Abstract
Malaria is a serious, complex disease caused by parasites of the genus Plasmodium. Plasmodium parasites affect multiple tissues as they evade immune responses, replicate, sexually reproduce, and transmit between vertebrate and invertebrate hosts. The explosion of omics technologies has enabled large-scale collection of Plasmodium infection data, revealing systems-scale patterns, mechanisms of pathogenesis, and the ways that host and pathogen affect each other. Here, we provide an overview of recent efforts using systems biology approaches to study host-Plasmodium interactions and the biological themes that have emerged from these efforts. We discuss some of the challenges in using systems biology for this goal, key research efforts needed to address those issues, and promising future malaria applications of systems biology.
Keywords: Plasmodium, malaria, systems biology, omics, host-pathogen interaction
Host-Parasite Interactions: A Key to Understanding Malaria
Malaria is caused by protozoan parasites of the genus Plasmodium. The Plasmodium life cycle involves two hosts: 1) a vertebrate host in which parasites reproduce asexually, begin sexual development, and cause the disease malaria, and 2) an invertebrate host that acts as a vector for transmitting the disease between vertebrate hosts, and in which sexual reproduction occurs (Figure 1). Mosquitoes, mainly of the genus Anopheles, are the invertebrate hosts. Plasmodium vertebrate hosts include reptiles, birds, rodents, and primates [1] (Table 1). In their vertebrate hosts, infection by Plasmodium parasites can lead to serious illness and even death [2]. Plasmodium parasites also affect the survival, behavior, and reproductive success of their invertebrate hosts in the course of completing the sexual stage of their life cycle and transmitting to new vertebrate hosts [2].
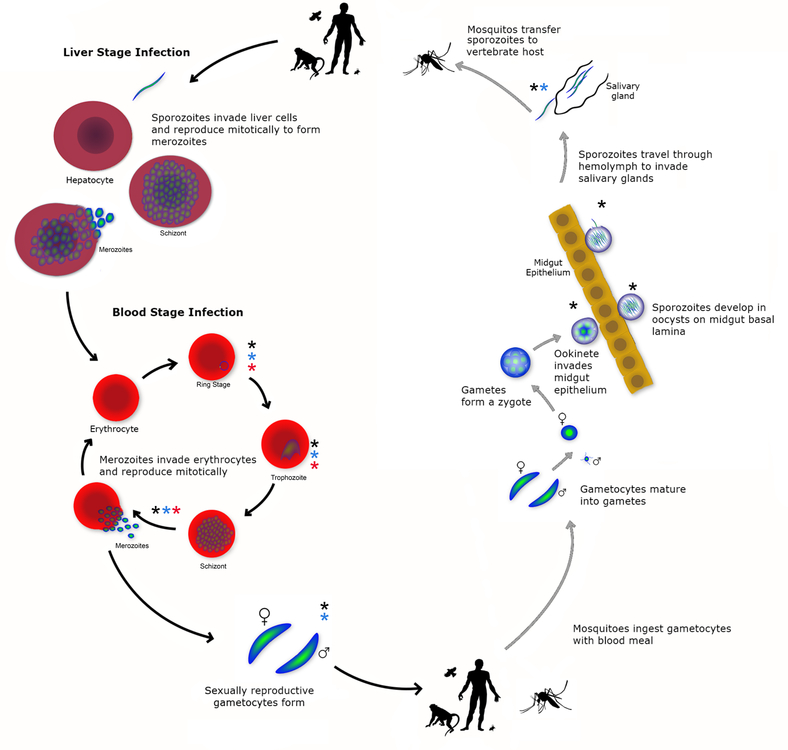
Figure 1:
Life cycle of Plasmodium parasites including developmental stages in vertebrate and invertebrate hosts and transmission between hosts. Asterisks indicate the omic studies (see Table 3 for specific references) that have been performed for the different life cycle stages: black asterisks indicate transcriptomics studies; blue asterisks indicate proteomics studies; and red asterisks indicate metabolomics and/or lipidomics studies. The crescent-shaped gametocytes depicted represent the morphology in Plasmodium falciparum; other Plasmodium species have rounded/brick-shaped gametocytes.
Table 1:
Representative Plasmodium Species, their hosts, and selected references.
| Host | Plasmodium species | Omics type | References |
|---|---|---|---|
| Human | P. falciparum, P. vivax, P. knowlesi | Transcriptomics | [10, 11, 16, 19–22, 42–44, 51, 52, 57, 58] |
| Proteomics | [31, 56, 60, 98] | ||
| Metabolomics/Lipidomics | [18, 34, 36, 39–41, 48, 80, 99] | ||
| Nonhuman Primate | P. cynomolgi, P. knowlesi | Transcriptomics | [100, 101] |
| Proteomics | [13, 14] | ||
| Rodent | P. berghei, P. chabaudi, P. yoelii | Transcriptomics | [25–28, 30, 49] |
| Metabolomics/Lipidomics | [15, 37–39] | ||
| Avian | P. ashfordi, P. gallinaceum | Transcriptomics | [12, 61, 62] |
| Insect | *all Plasmodium species | Transcriptomics | [66–70, 78–84, 86, 102–104] |
| Proteomics | [87–92] | ||
| Metabolomics/Lipidomics | [71] |
Broadly, the term “host-parasite interaction” refers to the relationship between a host and an organism that lives at its expense. These interactions may be direct, physical binding events at the molecular or cellular level, or they may be more indirect effects of the parasite on the host or of the host on the parasite. In this review, we define host-parasite interactions as any molecular, cellular, or even behavioral changes that occur in a parasite or host due to the influence of one organism on the other, including secondary and higher-order effects. Our focus is specifically on the complex interactions between malaria-causing parasites and their hosts, and how systems biology techniques can be used to elucidate and understand these interactions. Importantly, these interactions are typically best studied in an in vivo context, as in vitro studies often cannot capture the immune and other systemic host responses with major impacts on the parasite and on disease progression. As a result, in this review we focus whenever possible on in vivo studies most likely to capture the full breadth of host-Plasmodium interactions.
Due to the major impact of malaria on human health, elucidating the complex interactions between Plasmodium and its hosts is an area of intense research interest. Recent technological and analytical advances are enabling us to move towards this goal at an unprecedented rate.
Systems Biology: An Emerging Approach to Studying Malaria
Definitions of systems biology (see Glossary) can vary widely; here, we use a definition consistent with that used by the United States National Institutes of Health [3]. Systems biology approaches entail the study of a biological system via a near-comprehensive examination of a specific class of biomolecules, in contrast to a reductionist approach which looks at small subsets of a class of biomolecules. For example, a reductionist approach might entail studying the transcriptional levels of the genes in a small, well-defined pathway, while a systems approach would entail measuring genome-wide transcription levels. Computational modeling and analysis are also important aspects of systems biology, as the scale of the system being considered and the large datasets generated by experimental techniques associated with systems biology are often not amenable to standard data analyses. Systems biology also ideally involves mechanistic mathematical models of a system beyond the aforementioned computational models and analyses, allowing biological insight and the ability to predict system behavior. In the field of malaria research, mathematical modeling is most commonly used in population modeling, to track and predict the transmission of malaria through host populations [4] – an approach not quite in the vein of systems biology per se, and beyond the scope of this review. While specific aspects such as immune response and even the distribution and timing of parasite sequestration in different body tissues have recently been studied using modeling approaches [5], mechanistic mathematical modeling at the systems scale or otherwise has not been undertaken nearly as broadly or as effectively in malaria host-pathogen interactions as it has in other diseases, like cancer. This is in part due to the fact that a large majority of the systems-scale data in the literature to date has been in vitro, rather than in vivo. As a result, this review will focus more on the results of the diverse systems-scale experimental analyses performed in recent years and the biological themes that have emerged from the computational analyses of these datasets.
Advanced analytical techniques are necessary to generate the expansive, systems-scale experimental datasets characteristic of systems biology, which are sometimes collectively and generically referred to as “omics”. Transcriptomics entails the use of RNA-sequencing and microarrays for the systems-scale measurement and study of gene expression. Proteomics, metabolomics, and lipidomics typically entail the use of mass spectrometry or nuclear magnetic resonance spectroscopy for measuring protein, metabolite, and lipid levels, respectively; advanced immunoassays can also be used for measuring protein levels. Collectively, these techniques enable large-scale collection of molecular-level data involving diverse classes of biomolecules. In the field of malaria research, these omics technologies are increasingly being used to study how Plasmodium parasites affect their hosts and how the host environment affects the parasite [6].
Critical to turning such complex datasets into biological insight is the suite of computational methods used for their analysis and interpretation. Beyond traditional statistical analyses, computational approaches such as network modeling, ontological analysis, and phenotype association are used for the analysis and interpretation of these data. Network modeling involves using a structural or graphical model to represent relationships between constituent elements of a dataset [7]. These relationships may come from the experimental dataset itself, such as significant correlation or mutual information between two measured variables, or from other preexisting knowledge such as sequence data or previously reported relationships. Ontological analysis involves associating individual measured variables with groups, sets, or classes to which they belong and then assessing statistical trends, such as enrichment in significantly changing variables for each class based on the dataset [8]. Phenotype association refers to identifying relationships between abundances of a biomolecule, such as a protein or RNA product, to a trait of interest in order to identify which biomolecules may affect the trait [9]. Taken together, these computational approaches can help lead to a systems-scale understanding of the interactions between host and parasite that will be key for disrupting them via the development of new drugs and vaccines in the fight against malaria.
Malaria Systems Biology Studies in Vertebrate Hosts
Plasmodium parasites have evolved to infect a wide range of vertebrate hosts including reptiles, birds, and a variety of mammals, from rodents to primates [1]. The most commonly used animal models in malaria research are mice, birds, and nonhuman primates, with each selected for a variety of reasons including availability, ease of handling, and evolutionary relationship to humans [1]. Systems-scale studies of both human and nonhuman hosts are playing an increasingly important role in understanding the interplay between Plasmodium parasites and their vertebrate hosts.
Immune Response to Plasmodium Infection
One of the most central aspects of host-Plasmodium interactions is the host response to infection by Plasmodium parasites, which has been studied in multiple host/pathogen model systems using a variety of techniques. Transcriptomics is by far the most commonly used approach in malaria omics studies, and has been of particular use in this research area when applied to host cells, such as those collected from a blood sample. Transcriptomics studies of malaria have been performed in mice, birds, nonhuman primates, and humans. Across these studies, a picture of host response begins to emerge with the unsurprising theme of pathways involved in immune response playing a significant role. Human transcriptomics studies revealed cytokine activation, regulation of apoptosis, co-expression of Toll-like receptor and type 1 interferon genes as a group, and correlation to parasitological and/or clinical factors such as parasitemia as important biological themes [10, 11]. A longitudinal study of Plasmodium ashfordi infection in Eurasian siskins also revealed disruption of T-cell and B-cell mediated immunity, oxidative stress response, and telomerase activity in the host. This study also compared the transcriptome response to malaria in birds to that of humans and mice, and found significant overlap, particularly in genes involved in T-cell activation [12]. Broadening the scope to look also at proteomics work in this area, a study performed using blood samples from the nonhuman primate host Saimiri boliviensis also indicated upregulation during P. vivax blood-stage infection of host immune-associated pathways such as oxidative stress, vesicular trafficking, and cytoskeletal proteins. Exploration of the parasite proteome also detected in these samples revealed upregulated P. vivax proteins in pathways including glycolysis and pyruvate metabolism, translation initiation, elongation, unfolded protein response, and intracellular vesicle trafficking [13, 14].
Metabolomics and lipidomics, usually performed on blood samples when used in in vivo studies, are some of the newest forms of analysis being used in the study of malaria, allowing analysis of metabolism to move beyond inferences based on genome, transcriptome, or proteome [15, 16]. These efforts may not always directly implicate specific cellular processes, since any given metabolite is involved in multiple cellular processes and measured changes in blood metabolites may be due to contributions of multiple tissues. Nonetheless, they often provide supporting evidence for previous findings and valuable metabolic context. One such study in mouse models revealed increased energy demand and impaired glycolysis [15]; energy metabolism is potentially related to immune function [17]. A metabolomic study of human subjects infected with P. vivax also identified associations between parasitemia and metabolism, where metabolites with significant, and usually inverse, associations with parasitemia were enriched for heme and glycerophosopholipid metabolism [18]. The strongest association identified in this study was the presence of increased biliverdin and bilirubin levels in patients with high parasitemia, each of which have direct immunomodulatory properties. Heme oxygenase-1, which breaks down heme into biliverdin, is known to be upregulated in patients infected with P. vivax; it is also known to have immunomodulatory and antiinflammatory properties. Moreover, Plasmodium parasites metabolize heme into bilirubin, which could impact leukocyte function and enable parasite invasion of the host immune response [18]. Taken together, these studies show how, across omics levels and across host species, the importance of the host immune response can be observed and characterized at systems scale.
Differential Host and Parasite Biomolecular Profiles Associated with Malaria Severity
Changes that occur in both host and parasite during severe versus non-severe malaria infections is another area of intense research interest, as human malaria can range in severity from asymptomatic to lethal [1, 2]. There are several open questions in this area of study receiving significant attention. For example, do differences in host response determine the severity of malarial illness? Do the parasites have different biomolecular profiles in severely and mildly ill hosts? Is there is any interaction between the biological states of host and parasite that may affect infection severity? Systems biology studies have enabled broader investigations into these and other questions than was previously possible.
Transcriptomics has been among the more widely-used and productive approaches to study this aspect of malaria, in terms of both host and parasite gene expression. For example, the blood transcriptomes of individuals who have experienced multiple P. falciparum infections was compared to those of malaria-naive individuals [19]. This study revealed that, despite differences in symptom presentation, febrile malaria-experienced individuals and asymptomatic malaria-experienced individuals had more similar transcriptome profiles to each other than to malaria-naive individuals. Genes differentially expressed between the groups were enriched for pro-inflammatory cytokines [19]. A similar study comparing P. vivax malaria-naïve and malaria-experienced individuals also identified differences in expression in pro-inflammatory cytokines, as well as interferons [20].
Human transcriptomic studies of cerebral malaria, perhaps the most severe manifestation of Plasmodium infection, have shown upregulation of known neurodegeneration pathways as well as pathways involved in protein transport in blood samples, although the parasite transcriptional profiles were not found to be different [21, 22]. In other studies, parasite transcriptomic signatures in blood samples from patients with cerebral and uncomplicated malaria revealed the expression of surface antigens of the var gene family to be highly associated with malaria severity [23, 24]. Gene expression differences in the host brain between cerebral and uncomplicated malaria have also been identified in mice, with biological processes such as chromatin remodeling, apoptosis, interferon signaling, and regulation of muscle contraction upregulated in mice with cerebral compared to uncomplicated malaria [25–27]. Mouse studies of severe malaria also showed earlier dysregulation of erythropoiesis and increased pulmonary inflammation in severe malaria compared to non-severe, indicating distinct gene expression profiles in different tissue types during severe malaria [28–30].
Proteomics studies have also identified differences in parasite protein expression profiles between these two groups. In particular, significantly higher expression of Plasmodium MESA/PfEMP2 protein was seen in patients with cerebral malaria. This protein is an antigen that is exported from mature Plasmodium parasites and interacts with the host erythrocyte cytoskeleton and surface membrane [31–33]. This result highlights the importance of proteomics data in supporting the results of transcriptomics data, especially when previously published studies may have reported conflicting results. Accordingly, efforts to integrate multiple data types, including proteomics, in future systems biology studies of host-Plasmodium interaction are thus particularly important to allow for proper and holistic understanding of the system.
Other omics techniques have also been brought to bear on the study of severe versus uncomplicated malaria. Proteomics and metabolomics studies in both humans [34–36] and mice [37–39] have been used to further explore host physiology during cerebral malaria, potentially identifying early markers of cerebral malaria that would allow for more prompt medical intervention [40, 41]. One such study found significant correlation between brain swelling in cerebral malaria patients and upregulation of PLA2 pathway-associated metabolites in blood plasma, including arachidonic and pentadecanoic acid. These findings were then confirmed using enzyme assays that confirmed positive correlation between PLA2 activity and brain swelling [35]. Other significant differences found in the plasma metabolic profile of patients during cerebral malaria compared to during convalescence post-illness include amino acid depletion and broad enrichment for fatty acids [36]. These analyses have not explored whether the metabolic profiles associated with cerebral malaria are the result of a general immune response to infection, or whether interactions between the host and Plasmodium parasites have a unique effect on the host metabolic profile. Moreover, the involvement of, for example, fatty acids and amino acids in a broad range of physiological processes makes the conclusions and hypotheses that can come from these studies typically less specific than those from transcriptomics and proteomics studies, which can implicate specific genetic targets; nonetheless, they are still quite informative.
Plasmodium Vertebrate Life Cycle Stages and Comparative Analysis
Systems biology analyses from in vitro studies have already provided a significant baseline of understanding of the biomolecular changes that happen during the stages of the Plasmodium life cycle that occur in vertebrate hosts (Figure 1). Stage-specific gene expression has long been observed in P. falciparum in vitro, with a large number of genes upregulated during the intraerythrocytic development cycle (IDC) compared to, for example, the gametocyte stage [42–47]. Metabolomics analysis of in vitro cultures has even revealed specific host-parasite interactions, such as the fact that Plasmodium incorporates host arginine during the IDC [48].
Systems biology analyses from in vivo infections have further deepened our understanding of the Plasmodium life cycle, with significant progress in characterizing the dynamic transcriptional programs in the IDC. Transcriptomic studies of Plasmodium gene expression in mice and human subjects have identified life cycle-specific clusters of co-expressed genes representing host cell invasion, cell gliding, fatty acid processing, transcriptional regulation, and cellular proliferation during blood stages of the Plasmodium life cycle [11, 49–53]. Parasite transcriptional profiling studies from P. falciparum-infected humans have shown that while some aspects of in vitro IDC profiles can be observed easily, others cannot; the characterized patterns suggested three different transcriptional states: active growth phase, starvation response, and environmental stress response [54]. In another study, network analysis was used to identify clusters of co-expressed genes during the IDC enriched for erythrocyte and reticulocyte variant antigens, particularly those in the var, rif and stevor gene families [55]. This study also identified clusters enriched in Plasmodium genes that have previously been associated with gametocyte development and microtubule function. Based on these findings, the authors hypothesized that regulation of exflagellation of male gametocytes begins in vertebrate hosts before maturation to gametes in mosquitoes. This study was particularly noteworthy and strong in that it harnessed existing published data from both in vitro experiments and in vivo infections while also including new samples and analyses from both in vitro experiments and human samples. Moreover, the authors explicitly sought to characterize the differences between the in vitro and in vivo samples, thus directly investigating the impacts of the host-pathogen interactions that would cause molecular profile differences between those types of samples [55].
Proteome signatures of specific Plasmodium life cycle stages, particularly those of Plasmodium species that infect humans, have further enriched transcriptional studies. In particular, a study by Florens et al. found that only 6% of proteins expressed during blood stages were also expressed in sporozoites collected from mosquito salivary glands [56]. Proteins expressed in sporozoites included known sporozoite markers that are involved in host cell invasion such as circumsporozoite protein (CSP) and sporozoite surface protein 2 (SSP2). However, protein products from the var and rif gene families that had not been previously associated with sporozoites were also identified. Furthermore, this study also found that only a few var and rif protein products expressed in sporozoites were also expressed during the IDC. This finding was one of the first times that evidence for antigenic variation, a known host evasion response during IDC malaria, was observed in the sporozoite stage. This study also showed that host cell invasion proteins expressed in sporozoites were different than those expressed in merozoites [56], supporting the idea that the processes by which Plasmodium invades host cells are very specific to each phase of its life cycle.
In addition to revealing the biological programming occurring at each life cycle stage, systems biology analyses have also been used to gain insight into the similarities and differences between Plasmodium species. Significant effort has been focused on studying the two most common human malaria species, P. falciparum and P. vivax. Network modeling using existing ontology data has been used to identify clusters of co-expressed blood-stage genes from P. vivax transcriptome data that overlap with similar data from P. falciparum. Genes that overlap with those expressed in P. falciparum during vertebrate life cycle stages are known to be involved in liver-stage infection, antigenic variation, and malaria drug resistance [51, 52]. Applying statistical and modeling techniques to gene expression studies of P. vivax in ex vivo cultures [57] and patient blood samples [58] has revealed highly correlated expression during blood stages between P. vivax and P. falciparum. In spite of their similarities, however, these two species show differences in timing of life cycle stages as well as host clinical presentation, including parasitemia in vertebrate hosts, ability to cause relapse, and likelihood of serious complications [59]. Genes coding for metabolic enzymes or of conserved function such as dhfr-ts and msp1 showed nearly identical expression patterns in P. vivax and P. falciparum during the IDC, but 22% of identified transcripts showed significant differences in expression patterns [57]. One example was msp8, a gene with high expression during early ring stages in P. falciparum that continues through late ring and trophozoite stages only in P. vivax (Figure 1). The Plasmodium gene pfkahrp also showed differences in expression timing between these species; it is known to be involved in sequestration during P. falciparum infection by contributing to the formation of protein protrusions on the surface of infected erythrocytes [57]. This process does not occur in P. vivax, but the increased expression of pfkahrp during the late schizont stage in P. vivax, but not in P. falciparum, suggests this gene may play a yet-unknown role in the late IDC in P. vivax [57]. Transcriptional profile differences such as these have been hypothesized to potentially underlie the preference of P. vivax for infection of early reticulocytes and, possibly, the transition to the dormant hypnozoite stage [57]. A proteomics study of P. vivax clinical isolates also identified five expressed proteins, of varying putative function, with no P. falciparum orthologs [60].
Analyses of systems-scale datasets have also identified gene expression patterns that may underlie vertebrate host specificity (Table 1) and differences in the infective behavior of different Plasmodium species. For example, an analysis of P. gallinaceum gene expression in blood samples from infected chickens showed significant differences in the regulation of genes from erythrocyte invasion pathways compared to human malaria parasites [61]. This finding from avian models supports the idea that different Plasmodium species have evolved different gene expression patterns based on their preferred vertebrate host [62]. Molecular mechanisms underlying host specificity are one aspect of Plasmodium-host interaction research that is ripe for future study, since most Plasmodium species do not transmit between vertebrate clades (Table 1) [1].
Malaria Systems Biology Studies in Invertebrate Hosts
While focusing on the human host may be an obvious step in understanding malaria, it is important to remember that Plasmodium transmission also requires an invertebrate insect host where the parasite completes the sexual stage of its life cycle [1]. The main contemporary invertebrate hosts are Anopheles mosquitoes. The insect host stage of the Plasmodium life cycle begins when a mosquito takes a blood meal from an infected animal. Gametocytes in the blood meal form a zygote in the gut, and the zygote then develops into an ookinete that invades the midgut wall to begin the process of developing into sporozoites. Sporozoites then travel through the hemolymph and invade the salivary glands in order to transmit to the next vertebrate host (Figure 1) [1]. Although Plasmodium parasites do not affect insect host health as dramatically as they do in vertebrate hosts, there are definite impacts on invertebrate hosts, from behavioral changes to reduced lifespan [63, 64]. Perhaps more importantly, enhanced characterization of molecular profiles in invertebrate hosts could help shape our understanding of transmission and spur new ways to limit it.
Mechanisms Underlying the Host-Plasmodium Evolutionary Arms Race
Plasmodium parasites and their insect hosts have engaged in a long-standing evolutionary arms race between the insect’s defenses to fight off Plasmodium invaders and Plasmodium’s mechanisms for evading the insect’s immune system [65]. This biological phenomenon is another complex process that systems-scale data analysis has begun to elucidate. For example, a study of Anopheles stephensi mosquitoes using supervised learning and network modeling identified a network of invertebrate host oxidative stress-responsive genes that are disrupted by Plasmodium infection during the oocyst development stage (Figure 1) [66]. Transcriptomics data have also been used to examine hemocyte immune response to Plasmodium sporozoites, revealing a distinct pattern of gene expression when compared to the insect immune response to bacterial pathogens. Pathways regulated in response to Plasmodium sporozoite presence in the hemolymph include FBN family immunolectins and Imd/REL2 pathway genes [67].
While these host-Plasmodium interactions are noteworthy, some of the most interesting findings have more directly identified the importance of evolutionary pressures on both the Plasmodium species and the invertebrate host. One of the most surprising findings from transcriptomic studies is that Anopheles mosquitoes mount an immune response against Plasmodium after taking a blood meal, regardless of whether any parasites are actually present in the blood bolus or not [65, 68]. The putative selective advantage provided by automatically mounting an immune response that will often be unnecessary is indicative of the significant influence Plasmodium species have had on insect evolution. Another study found that exposure to P. berghei for several generations leads to stronger upregulation of specific immune response genes in response to Plasmodium infection compared to mosquitoes from a malaria-naïve line. These genes included the known malaria response genes TEP1, LRIM1 and SPCLIP1. This finding indicates a targeted immune response may be acquired over generations with the selective pressure of constant Plasmodium exposure [69]. Moreover, transcriptomics analysis has also exposed host expression differences in A. stephensi infected with drug-resistant Plasmodium yoelii compared to strains that are not drug-resistant, particularly in genes involved in phagosome activity, melanization, and complement activation. This indicates that the selective pressure of anti-malarial drugs on Plasmodium species has indirect impacts on the invertebrate host as well [70].
Metabolomics analysis of insects, called entometabolomics, is a relatively new area of inquiry with interesting potential for the study of the co-evolved competitive interactions between Plasmodium species and their invertebrate hosts. An untargeted metabolomics study of Anopheles gambiae midgut tissue after feeding with P. falciparum-infected and uninfected blood was recently reported; while analysis and biological interpretation of this dataset was minimal, it nonetheless represents one of the first attempts to track the full metabolic response of a mosquito tissue to Plasmodium infection [71]. Metabolomics studies of other mosquito-borne diseases suggest that lipidome disruption is common in pathogen-infected mosquitoes [72–74], and as such may also be found in mosquitoes infected with Plasmodium parasites even though it has not yet been identified in the literature.
Plasmodium Transmission between Insect and Vertebrate Hosts
The molecular mechanisms underlying sporozoite transmissibility from insect to vertebrate host are of great interest to the malaria research community because transmission is one possible point of intervention to reduce the spread of malaria. Interaction between host and parasite in the form of protein-ligand binding and glycoprotein recognition [75–77] has been well-documented. Recently, systems biology studies have also used transcriptional profiling and other systems-scale screening techniques [78] to quantify changes in the insect host or parasite during sporozoite maturation and development of virulence. These analyses also successfully uncovered some of the mechanisms by which ookinetes traverse the mosquito midgut [79, 80], attach to the basal lamina as oocysts [81], and by which sporozoites invade the salivary glands [82], including identifying new potential ligands for salivary gland invasion [83, 84]. Proteomics approaches have also contributed to this line of inquiry by cataloging protein expression at various stages of the Plasmodium life cycle [56, 85, 86]. Protein expression differences between oocyst and salivary gland sporozoites have been used to identify putative ligands involved in mosquito salivary gland [87–89] and vertebrate hepatic cell invasion [90]. These analyses have also uncovered similarities in protein expression [88] and protein modification [89] in the two major human malaria species P. vivax and P. falciparum, a finding with important implications for the development of transmission-blocking interventions. Recent work also found that lipid rafts from mosquito midgut cells were indeed enriched for known ookinete interacting proteins [91]. Another study integrated findings from several types of previous analyses to identify a protein of previously unknown function, AgSGU, that is concentrated in lipid rafts in the mosquito midgut and whose expression significantly changes after blood feeding. Follow-up in vitro experiments suggested that AgSGU activity may inhibit ookinete midgut invasion and, thus, oocyst formation [92].
Concluding Remarks
Experimental and computational techniques for systems-scale analysis have allowed researchers to, in a previously unimaginable way, characterize regulation of the Plasmodium life cycle, host immune response to the presence of Plasmodium, and ways in which host and parasite influence each other. However, even as these approaches have deepened our understanding of host-parasite interaction, many unanswered questions remain about how the host’s biochemistry and immune system influence the biochemical, cellular, and behavioral responses of Plasmodium parasites and vice-versa.
These are multifaceted questions whose answers will require analysts to combine, or integrate, information from different omics data types. Integration of multiple types of omics data will allow us to study and understand the coordinated changes in the cellular environment that occur across molecular scales in response to parasite invasion. Integration techniques have been widely used in other fields to integrate genomic, transcriptomics, and phenotype data, often for the purpose of identifying genomic sequences that contribute to a specific trait or disease via population-scale analyses [93]. However, truly integrative analysis will also undoubtedly require development of new computational and analytical techniques for efficient exploitation of these large datasets with complex interrelatedness (Figure 2). Towards this goal, two classes of techniques will likely find great use in the integration of diverse data types: approaches that map multiple data types to known pathways and network topologies, and approaches that identify network topologies between data types strictly based on the datasets themselves. The first approach links data to biological knowledge and thus increases confidence in resulting biological inferences, while the second is more likely to reveal currently unknown relationships, yielding unexpected and potentially more impactful insights. While some tools do exist for these tasks, there is still an overwhelming need for improved, advanced methods in this area, which would have an outsize impact on our ability to interpret systems biology data.
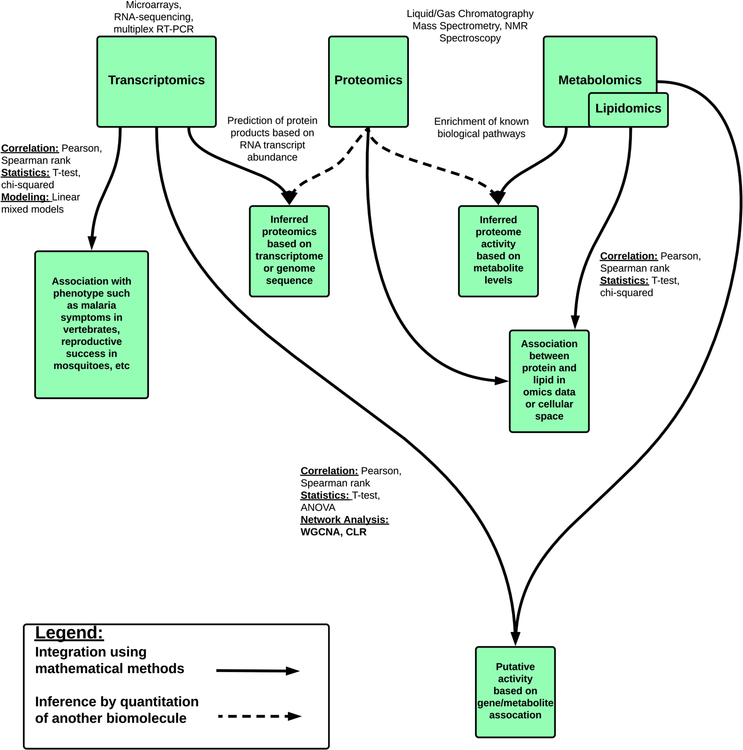
Figure 2:
Overview of omics data types, methods for generation and analysis of the data, and strategies for integration across data types and the expected information to be learned from such analyses. Solid lines represent direct integration of data types; dotted lines represent inference of one data type from another, with potential validation using experimental measurements of the inferred values.
Another challenge to the effective use of systems biology in studying malaria is that sample sizes in both human and animal in vivo model studies are often undesirably small. This occurs for a variety of reasons including cost, restrictions or difficulties in sample collection, and ethical concerns that outweigh the benefit of greater statistical power that comes with larger sample sizes. Both new studies with bigger cohorts and the use of meta-analysis methods to combine data from several independent studies will likely be needed to overcome these limitations [94]. Systems biology approaches to meta-analysis, however, are still a relatively new research focus in need of development of new techniques and testing of existing methods in order to determine their validity and effectiveness. P-value combination, where each study is considered an independent test and p-values are then combined into one statistic, is one promising option based on a recent application to a large number of RNA-seq studies [95].
Finally, mathematical modeling is relatively under-utilized in the study of host-Plasmodium interactions. Malaria systems biology in its most ideal form would include the development of predictive mathematical models that both codify and enhance our understanding of the disease. Beyond the well-trodden field of mathematical modeling of population-scale disease transmission, modeling approaches have also been used to gain greater insights into, for example, the timing of anemia compensation during the course of Plasmodium infection [96], the timing of parasite infection and release before parasitemia can be detected by current methods [5, 55], and how models of metabolic pathways may be used to interpret transcriptomic datasets [97]. Efforts like these range from focused models to fit only a few types of physiological measurements to broader pathway-level models, and they have provided noteworthy insight. Nonetheless, the extent of truly systems-scale mathematical modeling in malaria to date has been limited, though that is slowly changing. Such systems-scale models could potentially be used for the identification of therapeutic targets that are most likely to affect Plasmodium parasites while minimizing impact on host cells.
Taken together, research to date has shown systems biology to be a valuable tool to uncover host-parasite interactions at the molecular level between Plasmodium parasites and their hosts, whether at the level of RNA transcripts, proteins, metabolites, or lipids. These approaches have been effective in uncovering biological insights across a wide variety of host-parasite model systems. They hold great promise to help develop our understanding of emerging areas of host-parasite interactions, such as modulation of host behavior that facilitates interaction between vertebrate and invertebrate hosts, the effects of the host’s microbiome, and even physical interactions between Plasmodium parasites and infected tissues in the host’s body [5]. Further use of systems biology analysis to uncover the interactive response between host and parasite will undoubtedly lead to deeper understanding of malaria-related pathology and transmission and provide valuable insight toward the identification of new therapeutic targets.
Table 2:
Major findings from vertebrate and invertebrate hosts and relevant references
| Vertebrate Hosts | ||
|---|---|---|
| Major findings | References | |
| Immune response | • Disruption of oxidative stress response by Plasmodium infection identified by transcriptomics in humans and birds, and proteomics in nonhuman primates. • T-cell activation during Plasmodium infection at transcriptome level in birds and mice. • Metabolomics data from rodents suggests immune response to Plasmodium infection affects cellular metabolic processes. |
[10–15] |
| Malaria severity | • Transcriptome profiles of Plasmodium experienced or naïve individuals suggests interferon and cytokine mediated immune response differs depending on previous malaria exposure. • Differences in host expression seen at the transcriptome level between cerebral and uncomplicated malaria, but not in Plasmodium gene expression. • Differences in Plasmodium surface antigen expression between cerebral and uncomplicated malaria. |
[19, 20, 22, 25–27, 31] |
| Vertebrate life cycle stages and host specificity | • Stage-specific gene expression identified in in vitro and ex vivo Plasmodium parasites. • Immune evasion proteins expressed at all Plasmodium life cycle stages. • Significant differences between avian Plasmodium and the human Plasmodium species P. vivax and P. falciparum erythrocyte invasion pathways. |
[13, 14, 42–45, 56–58, 60, 62]. |
| Invertebrate Hosts | ||
| Major findings | References | |
| Insect-Plasmodium evolutionary arms race | • Insect oxidative stress response disrupted by oocyst development. • Significantly different insect immune response to Plasmodium compared to bacterial or viral infection found at the transcriptome and metabolome level. • Transcriptome evidence of an immune response to Plasmodium after blood meal, even when Plasmodium parasites are not present. • Significantly different immune response in phagosome activity, melanization, and complement activation during infection with drug resistant compared to non-drug resistant Plasmodium. |
[66, 67, 69, 70] |
| Transmission between insect and vertebrate hosts | • Similar expression between P. vivax and P. falciparum in proteins involved in salivary gland and hepatocyte invasion. • Ookinete interacting proteins found in lipid rafts in insect midgut cells. • Differences in red blood cell and skin bacteria metabolomes identified in Plasmodium infected vertebrates, correlating to increased insect attraction. |
[63, 87–89, 91, 92] |
Table 3:
Selected references from specific Plasmodium life cycle stages organized by omics type
| Sporozoite | Ring | Trophozoite | Merozoite and schizont (blood and liver stage) | Gametocyte (vertebrate and invertebrate) | Ookinete | Oocyst | |
|---|---|---|---|---|---|---|---|
| Transcriptomics | [45, 79, 102, 103, 105] | [22, 43, 45, 50, 85, 106] | [23, 42–45, 49, 55, 85, 106–108] | [24, 33, 42, 43, 45, 49, 55, 85, 106, 108] | [44, 45, 49, 55, 85, 107, 109] | [49, 79] | [79] |
| Proteomics | [56, 85, 87–90] | [85, 98] | [13, 14, 31, 56, 60, 85, 98] | [13, 31, 56, 60, 85, 98] | [56] | * | * |
| Metabolomics and Lipidomics | * | [110, 111] | [48, 110–112] | [48, 110, 111] | * | * | * |
To the best of our knowledge, no relevant omic studies have been performed for these life cycle stages.
Acknowledgements
This work has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [Contract No. HHSN272201200031C]. The authors thank the MaHPIC consortium for their support, and thank Mary R. Galinski, Jessica C. Kissinger, Chester J. Joyner, Regina Joice Cordy, and Jeremy D. DeBarry for their critical reviews of the manuscript.
Glossary
- Antigenic Variation
The process by which Plasmodium parasites vary expression of surface molecules in order to evade the host immune system
- Cell gliding
Movement of a microorganism on the surface of, or through, a medium without the aid of propulsive organelles such as flagella
- Cerebral malaria
A severe form of malaria characterized by encephalopathy
- Chromatin remodeling
The modification of chromatin architecture between DNA condensed states and open states to make DNA more or less accessible for transcription
- Context likelihood relatedness (CLR)
A network analysis method based on mutual information between variables
- Erythrocyte
A vertebrate red blood cell
- Erythropoiesis
The process by which red blood cells are produced
- Gametocyte
The sexual stage of the Plasmodium life cycle that occurs in vertebrate host blood and is taken up by the bite of an invertebrate host
- Hemocyte
An invertebrate blood cell
- Interferon
A class of several proteins produced by the immune system in response to the presence of pathogens
- Lipidomics
The study of the complete set of lipids that are produced by a cell or population of cells under specific circumstances
- Mass Spectrometry (MS)
An analytical technique that measures the masses of molecules in a sample
- Merozoite
The asexual stage of the Plasmodium life cycle that begins in the liver and is responsible for beginning and perpetuating blood stage infection
- Metabolomics
The study of the complete set of metabolites that are produced by a cell or population of cells under specific circumstances
- Microarray
Microscope slide with attached probes that are used to determine the levels of thousands of cDNAs from RNA transcripts at once
- Nuclear Magnetic Resonance Spectroscopy (NMR)
An analytical technique that characterizes the molecules in a sample by exploiting the magnetic properties of their atomic nuclei
- Omics
a generic term referring to genomics, transcriptomics, metabolomics, proteomics, or other systems-scale analyses of biomolecules
- Ontology
a set of concepts within a certain subject area that describe properties or relationships between them. In biology tends to refer to a set of genes, proteins, or other biomolecule that are involved in a known biological process
- Proteomics
The study of the complete set of proteins that are produced by a cell or population of cells under specific circumstances
- RNA-sequencing (RNA-seq)
A technique to determine the levels of thousands of cDNAs from RNA transcripts at once using high-throughput sequencing methods
- Sequestration
A phenomenon observed with Plasmodium falciparum parasites whereby parasites adhere to the endothelial lining of blood vessels. Considered a marker of severe malaria
- Sporozoite
The motile stage of the Plasmodium life cycle that invades insect salivary glands, is transmitted by bite to a vertebrate host, and then invades vertebrate liver cells
- Supervised learning
A task in the field of machine learning where a training dataset, with the class membership of each training data point known, is used to develop mathematical predictors to classify new input data
- Systems Biology
The comprehensive study of a biological system on a large scale rather than with a focus on only a few constituent parts. Approaches include bioinformatic analysis and network modeling of high-throughput data (see omics), and mathematical modeling of biological systems
- Transcriptomics
The study of the complete set of RNA transcripts that are produced by a cell or population of cells under specific circumstances
- Uncomplicated malaria
Malaria manifestation where symptoms are present but there is no indication of organ dysfunction
- Weighted Gene Correlated Network Analysis (WGCNA)
A network analysis method based on correlation between variables
References
- 1.Baird JK (2009) Malaria zoonoses. Travel Med Infect Dis 7 (5), 269–77. [DOI] [PubMed] [Google Scholar]
- 2.WHO (2016) Malaria World Report.
- 3.Wanjek C (2011) Systems Biology as Defined by the NIH. The NIH Catalyst 19 (6). [Google Scholar]
- 4.Gutierrez JB et al. (2015) From Within Host Dynamics to the Epidemiology of Infectious Disease: Scientific Overview and Challenges. Mathematical biosciences 270 (0 0), 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonseca LL et al. (2017) A model of Plasmodium vivax concealment based on Plasmodium cynomolgi infections in Macaca mulatta. Malar J 16 (1), 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aderem A et al. (2011) A Systems Biology Approach to Infectious Disease Research: Innovating the Pathogen-Host Research Paradigm. mBio 2 (1), e00325–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabasi AL and Oltvai ZN (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5 (2), 101–13. [DOI] [PubMed] [Google Scholar]
- 8.The Gene Ontology, C. et al. (2000) Gene Ontology: tool for the unification of biology. Nature genetics 25 (1), 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison PW et al. (2012) The evolution of gene expression and the transcriptome–phenotype relationship. Seminars in cell & developmental biology 23 (2), 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colborn JM et al. (2015) Human Gene Expression in Uncomplicated Plasmodium falciparum Malaria. J Immunol Res 2015, 162639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagishi J et al. (2014) Interactive transcriptome analysis of malaria patients and infecting Plasmodium falciparum. Genome Res 24 (9), 1433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Videvall E et al. (2015) The Avian Transcriptome Response to Malaria Infection. Molecular Biology and Evolution 32 (5), 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson DC et al. (2017) A large scale Plasmodium vivax-Saimiri boliviensis trophozoite-schizont transition proteome. PLoS ONE 12 (8), e0182561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson DC et al. (2015) Plasmodium vivax trophozoite-stage proteomes. Journal of proteomics 115, 157–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JV et al. (2008) Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. J Proteome Res 7 (9), 3948–56. [DOI] [PubMed] [Google Scholar]
- 16.Carlton JM et al. (2008) Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455 (7214), 757–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaber T et al. (2017) Metabolic regulation of inflammation. Nat Rev Rheumatol 13 (5), 267–279. [DOI] [PubMed] [Google Scholar]
- 18.Gardinassi LG et al. (2017) Metabolome-wide association study of peripheral parasitemia in Plasmodium vivax malaria. Int J Med Microbiol 307 (8), 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran TM et al. (2016) Transcriptomic evidence for modulation of host inflammatory responses during febrile Plasmodium falciparum malaria. Sci Rep 6, 31291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rojas-Pena ML et al. (2015) Transcription Profiling of Malaria-Naive and Semi-immune Colombian Volunteers in a Plasmodium vivax Sporozoite Challenge. PLoS Negl Trop Dis 9 (8), e0003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabantous S et al. (2017) Gene Expression Analysis Reveals Genes Common to Cerebral Malaria and Neurodegenerative Disorders. J Infect Dis 216 (6), 771–775. [DOI] [PubMed] [Google Scholar]
- 22.Almelli T et al. (2014) Differences in gene transcriptomic pattern of Plasmodium falciparum in children with cerebral malaria and asymptomatic carriers. PLoS One 9 (12), e114401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milner DA Jr. et al. (2012) Transcriptional Profiling of Plasmodium falciparum Parasites from Patients with Severe Malaria Identifies Distinct Low vs. High Parasitemic Clusters. PLOS ONE 7 (7), e40739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonkin-Hill GQ et al. (2018) The Plasmodium falciparum transcriptome in severe malaria reveals altered expression of genes involved in important processes including surface antigen–encoding var genes. PLOS Biology 16 (3), e2004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desruisseaux MS et al. (2010) Alterations in the Brain Transcriptome in Plasmodium Berghei ANKA Infected Mice. J Neuroparasitology 1. [PMC free article] [PubMed] [Google Scholar]
- 26.Delahaye NF et al. (2007) Gene expression analysis reveals early changes in several molecular pathways in cerebral malaria-susceptible mice versus cerebral malaria-resistant mice. BMC Genomics 8, 452–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miu J et al. (2008) Predominance of Interferon-Related Responses in the Brain during Murine Malaria, as Identified by Microarray Analysis. Infection and Immunity 76 (5), 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin JW et al. (2017) Signatures of malaria-associated pathology revealed by high-resolution whole-blood transcriptomics in a rodent model of malaria. Sci Rep 7, 41722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaecher K et al. (2005) Genome-Wide Expression Profiling in Malaria Infection Reveals Transcriptional Changes Associated with Lethal and Nonlethal Outcomes. Infect Immun 73 (9), 6091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovegrove FE et al. (2006) Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics 7, 295–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertin GI et al. (2016) Proteomic analysis of Plasmodium falciparum parasites from patients with cerebral and uncomplicated malaria. Sci Rep 6, 26773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waller KL et al. (2003) Mature parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum binds to the 30-kDa domain of protein 4.1 in malaria-infected red blood cells. Blood 102 (5), 1911–4. [DOI] [PubMed] [Google Scholar]
- 33.Vignali M et al. (2011) NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J Clin Invest 121 (3), 1119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta A et al. (2016) Host metabolic responses to Plasmodium falciparum infections evaluated by 1H NMR metabolomics. Mol Biosyst 12 (11), 3324–3332. [DOI] [PubMed] [Google Scholar]
- 35.Pappa V et al. (2015) Lipid metabolites of the phospholipase A(2) pathway and inflammatory cytokines are associated with brain volume in paediatric cerebral malaria. Malaria Journal 14, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta S et al. (2017) Extensive alterations of blood metabolites in pediatric cerebral malaria. PLoS One 12 (4), e0175686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penet MF et al. (2007) Magnetic resonance spectroscopy reveals an impaired brain metabolic profile in mice resistant to cerebral malaria infected with Plasmodium berghei ANKA. J Biol Chem 282 (19), 14505–14. [DOI] [PubMed] [Google Scholar]
- 38.Rae C et al. (2004) Brain gene expression, metabolism, and bioenergetics: interrelationships in murine models of cerebral and noncerebral malaria. Faseb j 18 (3), 499–510. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh S et al. (2012) Metabolic fingerprints of serum, brain, and liver are distinct for mice with cerebral and noncerebral malaria: a (1)H NMR spectroscopy-based metabonomic study. J Proteome Res 11 (10), 4992–5004. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh S et al. (2016) Early prediction of cerebral malaria by (1)H NMR based metabolomics. Malar J 15, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surowiec I et al. (2015) Metabolic Signature Profiling as a Diagnostic and Prognostic Tool in Pediatric Plasmodium falciparum Malaria. Open Forum Infect Dis 2 (2), ofv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben Mamoun C et al. (2001) Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol Microbiol 39 (1), 26–36. [DOI] [PubMed] [Google Scholar]
- 43.Bozdech Z et al. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1 (1), E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayward RE et al. (2000) Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol Microbiol 35 (1), 6–14. [DOI] [PubMed] [Google Scholar]
- 45.Le Roch KG et al. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301 (5639), 1503–8. [DOI] [PubMed] [Google Scholar]
- 46.Reid AJ et al. (2018) Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. eLife 7, e33105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorber K et al. (2011) RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Research 39 (9), 3820–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olszewski KL et al. (2009) Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe 5 (2), 191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashano E et al. (2016) Cluster analysis of Plasmodium RNA-seq time-course data identifies stage-specific co-regulated biological processes and regulatory elements. F1000Research 5, ISCB Comm J-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoo R et al. (2016) Integrated analysis of the Plasmodium species transcriptome. EBioMedicine 7, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu L et al. (2016) New insights into the Plasmodium vivax transcriptome using RNA-Seq. Sci Rep 6, 20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westenberger SJ et al. (2010) A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis 4 (4), e653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young JA et al. (2008) In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daily JP et al. (2007) Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450 (7172), 1091–5. [DOI] [PubMed] [Google Scholar]
- 55.Pelle KG et al. (2015) Transcriptional profiling defines dynamics of parasite tissue sequestration during malaria infection. Genome Medicine 7 (1), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florens L et al. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419 (6906), 520–6. [DOI] [PubMed] [Google Scholar]
- 57.Bozdech Z et al. (2008) The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A 105 (42), 16290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim A et al. (2017) Characterization of P. vivax blood stage transcriptomes from field isolates reveals similarities among infections and complex gene isoforms. Sci Rep 7 (1), 7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q et al. (2014) The epidemiology of Plasmodium vivax and Plasmodium falciparum malaria in China, 2004–2012: from intensified control to elimination. Malaria Journal 13 (1), 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venkatesh A et al. (2017) Identification of Highly Expressed Plasmodium Vivax Proteins from Clinical Isolates Using Proteomics. Proteomics Clin Appl doi: 10.1002/prca.201700046. [Epub ahead of print] (1700046). [DOI] [PubMed] [Google Scholar]
- 61.Lauron EJ et al. (2015) De novo assembly and transcriptome analysis of Plasmodium gallinaceum identifies the Rh5 interacting protein (ripr), and reveals a lack of EBL and RH gene family diversification. Malaria Journal 14, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Videvall E et al. (2017) The transcriptome of the avian malaria parasite Plasmodium ashfordi displays host-specific gene expression. Mol Ecol 26 (11), 2939–2958. [DOI] [PubMed] [Google Scholar]
- 63.Busula AO et al. (2017) Mechanisms of Plasmodium-Enhanced Attraction of Mosquito Vectors. Trends Parasitol 33 (12), 961–973. [DOI] [PubMed] [Google Scholar]
- 64.Cator LJ et al. (2014) Alterations in mosquito behaviour by malaria parasites: potential impact on force of infection. Malar J 13, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong Y et al. (2006) Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog 2 (6), e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shrinet J et al. (2014) Inference of the oxidative stress network in Anopheles stephensi upon Plasmodium infection. PLoS One 9 (12), e114461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baton LA et al. (2009) Genome-wide transcriptomic profiling of Anopheles gambiae hemocytes reveals pathogen-specific signatures upon bacterial challenge and Plasmodium berghei infection. BMC Genomics 10, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Upton LM et al. (2015) Anopheles gambiae blood feeding initiates an anticipatory defense response to Plasmodium berghei. J Innate Immun 7 (1), 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aguilar R et al. (2007) Continuous exposure to Plasmodium results in decreased susceptibility and transcriptomic divergence of the Anopheles gambiae immune system. BMC Genomics 8, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J et al. (2017) Differential gene expression in Anopheles stephensi following infection with drug-resistant Plasmodium yoelii. Parasit Vectors 10 (1), 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Champion CJ et al. (2017) Anopheles gambiae: Metabolomic Profiles in Sugar-Fed, Blood-Fed, and Plasmodium falciparum-Infected Midgut. Dataset Papers in Science 2017, 49. [Google Scholar]
- 72.Hoxmeier JC et al. (2015) Analysis of the metabolome of Anopheles gambiae mosquito after exposure to Mycobacterium ulcerans. Sci Rep 5, 9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Melo CF et al. (2016) A Lipidomics Approach in the Characterization of Zika-Infected Mosquito Cells: Potential Targets for Breaking the Transmission Cycle. PLoS One 11 (10), e0164377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perera R et al. (2012) Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog 8 (3), e1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lavazec C and Bourgouin C (2008) Mosquito-based transmission blocking vaccines for interrupting Plasmodium development. Microbes Infect 10 (8), 845–9. [DOI] [PubMed] [Google Scholar]
- 76.Sreenivasamurthy SK et al. (2013) A compendium of molecules involved in vector-pathogen interactions pertaining to malaria. Malar J 12, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brennan JDG et al. (2000) Anopheles gambiae salivary gland proteins as putative targets for blocking transmission of malaria parasites. Proceedings of the National Academy of Sciences of the United States of America 97 (25), 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosinski-Chupin I et al. (2007) SAGE analysis of mosquito salivary gland transcriptomes during Plasmodium invasion. Cell Microbiol 9 (3), 708–24. [DOI] [PubMed] [Google Scholar]
- 79.Raibaud A et al. (2006) Differential gene expression in the ookinete stage of the malaria parasite Plasmodium berghei. Mol Biochem Parasitol 150 (1), 107–13. [DOI] [PubMed] [Google Scholar]
- 80.Abraham EG et al. (2004) Analysis of the Plasmodium and Anopheles transcriptional repertoire during ookinete development and midgut invasion. J Biol Chem 279 (7), 5573–80. [DOI] [PubMed] [Google Scholar]
- 81.Srinivasan P et al. (2004) Analysis of the Plasmodium and Anopheles transcriptomes during oocyst differentiation. J Biol Chem 279 (7), 5581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vlachou D et al. (2005) Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol 15 (13), 1185–95. [DOI] [PubMed] [Google Scholar]
- 83.Pinheiro-Silva R et al. (2015) Gene expression changes in the salivary glands of Anopheles coluzzii elicited by Plasmodium berghei infection. Parasit Vectors 8, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dixit R et al. (2009) Salivary gland transcriptome analysis during Plasmodium infection in malaria vector Anopheles stephensi. Int J Infect Dis 13 (5), 636–46. [DOI] [PubMed] [Google Scholar]
- 85.Hall N et al. (2005) A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307 (5706), 82–6. [DOI] [PubMed] [Google Scholar]
- 86.Rosinski-Chupin I et al. (2007) Serial Analysis of Gene Expression in Plasmodium berghei salivary gland sporozoites. BMC Genomics 8, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lindner SE et al. (2013) Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol Cell Proteomics 12 (5), 1127–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Swearingen KE et al. (2017) Proteogenomic analysis of the total and surface-exposed proteomes of Plasmodium vivax salivary gland sporozoites. PLoS Neglected Tropical Diseases 11 (7), e0005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swearingen KE et al. (2016) Interrogating the Plasmodium Sporozoite Surface: Identification of Surface-Exposed Proteins and Demonstration of Glycosylation on CSP and TRAP by Mass Spectrometry-Based Proteomics. PLoS Pathog 12 (4), e1005606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lasonder E et al. (2008) Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog 4 (10), e1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parish LA et al. (2011) Ookinete-interacting proteins on the microvillar surface are partitioned into detergent resistant membranes of Anopheles gambiae midguts. J Proteome Res 10 (11), 5150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mathias DK et al. (2014) Differential roles of an Anopheline midgut GPI-anchored protein in mediating Plasmodium falciparum and Plasmodium vivax ookinete invasion. Infect Genet Evol 28, 635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun YV and Hu YJ (2016) Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv Genet 93, 147–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hasin Y et al. (2017) Multi-omics approaches to disease. Genome Biol 18 (1), 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rau A et al. (2014) Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinformatics 15, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fonseca LL et al. (2016) Quantifying the removal of red blood cells in Macaca mulatta during a Plasmodium coatneyi infection. Malar J 15 (1), 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang Y et al. (2017) Metabolic modeling helps interpret transcriptomic changes during malaria. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease pii:S0925-4439 (17), 30387–3. [DOI] [PMC free article] [PubMed]
- 98.Acharya P et al. (2009) A glimpse into the clinical proteome of human malaria parasites Plasmodium falciparum and Plasmodium vivax. Proteomics Clin Appl 3 (11), 1314–25. [DOI] [PubMed] [Google Scholar]
- 99.Sengupta A et al. (2011) Liver Metabolic Alterations and Changes in Host Intercompartmental Metabolic Correlation during Progression of Malaria. J Parasitol Res 2011, 901854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang Y et al. (2017) Integrative analysis associates monocytes with insufficient erythropoiesis during acute Plasmodium cynomolgi malaria in rhesus macaques. Malaria Journal 16, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ylostalo J et al. (2005) Transcriptome profiles of host gene expression in a monkey model of human malaria. J Infect Dis 191 (3), 400–9. [DOI] [PubMed] [Google Scholar]
- 102.Mikolajczak SA et al. (2008) Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol Cell Biol 28 (20), 6196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kappe SH et al. (2001) Exploring the transcriptome of the malaria sporozoite stage. Proc Natl Acad Sci U S A 98 (17), 9895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu X et al. (2005) Transcriptome analysis of Anopheles stephensi-Plasmodium berghei interactions. Mol Biochem Parasitol 142 (1), 76–87. [DOI] [PubMed] [Google Scholar]
- 105.Daily JP et al. (2004) In vivo transcriptional profiling of Plasmodium falciparum. Malar J 3, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Llinas M et al. (2006) Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34 (4), 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Otto TD et al. (2014) A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biology 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Broadbent KM et al. (2011) A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12 (6), R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Young JA et al. (2005) The Plasmodium falciparum sexual development transcriptome: A microarray analysis using ontology-based pattern identification. Molecular and Biochemical Parasitology 143 (1), 67–79. [DOI] [PubMed] [Google Scholar]
- 110.Park YH et al. (2015) High-resolution metabolomics to discover potential parasite-specific biomarkers in a Plasmodium falciparum erythrocytic stage culture system. Malaria Journal 14 (1), 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vo Duy S et al. (2012) A quantitative liquid chromatography tandem mass spectrometry method for metabolomic analysis of Plasmodium falciparum lipid related metabolites. Analytica Chimica Acta 739, 47–55. [DOI] [PubMed] [Google Scholar]
- 112.Teng R et al. (2009) Metabolite profiling of the intraerythrocytic malaria parasite Plasmodium falciparum by (1)H NMR spectroscopy. NMR Biomed 22 (3), 292–302. [DOI] [PubMed] [Google Scholar]