Abstract
As it is likely that both common and rare genetic variation are important for complex disease risk, studies that examine the full range of the allelic frequency distribution should be utilized to dissect the genetic influences on mental illness. The rate limiting factor for inferring an association between a variant and a phenotype is inevitably the total number of copies of the minor allele captured in the studied sample. For rare variation, with minor allele frequencies of 0.5% or less, very large samples of unrelated individuals are necessary to unambiguously associate a locus with an illness. Unfortunately, such large samples are often cost prohibitive. However, by using alternative analytic strategies and studying related individuals, particularly those from large multiplex families, it is possible to reduce the required sample size while maintaining statistical power. We contend that using whole genome sequence (WGS) in extended pedigrees provides a cost-effective strategy for psychiatric gene mapping that complements common variant approaches and WGS in unrelated individuals. This was our impetus for forming the “Pedigree-Based Whole Genome Sequencing of Affective and Psychotic Disorders” consortium. In this review, we provide a rationale for the use of WGS with pedigrees in modern psychiatric genetics research. We begin with a focused review of the current literature, followed by a short history of family-based research in psychiatry. Next, we describe several advantages of pedigrees for WGS research, including power estimates, methods for studying the environment, and endophenotypes. We conclude with a brief description of our consortium and its goals.
Large-scale genome wide association studies (GWAS) comprise the dominant paradigm in psychiatric genetics research today [1]. Case/control GWAS, that compare the frequency of minor alleles from common polymorphisms between unrelated individuals [2], have provided numerous insights into the genetic architecture [1] and the interrelatedness [3, 4] of psychiatric disorders. However, like any experimental approach, the case/control GWAS design has relative strengths and weaknesses. Unfortunately, it is unlikely that any single design will be able to dissect all of the genetic influences on multifactorial traits [5, 6] such as mental illnesses [7, 8]. Rather, diverse complementary approaches may be necessary to garner the full spectrum of biological insights that genetics could provide neuropsychiatry [5, 9–12]. Chief among these approaches is the use of whole genome sequencing (WGS) which catalogues almost all genomic DNA sequence variation within an organism [13]. Early sequencing efforts confirmed that the substantial majority of human genetic variation is rare (occurring in less than 1% of the population) or private (only occurring in a single individual and their close relatives) [13, 14]. There is a growing appreciation of the impact of rare variation on human disease [11, 15, 16], particularly given the excess of rare functional variants resulting from recent accelerated population growth and relatively weak purifying selection [17]. Rare variants, especially loss of function variants or those deleterious to protein expression, are far more amenable to biological experimentation, and subsequent molecular insights, than common loci [18–22], which are often localized outside of transcribed regions [23, 24]. As it is likely that both common and rare variation are relevant for complex diseases [11], both GWAS and WGS methods should be utilized in a complementary manner to dissect the genetic influences on mental illness.
The rate limiting factor for inferring an association between a particular rare variant and a phenotype is inevitably the total number of copies of that variant captured in the sample [25–27]. Typically, to have enough copies of a rare variant for statistical analysis, one must sequence very large samples of unrelated individuals (e.g., ~700,000 in the recent human height exome study [20]). Consistent with this notion, the Whole Genome Sequencing of Psychiatric Disorders (WGSPD) consortium estimated that sequences from at least 20,000 unrelated cases and controls are needed to adequately power a gene burden-type analysis [8], though far larger samples are necessary to identify specific risk variants for mental illness. However, by using alternative analytic strategies and studying related individuals, particularly those from large multiplex families, it is possible to reduce the required sample size while maintaining statistical power [28–31]. Given this and other benefits discussed below, we contend that WGS in extended pedigrees provides a cost-effective strategy for psychiatric gene mapping that complements GWAS and WGS in unrelated individuals. In fact, family-based methods may be the only feasible study design for specifically identifying the rarest functional variants that are private to family lineages. This was our impetus for forming the “Pedigree-Based Whole Genome Sequencing of Affective and Psychotic Disorders” consortium, an international group of scientists using family-based designs to identify rare variants that increase risk for psychiatric disorders.
In this review, we provide a rationale for the use of WGS with pedigrees in modern psychiatric genetics research. We begin with a focused review of the current literature, followed by a short history of family-based research in psychiatry. Next, we describe several advantages of pedigrees for WGS research, including power estimates, methods for studying the environment, and utilizing endophenotypes. We conclude with a brief description of our consortium and its goals.
The current state of psychiatric genetics
Large-scale GWAS meta-analyses have been successfully completed for schizophrenia (sample size: 36,989 cases/113,075 controls [32]), bipolar disorder (13,902/19,279 [33], 9,784/30,471 [34]), major depression (130,664/330,470 [35], 10,851/32,211 [36], 121,380/338,101 [37]), post-traumatic stress disorder (5131/15,092 [38]), attention deficit hyperactivity disorder (20,183/35,191 [39]), and autism (16,539/157,234 [40]). Together, these GWAS have localized over 200 genome-wide significant loci influencing mental illness risk [1]. Given the sample sizes listed above, it is quite possible that common loci with moderate to large effect sizes for the majority of mental illnesses have already been localized [10], at least among individuals of European ancestry. If so, this represents an important milestone for the field and provides an opportunity to explore alternate approaches for delineating the genetics of mental illness.
One lesson from GWAS is that mental illnesses, like other complex diseases, appear to be highly polygenic, involving large numbers of loci, most of which have a small or very small effect on risk [10, 41, 42]. This pattern of results is entirely consistent with Fisher’s multifactorial model [43], which predicts that as the number of risk loci grows, the contribution of each new locus correspondingly shrinks. Accordingly, results from meta-analyses have been used for individual risk prediction based on polygenic scores [44, 45] that include thousands to hundreds of thousands of variants to provide a risk index [46]. Additionally, loci from GWAS studies appear to be useful for selecting among potential therapeutic agents [47], a property which could have a significant impact in psychiatry [48] where novel drug development is at a near standstill [49].
A case for rare genetic variants in mental illness
Arguably, our understanding of the genetic underpinnings of autism spectrum disorders has advanced more than that of other mental illnesses because investigators have focused more on rare nonsynonymous variants [50] than common genetic variation [40]. These studies, which often search for exonic de novo mutations [51, 52], have identified at least 50 potential risk genes for the disorder that together with copy number variants (CNV) explain more than 30% of the genetic variance of the illness [53, 54]. While the relative contribution of largest-effect common variants and of higher-penetrance rare variants probably varies across mental illnesses [1, 55, 56], the genetic architecture of autism is likely not unique. For example, Singh and colleagues identified a set of rare, putative loss-of-function variants in an exon SETD1A that strongly increases risk for schizophrenia and intellectual disability [57]. Similarly, exome sequencing studies in schizophrenia have implicated genes expressed in neurons [58] and synapses [59] and shown that affected individuals have more rare protein-altering loss-of-function variants than unrelated controls [58].
Perhaps the strongest evidence that rare variation is important across mental illnesses [60] comes from findings that certain rare CNVs or insertion-deletions clearly influence risk for autism spectrum disorders [61], intellectual disability [62] and schizophrenia [63, 64], and may also contribute to bipolar disorder [65] and ADHD [66] risk. Indeed, the 22q11 CNV [67] is among the strongest genetic predictors of schizophrenia risk [63, 68].
Family studies in psychiatric genetics
Historically, the mapping of traits to genetic loci in humans depended almost exclusively on family studies. Early linkage studies posited simple, single major gene models of inheritance and utilized transmission of chromosome segments across generations in large pedigrees to map putative disease loci relative to a scaffold of a few hundred markers of known position. Later linkage approaches did not assume a Mendelian model and utilized identity-by-descent (IBD) allele sharing among relatives. Although these linkage methods successfully identified loci for some illnesses (e.g., Huntington’s disease, Alzheimer’s disease, macular degeneration, diabetes, and some forms of breast cancer), early attempts to localize the genetic influences on polygenic diseases were limited and often could not be replicated. Indeed, two early high profile reports of linkages for bipolar disorder, one on the X chromosome [69] and the other on 11p [70], could not be replicated [71, 72]. When reviewing this literature in 2008, Burmeister and colleagues [73] reported that no single locus was unequivocally replicated across multiple independent samples for any mental illness. This lack of results was likely due to underpowered studies that used suboptimal concordant sibling pair designs [73, 74] and were likely ineffectual where very rare or private mutations were causal. Nonetheless, discouraging progress with linkage analyses, combined with the simplicity of sampling unrelated cases and controls, undoubtedly added to the popularity of association methods and the field’s shift towards GWAS.
In an influential article, Risch and Merikangas [75] argued that linkage analysis has limited power to detect genes of modest effect (particularly in concordant sibling pair designs), but that family-based assocation methods have far greater power to detect the same loci, provided the locus is either directly genotyped or in strong linkage disequilibrium (LD) with a genotyped marker. The genome-wide application of this association strategy was made possible by the human genome project’s identification and mapping of hundreds of thousands of common genetic variants and the characterization of patterns of LD between them. It draws on shared population history rather than transmission among family members, to map loci of interest. This information, in turn, allowed investigators to estimate minor allele frequencies (MAF) and LD-structure for singletons, enabling GWAS in unrelated individuals [2]. Yet, the reliance on population level knowledge has drawbacks. For example, GWAS are population-specific. Most published GWAS have been in European-derived populations, where the LD structure is well defined and represented on GWAS arrays. Although work is ongoing, sample sizes in non-European populations are yet to reach levels that would support powerful GWAS [76]. Carefully ascertained, very large families do not require population level information (e.g., MAF or LD-structure), have the potential to provide sufficient copies of very rare alleles to identify their effects, and offer the opportunity to leverage both analytical approaches, combining genome-wide association and examination of familial transmission within the same analysis. Thus, while family-based designs were largely set aside in the GWAS era, the recurring focus on rare variants and functional genomics have renewed interests in pedigrees.
Rare variants and pedigrees
Pedigree-based studies represent an implicit enrichment strategy for identifying rare variants as transmission of a rare allele from parents to offspring follows Mendel’s laws, maximizing the chance that multiple copies of that allele exist in the pedigree. For example, 148 individuals from a single large pedigree sampled in our ongoing “Genetics of Brain Structure and Function” study [77, 78] are represented in Fig. 1. Based on the principles of Mendelian inheritance, the pedigree could maximally provide 105 copies of a rare or even private mutation originating in a single founder (founder and unilineal descendants). While the propagation of a particular variant within a pedigree is likely less extreme than this, the example provides an important heuristic for understanding how families enrich even the rarest of genetic variation where the segregation of rare variants in a pedigree provides multiple copies, facilitating their detection and effect estimation [29, 31, 79, 80]. For a known pedigree, each founding lineage can be directly assessed for the expected number of copies of a private variant originating at the top of the lineage using Mendelian transmission probabilities. The expected number of copies of a private variant originating in the focal founder of Fig. 1 is 13 (as is that of his founder spouse). While this founder pair exhibits the maximum number of potential copies, the founder female spouse of the third male sibling in generation II actually exhibits the highest expectation of potential copies with 14.125.
Fig. 1.
Demonstration of rare variant inheritance in a large extended pedigree. One hundred and forty-eight individuals from a single large pedigree sampled in our ongoing “Genetics of Brain Structure and Function” study are represented. Based on the principals of Mendelian inheritance, the pedigree could maximally provide 105 copies of a rare or even private mutation originating in a single founder (filled). The figure was created with CraneFoot [150]
For a fixed biological effect size, the power of pedigrees for capturing larger numbers of rare minor allele copies than that expected in an equivalent set of unrelated individuals is a direct function of pedigree structure. Basically, the variance of the number of minor allele copies (MACs) can be substantially larger (and therefore lead to potentially many more copies) in pedigrees than in a sample of unrelated individuals. Given that the expected correlation structure for the allelic dosages amongst family members is well represented by the coefficient of relationship matrix, R, standard covariance mathematics reveal that the expected excess in the variance of expected MACs in a pedigree can be approximated by a multiplicative variance inflation factor, where rij is the coefficient of relationship between the i-th and j-th individuals in the pedigree and n is the number of individuals in the pedigree. The larger the VIF for a pedigree, the greater the expected power is for capturing larger numbers of a private variant, which itself determines the expected power to detect an association of a rare variant conditional on biological effect size. A sibship yields a VIF equal to1 + (n – 1)(1/2), thus a large sibship of 10 siblings generates a VIF of 5.5 times that expected for 10 unrelated individuals. The pedigree shown in Fig. 1 generates a VIF of 8.6. Typically, large pedigrees with large lineages will yield the highest VIFs likely to be observed in humans. Thus, pedigrees are optimally suited for the examination of rare functional variants because in the limiting case of private variants, traditional epidemiological studies of unrelated individuals are highly unlikely to capture more than a single copy of such a variant (e.g., [58, 59, 81]). Pedigree-based studies could capture many more depending upon the size and structure of the pedigrees. However, a potential negative for such studies is the more limited number of genomes being observed over that of unrelated samples. For example, the pedigree in Fig. 1 represents independent genomes from 44 founders versus that 148 that would be observed if all these individuals were unrelated. Thus, while more copies of rare variants can be captured in pedigrees, we also expect fewer such variants overall than in samples of unrelated individuals.
For rare variants in the absence of inbreeding, the number of heterozygotes captured is a primary determinant of statistical power to detect association. In this case, the number of heterozygotes is equivalent to the number of minor allele copies captured in the sample. Following theory developed in Blangero and colleagues [82], the expected association test statistic for private variants in pedigrees can be approximated (for small relative effects) as:
where N is the sample size, is the heritability due to the variant in the sample, is the total heritability of the trait, H is the proportion of heterozygotes in the sample, and α is the displacement of the heterozygote mean trait value from the common homozygote in standard deviation units. The parameter, α, directly measures the biological effect size of the variant. The symbol c() represents a function of parameters within the parentheses and is used here as a correction that accounts for the non-independence amongst related individuals and is defined in detail elsewhere [82]. The value of c is generally small for most reasonable genetic effect sizes [82]. Thus, power is dominated by the biological effect size and NH that gives the observed number of heterozygotes (or the number of captured minor allele copies) in the sample.
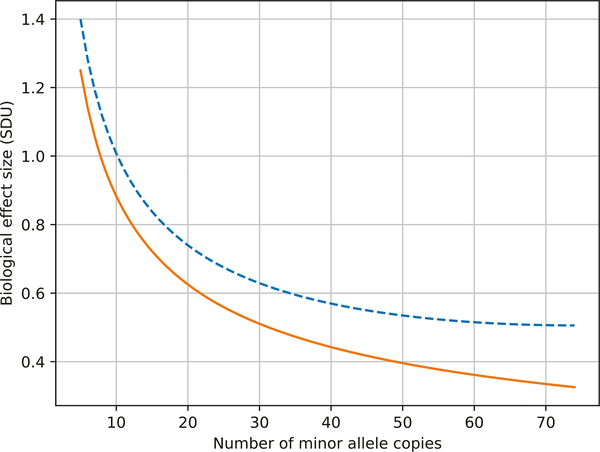
Figure 2 shows the biological effect size that can be detected at 80% power for a fixed number of observed heterozygotes in the pedigree in Fig. 1. We show the range of 5 to 70 heterozygotes/MACs. The lower bound of five minor allele copies required before testing is based on simulations that show that the resulting test distribution under the null hypothesis conforms with expectation (i.e., there is no excess type I error). As the number of captured MACs increases, power to detect moderate biological effect sizes improves. As a rough reference, a biological effect size of 4.5 SDU approaches nearly monogenic penetrance. Figure 2 also shows the effect of augmenting this pedigree with an additional 20,000 unrelated controls (the total sample size of the WGSPD consortium [8]). For the case of the rarest of variants (i.e., private variants), there is a relatively minor improvement in power with increased numbers of controls who are highly unlikely to harbor the rare variant. Thus, the recruitment of related individuals acts like an ascertainment bias to increase power by increasing the probability of capturing additional copies of rare variants that appear in the founders of the sampled lineages.
Fig. 2.
Biological effect size for rare variants as a function of minor allele copies (MAC). The blue dashed line shows the biological effect size that can be detected at 80% power for a fixed number of observed heterozygotes in the pedigree in Fig. 1. As the number of captured MACs increases, power to detect moderate biological effect sizes improves. The effect of augmenting this pedigree with an additional 20,000 unrelated controls is presented in the orange line. For the case of the rarest of variants, there is a relatively minor improvement in power with increased numbers of controls who are highly unlikely to harbor the rare variant
The prior discussion focuses on ascertainment of families simply through lineage size in order to maximize the capture of rare variants that originate in pedigree founders. However, additional power benefits accrue through additional ascertainment through disease or phenotype. For example, the co-segregation of rare variation and disease status in multiplex families can amplify association signals [31, 83, 84]. For the study of rare sequence variation, an implication of Mendelian transmission is that the required sample sizes can be orders of magnitude smaller for families than those required for designs based on unrelated subjects [85], particularly if sequence information is combined with linkage methods [28] in pedigrees of 20–25 individuals or larger [29], when comparing affected sibling pairs [30] or when searching for shared genomic segments [31]. For the rarest variants, large pedigrees have better power for detection of linkage/association when compared to equivalent-sized samples of smaller families [86] or unrelated subjects [80, 87, 88]. Family-based cohorts have substantially greater power than unrelated cases to detect rare genetic effects given an equivalent number of sampled individuals [89, 90].
An additional advantage to studying families is that, in contrast to unrelated individuals, the analysis of phenotypes among family members is constrained for genetic background (e.g., minimizes the impact of population admixture and stratification [91, 92]). Given that analytic techniques developed to correct for population stratification in common variant studies maybe less effective when the focus is on rare variants [93, 94], observations that pedigree-based experiments appear to be robust to population stratification are of particular importance [92]. In addition, reduced environmental variation among family members can reduce noise, improving statistical power to observe genotype-phenotype associations [95]. Shared familial environments also can alter the potential to observe signals resulting from gene-environment interactions. Pedigree-based designs allow for the investigation of de novo mutations, parent of-origin effects [96], transmission bias [97], phasing [98, 99], and compound heterozygosity [100, 101]. Finally, when pedigrees have multiple affected members it is often presumed that the same inherited mutation on a similar genetic background causes the illness in each case. This assumption appears to be better supported when a kindred includes at least three affected individuals [102, 103]. Although unambiguously demonstrating phenocopies is difficult in multifactorial phenotypes [104], it is possible that family-based studies provide a method for detecting phenocopies if a rare mutation appears to segregate with affection status in the pedigree[102]. To the extent that the segregating mutation also influences an illness endophenotype (see below), contrasting the endophenotype from the putative phenocopy and family members who carry the variant could provide further evidence of the non-genetic origin of the illness in that individual.
Rare variants, pedigrees and psychotic and affective disorders
Recently, Steinberg and colleagues [105] examined a single Icelandic pedigree with ten psychotic individuals (six schizophrenia, two schizoaffective disorder and two psychotic bipolar disorder) using WGS and long-range phasing. All affected individuals carried a rare nonsense mutation in RBM12 (RNA-binding-motif protein 12) resulting in a truncated protein lacking a predicted RNA-recognition motif while few unaffected had the mutation (p = 2.2 × 10−4). A Finnish family with a second loss of function RBM12 mutation replicated the finding (p = 0.020). Although the truncating mutation was not fully penetrant for psychosis, non-psychotic carriers were similar to their psychotic relatives in terms of neurocognitive endophenotypes, educational attainment and disability benefits received. Together, these data strongly associate RBM12 with psychosis risk and demonstrate the potential for gene identification using WGS and extended pedigrees.
Homann and colleagues [106] performed WGS on nine families with at least three members with schizophrenia. In one of these families, seven siblings with schizophrenia spectrum disorders carried a private missense variant within the SHANK2 gene. In a separate family, four affected siblings carried a novel private missense variant in the SMARCA1 gene. In a conceptually similar study, Timms and colleagues [107] used exome sequencing to examine rare nonsynonymous variants in five multiplex schizophrenia families. One pedigree carried a missense and frameshift substitution of GRM5, while another family had a missense substitution in PPEF2; both are genes that directly interact with the NMDA system [107]. Three pedigrees had missense substitutions within LRP1B, which is putatively related to the NMDA receptor. While these findings require replication and biological validation, nominated genes are reasonable empirical candidates for psychosis risk, warranting further research.
As can be seen in Table 1, an increasing number of family-based sequencing studies involving affective and psychotic disorders are being published, often with very small sample sizes. While findings from most of these studies have yet to be replicated, several of the more recent studies, particularly those conducted in population isolates [108] with larger sample sizes, provide strong candidate genes for these disorders.
Table 1.
Extended pedigree-based sequencing studies of psychotic or affective disorders
| Reference | Year published | Ascertainment | sequence | # of families | Individuals sequenced | Populationa | Findings replicated |
|---|---|---|---|---|---|---|---|
| Bouwkamp [109] | 2017 | Bipolar disorder | WGS | 1 | 1 | Dutch | No |
| Hornig [151] | 2017 | Schizophrenia | Exome | 1 | 8 | German | No |
| John [152] | 2017 | Schizophrenia | Exome | 1 | 4 | Indian | Partial |
| Rao [153] | 2017 | Bipolar disorder | Exome | 4 | 9 | USA | No |
| Steinberg [105] | 2017 | Schizophrenia | WGS | 1 | 7 | Icelandic | Yes |
| Zhang [154] | 2017 | Bipolar disorder | Exome | 1 | 6 | USA | No |
| Egawa [155] | 2016 | Schizophrenia | Exome | 1 | 3 | Japanese | No |
| Goes [156] | 2016 | Bipolar disorder | Exome | 8 | 36 | USA | No |
| Homann [106] | 2016 | Schizophrenia | WGS | 9 | 90 | USA | No |
| Kos [157] | 2016 | Schizophrenia | Exome | 8 | 134 | USA | No |
| Subaran [158] | 2016 | Major depression | Exome | 5 | 12 | USA | No |
| Watanabe [159] | 2016 | Schizophrenia | Exome | 3 | 12 | Japanese | Partial |
| Zhou [160] | 2016 | Schizophrenia | Exome | 1 | 4 | Chinese | No |
| Ament [161] | 2015 | Bipolar disorder | WGS | 41 | 200 | Amish | No |
| Kember [162] | 2015 | Bipolar disorder | WGS | 1 | 80 | Amish | Yes |
| Thygesen [163] | 2015 | Schizophrenia | WGS | 6 | 3 | Dane | No |
| Georgi [142] | 2014 | Bipolar disorder | WGS | 1 | 50 | Amish | No |
| Strauss [164] | 2014 | Bipolar disorder | Exome | 4 | 26 | Amish | No |
| Timms [107] | 2013 | Schizophrenia | Exome | 5 | 12 | USA | No |
Studies conducted in the USA involved subjects of European Ancestry
The foregoing discussion focused on identifying individual rare variants or CNVs strongly associated with risk for affective or psychotic disorders. The focus on a single variant or CNV is analytically consistent with method developed for monogenic disorders [109]. However, there is growing evidence that even in the case of a highly penetrant mutation, an individual’s genetic background contributes to illness risk [110]. For example, among individuals with a 22q11 deletion, rare CNVs outside of the 22q11 deletion region significantly contribute to schizophrenia risk [68]. Similarly, among members of a large multiplex pedigree with a balanced chromosomal translocation (1q42–11q14.3) associated with affective and psychotic disorders [111], common and rare variation in other areas of the genome appear to increase illness risk [112]. These finding are consistent with observations that genetic variation outside of the focal “causal” gene are often necessary for disease expression in monogenic disorders [103]. Together, these results serve as a reminder of the difficulty of making casual inferences in human genetics.
Cost-effectiveness of wgs in families
Family-based designs are cost effective. Given that genetic relationships between family members are known, WGS can be imputed [113] for individuals who have sparse genotype data, decreasing the effective cost per sample [114, 115]. This pedigree-based imputation or “pseudo-sequencing” is particularly effective for rarer, segregating variants [116]. Typically, this approach consists of two steps: (1) form optimal sub-pedigrees that maximize phase and IBD information and (2) pseudo-sequence each sub-pedigree. The resulting output will contain the expected number of copies for the tested allele (dosage), shown to yield the most power when used in association testing versus choosing most probable genotypes [90]. Livne and colleagues [114] applied similar methods (a combination of pedigree-based and LD-based imputation), reporting > 99% accuracy over the full range of allele frequencies. With data from the “Genetics of Brain Structure and Function” study, we found that pseudo-sequenced individuals show 97% accuracy for rare heterozygous variants and 99% for rare homozygotes compared to ExomeChip genotypes. Despite the accuracy of these “pseudo-sequencing” methods, once a rare variant is associated with a specific trait, we advocate directly genotyping that variant across the full sample to confirm the imputation.
Pedigree-based sequence data allows a level of quality control not available for population studies. Genotyping errors occur when the “true” genotype is not identical to the genotype determined after subsequent genotyping. These errors, can occur at every step of the genotyping process and cannot be fully eradicated as genotyping methods are not completely accurate [117]. Genotyping errors can lead to a number of possible biases, including an artificial excess of homozygotes [118], a false departure from Hardy–Weinberg equilibrium [119], an overestimation of inbreeding [118] or unreliable inferences about population substructures [120]. Incorporating evidence of Mendelian transmission of alleles between parents and offspring in pedigree data can dramatically reduce genotyping errors [121], even allowing for the detection of de novo mutation and the fact that 25% of typing errors may be Mendelian-compatible [122].
Using families to model environmental risk factors
Mental illness results from multiple genetic and environmental factors and, likely, from their interactions. In contrast to genetic data, the environment is ever changing and its impact can vary with developmental stage, making the study of non-genetic influences on mental illness risk particularly challenging. Yet, most studies of environmental risk factors for mental illness (e.g., [123–125.]) do not explicitly account for genetic background. For example, the incidence of schizophrenia is higher among individuals living in urban areas than to those living in rural areas [126, 127], which presumably reflects an environmental risk factor for psychotic disorders. However, even this classic environmental risk factor has an appreciable genetic component, where “urbanicity” is to some extent conditioned upon family history for schizophrenia [128] and individuals living in urban areas have higher polygenetic risk for the disorder than those living in rural areas [129]. Thus, Epidemiological studies designed to identify risk factors for mental illnesses should also include genetic information [124]. Pedigree-based designs, in addition to being of value for detecting genetic loci, enhance the study of environmental factors influencing mental illness as they provide a relatively straightforward method for optimally statistically controlling for genetic influences. Recently, we developed a best linear unbiased predictor estimation procedure to obtain individual-level estimates of genome-wide genetic effects [130]. This procedure uses all phenotypic information available for an individual and his or her relatives to infer the underlying genetic component of a phenotype. The estimated genetic value is then subtracted from the original phenotypic value to obtain an estimated environmental value devoid of the average additive genetic signal. Polygenic effect estimates derived in this way can be used to control for genetic influence when investigating non-genetic (environmental) contributions to mental illness.
Endophenotypes
An endophenotype is a trait influenced by some or all of the genes predisposing to an illness [131, 132]. As endophenotypes are measureable in both affected and unaffected individuals, they are theoretically capable of providing greater statistical power to localize and identify disease-related genes than affection status alone [26, 133]. Furthermore, as demonstrated by Steinberg and colleagues [105], endophenotypes can provide insight into unaffected carriers of putatively causal illness variants. Despite the consistent use of endophenotypes in other areas of human disease genetics (sometimes referred to as allied phenotypes or simply risk factors), their application in larger scale psychiatric genetics studies designed to identify novel risk loci has been limited [132]. However, methods for empirically selecting endophenotypes for specific illnesses based upon shared genetic covariance using related subjects [134, 135] or based upon common variants [46] have been developed and overlapping genetic influences for cognitive, electrophysiological, neuroimaging and transcriptional measures and various psychiatric disorders have been discovered [132]. Regardless of the genetic design employed, we strongly advocate deep phenotyping, including quantitative diagnostic/symptom measures and cognitive, imaging and molecular endophenotypes. While myriad of potential endophenotypes for psychotic and affective disorders exist, selecting those that are heritable, genetically correlated with illness risk and amenable to large scale data collection is critical [132]. Tools for such deep phenotyping are now available in the public domain (e.g., PhenX Early Psychosis Translational Research Collection https://www.phenxtoolkit.org/index.php?pageLink=browse.nimh.eptr).
Effects of ascertainment
How families are selected for study may influence both the phenotypic spectrum of the sample and the underlying genetic contributors. Probands recruited as part of families may differ from those recruited as singleton cases, presumably as an effect of selecting individuals with intact family relationships. For example, the Consortium on the Genetics of Schizophrenia (COGS) examined neurocognitive measures and other endophenotypes in families selected through a proband (COGS-1) and in a case-control (COGS-2) study [136]. Patients ascertained through the family-based design, compared to case-control, were younger, had higher educational attainment, better educated parents and superior performance on some neurocognitive tests. Thus, studies that use case-control ascertainment may tap into populations with more severe forms of illness that are exposed to less favorable factors compared to those ascertained through designs that require family participation. However, designs that require multiple affected individuals in a family may result in a more severe phenotypic profile and a different underlying genetic architecture as compared to simplex families. For example, a comparison of multiplex and simplex ASD families found an enrichment of CNVs in ASD risk loci in both but a lower rate of de novo CNVs in the multiplex families [137]. Family selection also impacts the distribution of phenotypes among unaffected family members, with members of multiplex families generally having greater endophenotype impairment than simplex family members [138–140]. In addition to enriching for inherited, as opposed to de novo, risk alleles selection on multiplex families may enrich for loci of larger effect which are presumably rarer [102, 107, 141].
Pedigree-based whole genome sequencing of affective and psychotic disorders consortium
To capitalize on the benefits of family-based designs for variant localization and gene identification, we formed an eight-site international consortium to use whole genome sequence data and novel analytic methods to identify rare variants that increase risk for affective and/or psychotic illness. Initially, participating studies included: individuals from large families of Amish and Mennonite descent ascertained for bipolar disorder and living in Pennsylvania, Ohio, and Indiana [142, 143]; individuals from 88 multiplex families living in Western Australia [144]; persons from extended families living in Costa Rica’s central valley who were identified via a sibling pair concordant for either schizophrenia or bipolar disorder [145]; large multiplex multigenerational families from Pennsylvania selected for schizophrenia [146]; individuals with from Scottish families multiply affected with bipolar disorder or schizophrenia [147, 148]; and large extended Mexican–American pedigrees living in Texas and selected without regard to phenotype [77]. Our cost-effective approach leverages existing DNA, phenotypic data and some existing sequence data from extended pedigrees with at least three affected family members. Together, we have marshaled over 4000 individuals in approximately 269 families (see Table 2). Other research groups who have generated WGS in additional well-characterized families are encouraged to join us.
Table 2.
Studies currently participating in the consortium
| Study | Sample (n/families) | Existing sequence | Ascertainment | Race/ethnicity | SCZ | SCA | BPD | MDD | Cognitive |
|---|---|---|---|---|---|---|---|---|---|
| AMBiGena | 750/3 | Yes | BPD | EA | 2 | 13 | 198 | 52 | 156 |
| Australia | 316/88 | Yes | SCZ | EA | 76 | 17 | 7 | 76 | 316 |
| Costa Rica | 488/7 | No | BPD/SCZ | LA | 36 | 14 | 72 | 83 | 475 |
| Pennsylvania | 725/45 | No | SCZ | EA | 87 | 24 | 11 | 147 | 656 |
| Scotland | 569/76 | No | BPD/SCZ | EA | 60 | 0 | 130 | 165 | – |
| Texas | 1329/50 | Yes | Random | LA | 5 | 13 | 21 | 418 | 1256 |
| Total | 4097/269 | 266 | 81 | 439 | 941 | 2703 |
SCZ Schizophrenia, SCA Schizoaffective, BPD bipolar disorder, MDD major depression, EA European ancestry, LA Latino ancestry
Amish Mennonite Bipolar Genetics Study
Conclusion
It is clear that common and rare variants, as well as environmental factors, play a role in risk for mental illness. Large meta analytic GWAS have likely localized most or all of the common variants with moderate to large effect sizes for the major psychiatric disorders [10]. Following this logic, Boyle, Li and Pritchard [10] recently suggested that after the biggest hits from GWAS have been identified, “the next most promising step is to hunt for lower-frequency variants of larger effects” (page 1184). Given the recent progress with common variation, it would seem that the field of psychiatric genetics should now capitalize on those successes by identifying and characterizing analogous rare variation and confirming those previously identified [149]. Extended pedigrees represent an implicit enrichment strategy for identifying rare variants since Mendelian transmission maximizes the chance that multiple copies will exist in the family. Given this enrichment, the associated improvement in statistical power, plus the economic advantages of pseudo-sequencing through genotype imputation, we formed the “Pedigree-Based Whole Genome Sequencing of Affective and Psychotic Disorders” consortium, a group of international scientists dedicated to using family-based designs to identify rare variants that increase risk of psychiatric disorders. WGS in multiplex pedigrees provides an important complementary experimental approach for identifying genes that confer risk for mental illness.
Acknowledgements
This research was supported by National Institute of Mental Health grants U01 MH105630 (DCG), U01 MH105634 (REG), U01 MH105632 (JB), R01 MH078143 (DCG), R01 MH083824 (DCG & JB), R01 MH078111 (JB), R01 MH061622 (LA), R01 MH042191 (REG), and R01 MH063480 (VLN). We thank Dr. Steve Hyman for his continued support for psychiatric genetics.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2017;175:15–27. appiajp201717030283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6: 95–108. [DOI] [PubMed] [Google Scholar]
- 3.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381: 1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint J, Mott R. Finding the molecular basis of quantitative traits: successes and pitfalls. Nat Rev Genet. 2001;2:437–45. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11:2417–23. [DOI] [PubMed] [Google Scholar]
- 7.McClellan J, King MC. Genomic analysis of mental illness: a changing landscape. JAMA. 2010;303:2523–4. [DOI] [PubMed] [Google Scholar]
- 8.Sanders SJ, Neale B, Huang H, Werling D, An J-Y, Dong S, et al. Whole genome sequencing in psychiatric disorders: the WGSPD consortium. Nat Neurosci. 2017;12: 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–48. [DOI] [PubMed] [Google Scholar]
- 10.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rare Gibson G. and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freimer N, Sabatti C. The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat Genet. 2004;36:1045–51. [DOI] [PubMed] [Google Scholar]
- 13.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–25. [DOI] [PubMed] [Google Scholar]
- 14.Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–7. [DOI] [PubMed] [Google Scholar]
- 16.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–75. [DOI] [PubMed] [Google Scholar]
- 19.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335: 823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sveinbjornsson G, Albrechtsen A, Zink F, Gudjonsson SA, Oddson A, Masson G, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet. 2016;48:314–7. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarti A, Clark AG, Mootha VK. Distilling pathophysiology from complex disease genetics. Cell. 2013;155:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11:773–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Localization Blangero J. and identification of human quantitative trait loci: king harvest has surely come. Curr Opin Genet Dev. 2004;14:233–40. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey-Wilson JE, Wilson AF. Linkage analysis in the next-generation sequencing era. Hum Hered. 2011;72:228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Hum Genet. 2012;131:1555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epstein MP, Duncan R, Ware EB, Jhun MA, Bielak LF, Zhao W, et al. A statistical approach for rare-variant association testing in affected sibships. Am J Hum Genet. 2015;96:543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight S, Abo RP, Abel HJ, Neklason DW, Tuohy TM, Burt RW, et al. Shared genomic segment analysis: the power to find rare disease variants. Ann Hum Genet. 2012;76:500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charney AW, Ruderfer DM, Stahl EA, Moran JL, Chambert K, Belliveau RA, et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiatry. 2017;7: e993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landen M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depressive disorder. Nat Genet. 2018;50:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall L, Adams M, Arnau-Soler A, Clarke T, Howard D, Zeng Y, et al. Genome-wide meta-analyses of stratified depression in generation Scotland and UK biobank. bioRxiv. 2017;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, et al. Largest GWAS of PTSD (N = 20,070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2018;23:666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demontis D, Walters R, Martin J, Mattheisen M, Als T, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for ADHD. bioRxiv 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Autism Spectrum Disorders Working Group of The Psychiatric Genomics C. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism. 2017;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. [DOI] [PubMed] [Google Scholar]
- 42.Manolio T, Collins F, Cox N, Goldstein D, Hindorff L, Hunter D, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- 44.Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet. 2009;18:3525–31. [DOI] [PubMed] [Google Scholar]
- 45.Purcell S, Wray N, Stone J, Visscher P, O’Donovan M, Sullivan P, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. [DOI] [PubMed] [Google Scholar]
- 48.Breen G, Li Q, Roth BL, O’Donnell P, Didriksen M, Dolmetsch R, et al. Translating genome-wide association findings into new therapeutics for psychiatry. Nat Neurosci. 2016;19:1392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyman SE. Revolution stalled. Sci Transl Med. 2012;4: 155cm111. [DOI] [PubMed] [Google Scholar]
- 50.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders SJ. First glimpses of the neurobiology of autism spectrum disorder. Curr Opin Genet Dev. 2015;33:80–92. [DOI] [PubMed] [Google Scholar]
- 55.Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. 2017;49:978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi H, Kichaev G, Pasaniuc B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am J Hum Genet. 2016;99:139–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17: 9–18. [DOI] [PubMed] [Google Scholar]
- 63.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. [DOI] [PubMed] [Google Scholar]
- 65.Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92:7612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bassett AS, Lowther C, Merioo D, Costain G, Chow EWC, van Amelsvoort T, et al. Rare genome-wide copy number variation and expression of Schizophrenia in 22q11.2 deletion syndrome. Am J Psychiatry. 2017;174:1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baron M, Risch N, Hamburger R, Mandel B, Kushner S, Newman M, et al. Genetic linkage between X-chromosome markers and bipolar affective illness. Nature. 1987;326:289–92. [DOI] [PubMed] [Google Scholar]
- 70.Egeland JA, Gerhard DS, Pauls DL, Sussex JN, Kidd KK, Allen CR, et al. Bipolar affective disorders linked to DNA markers on chromosome 11. Nature. 1987;325:783–7. [DOI] [PubMed] [Google Scholar]
- 71.Kelsoe JR, Ginns EI, Egeland JA, Gerhard DS, Goldstein AM, Bale SJ, et al. Re-evaluation of the linkage relationship between chromosome 11p loci and the gene for bipolar affective disorder in the Old Order Amish. Nature. 1989;342:238–43. [DOI] [PubMed] [Google Scholar]
- 72.Gershon ES. Marker genotyping errors in old data on X-linkage in bipolar illness. Biol Psychiatry. 1991;29:721–9. [DOI] [PubMed] [Google Scholar]
- 73.Burmeister M, McInnis MG, Zollner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–40. [DOI] [PubMed] [Google Scholar]
- 74.Risch N Genetic linkage and complex diseases, with special reference to psychiatric disorders. Genet Epidemiol. 1990;7:3–16. discussion 17–45 [DOI] [PubMed] [Google Scholar]
- 75.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. [DOI] [PubMed] [Google Scholar]
- 76.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, et al. Common genetic influences on depression, alcohol, and substance use disorders in Mexican-American families. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKay DR, Knowles EE, Winkler AA, Sprooten E, Kochunov P, Olvera RL, et al. Influence of age, sex and genetic factors on the human brain. Brain Imaging Behav. 2014;8:143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hinrichs AL, Suarez BK. Incorporating linkage information into a common disease/rare variant framework. Genet Epidemiol. 2011;35(Suppl 1):S74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson AF, Ziegler A. Lessons learned from Genetic Analysis Workshop 17: transitioning from genome-wide association studies to whole-genome statistical genetic analysis. Genet Epidemiol. 2011;35(Suppl 1):S107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blangero J, Diego VP, Dyer TD, Almeida M, Peralta J, Kent JW,et al. A kernel of truth: statistical advances in polygenic variance component models for complex human pedigrees. Adv Genet. 2013;81:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Teng J, Risch N. The relative power of family based and case-control designs for linkage disequilibrium studies of complex human diseases. II. Individual genotyping. Genome Res. 1999;9:234–41. [PubMed] [Google Scholar]
- 84.Zöllner S. Sampling strategies for rare variant tests in case-control studies. Eur J Hum Genet. 2012;20:1085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wijsman EM. Family-based approaches: design, imputation, analysis, and beyond. BMC Genet. 2016;17(Suppl 2):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wijsman E, Amos C. Genetic analysis of simulated oligogenic traits in nuclear families and extended pedigrees: summary of GAW10 contributions. Genet Epidemiol. 1997;14:719–35. [DOI] [PubMed] [Google Scholar]
- 87.Gagnon F, Roslin NM, Lemire M. Successful identification of rare variants using oligogenic segregation analysis as a prioritizing tool for whole-exome sequencing studies. BMC Proc. 2011;5(Suppl 9):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simpson CL, Justice CM, Krishnan M, Wojciechowski R, Sung H, Cai J, et al. Old lessons learned anew: family-based methods for detecting genes responsible for quantitative and qualitative traits in the Genetic Analysis Workshop 17 mini-exome sequence data. BMC Proc. 2011;5(Suppl 9):S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li M, Boehnke M, Abecasis GR. Efficient study designs for test of genetic association using sibship data and unrelated cases and controls. Am J Hum Genet. 2006;78:778–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saad M, Wijsman EM. Power of family-based association designs to detect rare variants in large pedigrees using imputed genotypes. Genet Epidemiol. 2014;38:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–94. [DOI] [PubMed] [Google Scholar]
- 92.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Family-based association tests for sequence data, and comparisons with population-based association tests. Eur J Hum Genet. 2013;21:1158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Q, Nicolae DL, Chen LS. Marbled inflation from population structure in gene-based association studies with rare variants. Genet Epidemiol. 2013;37:286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Borecki IB, Province MA. Genetic and genomic discovery using family studies. Circulation. 2008;118:1057–63. [DOI] [PubMed] [Google Scholar]
- 96.Haghighi F, Hodge SE. Likelihood formulation of parent-of-origin effects on segregation analysis, including ascertainment. Am J Hum Genet. 2002;70:142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993;52:506–16. [PMC free article] [PubMed] [Google Scholar]
- 98.Gao G, Allison DB, Hoeschele I. Haplotyping methods for pedigrees. Hum Hered. 2009;67:248–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schouten MT, Williams CK, Haley CS. The impact of using related individuals for haplotype reconstruction in population studies. Genetics. 2005;171:1321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giudicessi JR, Ackerman MJ. Prevalence and potential genetic determinants of sensorineural deafness in KCNQ1 homozygosity and compound heterozygosity. Circ Cardiovasc Genet. 2013;6:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhong K, Zhu G, Jing X, Hendriks AEJ, Drop SLS, Ikram MA, et al. Genome-wide compound heterozygote analysis highlights alleles associated with adult height in Europeans. Hum Genet. 2017;136:1407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dudbridge F, Brown SJ, Ward L, Wilson SG, Walsh JP. How many cases of disease in a pedigree imply familial disease? Ann Hum Genet. 2017;82:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chakravarti A, Turner TN. Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. Bioessays. 2016;38:578–86. [DOI] [PubMed] [Google Scholar]
- 104.Lescai F, Franceschi C. The impact of phenocopy on the genetic analysis of complex traits. PLoS ONE. 2010;5:e11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Steinberg S, Gudmundsdottir S, Sveinbjornsson G, Suvisaari J, Paunio T, Torniainen-Holm M, et al. Truncating mutations in RBM12 are associated with psychosis. Nat Genet. 2017;49: 1251–4. [DOI] [PubMed] [Google Scholar]
- 106.Homann OR, Misura K, Lamas E, Sandrock RW, Nelson P, McDonough SI, et al. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Mol Psychiatry. 2016;21:1690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Timms AE, Dorschner MO, Wechsler J, Choi KY, Kirkwood R, Girirajan S, et al. Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry. 2013;70: 582–90. [DOI] [PubMed] [Google Scholar]
- 108.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–90. [DOI] [PubMed] [Google Scholar]
- 109.Bouwkamp CG, Kievit AJ, Olgiati S, Breedveld GJ, Coesmans M, Bonifati V, et al. A balanced translocation disrupting BCL2L10 and PNLDC1 segregates with affective psychosis. Am J Med Genet B Neuropsychiatr Genet. 2017;174:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tansey KE, Rees E, Linden DE, Ripke S, Chambert KD, Moran JL, et al. Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry. 2016;21:1085–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomson PA, Duff B, Blackwood DH, Romaniuk L, Watson A, Whalley HC, et al. Balanced translocation linked to psychiatric disorder, glutamate, and cortical structure/function. NPJ Schizophr. 2016;2:16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ryan NM, Lihm J, Kramer M, McCarthy S, Evans KL, Ghiban E, et al. Beyond the translocation: whole genome sequencing analysis of the Scottish t(1;11) family. Orlando, FL: World Congress of Psycahtric Genetics; 2017. [Google Scholar]
- 113.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Livne OE, Han L, Alkorta-Aranburu G, Wentworth-Sheilds W, Abney M, Ober C, et al. PRIMAL: fast and accurate pedigree-based imputation from sequence data in a founder population. PLoS Comput Biol. 2015;11:e1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meuwissen T, Goddard M. The use of family relationships and linkage disequilibrium to impute phase and missing genotypes in up to whole-genome sequence density genotypic data. Genetics. 2010;185:1441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheung CY, Thompson EA, Wijsman EM. GIGI: an approach to effective imputation of dense genotypes on large pedigrees. Am J Hum Genet. 2013;92:504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Mol Ecol. 2004;13: 3261–73. [DOI] [PubMed] [Google Scholar]
- 118.Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, et al. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24: 3189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu J, Turner A, Little J, Bleecker ER, Meyers DA. Positive results in association studies are associated with departure from Hardy-Weinberg equilibrium: hint for genotyping error? Hum Genet. 2002;111:573–4. [DOI] [PubMed] [Google Scholar]
- 120.Miller CR, Joyce P, Waits LP. Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics. 2002;160: 357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Douglas JA, Skol AD, Boehnke M. Probability of detection of genotyping errors and mutations as inheritance inconsistencies in nuclear-family data. Am J Hum Genet. 2002;70:487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McGrath J, Saha S, Welham J, El Saadi O, MacCauley C, Chant D. A systematic review of the incidence of schizophrenia: the distribution of rates and the influence of sex, urbanicity, migrant status and methodology. BMC Med. 2004;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Modabbernia A, Velthorst E, Reichenberg A. Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol Autism. 2017;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D, et al. Advancing paternal age and the risk of schizophrenia. Arch Gen Psychiatry. 2001;58:361–7. [DOI] [PubMed] [Google Scholar]
- 126.Vassos E, Pedersen CB, Murray RM, Collier DA, Lewis CM. Meta-analysis of the association of urbanicity with schizophrenia. Schizophr Bull. 2012;38:1118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Os J, Kenis G, Rutten BP. The environment and schizophrenia. Nature. 2010;468:203–12. [DOI] [PubMed] [Google Scholar]
- 128.Krabbendam L, van Os J. Schizophrenia and urbanicity: a major environmental influence--conditional on genetic risk. Schizophr Bull. 2005;31:795–9. [DOI] [PubMed] [Google Scholar]
- 129.Colodro-Conde L, Couvy-Duchesne B, Whitfield JB, Streit F, Gordon S, Rietschel M, et al. Higher genetic risk for schizophrenia is associated with living in urban and populated areas. bioRxiv 2017. [Google Scholar]
- 130.Quillen EE, Voruganti VS, Chittoor G, Rubicz R, Peralta JM, Almeida MA, et al. Evaluation of estimated genetic values and their application to genome-wide investigation of systolic blood pressure. BMC Proc. 2014;8:S66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45. [DOI] [PubMed] [Google Scholar]
- 132.Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventós H, Blangero J, et al. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Almasy L, Blangero J. Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. Am J Med Genet. 2001;105:42–44. [PubMed] [Google Scholar]
- 134.Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW, Charlesworth JC, et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012;71:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Glahn DC, Williams JT, McKay DR, Knowles EE, Sprooten E, Mathias SR, et al. Discovering schizophrenia endophenotypes in randomly ascertained pedigrees. Biol Psychiatry. 2015;77: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gur RC, Braff DL, Calkins ME, Dobie DJ, Freedman R, Green MF, et al. Neurocognitive performance in family-based and case-control studies of schizophrenia. Schizophr Res. 2015;163:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Leppa VM, Kravitz SN, Martin CL, Andrieux J, Le Caignec C, Martin-Coignard D, et al. Rare inherited and de novo CNVs reveal complex contributions to ASD risk in multiplex families. Am J Hum Genet. 2016;99:540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Virkud YV, Todd RD, Abbacchi AM, Zhang Y, Constantino JN. Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oerlemans AM, Hartman CA, de Bruijn YG, Franke B, Buitelaar JK, Rommelse NN. Cognitive impairments are different in single-incidence and multi-incidence ADHD families. J Child Psychol Psychiatry. 2015;56:782–91. [DOI] [PubMed] [Google Scholar]
- 140.Donaldson CK, Stauder JEA, Donkers FCL. Increased sensory processing atypicalities in parents of multiplex ASD families versus typically developing and simplex ASD families. J Autism Dev Disord. 2017;47:535–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bureau A, Parker MM, Ruczinski I, Taub MA, Marazita ML, Murray JC, et al. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics. 2014;197:1039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Georgi B, Craig D, Kember RL, Liu W, Lindquist I, Nasser S, et al. Genomic view of bipolar disorder revealed by whole genome sequencing in a genetic isolate. PLoS Genet. 2014;10: e1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hou L, Faraci G, Chen DT, Kassem L, Schulze TG, Shugart YY, et al. Amish revisited: next-generation sequencing studies of psychiatric disorders among the Plain people. Trends Genet. 2013;29:412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McCarthy NS, Melton PE, Ward SV, Allan SM, Dragovic M, Clark ML, et al. Exome array analysis suggests an increased variant burden in families with schizophrenia. Schizophr Res. 2017;185:9–16. [DOI] [PubMed] [Google Scholar]
- 145.Carmiol N, Peralta JM, Almasy L, Contreras J, Pacheco A, Escamilla MA, et al. Shared genetic factors influence risk for bipolar disorder and alcohol use disorders. Eur Psychiatry. 2014;29:282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gur R, Nimgaonkar V, Almasy L, Calkins M, Ragland J, Pogue-Geile M, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164:813–9. [DOI] [PubMed] [Google Scholar]
- 147.Whalley HC, Sussmann JE, Chakirova G, Mukerjee P, Peel A, McKirdy J, et al. The neural basis of familial risk and temperamental variation in individuals at high risk of bipolar disorder. Biol Psychiatry. 2011;70:343–9. [DOI] [PubMed] [Google Scholar]
- 148.Christoforou A, McGhee KA, Morris SW, Thomson PA, Anderson S, McLean A, et al. Convergence of linkage, association and GWAS findings for a candidate region for bipolar disorder and schizophrenia on chromosome 4p. Mol Psychiatry. 2011;16:240–2. [DOI] [PubMed] [Google Scholar]
- 149.Auer PL, Reiner AP, Wang G, Kang HM, Abecasis GR, Altshuler D, et al. Guidelines for large-scale sequence-based complex trait association studies: lessons learned from the NHLBI exome sequencing project. Am J Hum Genet. 2016;99:791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Makinen VP, Parkkonen M, Wessman M, Groop PH, Kanninen T, Kaski K. High-throughput pedigree drawing. Eur J Hum Genet. 2005;13:987–9. [DOI] [PubMed] [Google Scholar]
- 151.Hornig T, Gruning B, Kundu K, Houwaart T, Backofen R, Biber K, et al. GRIN3B missense mutation as an inherited risk factor for schizophrenia: whole-exome sequencing in a family with a familiar history of psychotic disorders. Genet Res. 2017;99:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.John J, Kukshal P, Bhatia T, Chowdari KV, Nimgaonkar VL, Deshpande SN, et al. Possible role of rare variants in trace amine associated receptor 1 in schizophrenia. Schizophr Res. 2017;189: 190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rao AR, Yourshaw M, Christensen B, Nelson SF, Kerner B. Rare deleterious mutations are associated with disease in bipolar disorder families. Mol Psychiatry. 2017;22:1009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang T, Hou L, Chen DT, McMahon FJ, Wang JC, Rice JP. Exome sequencing of a large family identifies potential candidate genes contributing risk to bipolar disorder. Gene. 2017;645:119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Egawa J, Hoya S, Watanabe Y, Nunokawa A, Shibuya M, Ikeda M, et al. Rare UNC13B variations and risk of schizophrenia: whole-exome sequencing in a multiplex family and follow-up resequencing and a case-control study. Am J Med Genet B Neuropsychiatr Genet. 2016;171:797–805. [DOI] [PubMed] [Google Scholar]
- 156.Goes FS, Pirooznia M, Parla JS, Kramer M, Ghiban E, Mavruk S, et al. Exome sequencing of familial bipolar disorder. JAMA Psychiatry. 2016;73:590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kos MZ, Carless MA, Peralta J, Blackburn A, Almeida M, Roalf D, et al. Exome sequence data from multigenerational families implicate AMPA receptor trafficking in neurocognitive impairment and schizophrenia risk. Schizophr Bull. 2016;42:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Subaran RL, Odgerel Z, Swaminathan R, Glatt CE, Weissman MM. Novel variants in ZNF34 and other brain-expressed transcription factors are shared among early-onset MDD relatives. Am J Med Genet B Neuropsychiatr Genet. 2016;171B:333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Watanabe Y, Nunokawa A, Shibuya M, Ikeda M, Hishimoto A, Kondo K, et al. Rare truncating variations and risk of schizophrenia: whole-exome sequencing in three families with affected siblings and a three-stage follow-up study in a Japanese population. Psychiatry Res. 2016;235:13–18. [DOI] [PubMed] [Google Scholar]
- 160.Zhou Z, Hu Z, Zhang L, Hu Z, Liu H, Liu Z, et al. Identification of RELN variation p.Thr3192Ser in a Chinese family with schizophrenia. Sci Rep. 2016;6:24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ament SA, Szelinger S, Glusman G, Ashworth J, Hou L, Akula N, et al. Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci USA. 2015;112:3576–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kember RL, Georgi B, Bailey-Wilson JE, Stambolian D, Paul SM, Bucan M. Copy number variants encompassing Mendelian disease genes in a large multigenerational family segregating bipolar disorder. BMC Genet. 2015;16:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Thygesen JH, Zambach SK, Ingason A, Lundin P, Hansen T, Bertalan M, et al. Linkage and whole genome sequencing identify a locus on 6q25–26 for formal thought disorder and implicate MEF2A regulation. Schizophr Res. 2015;169:441–6. [DOI] [PubMed] [Google Scholar]
- 164.Strauss KA, Markx S, Georgi B, Paul SM, Jinks RN, Hoshi T, et al. A population-based study of KCNH7 p.Arg394His and bipolar spectrum disorder. Hum Mol Genet. 2014;23:6395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]