Chem 288, 14046–14058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goudriaan JR et al. The VLDL receptor plays a major role in chylomicron metabolism by enhancing LPL-mediated triglyceride hydrolysis. J. Lipid Res. 45, 1475–1481 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Obunike JC et al. Transcytosis of lipoprotein lipase across cultured endothelial cells requires both heparan sulfate proteoglycans and the very low density lipoprotein receptor. J. Biol. Chem 276, 8934–8941 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Alva JA et al. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn 235, 759–767 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Brüning JC et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559–569 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Andersen OM et al. Essential role of the apolipoprotein E receptor-2 in sperm development. J. Biol. Chem 278, 23989–23995 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Frykman PK, Brown MS, Yamamoto T, Goldstein JL & Herz J Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 92, 8453–8457 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goudriaan JR et al. Protection from obesity in mice lacking the VLDL receptor. Arterioscler. Thromb. Vasc. Biol 21, 1488–1493 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Choi J et al. The E3 ubiquitin ligase Idol controls brain LDL receptor expression, ApoE clearance, and Aβ amyloidosis. Sci Transl Med 7, 314ra184–314ra184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J et al. The E3 ubiquitin ligase IDOL regulates synaptic ApoER2 levels and is important for plasticity and learning. Elife 6, 1741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mina AI et al. CalR: A Web-based Analysis Tool for Indirect Calorimetry Experiments. (2017). doi: 10.1101/213967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cannon B & Nedergaard J Brown adipose tissue: function and physiological significance. Physiol. Rev 84, 277–359 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Berbée JFP et al. Brown fat activation reduces hypercholesterolaemia and protects from atherosclerosis development. Nat Commun 6, 6356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautron L, Elmquist JK & Williams KW Neural control of energy balance: translating circuits to therapies. Cell 161, 133–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tschöp MH et al. A guide to analysis of mouse energy metabolism. Nat Meth 9, 57–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caron A, Lee S, Elmquist JK & Gautron L Leptin and brain-adipose crosstalks. Nat. Rev. Neurosci 19, 153–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong D et al. GABAergic RIP-Cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell 151, 645–657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan H-B et al. O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell 159, 306–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Loon NM et al. Inactivation of the E3 Ubiquitin Ligase IDOL Attenuates Diet-Induced Obesity and Metabolic Dysfunction in Mice. Arterioscler. Thromb. Vasc. Biol 38, 1785–1795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterson MJ & Horvath TL Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell Metab. 22, 962–970 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Nectow AR et al. Identification of a Brainstem Circuit Controlling Feeding. Cell 170, 429–442.e11 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Williams KW & Elmquist JK From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat Neurosci 15, 1350–1355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chechi K, Carpentier AC & Richard D Understanding the brown adipocyte as a contributor to energy homeostasis. Trends Endocrinol. Metab 24, 408–420 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Bamshad M, Song CK & Bartness TJ CNS origins of the sympathetic nervous system outflow to brown adipose tissue. Am. J. Physiol 276, R1569–78 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Friedman J 20 years of leptin: leptin at 20: an overview. J. Endocrinol 223, T1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Owen BM et al. FGF21 Acts Centrally to Induce Sympathetic Nerve Activity, Energy Expenditure, and Weight Loss. Cell Metab. 20, 670–677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu M et al. Brain PPAR-γ promotes obesity and is required for the insulin-sensitizing effect of thiazolidinediones. Nat. Med 17, 618–622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, O’Malley BW & Elmquist JK Brain nuclear receptors and body weight regulation. J. Clin. Invest 127, 1172–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.López M et al. Hypothalamic AMPK and fatty acid metabolism mediate thyroid regulation of energy balance. Nat. Med 16, 1001–1008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez-Crespo M et al. Essential role of UCP1 modulating the central effects of thyroid hormones on energy balance. Mol Metab 5, 271–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mancini G & Horvath TL Viral Vectors for Studying Brain Mechanisms that Control Energy Homeostasis. Cell Metab. 27, 1168–1175 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Schwartz MW, Woods SC, Porte D, Seeley RJ & Baskin DG Central nervous system control of food intake. Nature 404, 661–671 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Leeb C, Eresheim C & Nimpf J Clusterin is a ligand for apolipoprotein E receptor 2 (ApoER2) and very low density lipoprotein receptor (VLDLR) and signals via the Reelin-signaling pathway. J. Biol. Chem 289, 4161–4172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pohlkamp T, Wasser CR & Herz J Functional Roles of the Interaction of APP and Lipoprotein Receptors. Front Mol Neurosci 10, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zisman S et al. Proteolysis and membrane capture of F-spondin generates combinatorial guidance cues from a single molecule. J. Cell Biol. 178, 1237–1249 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bajari TM, Strasser V, Nimpf J & Schneider WJ A model for modulation of leptin activity by association with clusterin. FASEB J. 17, 1505–1507 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Oomura Y, Nakamura T, Sugimori M & Yamada Y Effect of free fatty acid on the rat lateral hypothalamic neurons. Physiol. Behav 14, 483–486 (1975). [DOI] [PubMed] [Google Scholar]
- 58.Lam TKT et al. Hypothalamic sensing of circulating fatty acids is required for glucose homeostasis. Nat. Med 11, 320–327 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Lam TKT, Schwartz GJ & Rossetti L Hypothalamic sensing of fatty acids. Nat Neurosci 8, 579–584 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Yue JTY et al. A fatty acid-dependent hypothalamic-DVC neurocircuitry that regulates hepatic secretion of triglyceride-rich lipoproteins. Nat Commun 6, 5970 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Scotti E et al. Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Mol. Cell. Biol 31, 1885–1893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Postic C et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem 274, 305–315 (1999). [DOI] [PubMed] [Google Scholar]
- 63.Swayze EE et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seth PP et al. Synthesis and biophysical evaluation of 2’,4’-constrained 2’O-methoxyethyl and 2’,4’-constrained 2’O-ethyl nucleic acid analogues. J. Org. Chem 75, 1569–1581 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Østergaard ME et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 41, 9634–9650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brewer GJ & Torricelli JR Isolation and culture of adult neurons and neurospheres. Nat. Protoc 2, 1490–1498 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Poon K, Barson JR, Fagan SE & Leibowitz SF Developmental changes in embryonic hypothalamic neurons during prenatal fat exposure. Am. J. Physiol. Endocrinol. Metab 303, E432–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waltman L & van Eck NJ A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 75 (2013). [Google Scholar]
- 70.Chung NC & Storey JD Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romanov RA et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci 20, 176–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen R, Wu X, Jiang L & Zhang Y Single-Cell RNA-Seq Reveals Hypothalamic Cell Diversity. Cell Rep 18, 3227–3241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell JN et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci 20, 484–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request, furthermore the Reporting Summary is available from the Nature Metabolism website. We deposited the single-cell RNA sequencing data in the NCBI Gene Expression Omnibus - accession number GSE119960.