SUMMARY
Oxidized phospholipids (OxPLs), which arise due to oxidative stress, are pro-inflammatory and pro-atherogenic, but their roles in non-alcoholic steatohepatitis (NASH) are unknown. Here, we show that OxPLs accumulate in human and mouse NASH. Using a transgenic mouse that expresses a functional single chain variable fragment of E06, a natural antibody that neutralizes OxPLs, we demonstrate the casual role of OxPLs in NASH. Targeting OxPLs in hyperlipidemic Ldlr−/− mice improved multiple aspects of NASH, including steatosis, inflammation, fibrosis, hepatocyte death and progression to hepatocellular carcinoma. Mechanistically, we found that OxPLs promote ROS accumulation to induce mitochondrial dysfunction in hepatocytes. Neutralizing OxPLs in AMLN diet-fed Ldlr−/− mice reduced oxidative stress, improved hepatic and adipose tissue mitochondrial function and fatty acid oxidation. These results suggest targeting OxPLs may be an effective therapeutic strategy for NASH.
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is a spectrum ranging from fatty liver (NAFL) to non-alcoholic steatohepatitis (NASH) to cirrhosis. In the US alone, the number of NAFLD cases is expected to grow from 83 million in 2015 to 101 million in 2030, with about 27% of cases meeting the criteria for NASH (Estes et al., 2018; Friedman et al., 2018). The rising disease prevalence is accompanied by an increased number of individuals with both cirrhosis and end-stage liver disease, needing liver transplantation (Wong et al., 2015). Currently, the accurate diagnosis of NASH still requires a liver biopsy that is invasive, costly and associated with potential complications. Further, there is no effective treatment to reverse NASH (Younossi et al., 2018). Therefore, identifying the key events that underlie and control NASH progression is of utmost importance in clinical research.
NASH is defined as hepatic steatosis with inflammation and hepatocyte death (Antunes and Bhimji, 2018). Typically, NASH is accompanied by hepatocellular ballooning, varying degrees of pericellular fibrosis and elevated serum transaminase levels (Friedman et al., 2018; Hernandez-Gea and Friedman, 2011). Left untreated, NASH may progress to cirrhosis and hepatocellular carcinoma (HCC). While hepatic steatosis predisposes the liver to NASH, other factors, such as genetics, oxidative stress, inflammation, mitochondrial dysfunction, gut dysbiosis and endotoxin, all contribute in varying degrees to the severity of NASH and its consequences (Barbara et al., 2018; Kim et al., 2017; Rong et al., 2017; Tiniakos et al., 2010; Wang et al., 2016; Yu et al., 2016). Lipotoxic factors such as oxidized lipids, generated by lipid peroxidation, are ubiquitous and found in many inflammatory settings, including atherosclerosis (Lee et al., 2012; Navab et al., 2004; Senders et al., 2018), infectious and chemical pulmonary diseases (Baldan et al., 2014; Imai et al., 2008) and NAFLD (Fujii et al., 2009; Ikura et al., 2006). Phosphocholine (PC)-containing phospholipids are major components of LDL, as well as cell membranes and lipid droplets. They usually contain polyunsaturated fatty acids at the sn-2 position, which are highly susceptible to free radical-induced lipid peroxidation, resulting in the exposure of hydrophilic PC headgroups and formation of highly reactive PC-containing oxidized phospholipids (OxPLs), which present as OxPLs or OxPL protein adducts (Binder et al., 2002; Binder et al., 2016). OxPLs are known to accumulate not only in OxLDL but also in apoptotic cells and microparticles released by activated and dying cells (Binder et al., 2016). OxPLs exert variable effects on cells by modulating activity of intracellular signal transduction and gene expression, inducing cellular stress and apoptosis (Li et al., 2007; Rouhanizadeh et al., 2005; Van Lenten et al., 2001). However, because OxPLs are primarily products of non-enzymatic lipid peroxidation, methods to neutralize them have not been available. Therefore, their actual pathologic roles in vivo are largely unknown.
We previously cloned the IgM natural antibody E06, which binds the PC headgroup of OxPLs, but does not bind unoxidized PLs. E06 blocks the uptake of OxLDL by macrophages and inhibits proinflammatory properties of OxPLs (Friedman et al., 2002; Shaw et al., 2000). To determine the role of OxPLs in vivo in atherosclerosis, we recently generated transgenic mice that expressed a single-chain variable fragment of E06 (E06-scFv) (Que et al., 2018). Because the E06-scFv lacks the Fc effector functions of antibodies, biological effects observed are predicted to be due solely to blocking biological effects of OxPLs. The E06-scFv is driven by the Apoe promoter and secreted from the liver and macrophages and present in plasma at a concentration of 20–30 ug/ml. E06-scFv binds to OxLDL and OxPL epitopes and inhibits both inflammation and atherosclerosis (Que et al., 2018). Shiri-Sverdlov and colleagues reported an inverse correlation of IgM to OxPLs in individuals with NASH (Hendrikx et al., 2016). However, it is unknown if OxPLs might be pathogenically involved in NASH and its complications, such as fibrosis and liver damage. Therefore, we sought to take advantage of our E06-scFv transgenic mice to determine the roles of OxPLs in NASH.
There are a variety of mouse models that have been used to study NASH, each of which displays one or more features of human NASH and consequences such as fibrosis, cirrhosis and development of HCC (Friedman et al., 2018; Tsuchida et al., 2018). We generated NASH models in the Ldlr−/− mouse fed the AMLN diet (40 kcal% Fat, 20 kcal% Fructose and 2% Cholesterol), a diet widely used as a preclinical model for identifying pharmacological interventions with greater likelihood of translating to the clinic (Clapper et al., 2013; Friedman et al., 2018). Ldlr−/− mice fed the AMLN diet develop both histologic and metabolic features of human NASH (Clapper et al., 2013). By combining the AMLN diet with the Ldlr−/− background, we intend to build a model with hypercholesterolemia, inflammation, oxidant stress, increased fatty acid production and fibrosis to study the roles of OxPLs in NASH (Friedman et al., 2018).
Here we report that OxPLs accumulate in the liver and serum of diverse mouse NASH models and human subjects. Targeting OxPLs by E06-scFv ameliorated multiple aspects of NASH, including steatosis, inflammation, fibrosis, cell death and its progression to HCC. Further, in vivo studies indicated that targeting OxPLs improved AMLN diet-induced mitochondrial dysfunction, which reduced hepatic lipid accumulation and attenuated obesity. OxPLs induced mitochondrial damage and ROS accumulation, in part through covalent modification of manganese superoxide dismutase (MnSOD/SOD2), inactivating the enzyme. Neutralization of OxPLs decreased hepatic inflammation as indicated by decreased numbers of recruited macrophages, decreased inflammatory gene expression and reduced serum cytokine levels. Taken together, these findings reveal a causal role of OxPLs in the pathogenesis of NASH and that targeting OxPLs may be an effective therapeutic strategy to ameliorate NASH.
RESULTS
OxPLs accumulate in liver and serum of human and mouse NASH
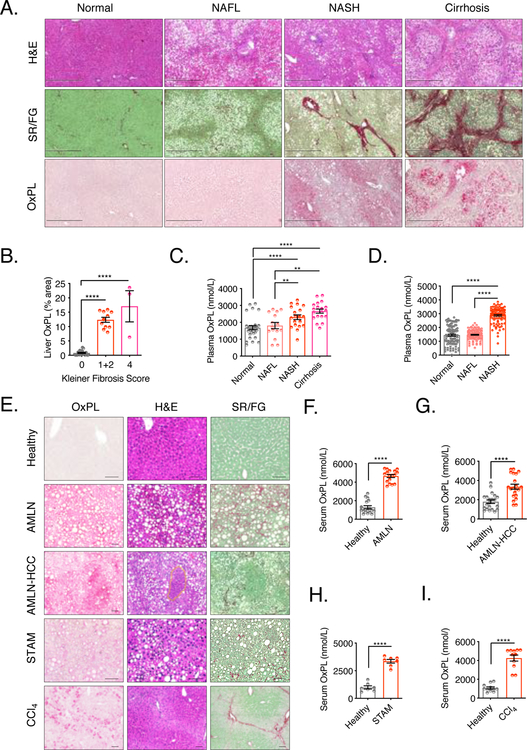
To determine if OxPLs accumulate during the pathogenesis of human NASH, we utilized E06 to stain OxPLs in liver sections from clinical samples, which were diagnosed blindly by a pathologist into the following categories: normal, steatosis (NAFL), and NASH-induced liver fibrosis stages 1, 2, and 4. H&E and Sirius Red/Fast Green (SR/FG) staining were conducted to indicate the extent of hepatic steatosis and fibrosis, respectively. OxPL content was higher in liver sections from subjects with NASH and NASH-associated cirrhosis compared with subjects with normal and NAFL tissue (Fig. 1A). Of note, OxPLs distributed in locales surrounding the extensive fibrosis. The quantified liver OxPL staining area showed a positive correlation to fibrosis scores (Fig. 1B). Moreover, we analyzed plasma of subjects in whom liver status had been previously determined by liver biopsy (Gorden et al., 2015). Plasma OxPLs were measured by a newly developed competitive ELISA. Elevated plasma total OxPL levels were observed in both NASH and cirrhosis subjects, and both groups were distinguished from normal or NAFL groups (Fig. 1C). In a third study, we analyzed plasma of individuals from an outpatient clinic who were diagnosed as normal, NAFL or NASH. In this cohort, plasma total OxPL levels were significantly elevated in NASH subjects compared to normal or NAFL subjects (Fig. 1D). These preliminary data suggest that OxPLs accumulate in both liver and plasma of human subjects with NASH.
Figure 1. OxPLs accumulate in liver and serum of human and mouse models of NASH.
(A) Human liver sections were classified blindly by a liver pathologist and determined levels of OxPL accumulation, histology and collagen fiber deposition respectively in different stages of liver disease. Normal (no steatosis, Kleiner fibrosis score 0), NAFL (steatosis, Kleiner fibrosis score 0), NASH (steatosis, Kleiner fibrosis score 1–2), cirrhosis (steatosis, Kleiner fibrosis score 4 and plural pseudo-lobules). n = 3–11. Scale bar = 500μm.
(B) Quantification of liver OxPL staining in (A), plotted against Kleiner fibrosis scores.
(C) Plasma OxPL levels were determined in 82 subjects previously characterized for extent of liver disease by liver biopsy. n = 15–29.
(D) Plasma OxPL levels were determined in 322 outpatient subjects diagnosed as Normal (no steatosis by ultrasound with normal liver ALT and AST levels), NAFL (steatosis by ultrasound with normal liver ALT and AST levels) or NASH (steatosis and both elevated ALT and AST). n = 100–118.
(E) Healthy mouse model: 20 weeks old Ldlr−/− mice on chow diet; AMLN Model: Ldlr−/− mice were fed AMLN diet for 30 weeks; AMLN-HCC Model: Ldlr−/− mice were fed AMLN diet for 48 weeks; STAM Model: male Ldlr−/− mice were subcutaneously injected with 200μg streptozotocin (STZ) or vehicle within 48 hours after birth and fed with HFD for 4 weeks stating at 4 weeks of age; CCl4 Model: Ldlr−/− mice were injected intraperitoneally with CCl4 (0.5ml/kg body weight, 1:5 diluted in corn oil) or vehicle (corn oil) twice a week for 4 weeks. Paraffin-embedded mouse liver sections were stained with biotinylated-E06 IgM, H&E and SR/FG to determine OxPL deposition, histology and collagen fiber deposition respectively. n = 6. Scale bar = 50μm.
(F) OxPL in serum of healthy (littermate control on chow diet) and AMLN-NASH mice. n = 17.
(G) Serum OxPL in healthy (littermate control on chow diet) and AMLN-HCC mice. n = 17–21.
(H) Serum OxPL in healthy (littermate control injected with vehicle on chow diet) and STAM-NASH mice. n = 7–17.
(I) Serum OxPL in healthy (littermate control injected with vehicle) and CCl4-liver fibrosis mice. n = 11–17.
Data are mean ± SEM. **, P < 0.01; ****, P < 0.0001. See also Figure S1.
To determine if OxPLs accumulate in mouse NASH models, we examined OxPL levels in healthy mice and different NASH/liver damage models (the timeframe for the NASH/liver damage models are shown in Fig. S1, A–D). We stained serial hepatic sections with E06, H&E and SR/FG to indicate the extent of hepatic OxPL accumulation, steatosis, and fibrosis, respectively, in the same liver areas. Serum total OxPLs were also measured by the competitive ELISA. Ldlr−/− mice fed the AMLN diet for 30 weeks developed human NASH features, including steatosis, inflammation and peri-cellular fibrosis. Of note, there was marked accumulation of OxPLs in the liver (Fig. 1E–AMLN). Total-OxPLs were about 3-fold higher in the blood of AMLN-fed mice compared to chow-fed mice (Fig. 1F). Moreover, these mice developed HCC with prolonged feeding of AMLN diet (Fig. 1E–AMLN-HCC). After 48 weeks of feeding, there was marked accumulation of OxPLs in the liver, especially at the tumor sites shown by H&E staining and enhanced OxPL levels in serum (Fig. 1E–AMLN-HCC, Fig. 1G). Similarly, high-fat diet (HFD)-fed streptozotocin-treated Ldlr−/− mice (STAM) developed NASH with mild fibrosis after only 4 weeks of feeding, and these mice also exhibited substantially higher level of liver and blood OxPLs compared to chow-fed mice (Fig. 1E–STAM, H). Intraperitoneal injection of CCl4 enhances free radical formation and rapidly induces hepatic damage (Muriel, 2017; Shrestha et al., 2016). In this model of repeated CCl4 administration over 4 weeks, there was OxPL accumulation in liver in areas coinciding with damaged hepatocytes and adjacent to the collagen fibers as shown by both H&E and SR/FG staining and serum OxPL levels were also ~ 3-fold higher than mice without CCl4 treatment (Fig. 1E– CCl4, I).
Neutralization of OxPLs protects against NASH in multiple models
We next sought to determine if the increased serum and liver OxPLs promoted the pathogenesis of NASH. We used our recently developed transgenic mice that express E06-scFv to neutralize OxPLs in vivo (Que et al., 2018). Immunohistological staining with IgM E06 of livers from AMLN-fed E06-scFvLdlr−/− mice showed less content of immunological recognized OxPLs compared to Ldlr−/− mice (Fig. 2A–OxPL). (A caveat is that hepatocytes are a major source of the E06-scFv). E06-scFvLdlr−/− and Ldlr−/− mice have similar levels of serum cholesterol (~1300mg/dL) and triglyceride (~400mg/dL) (Fig. S2A and B). There was a substantial reduction in steatosis, which was confirmed by a significant reduction in liver weight, and by ~50% reduction in hepatic triglyceride and ~45% reduction in cholesterol content (Fig. 2A–D). E06-scFv substantially ameliorated hepatic inflammation as evidenced by decreased macrophage F4/80 staining (Fig. 2A). Of particular note, there was ~ 70% reduction of liver fibrosis as measured by SR/FG staining and hydroxyproline content (Fig. 2A, 2E and S2C). Moreover, decreased TUNEL staining observed in liver of E06-scFvLdlr−/− mice supported reduced hepatocyte death, which is reflected in lower serum ALT, AST and ALP levels (Fig. 2A, F–H). The histological features were blindly assessed by an experienced pathologist according to the Kleiner Scoring System (Kleiner et al., 2005). Steatosis, inflammation, hepatocellular injury and fibrosis were all substantially less in AMLN-fed E06-scFvLdlr−/− mice compared to Ldlr−/− mice (Table S1). Neutralization of OxPLs also reduced the progression of NASH to HCC, as evidenced by a marked decrease of big tumor (T) incidence (> 4 mm), tumor number and volume in the E06-scFvLdlr−/− mice fed AMLN diet for 48 weeks (Fig. 2I–K).
Figure 2. Neutralization of OxPL restrains AMLN diet-induced hepatic steatosis, inflammation and fibrosis, increases energy expenditure and attenuates AMLN diet-induced obesity.
(A) Levels of OxPLs; neutral lipid deposition, histology, macrophage accumulation, collagen fiber deposition and apoptosis of liver sections from Ldlr−/− and E06-scFvLdlr−/− mice fed AMLN diet for 30 weeks. n = 6. Scale bar = 100μm. (for TUNEL staining, scale bar = 20μm).
(B) Liver weight of indicated mice fed AMLN diet for 30 weeks. n = 10.
(C-E) Content of liver triglyceride (C, TG), total cholesterol (D, TC) and hydroxyproline (E) of indicated mice fed AMLN diet for 30 weeks. n = 5–8.
(F-H) Serum Alanine Aminotransferase (F, ALT), Aspartate Aminotransferase (G, AST) and Alkaline Phosphatase (H, ALP) of indicated mice fed AMLN diet for 30 weeks. n = 8–9.
(I) Representative gross liver morphology and big tumor (big T, >0.4cm) incidence in indicated mice after 48 weeks of AMLN diet. n = 14–16. Scale bar = 1cm.
(J, K) Tumor numbers (J) and volumes (K) of same mice as in (I).
(L) Body weights of indicated mice at baseline (8weeks old) and after 30 weeks of indicated diet feeding. n = 6–10 per group.
(M) Photo of representative indicated mice after 30 weeks of AMLN diet and H&E staining of respective IWAT. Scale bar = 100μm.
(N) Percentage of fat mass of mice as determined by DEXA imaging. n = 4–6.
(O-Q) IWAT (Q), EWAT (R) or BAT (S) mass in indicated mice fed AMLN diet for 30 weeks. n = 6–10.
(R, S) Oxygen consumption rate over time (R, VO2) and ANCOVA analyzed VO2 statistics (S, normalized to body weight) of respective mice of indicated groups fed AMLN diet for 30 weeks. n = 4–5.
(T) ANCOVA analyzed energy expenditure (normalized to body weight) of mice in experiment shown in panel R and S.
Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. See also Figure S2.
Importantly, the protective effects on E06-scFv were validated in AMLN-fed C57BL/6 and E06-scFvC57BL/6 mice that had normal low LDL cholesterol levels (Fig. S2D and E). In a similar intervention study in STAM mice, expression of E06-scFv improved both hepatic steatosis and fibrosis (Fig. S2F). These observations indicate that efficient neutralization of OxPLs attenuates NASH in multiple mouse models.
Neutralization of OxPLs increases energy expenditure and attenuates diet-induced obesity
Neutralizing OxPLs had no effect on body weight in chow-fed mice. However, when fed the AMLN diet, E06-scFvLdlr−/− mice gained less body weight compared to Ldlr−/− mice (Fig. 2L, M). Adipocyte size was smaller in AMLN diet-fed E06-scFvLdlr−/− mice (Fig. 2M, right), suggesting that OxPL neutralization attenuates AMLN diet-induced adipocyte hypertrophy. Dual energy X-ray absorptiometry (DEXA) scanning revealed that E06-scFvLdlr−/− mice had significantly less fat mass, which was confirmed by decreased weight of inguinal White Adipose Tissue (IWAT), epididymal White Adipose Tissue (EWAT), and Brown Adipose Tissue (BAT) in E06-scFvLdlr−/− mice (Fig. 2N–Q, S2G). ANCOVA analysis with total body weight as covariant, or lean body mass as covariant, both indicated higher oxygen consumption rate and energy expenditure in AMLN-fed E06-scFvLdlr−/− mice compared to Ldlr−/− mice (Fig. 2R–T, S2H–I). Note that no difference in oxygen consumption rate was observed when the mice were on chow diet (Fig. S2J). Neutralization of OxPLs did not affect respiratory exchange rate (RER), physical activity or food intake (Fig. S2K–M). Neutralization of OxPLs also did not affect serum non-esterified fatty acid (NEFA) or glycerol concentrations, nor did it alter glucose homeostasis, as indicated by similar fasting glucose and insulin levels and similar glucose tolerance and insulin sensitivity (Fig. S2N–S). These data suggest that insulin sensitivity was not affected by E06-scFv under an AMLN dietary regimen.
Neutralization of OxPLs protects mitochondria and promotes mitochondrial biogenesis
To assess putative mechanisms by which targeting OxPLs restrains NASH, we performed RNA-seq of livers from AMLN-fed Ldlr−/− and E06-scFvLdlr−/− mice. 1628 genes were significantly up-regulated (fold change>1.5, P-adj < 0.05) in E06-scFvLdlr−/− livers (Fig. 3A, blue dots). Gene ontology analysis revealed that top enriched pathways in E06-upregulated genes were related to mitochondrial functions, such as oxidative phosphorylation, respiratory chain complex assembly, fatty acid metabolism, and fatty acid transport (Fig. 3B). The relative fold increase of 72 genes in the E06-scFvLdlr−/− mice most closely related to mitochondrial function (Mito function) is depicted relative to overall liver mRNA fold change (Fig. 3A, red dots). These 72 genes include 63 oxidative phosphorylation genes, 6 mitochondrial assembly genes, and 4 fatty acid transportation genes (Fig. 3C). Note that these genes were not regulated by E06 in chow diet-fed mice (Fig. S3). Collectively, these findings indicate that under AMLN diet, OxPL significantly down-regulates genes regulating mitochondrial function, which E06-scFv effectively counteracts.
Figure 3. Neutralization of OxPL promotes mitochondrial biogenesis at the transcriptional level.
(A) Comparison of RNA-seq for poly A transcripts in livers of Ldlr−/− (L) and E06-scFvLdlr−/− (EL) mice on AMLN diet for 30 weeks. Mean log2 (TPM + 1) values (y axis) are plotted versus log2 Fold Change (x axis) of the transcripts of livers between L and EL mice (TPM, transcripts per kilobase million). All transcripts with mean TPM greater than 4 are in grey. Transcripts upregulated in EL mice (EL > L, > 1.5-fold change, P-adj < 0.05) are in blue. Genes highlighted in red are the 72 upregulated genes most closely related to mitochondrial functions.
(B) Functional annotations associated with genes expressed more highly in EL mice (> 1.5-fold change, P-adj < 0.05).
(C) Relative expression values (Z-scaled log2 (TPM + 1)) for the 72 genes highlighted in red in (A) are illustrated, including 62 oxidative phosphorylation genes, 6 mitochondrial assembly machinery genes and 4 fatty acid transportation genes.
See also Figure S3.
Oxidative stress plays an important role in NASH (Mansouri et al., 2018). The balance between oxidant and antioxidant agents controls redox state. ROS generation during chronic hepatic steatosis and inflammation leads to oxidative damage to mitochondrial proteins, membranes and impaired oxidative phosphorylation (Ucar et al., 2013). In this study, we demonstrated that OxPLs directly induced ROS accumulation. We treated Ldlr−/− and E06-scFvLdlr−/−primary hepatocytes with oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (OxPAPC, a mixture of OxPLs). OxPAPC significantly increased total ROS (tROS) accumulation in Ldlr−/− hepatocytes, but not in E06-scFvLdlr−/− hepatocytes (Fig. 4A), which secrete E06-scFv into the medium (after 12 hours, about 1/6 the concentration found in serum of E06-scFv mice) (Fig. S4A). Additionally, OxPAPC diminished mitochondrial membrane potential (Fig. 4B) and stimulated mitochondrial ROS (mtROS) accumulation (Fig. 4C) in Ldlr−/− but not in E06-scFv Ldlr−/− hepatocytes, indicating sustained mitochondrial damage induced by OxPLs. As a result, we found that Ldlr−/− hepatocytes exhibited significantly compromised fatty acid oxidation in response to OxPAPC, which was preserved in E06-scFvLdlr−/− hepatocytes (Fig. 4D). To confirm that OxPL-induced mitochondrial damage can be attributed in part to ROS, we pretreated hepatocytes with the ROS scavenger MnTBAP, a cell-permeable MnSOD/SOD2 mimetic. MnTBAP pretreatment restored OxPL-diminished mitochondrial membrane potential (Fig. 4E). MnTBAP abrogated both mitochondrial and total ROS accumulation in hepatocytes exposed to OxPAPC (Fig. 4F, Fig. S4B). These results indicate that OxPLs induce hepatocyte mitochondrial dysfunction and damage by increasing ROS accumulation.
Figure 4. Neutralization of OxPL protects mitochondria.
(A-D) Primary hepatocytes from Ldlr−/− and E06-scFvLdlr−/− mice were pretreated with Vehicle or OxPAPC (100μg/mL) for 12 hours. Total ROS (A, tROS), mitochondrial membrane potential (B, Ψm preservation), mitochondrial ROS (C, mtROS) and fatty acid oxidation (D) were measured. n = 3–5.
(E, F) Primary hepatocytes of Ldlr−/− mice were pretreated with Vehicle or 200μM of MnTBAP for 1 hour, then with OxPAPC (100μg/mL) for 4 hours. Mitochondrial membrane potential (E) and mitochondrial ROS (F) were measured. n = 4–5.
(G) Primary hepatocytes of Ldlr−/− mice treated with Vehicle, OxPAPC (20μg/mL) or OxPAPC (1-hour pre-incubation with 50μg/mL E06 IgM) for 1 hour were stained with E06 (green), Mitotracker (red) and DAPI (blue). Scale bar = 20μm. n = 3.
(H) Primary hepatocytes of Ldlr−/− mice were treated with Vehicle or OxPAPC (100μg/mL) for 1 hour. SOD activity in the cell lysate was measured. n = 3.
(I) ELISA detection of E06 IgM binding to recombinant native MnSOD or POVPC modified MnSOD. n = 3.
(J) MnSOD activity was measured in vitro for recombinant native MnSOD and POVPC modified MnSOD. n = 3.
(K) Western blot with E06 of liver MnSOD immunoprecipitated from livers of chow or AMLN diet fed Ldlr−/− and E06-scFvLdlr−/− mice.
(L) SOD activity measured in liver homogenates of chow or AMLN diet fed Ldlr−/− and E06-scFvLdlr−/− mice. n = 3–4.
(M) Thiobarbituric acid reactive substances (TBARS) in the plasma of 30 weeks AMLN diet fed mice. n = 3.
(N) Transmission electron micrographs of hepatic mitochondria of fresh liver tissue from indicated mice on AMLN diet. Asterisks indicate lipid droplets. Arrows indicates ballooned or rounded mitochondrial cristae. n = 3. Scale bar = 1μm. (Scale bar=0.3μm for zoomed images)
(O) NAD/NADH ratio in the livers of AMLN diet fed mice. n = 3.
(P) SIRT1 activity in the fresh liver tissue from indicated mice were measured. n = 4–5.
(Q) Normalized distribution of PGC1α ChIP-seq tag density, at promoters and enhancers within 3kb of the transcription start site of E06 up-regulated mitochondrial genes (Fig. 3C) in Ldlr−/− (L) and E06-scFvLdlr−/− (EL) mice on AMLN diet.
(R) UCSC genome browser images illustrating normalized tag counts for PGC1α at the indicated mitochondrial genes in same groups of mice described in (Q). The tick marks indicate peaks up-regulated (> 2-fold, P-adj < 0.05) in E06-scFvLdlr−/− mice determined by DESeq2 using duplicate experiments.
(S) Mitotracker staining of livers from indicated mice. Circles indicate lipid droplets. Scale bar=20μm.
(T) Fatty acid oxidation in the livers of indicated mice fed AMLN diet. n = 3.
(U) Cold tolerance test in 30 weeks AMLN diet-fed mice. Shown is the body temperature of indicated mice at indicated times after initiation of cold exposure (4 °C). n=6.
Data are mean ± SEM. * P < 0.05; ** P < 0.01; ***P < 0.001, ****P < 0.0001. See also Figure S4.
Given that MnTBAP abolished OxPAPC-induced mtROS accumulation, we asked if one mechanism by which OxPAPC might induce mtROS accumulation is by binding to MnSOD, thereby decreasing its antioxidant activity. To test this hypothesis, we acutely treated primary hepatocytes with OxPAPC for one hour, which significantly increased intracellular and mitochondrial OxPAPC levels, as detected by E06, whereas pre-incubation of OxPAPC with E06 IgM antibody significantly reduced the amount of OxPAPC entering the cells (Fig. 4G). The specificity of E06 neutralizing OxPL epitopes was further validated. Incubation of Dil-OxLDL with hepatocytes led to substantial uptake, whereas preincubation in the presence of E06 reduced its uptake (Fig. S4C, D). In contrast, E06 did not affect fatty acid (BODIPY-fatty acids) uptake or that of native LDL (Fig. S4E, F, G). These data indicate that E06 specifically blocks OxPL epitopes from entering hepatocytes. Further, OxPAPC substantially reduced SOD activity within 1 hour (Fig. 4H), indicating regulation of SOD by OxPLs. Reactive OxPLs, such as 1-palmitoyl-2-(5’-oxo-valeroyl)-sn-glycero-3-phosphocholine (POVPC), can form covalent adducts with proteins, which are recognized by E06 (Friedman et al., 2002). To test whether OxPLs can affect MnSOD activity through direct modification of MnSOD, we incubated recombinant MnSOD with POVPC to generate POVPC adducts with MnSOD. Compared to native MnSOD, POVPC-modified MnSOD was strongly bound by E06 in an ELISA format (Fig 4I). Moreover, POVPC-modified MnSOD had significantly decreased activity (Fig. 4J). To determine the relevance of these observations to livers from mice with NASH, we immunoprecipitated MnSOD from liver lysates of chow-fed and AMLN diet-fed Ldlr−/− and E06-scFvLdlr−/− mice, and performed western blots with E06. OxPL-modified MnSOD was not detectable in livers of chow-fed mice, but it was readily observed in AMLN-diet fed Ldlr−/− mice, and this modification was abrogated by E06-scFv (Fig. 4K). Measurement of SOD activity in liver homogenates indicated that SOD activity was substantially reduced in the liver of AMLN diet-fed Ldlr−/− mice compared to E06-scFvLdlr−/−mice, but not in chow-fed mice (Fig. 4L), even though expression of Sod2 was not altered (data not shown). In the STAM-NASH model, SOD activity in livers of E06-scFvLdlr−/− mice was also significantly higher compared to that in Ldlr−/− mice (Fig. S4H). Taken together, these data suggest that OxPLs can directly modify and inhibit MnSOD activity to promote accumulation of mitochondrial ROS. To confirm the redox status in vivo, we measured the amount of plasma malondialdehyde (MDA) epitopes (products of lipid peroxidation) using the thiobarbituric acid-reactive substances (TBARS) assay. Plasma MDA levels were significantly lower in E06-scFvLdlr−/− mice than in Ldlr−/− mice, indicative of reduced generalized lipid peroxidation and oxidative stress (Fig. 4M). Using electron microcopy (EM), we directly assessed liver mitochondrial morphology in Ldlr−/− and E06-scFvLdlr−/− mice fed AMLN diet. Mitochondria of Ldlr−/− mice exhibited disrupted outer membranes and ballooned or rounded cristae (arrows) compared with mitochondria in E06-scFvLdlr−/− mice (Fig. 4N), indicative of protection of mitochondrial damage by E06-scFv. Note also there were less lipid droplets (asterisks) in the liver of E06-scFvLdlr−/− mice compared to Ldlr−/− mice.
We next explored how prevention of mitochondrial damage leads to the increased expression of mitochondrial genes (Fig. 3). As improvement of mitochondrial function would be predicted to increase respiration (Wu et al., 1999), we examined NAD/NADH ratio in AMLN diet-fed mice, and demonstrated that the NAD/NADH ratio was significantly higher in livers from E06-scFvLdlr−/− mice (Fig. 4O). This was accompanied by increased activity of SIRT1, an NAD-dependent protein deacetylase, in E06-scFvLdlr−/− mice (Fig. 4P). SIRT1 deacetylates peroxisome proliferator initiated receptor gamma and coactivator 1 alpha (PGC1α), a central regulator promoting mitochondria biogenesis, to enhance its activity, thus promoting mitochondrial biogenesis (Canto et al., 2009; Katsyuba et al., 2018). To assess whether PGC1α exhibits enhanced activity to induce mitochondrial biogenesis, we performed ChIP-seq to analyze the local regulatory landscapes of mitochondrial genes that were more highly expressed in E06-scFvLdlr−/− livers. The analysis demonstrated that PGC1α recruitment onto promoters and enhancers of these E06-upregulated genes was significantly induced in E06-scFvLdlr−/− livers, exemplified by Ndufs8, Uqcr10, Cox6b1 and Atp5d, spanning mitochondrial respiratory chain complexes (Fig. 4Q–R, the tick marks show E06-upregulated peaks). Consistent with enhanced expression of mitochondrial genes (Fig. 3C), an increase of mitochondrial number in the liver of AMLN diet-fed E06-scFvLdlr−/− mice was confirmed by Mitotracker staining and mitochondrial DNA (mtDNA) copy number determined by mtDNA markers (D-loop and non-NUMT) to nuclear DNA markers (Tert and B2m) (Fig. 4S, S4I–J) (Medeiros, 2008). Moreover, ex vivo experiments demonstrated that fatty acid oxidation in E06-scFvLdlr−/− livers was significantly higher (Fig. 4T). Consequently, improvement of mitochondrial function and increased mitochondrial biogenesis contributed to reduced lipid accumulation in the livers of E06-scFvLdlr−/− mice compared to Ldlr−/− mice (Fig. 4S, circles; Fig. 2A–D; Table S1). Consistent with improved mitochondrial function and enhanced PGC1α activity, which plays an important role in defending against oxidative stress (St-Pierre et al., 2006), we observed higher expression of antioxidant enzymes in E06-scFvLdlr−/− livers (Fig. S4K). Taken together, our data suggest that under AMLN diet, E06-scFv reduced OxPLs mediated oxidative stress and mitochondrial damage, and led to an increased NAD/NADH ratio, which in turn activated SIRT1/PGC1α pathway to up-regulate mitochondrial biogenesis in a feedforward axis.
Since we observed reduced fat mass and increased energy expenditure in E06-scFvLdlr−/− mice on AMLN diet, we also measured mitochondrial oxidative phosphorylation in IWAT. Both fatty acid oxidation and the NAD/NADH ratio were significantly increased in IWAT of E06-scFvLdlr−/− mice (Fig. S4L–M). This was accompanied by increased SIRT1 activity in adipose tissue of E06-scFvLdlr−/− mice (Fig. S4N). Staining of IWAT with Mitotracker revealed a substantial increase of mitochondria in adipocytes of E06-scFvLdlr−/− mice (Fig. S4O).
The apparent increase in mitochondrial function in both hepatocytes and adipose tissue under AMLN diet suggested the possibility that E06-scFvLdlr−/− mice might also have improved thermogenic capacity despite the decreased white and brown adipose tissue mass due to less lipid accumulation. Indeed, the E06-scFvLdlr−/− mice demonstrated improved cold tolerance compared to Ldlr−/− mice (Fig. 4U). Collectively, these findings suggest that neutralizing OxPL improved mitochondrial function by reducing mitochondrial oxidative damage and increasing mitochondrial biogenesis.
Neutralization of OxPLs suppresses AMLN diet-induced liver and systemic inflammation
Enhanced inflammation is a key driver of NASH and infiltration of inflammatory leukocytes plays essential roles (Alisi et al., 2017). To understand a potential role of OxPLs in regulating inflammatory immune cell populations during NASH, we performed flow cytometry on hepatic non-parenchymal cell populations isolated from AMLN diet-fed Ldlr−/− and E06-scFvLdlr−/− mice. The final gating strategy to distinguish infiltrating blood monocytes and tissue macrophages is shown in Fig. S5. Importantly, staining for leukocytes with CD45, CD146, Fixvia-NIR, Ly6G, CX3CR1, as well as measuring Vitamin A autofluorescence excluded contamination with liver sinusoidal endothelial cells, dead cells, neutrophils and stellate cells respectively (Fig. S5).
Our data demonstrated that neutralization of OxPLs tended to reduce recruitment of monocyte-derived macrophage characterized as Ly6ChiCD45+F4/80−CD11bhiLy6G−CD146−Live and Ly6ClowCD45+F4/80−CD11bhiLy6G−CD146−Live cells (Fig. 5A, B). Circulating pro-inflammatory cytokines mediating Ly6Chi monocyte infiltration and differentiation (RANTES, M-CSF, MIP-1b, TNFα) were significantly decreased in E06-scFvLdlr−/− mice (Fig. 5C–F). Of interest, in parallel to the findings of decreased hepatic inflammation and apoptosis in the E06-scFvLdlr−/− mice (Fig. 2A), we observed a significantly lower proportion of Tim4− macrophages (Fig. 5G, H), which are thought to be a macrophage population derived from infiltrating monocytes during inflammation (Devisscher et al., 2017; Scott et al., 2018; Scott et al., 2016). Meanwhile, the proportion of resident Tim4+ Kupffer cells (KCs) that mediate engulfment of apoptotic cells (Miyanishi et al., 2007) moderately increased in E06-scFvLdlr−/− mice (Fig. 5G, H). The shift of Tim4− to Tim4+ macrophages suggests less inflammation in the E06-scFvLdlr−/− mice.
Figure 5. Neutralization of OxPL suppresses AMLN diet-induced liver and systemic inflammation.
(A) Flow cytometry of Ly6ChiCD45+F4/80−CD11bhiLy6G−CD146− Live and Ly6ClowCD45+ -F4/80−CD11bhiLy6G−CD146−Live recruited macrophages in the liver of Ldlr−/− (L) and E06-scFvLdlr−/− (EL) mice were fed with AMLN diet for 30 weeks. n = 5.
(B) Statistical analysis of (A). n = 5.
(C-F) Comparison of indicated blood cytokines of indicated mice fed with AMLN diet for 30 weeks. n = 5–10.
(G) Flow cytometry of Tim4+CD45+F4/80+CD11bint-CD146− Live and Tim4−CD45+F4/80+CD11bintCD146− Live macrophages in the liver of indicated mice fed with AMLN diet for 30 weeks. n = 5.
(H) Statistical analysis of (G). n = 5.
(I) Comparison of RNA-seq in livers of indicated mice fed with AMLN diet for 30 weeks. Mean log2 (TPM+1) values (y axis) are plotted versus log2 Fold Change (x axis) of the transcripts of livers between L and EL mice. Transcripts exhibiting EL < L (> 1.5-fold changes, P-adj < 0.05) are red. Genes highlighted in green are the 23 genes most closely related to inflammation (EL < L, > 1.5-fold change, P-adj < 0.05).
(J) Functional annotations associated with genes expressed lower in EL mice indicated in I (red dots).
(K) Relative expression values (Z-scaled log2 (TPM+1)) for the 23 genes highlighted in green in (I) are illustrated, including 7 macrophage marker genes, 15 cytokine/cytokine receptor (R)/inflammatory mediators and 1 apoptosis genes.
Data are mean ± SEM, * P < 0.05. See also Figure S5.
In support of a generally decreased inflammatory state in the E06-scFvLdlr−/− mice, RNA-seq analysis of whole liver revealed 1230 genes significantly down-regulated at a cutoff of 1.5-fold and a P-adj of 0.05 (Fig. 5I, red dots). Gene ontology analysis of reduced genes demonstrated that the top enriched pathways were linked to inflammation related categories including adhesion, cytokine production, leukocyte migration and myeloid leukocyte activation (Fig. 5J). The 23 genes downregulated in E06-scFvLdlr−/− mice corresponding to the inflammatory response terms are colored in green (Fig. 5I). Expression of 7 macrophage marker genes (Adgre1, Csf1r, Cybb, Fcgr1, Fcgr3, Itgax, Itgam), 15 cytokine/receptor/inflammatory mediator genes (Ccr2, Ccr5, Ccl6, Cx3cr1, Cxcl14, Il1a, Tnfrsf14, Tnfsf10, Tnfaip8, Tnfrsfl1a, Tnfrsfl9, Tlr1, Tlr4, Tlr8 and Casp1) and apoptotic gene Casp3 were reduced in liver of E06-scFvLdlr−/− mice (Fig. 5K). As noted above, a variety of inflammatory cytokines/chemokines were decreased in the E06-scFvLdlr−/− mice (Fig. 5C–F). Collectively, these findings suggest that OxPL neutralization decreased the inflammatory content of liver and improved systemic inflammation in AMLN diet-fed mice.
Targeting OxPL prevents hepatic fibrosis
The accumulation of extracellular matrix and collagen in the liver leads to fibrosis and cirrhosis and end stage liver disease, which are the most common fatal hepatic consequences of NASH (Friedman et al., 2018). Prevention of fibrosis is the major goal in therapeutic regimens being developed to treat NASH (Younossi et al., 2018). Consistent with the marked decrease in hepatic collagen deposition in the E06-scFvLdlr−/− mice (Fig. 2A, E), RNA-seq analysis of livers from mice on the AMLN diet revealed that OxPL neutralization caused a significant decrease in 28 genes related to fibrogenesis (Fig. 6A, purple dots), in comparison to overall liver mRNA gene changes that decreased (Fig. 6A, red dots) (fold change > 1.5, P-adj < 0.05). Gene ontology analysis of down-regulated genes of E06-scFvLdlr−/− liver also showed significant functional enrichment for fibrogenic related categories among the top enriched pathways, such as cell migration, extracellular matrix (ECM) organization, collagen formation and PDGF signaling (Fig. 6B). Relative expression values of 15 ECM/receptor (R) genes, 6 growth factor/receptor genes and 7 ECM remodeling gene from liver of both Ldlr−/− and E06-scFvLdlr−/− mice are shown in Fig. 6C.
Figure 6. Targeting OxPL inhibits hepatic fibrosis.
(A) Comparison of RNA-seq in livers of indicated mice fed with AMLN diet for 30 weeks. Mean log2 (TPM+1) values (y axis) are plotted versus log2 Fold Change (x axis) of the transcripts of livers between L and EL mice. Transcripts exhibiting EL < L (> 1.5-fold change, P-adj < 0.05) are red. Genes highlighted in purple are the downregulated genes closely related to fibrogenesis.
(B) Functional annotations associated with genes expressed lower in EL mice (> 1.5-fold changes, P-adj < 0.05).
(C) Relative expression values (Z-scaled log2 (TPM + 1)) of the 28 genes highlighted in purple in (A) are illustrated, including 15 extracellular matrix/receptor (ECM/R) genes, 6 growth factor/receptor (R) genes and 7 ECM remodeling genes.
(D) Ldlr−/− and E06-scFvLdlr−/− mice were injected with CCl4 (0.5ml/kg body weight, 1:5 diluted in corn oil) for 4 weeks. Paraffin-embedded mouse liver sections were stained with SR/FG to determine collagen fiber deposition and E06 IgM antibody to determine OxPL deposition. n = 6. Scale bar = 100μm.
(E-H) Serum triglyceride (E), cholesterol (F) and ALT (G) levels, as well as body weight loss (H) of the same groups of mice described in (D) are shown. n = 8–11.
(I) Proposed model for roles of OxPL in the pathophysiology of NASH.
Data are mean ± SEM, *, P < 0.05; **, P < 0.01. See also Figure S6.
Given that ROS production, an essential contributor to most types of liver fibrosis, was reduced in hepatocytes of E06-scFv expressing mice, we sought to determine whether neutralization of OxPLs could directly attenuate fibrogenesis in a liver damage model that was independent of hyperlipidemia or hepatic steatosis, but induces strong free radical formation and lipid peroxidation (Ayala et al., 2014; Muriel, 2017; Shrestha et al., 2016). Liver injury in Ldlr−/− and E06-scFvLdlr−/− mice was induced by intraperitoneal injection of CCl4. Fibrotic septa between parenchymal nodules were attenuated in E06-scFvLdlr−/− mice compared to Ldlr−/− mice, paralleling less OxPL accumulation along the necrotic area (Fig. 6D). Liver hydroxyproline content and expression of fibrogenic genes (Acta2, Col1a1, Col3a1, Tgfb1, Ddr2) were significantly reduced in E06-scFvLdlr−/− mice (Fig. S6A–B). The histological features of the respective CCl4 treated cohorts were assessed blindly by an experienced pathologist according to the Kleiner Scoring System (Kleiner et al., 2005). Inflammation, hepatocellular injury and fibrosis were all decreased in CCl4-treated E06-scFvLdlr−/− mice compared to Ldlr−/− mice (Table S2). Moreover, SOD activity in CCl4-treated E06-scFvLdlr−/− mice was higher than that in Ldlr−/− mice (Fig. S6C). The serum triglycerides and cholesterol were similar in both Ldlr−/− and E06-scFvLdlr−/− mice, e.g. triglyceride of 170 mg/dL and cholesterol levels of 50 mg/dL (Fig. 6E–F). Serum ALT was significantly lower in E06-scFvLdlr−/− mice compared to Ldlr−/− mice (Fig. 6G). Additionally, E06-scFvLdlr−/− mice lost less weight (Fig. 6H), indicating a relative protection from the toxic effects of the CCl4.
To investigate whether OxPLs have direct effects on hepatic stellate cells, the direct driver of liver fibrosis (Koyama and Brenner, 2017; Tsuchida and Friedman, 2017), we stimulated an immortalized human hepatic stellate cell line (hTERT-HSC) (Schnabl et al., 2002) with OxPAPC. Notably, OxPAPC treatment resulted in an increase in fibrogenic gene expression (Acta2, Col1a1, Tgfb1, Timp1) (Fig. S6D). Given that OxPLs promote ROS accumulation, hepatic inflammation and damage (Fig. 2, 4, 6), all of which activates stellate cells to induce fibrosis (Koyama and Brenner, 2017; Richter and Kietzmann, 2016; Richter et al., 2015), these data suggest that OxPLs contribute to liver fibrosis through both direct and indirect pathways.
DISCUSSION
In this study, we demonstrate that OxPLs accumulate in blood and liver of human subjects with NASH and cirrhosis as well as in three diverse models of NASH and liver fibrosis in mice. The accumulation of OxPLs during AMLN diet feeding of Ldlr−/− mice occurred in the context of exaggerated hyperlipidemia and steatosis that mimics the clinical setting most often found in human NASH. However, OxPLs also accumulated in the serum and liver of CCl4-treated Ldlr−/−mice, a model of liver fibrosis associated with neither hyperlipidemia nor hepatic steatosis, but is thought to be primarily driven by free radical formation. Moreover, in humans, the elevated plasma and liver OxPL content were closely associated with NASH rather than steatosis. In aggregate, these observations suggest that OxPL accumulation is closely associated with progression of NAFL to NASH and that measurement of plasma total OxPLs may represent a potential non-invasive approach to improve the clinical distinction between NAFL and NASH.
Second, and importantly, we demonstrate that the accumulation of OxPLs was causally related to NASH and its complications. Targeting OxPLs with the E06-scFv antibody restrained all measured manifestations of NASH in the AMLN-fed Ldlr−/− mouse models, including steatosis, inflammation, fibrosis, hepatocyte cell injury and death, and its progression to HCC. This model has been used in previous studies of NASH (Friedman et al., 2018). In our study, we took advantage of the marked hypercholesterolemia in the model, which provides a high inflammatory stimulus, and showed that even under these conditions, targeting OxPL decreased NASH and atherosclerosis (Que et al., 2018). In addition, we demonstrated the effectiveness of targeting OxPL in other models as well. OxPLs are a complex set of oxidized moieties that mediate pathological effects through multiple mechanisms, and it is difficult to separate out the relative importance of these myriad effects, many of which are interrelated. In our studies, we demonstrated that at least one major mechanistic effect of neutralizing OxPLs in the context of the AMLN diet was the promotion of improved mitochondrial function in both liver and adipose tissue. Overnutrition and metabolic stress promoted increases in OxPLs, which in turn promoted ROS accumulation and mitochondrial dysfunction. Both elevated ROS and mitochondrial dysfunction contribute to hepatic steatosis, inflammation, apoptosis and fibrosis in NASH. Moreover, in damaged mitochondria, deficiency of electron transport enhanced superoxide generation, which in turn would promote lipid peroxidation and enhanced OxPL formation. Therefore, OxPLs induced a pathogenic feedforward loop in NASH (Fig. 6I). Neutralizing OxPLs by E06 abolished the activation of this feedforward loop, resulting in the amelioration of hepatic steatosis, inflammation, apoptosis and fibrosis. Furthermore, neutralization of OxPLs also attenuated fibrogenesis in the CCl4 free-radical induced liver fibrosis model, which was devoid of hyperlipidemia or steatosis, and also decreased fibrosis in AMLN-fed C57BL/6 mice, which have low cholesterol levels. These observations suggest that OxPLs are one of the long postulated second hit “lipotoxic factors” that promote NASH and its complications, including factors possibly related to HCC formation. Because OxPLs are also pathogenically involved in atherogenesis, targeting OxPLs as a therapeutic strategy should decrease not only NASH and its complications but atherosclerosis as well.
Defects of mitochondrial function and biogenesis are essential reasons for exacerbated hepatic steatosis and liver damage in NASH pathogenesis (Aharoni-Simon et al., 2011; Finkel, 2012; Nassir and Ibdah, 2014; Rector et al., 2010; Win et al., 2018). In our studies, we provided extensive data in isolated hepatocytes that OxPLs induced profound mitochondrial damage and dysfunction and that neutralizing OxPLs abrogated these effects. Furthermore, we showed that besides improvement of mitochondrial function, neutralizing OxPLs also increased mitochondrial biogenesis in livers of AMLN-fed mice. Because of the difficulty in isolating high quality hepatocytes from livers of NASH mice, being that they are fibrotic and steatotic, we did not study specific mitochondrial respiratory complexes using the Seahorse XF Cell Mito Stress Test system. Nevertheless, we did show a similar improvement of mitochondrial function and biogenesis in white adipose tissue, leading to an enhanced whole-body oxygen consumption rate and increased energy expenditure, which resulted in decreased weight gain and enhanced cold tolerance despite decreased white and brown adipose tissue.
To investigate the mechanisms by which OxPLs could impair hepatocyte mitochondrial function, we demonstrated that OxPLs could covalently modify MnSOD and impair its activity. The in vivo relevance of this was supported by the demonstration of OxPL-modified MnSOD in the liver of NASH mice, but not in normal liver or in livers from AMLN fed E06-scFvLdlr−/− mice. The importance of enhanced mtROS generated by OxPLs was supported by evidence that the MnSOD mimetic MnTBAP abrogated the OxPL-induced loss of mitochondrial membrane potential and mtROS generation. In addition, these are complex events and it is likely that indirect effects are involved in regulation of SOD as well.
As noted above, mitochondrial damage would lead to aggravated ROS production in a feedforward loop (Fig. 6I). Therefore, neutralizing OxPL abolished the initiation of this feedforward loop to prevent ROS accumulation and mitochondrial dysfunction. Moreover, OxPL neutralization increased the NAD/NADH ratio because of improved oxidative phosphorylation, and consistent with the role of NAD to be a cofactor that directly activates SIRT1, we further showed that neutralizing OxPLs increased SIRT1 activity in AMLN diet-fed E06-scFvLdlr−/− mice. In turn, since SIRT1 deacetylates and activates PGC1α, the increased NAD/NADH ratio would be predicted to promote mitochondrial biogenesis in E06-scFvLdlr−/− mice. Indeed, gene expression supporting mitochondrial biogenesis was enhanced. Further, PGC1α ChIP-seq data demonstrated enhanced localization to enhancer/promoter regions of key mitochondrial genes. Together with electron microscopy, Mitotracker staining data, and direct measurement of mitochondrial DNA, these data support the pathway outlined leading to an increase of mitochondrial biogenesis in livers from E06-scFvLdlr−/− mice. Taken together, improvement of mitochondrial function by neutralizing OxPLs in the context of the AMLN diet led to increased mitochondrial biogenesis in an NAD/SIRT1/PGC1α-mediated feedforward axis.
Numerous studies have indicated that OxPLs induce inflammatory gene expression and pro-inflammatory cytokine production (Huber et al., 2002; Que et al., 2018; Romanoski et al., 2011; Serbulea et al., 2018; Van Lenten et al., 2001). We show here that another major effect of neutralization of OxPLs was a decreased proportion of pro-inflammatory Ly6Chi monocytes-derived macrophage in the liver, reduced pro-inflammatory gene expression and reduced levels of circulating inflammatory cytokines, including RANTES, M-CSF, MIP-1b, and TNFα. These observations indicate that OxPLs directly (or indirectly) mediate both systemic and hepatic inflammation during NASH. Moreover, we found decreased apoptotic cell in livers of E06-scFvLdlr−/− mice, along with decreased serum transaminases in blood, all consistent with improved NASH. Neutralization of OxPLs, which are known as potent inducers of cell injury and apoptosis (Seimon et al., 2010), was partially responsible for this but in addition, we found a moderately increased proportion of resident Tim4+ KCs in E06-scFvLdlr−/− mice, which mediate engulfment of apoptotic cells (Miyanishi et al., 2007). In turn, there was a lower proportion of Tim4−macrophages, which are thought to be a macrophage population derived from infiltrating monocytes during inflammation (Devisscher et al., 2017; Scott et al., 2018; Scott et al., 2016). The decrease in cell death likely further leads to decreased inflammation. Injured and dying cells are known to release microparticles and extracellular vesicles that are enriched in OxPLs (Tsiantoulas et al., 2015; Yang et al., 2018; Zanoni et al., 2017), which in turn can promote ROS production leading to more OxPL production. As a result, OxPLs activated a feedforward loop to induce hepatocyte apoptosis and inflammation.
Development of fibrosis leading to cirrhosis and liver failure is a critical complication of NASH and a major target of all therapeutic regimens. We demonstrate that another major effect of targeting OxPLs was to inhibit fibrosis in the AMLN diet-induced NASH model, in the CCl4-induced liver fibrosis model not associated with hyperlipidemia, and in AMLN-fed C57BL/6 mice with low LDL values. Fibrogenesis is thought to be driven by activated hepatocytes and KCs, leading to differentiation of resident stellate cells into myofibroblasts, which in turn overproduce matrix proteins. We show that stimulation of an HSC cell line by OxPAPC in fact activates the fibrogenic pathway. As noted from our RNA-seq data, there was a marked decrease in the livers of E06-scFv mice of key fibrogenic genes, as well as in growth factors/receptors. In addition, OxPLs can likely indirectly induce immune cell-mediated fibrosis by recruitment of leukocytes, especially inflammatory macrophages, which can further induce fibrosis via TGFβ and PDGF (Wynn and Barron, 2010) and ROS production (Richter and Kietzmann, 2016). OxPLs accumulate within alveolar macrophages in bleomycin-induced lung fibrosis, which then display enhanced production of TGFβ (Romero et al., 2015). Direct instillation of OxPAPC into the mouse lung induce foam cell formation (Romero et al., 2015). In addition to direct activation of HSCs, OxPL-activation of KCs or hepatocytes leads to enhanced TGFβ production, which also initiates HSC activation and the promotion of fibrosis. OxPLs might thus contribute to the pulmonary toxicity of bleomycin that promotes fibrosis. Future studies will be needed to test these hypotheses. Understanding the cellular and molecular mechanisms by which OxPLs promote fibrosis may have widespread therapeutic implications aside from liver fibrosis; for example, in diseases such as radiation- and bleomycin-induced lung fibrosis.
We emphasize that the cellular and molecular mechanisms by which OxPLs mediate the pathological effects demonstrated here are complex and multifactorial, and likely differ with different cell types involved. OxPLs have been shown to induce cellular responses in immune cells including activation of CD36, TLR2/1 and TLR2/6, TLR4, CD14 and combinations of these receptors (Bieghs et al., 2012; Binder et al., 2016; Imai et al., 2008; Kadl et al., 2011; Lee et al., 2012; Miller et al., 2011; Podrez et al., 2002; Seimon et al., 2010; Zanoni et al., 2017). We now show yet another mechanism by which reactive OxPLs can alter normal cellular metabolism in hepatocytes; namely, by the direct covalent modification of proteins such as MnSOD. The distinct cellular pathways mediating individual effects will need to be studied by combinations of in vitro and in vivo studies in relevant cell populations of E06-scFv mice. These effects may also differ with different OxPL species, as, for example, Leitinger and colleagues have shown that fragmented OxPLs are responsible for reprogramming of macrophage metabolism, whereas full-length OxPLs were more responsible for proinflammatory gene expression (Serbulea et al., 2018; Serbulea et al., 2018). Further studies will also be needed to address the etiology of the increased OxPLs that accumulate in both liver and blood of both the mouse models and the human subjects with NASH. Enhanced inflammation and lipid peroxidation occur with the marked hyperlipidemia associated with the AMLN diet (Dhibi et al., 2011), and this no doubt leads to enhanced non-enzymatic lipid peroxidation and OxPL generation and accumulation. On the other hand, CCl4 is known to initiate free radical formation and also led to OxPL accumulation even in the absence of steatosis or hyperlipidemia. Of further relevance, it has been shown that activation of TLR4 on macrophages in culture leads to secretion of OxPLs into the media (Popat et al., 2017) and similarly, we have shown that TLR2 activation also leads to macrophage release of OxPLs (data not shown). In turn, as shown in our studies, OxPLs stimulated both total and mtROS production and accumulation in hepatocytes, which would promote lipid peroxidation and further OxPL generation. Thus, it is likely that there are a series of interacting “vicious cycles” that feedforward to lead to sustained enhanced OxPL accumulation, which in turn promote NASH and its complications (Fig. 6I).
In summary, our data demonstrate that OxPLs are increased in plasma and liver of humans with NASH and that targeting OxPLs in relevant mouse models restrains NASH and its complications of steatosis, inflammation, hepatocyte injury and cell death, fibrosis and possibly HCC formation. Targeting OxPL by the E06-scFv reduced all of these adverse events. The E06 antibody used in these studies is an innate natural antibody present in mice. Similar natural antibodies that target OxPLs exist in humans (Ravandi et al., 2011; Senders et al., 2018; Tsimikas et al., 2007), suggesting the feasibility of translating this approach to humans.
Limitations of Study
Our studies to date have used transgenic mice that constitutively express the E06-scFv antibody at a concentration of 20–30ug/ml or less, which seems sufficient to prevent many of the consequences of NASH observed in these varying mouse models. Importantly, translational studies in which an appropriate concentration of an IgG version of E06 is infused into mice to show prevention of NASH, as well as regression of NASH in models with existing disease, will be required to show the importance of these studies for therapeutic purposes.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph Witztum (jwitztum@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Ldlr−/− (L) mice in the C57BL/6J background were purchased from Jackson laboratory. E06-scFv Ldlr−/− (EL) mice were generated by crossing E06-scFv (Que et al., 2018) to Ldlr−/− mice all on C57BL/6 background. All mice were bred and maintained at the UCSD pathogen-free animal facility and were used in accordance with the Guide for Care and Use of Laboratory Animals of the National Institute of Health. The protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of UCSD. All mice were housed in a temperature-controlled environment at 22 °C with 12-h dark, 12-h light cycles and given free access to water and food, except for fasting period. All mice were in good health conditions. Only male mice were used for experiments. When indicated, mice (Ldlr−/− or C57BL/6) were fed an AMLN diet consisting of 40 kcal% Fat /20kcal% Fructose /2% Cholesterol from Research Diet (D09100301 Research Diets Inc) starting at 8 weeks old of age for 20–30 weeks to generate NASH model and for 48 weeks for HCC model. For experimental neoplasia assessment, which includes tumor size limits (not exceeding 2cm for a single tumor) and monitoring parameters, tumor volumes were calculated as (width2 × length)/2, and for multiple liver tumors the volumes of single tumor were added for a total tumor volume (Shalapour et al., 2017). In the CCl4 model, indicated mice were injected intraperitoneally with CCl4 (0.5ml/kg body weight, 1:5 diluted in corn oil) twice a week for 4 weeks, and mice were sacrificed 72 hours after the last injection. In the STAM model, male L and EL mice were subcutaneously injected with 200μg streptozotocin (STZ) 2 days after birth and fed with high fat diet consisting of 60% of calories from fat (D12492 Research Diets Inc.) starting at 4 weeks of age for 4 weeks.
For all experiments, Ldlr−/− and E06-scFvLdlr−/−, C57BL/6 and E06-scFvC57BL/6 mice were littermates and cage mates. The E06-scFv were all heterozygotes. Animals in each cohort were produced from 20 breeding pairs to minimize the birthdate range. Identification codes were assigned to each mouse and the investigators were blinded to treatment or genotype during experiments. For metabolic study, mice were subject to CLAMs indirect calorimetry and dual energy DEXA scanning at ACP phenotyping core of UCSD. Oxygen consumption rate (OCR) and energy expenditure (EE) per kilogram of body weight were determined. Analysis of covariance (ANCOVA) (Tschop et al., 2011) was performed to test the difference of OCR and EE between groups (body weight as covariant) with IBM SPSS Statistics. EE was calculated as a function of OCR and carbon dioxide production according to the following formula: energy expenditure (kcal/hr)=(3.941xVO2(ml/hr)+1.106xVCO2(ml/hr))/1000 (Owen et al., 2014). Rectal temperature was measured by Model 4600 Thermometer (Alpha Technics). Serum insulin was measured with Ultra-Sensitive Mouse Insulin ELISA kit (Crystal Chem).
Human liver sections and plasma
Human liver samples used in the study were obtained by Dr. Kisseleva via collaboration with Lifesharing (www.lifesharing.org). Classification of liver histology was performed by an experienced liver pathologist in a double blinded manner, and categorized as normal, steatosis, and NASH induced liver fibrosis stage 1, 2, and 4. Plasma samples for measurement of OxPLs were obtained from human outpatient subjects and from a previously published clinical study (Gorden et al., 2015). All human samples were collected under protocols approved by the UCSD Human Research Protections Program under informed patients’ consent prior to inclusion in this study.
METHOD DETAILS
Primary hepatocyte isolation:
Primary hepatocytes were isolated from 6-week-old Ldlr−/− and E06-scFvLdlr−/− male mice by a 2-step collagenase perfusion method. Briefly, HBSS (Life Technologies Co.; no Calcium and Magnesium, 0.5mM EDTA, 25mM HEPES) was used to perfuse liver at 10mL/min speed until the liver turns into pale. Afterward, the liver was perfused with HBSS digestion buffer (Life Technologies Co.; 30mg/100mL collagenase I, 2 tablet/100mL protease inhibitor) at 15mL/min speed for 18 min. After sequential flows, cells were smashed through 100μm strainer and washed with Williams’ Medium E (Gibco, Grand Island, NY). Hepatocytes were isolated by density gradient centrifugation using percoll (Pharmacia, Sweden). Hepatocytes with 95% viability were cultured in Williams’ Medium E supplemented with 5% serum, 0.5% penicillin/streptomycin and 15mM HEPES at 37°C in a 5% CO2 incubator overnight before use in experimentation.
Non-parenchymal cells (NPCs) isolation from the mouse liver:
Liver NPCs were processed for fluorescence activate cell sorting of Kupffer cells, with modifications from previous published methodology(Mederacke et al., 2015; Muse et al., 2018; Seki et al., 2007). In brief, liver was perfused with pre-warmed HBSS (no calcium and magnesium, 0.5mM EGTA, 0.75mM EDTA, 20mM HEPES, 1µM flavopiridol) for 3 min at a speed of 7mL/min through inferior vena cava. This was followed by 60ml of digestion buffer (HBSS, 0.1 mg/ml Liberase TM, 20 μg/ml DNaseI, 20mM HEPES, 1 µM flavopiridol) at the speed of 7ml/min for 8 min. Liver was then dissected and incubated in 50ml conical tube containing 20ml of digestion buffer for 20 additional minutes at 37 °C with gentle rotation using a Miltenyi MACSmix tube rotator. Cells were then smashed through 70 μm cell strainer. Hepatocytes were removed by a 2 low-speed centrifugation steps at 50g for 2 min. Cells were then washed with wash buffer (HBSS containing 20 µg/ml DNase1, 2% FBS, 20mM HEPES). NPCs were separated from debris by pelleting for 15 min at 600g by density gradient centrifugation using 20% isotonic Percoll (Pharmacia, Sweden). Cells were then washed with 28% OptiPrep (Sigma Aldrich) and carefully underlaid beneath 3mL of wash buffer. The gradient was centrifuged a 1400g for 25min and cells enriched at the interface were saved and subjected to isotonic erythrocyte lysis. Later, enriched NPCs were washed, suspend in PBS and then stained for indicated antibodies for flow cytometer.
Histology:
Mice were euthanized by CO2 inhalation. Tissues were dissected, and then fixed in sucrose fix working solution (4% paraformaldehyde, 20mM sodium phosphate buffer, 2mM EDTA, 7.5% sucrose). Paraffin/OCT-embedding tissues were sectioned and subject to H&E staining in the La Jolla Atherosclerosis Morphology Core. Fast Green (Fisher Scientific)/Sirius Red (Sigma Aldrich Inc.) staining was carried out on paraffin sections to assess liver fibrosis. Oil Red O staining was conducted on frozen-sections embedded in OCT to determine hepatic steatosis. Stained tissue was visualized with NanoZoomer Slide Scanner. Signal intensity was determined by ImageJ (NIH, Maryland, USA) analysis of H&E, Oil Red O and Fast Green/Sirius Red stained tissues. As described in the text, in some studies, formal histological analyses of features of NASH in various models were blindly assessed by an experienced pathologist according to Kleiner Scoring System (Kleiner et al., 2005) and results of those analyses provided in Tables S1 and S2.
Immunostaining and TUNEL staining:
Paraffin embedded tissue sections were subjected to de-paraffinization and rehydration, and then were immersed in 95 °C antigen retrieval buffer (10mM sodium citrate, 0.05% Tween 20, pH6.0) for 30 min. Cells were fixed with 10% buffered formalin for 10 min at room temperature and permeabilized with 0.02% Triton X-100 for 5 min. Tissue sections or cells were blocked with 1% normal donkey serum for 30 min. For E06 staining, the sections or cells were blocked sequentially by donkey serum and biotin/avidin blocking. Sections were incubated with primary antibodies for 12h at 4 °C. F4/80 (AbD Serotec) and biotinylated E06 staining (Que et al., 2018) were conducted on indicated liver/adipose paraffin sections to analyze macrophage infiltration and OxPL accumulation. Mitochondria in the liver and adipose tissue were stained with 200nM Mitotracker Red (Life Technologies) for 45 min at room temperature. Hepatic apoptosis was determined by TUNEL staining of liver sections from mice on AMLN diet with ApoBrdU DNA Fragmentation Assay Kit (K401 BioVision Inc) according to the manufacturer’s instruction. Nuclei were stained with DAPI. IHC stained tissue was visualized with NanoZoomer Slide Scanner. Fluorescence stained sections were examined using Zeiss LSM 880 with FAST Airyscan (Zessi, Germany).
Hepatic hydroxyproline measurement:
Liver tissue was homogenized in distilled water (100μl/10mg tissue). 100μl of 10 mol/L concentrated NaOH was added to each 100 μl samples and hydrolyzed at 120 °C for 1 hour. Supernatants were cooled on ice and neutralized with 10 mol/L concentrated HCl, followed by 10000g centrifugation for 5 min. The supernatants were then collected without lipid content. Hepatic hydroxyproline was measured with Hydroxyproline Assay Kit (Abcam) according to the manufacturer’s instruction.
Measurement of E06-scFv titers in culture supernatants:
E06-scFv titers in the culture supernatants of primary hepatocytes were determined by chemiluminescent ELISA assays. The principle of assay is that the E06-scFv has an His tag. In brief, 96-well round-bottom MicroFluor plates (Phenix Research, NC) were coated with PC-KLH (PC-1013–5, Biosearch Tech) at 5μg/ml (50μl per well) in PBS overnight at 4 °C. Culture media of primary hepatocytes were collected after 12 hours of culture, centrifuge at 1000g for 10 min and the supernatant collected for E06-scFv titer test. After the plates were washed and blocked with 1% BSA in Tris-buffered saline (TBS) for 60 min, 40μl of culture media were added to the wells, and incubated for 60 min at room temperature. Bound E06-scFv was detected with anti-His6-tag antibody conjugated to alkaline phosphatase (Sigma-Aldrich), in TBS buffer containing 1% BSA, followed by three rinses with TBS and the addition of 25μl of 50% LumiPhos 530 (Lumigen) as luminescent substrate. The light emissions were measured, and counts expressed as relative light units over 100ms (RLU/100ms) using a Dynex Luminometer (BioTek, VT). All determinations were done in triplicate.
Measurement of Total-OxPLs in serum/plasma by ELISA:
A competitive ELISA was established to measure the concentration of total immunodetectable OxPLs in mouse and human subject serum. Limiting amounts of E06-IgM (330001 Avanti Polar Lipids Inc) was coated in wells of microtiter plates and then a fixed concentration of PC-KLH (PC-1013–5, Biosearch Tech) was added in the absence and presence of serum/plasma and the extent of PC-KLH binding to plated E06 determined using anti-KLH antibody. The presence of OxPLs in serum/plasma will compete with PC-KLH for binding to E06. In brief, a limited amount of E06-IgM antibody (2μg/mL in PBS) was used to coat immunograde White U Bottom 96 Well Plates (Phenix Research, NC) overnight at 4°C. The plates were blocked by 1% fatty acid free BSA for 1h at room temperature. Human or mouse serum/plasma (1:150) were incubate for 1h at room temperature in the E06 coated plates to allow pre-binding. This was followed by PC-KLH (1μg/mL) addition to the wells for 1h at room temperature. After each 1 hour of incubation, the wells were washed 3 times with TBS. Alkaline phosphatase-conjugated anti-KLH antibody (source 600–405-466 Rockland Inc) was then applied to detect bound PC-KLH using Lumi-Phos 530. Data were collected as RLU/100ms in a Synergy HTX Multi-Mode Reader (BioTek, VT). A standard curve of PC-KLH without serum/plasma competitor was run in parallel on each plate to allow calculation of concentration of immunodetectable OxPLs in the samples. All determinations were done in triplicate.
Triglyceride and Cholesterol measurement:
Blood/Tissue triglyceride and cholesterol levels were determined using the Triglyceride Quantification Colorimetric/Fluorometric Kit (k622 Biovision Inc.) and total Cholesterol and Cholesterol Ester Colorimetric/Fluorometric Kit (k603 Biovision Inc.) according to the manufacturer’s instruction respectively. All values were analyzed from 12 hours fasted mice.
Free fatty acid and glycerol measurement:
Blood free fatty acid and glycerol levels were measured with NEFA HR color reagent (Wako Life Sciences) and Free Glycerol Determination kit (Sigma) respectively, according to the manufacturer’s instructions. All mice sera were from mice fasted for 12 hours.
Glucose and Insulin tolerance tests:
Fasting blood glucose was measured after 12 hours fast, using Easy Step Blood Glucose Monitoring System. Mice were then intraperitoneally injected with D-[+]-glucose (Sigma) at a dose of 1g/kg BW for ALMN diet-fed mice. Blood glucose levels were measured at 15, 30, 45, 60, 90 and 120 min after injection. For insulin tolerance test, fasting blood glucose was measured after 4 hours fast, using Easy Step Blood Glucose Monitoring System. Mice were then intraperitoneally injected with insulin (Humulin R) at a dose of 1.2U/kg BW for AMLN diet-fed mice. Blood glucose levels were measured at 15, 30, 45, 60, 90 and 120 min after injection (Zhao et al., 2018).
RNA-seq library preparation:
Total RNA was isolated from mice livers homogenized with TRIzol reagent and purified using Quick RNA mini prep columns and RNase-free DNase digestion according to the manufacturer’s instructions (Life Technologies). RNA quality was assessed by an Agilent 2100 Bioanalyzer. Sequencing libraries were prepared in biological replicates from polyA enriched mRNA. RNA-seq libraries were prepared from poly(A)-enriched mRNA as previously described (Oishi et al., 2017). Libraries were size selected by gel extraction, quantified using a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) and sequenced on a Hi-seq 4000 (Illumina, San Diego, CA) according to the manufacturer’s instructions.
RNA-seq analysis:
RNA-seq analysis was conducted as previously described (Link et al., 2018). FASTQ files from sequencing experiments were mapped to the mouse mm10 genome. STAR with default parameters was used to map RNA-seq experiments (Dobin et al., 2013). To compare differential gene expression between indicated groups, HOMER’s analyzeRepeats with the option rna and the parameters -condenseGenes, -noadj, and -count exons was used on two-three replicates per condition (Heinz et al., 2010). Each sequencing experiment was normalized to a total of 107 uniquely mapped tags by adjusting the number of tags at each position in the genome to the correct fractional amount given the total tags mapped. Sequence experiments were visualized by preparing custom tracks for the UCSC genome browser. Differential gene expression was assessed with DESeq2 using HOMER’s getDiffExpression.pl with the parameters -p-adj 0.05 and -log2 fold 0.585 (for 1.5-fold differently expressed genes) (Love et al., 2014). For all genes the TPM (transcript per kilobase million) values were plotted and colored according to fold change. For various ontology analyses, either HOMER or Metascape was used (Tripathi et al., 2015).
PGC-1α ChIP-seq of hepatic nuclei:
Livers from AMLN diet-fed Ldlr−/− and E06-scFvLdlr−/−mice were crosslinked by a two-step perfusion method. Briefly, 1mg/mL disuccinimidyl glutarate (DSG) in PBS was used to perfuse the liver for 30 min, followed by 1% PFA in PBS for 10 min. Afterward, the livers were perfused with 20mL 0.125M glycine to quench the crosslinking. After sequential flows, livers were Dounce homogenized and filtered through 70 μm strainer and washed with NF buffer (10mM Tris-HCl, pH8.0, 1mM EDTA, 5mM MgCl2, 0.1M Sucrose, 0.5% Triton X-100). Liver homogenate were then centrifugation at 1200g for 7min, then washed with 10mL HBSS (adding 1%BSA, 1mM EDTA) and filtered through 30 μm strainer. Hepatic nuclei were then washed again with 15mL HBSS (adding 1%BSA, 1mM EDTA) followed by centrifugation and the pellet was saved for ChIP-seq. ChIP from hepatic nuclei was performed as described previously (Oishi et al., 2017). Briefly, nuclei were suspended in 130ul RIPA-NR lysis buffer (20mM Tris/HCl pH7.5, 1mM EDTA, 0.5mM EGTA, 0.1% SDS, 0.4% Na-Deoxycholate, 1% NP-40 alternative, 0.5mM DTT, 1X protease inhibitor cocktail (Sigma), 1mM PMSF) and chromatin was sheared by sonication using a Covaris E220 for 18 cycles with the following setting: time, 60s; duty, 5.0; PIP, 140; cycles, 200; amplitude, 0.0; velocity, 0.0; dwell, 0.0. Immunoprecipitation was carried out with 2.5μg each of the indicated PGC1α antibodies (sc-517380 from Santa Cruz Biotechnology and NBP1–04676 from Novus Biologicals) with slow rotation at 4 °C overnight. Libraries were PCR amplified for 12–15 cycles, size selected by gel extraction, and sequenced on a NextSeq 500 to a depth of 10–20 million reads.
ChIP-seq analysis:
ChIP-seq analysis was conducted as previously described (Link et al., 2018). FASTQ files from sequencing experiments were mapped to the mouse mm10 genome using Bowtie2 with default parameters (Langmead and Salzberg, 2012). HOMER was used to convert aligned reads into “tag directories” for further analysis (Heinz et al., 2010). ChIP-seq experiments were performed in replicate with corresponding input experiments. Peaks were called with HOMER for each tag directory with relaxed peak finding parameters -L 0 -C 0 -fdr 0.9 against the corresponding input directory. IDR (Li et al., 2011) was used to test for reproducibility between replicates, and only peaks with IDR < 0.05 were used for downstream analysis. The pooled tag directory from two replicates was used for track visualization. To quantify transcription factor (TF) binding, peak files were merged with HOMER’s mergePeaks and annotated with raw tag counts with HOMER’s annotatePeaks using parameters -noadj, -size given. Subsequently, DESeq2 (Love et al., 2014) was used to identify the differentially bound TF with > 2 fold-change and P-adj < 0.05, unless stated otherwise in the text. The UCSC genome browser (Kent et al., 2002) was used to visualize ChIP-seq data.
Each sequencing experiment was normalized to a total of 107 uniquely mapped tags by adjusting the number of tags at each position in the genome to the correct fractional amount given the total tags mapped. Sequence experiments were visualized by preparing custom tracks for the UCSC genome browser.
Transmission electron microscopy:
Mice were perfused with 10ml of modified Karnovsky’s fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.15 M sodium cacodylate buffer, pH 7.4) carefully. Liver were then dissected and fixed for at least 4 hours, postfixed in 1% osmium tetroxide in 0.15 M cacodylate buffer for 1 hour and stained en bloc in 2% uranyl acetate for 1 hour. Samples were dehydrated in ethanol, embedded in Durcupan epoxy resin (Sigma-Aldrich), sectioned at 50 to 60 nm on a Leica UCT ultramicrotome, and picked up on Formvar and carbon-coated copper grids. Sections were stained with 2% uranyl acetate for 5 minutes and Sato’s lead stain for 1 minute. Grids were viewed using a JEOL 1200EX II (JEOL, Peabody, MA) transmission electron microscope and photographed using a Gatan digital camera (Gatan, Pleasanton, CA), or viewed using a Tecnai G2 Spirit BioTWIN transmission electron microscope equipped with an Eagle 4k HS digital camera (FEI, Hilsboro, OR).
ROS and Mitochondrial membrane potential measurement:
Total ROS was determined with Total ROS detection Kit (ENZ-51011 Enzo Life Sciences Inc.) and mitochondrial ROS levels using MitoSOX (Invitrogen) following the manufacturers’ instructions. Mitochondrial membrane potential (Ψm) was measured using TMRM as previously described (Shimada et al., 2012).
Ex vivo/in vitro fatty acid oxidation assay:
Liver or adipose tissues were dissected, weighted, quickly rinsed in PBS. Minced tissues were placed in 96-well tissue culture plate. For cultured cells, PBS was used to rinse the cells. Fatty Acid Oxidation was measured with combination of Fatty Acid Oxidation Assay Kit (Abcam) and Oxygen Consumption Assay Kit (Abcam) according to the manufacturer’s instruction. 150μl reaction medium and 10μl oxygen consumption reagent were added to each well. Wells were sealed with pre-warmed high sensitivity mineral oil. Fluorescence was measured at 37°C for 30min by Tecan Infinite M200 Pro.
Immunoprecipitation:
Liver tissue was homogenized in PBS with freshly added protease inhibitors tablet (Roche). Immunoprecipitation was performed with anti-SOD2 antibody (Abcam) overnight at 4C followed by incubation with Protein A/G agarose beads for 4h at 4C. Protein A/G beads were washed with PBS for 6 times. Protein was eluted with SDS loading buffer.
Western blot:
Immunoprecipated MnSOD complex was resolved by SDS–PAGE and transferred to nitrocellulose membranes (Bio-Rad). Nitrocellulose membranes were sequentially blocked by 5% milk and Avidin/Biotin blocking buffer. Biotinlyzed-E06 antibody was used to detect OxPL modification on MnSOD. E06 signal was visualized on film using horseradish peroxidase-conjugated NeutrAvidin (Life Technologies) and SuperSignal West Pico Chemilunminescent Substrate (Thermo Scientific).
In vitro modification of SOD and SOD activity:
Recombinant human MnSOD (Abcam, ab82656) was incubated with POVPC (Avanti Polar Lipids, 870606) (molar ration 1: 40) or vehicle at 37 °C overnight. 300μg/mL NaBH3CN was added to stabilize reversible OxPL adducts. Both native and POVPC modified MnSOD were dialyzed extensively against PBS. SOD activity of recombinant and modified MnSOD and liver and cell lysates were measured with Superoxide Dismutase Activity Assay Kit (Abcam) according to the manufacturer’s instruction.
NAD/NADH ratio measurement:
Tissue NAD/NADH ratio was measured with NAD/NADH Assay kit (Abcam) according to manufacturer’s instruction. Tissues were dissected, rinsed in PBS, and homogenized in NAD/NADH extraction buffer from the kit. After centrifugation, supernatants were collected for measurements.
Serum cytokine analysis:
Serum cytokine levels of mice, including TNF-α, GM-CSF, MIP-1β and RANTES, were measured used a Bio-Plex Pro™ Mouse Cytokine 23-Plex panel using the Bio-Plex® Protein Array system (Bio-Rad Laboratories, Hercules, CA, USA) following manufacturer instructions.
LDL uptake assay.
Fluorescence-labelled LDL (Dil-OxLDL/Dil-nLDL) were used. HepG2 cells were serum starved for four hours. 10μg/mL Dil-OxLDL or Dil-nLDL were incubated with HepG2 cells for 3 hours. Cells were washed 3 times with cold PBS and fixed with 10% buffered formalin for 10 min at room temperature. Cellular uptake of OxLDL/nLDL were measured by fluorescence intensity or confocal microscopy. Fluorescence intensity was measured (absorption/emission: 554/571 nm). In some experiments, nuclei were co-stained with DAPI, and confocal imaging was conducted to capture images of indicated groups of HepG2 cells.
Fatty acid uptake assay.
Fluorescence-labelled fatty acid (BODIPY- fatty acids) was used. HepG2 cells were serum starved for four hours. 1μM BODIPY-fatty acids (BODYPI-FA) was pre-conjugated with 1% BSA at 37C for 1 hour. BODIPY-FA/BSA pre-incubated with IgM isotype control or E06 IgM were incubated with HepG2 cells for 30 min. Cells were washed 3 times with cold PBS and fixed with 10% buffered formalin for 10 min at room temperature. Cell uptake of BODIPY-FA were measured by fluorescence intensity or confocal microscopy. Fluorescence intensity was measured (absorption/emission: 500/510 nm). In some experiments, nuclei were co-stained with DAPI, and confocal imaging was conducted to capture images of indicated groups of HepG2 cells.
Measurement of mtDNA copy number.
Total liver DNA of AMLN diet-fed Ldlr−/− and E06-scFv Ldlr−/− mice was isolated using PureLink DNA Mini Kit (K182002, Life Technologies) according to manufacturer’s instruction. mtDNA was quantified by qPCR using primers specific for the mitochondrial D-loop region or a specific region of mtDNA that is not inserted into nuclear DNA (non-NUMT) (Malik et al., 2016). Nuclear DNA encoding Tert and B2m was used for normalization. Primer sequences are provided in Key Resource Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| F4/80 antibody | Bio-Rad Laboratories | MCA497R |
| E06 IgM antibody | Avanti Polar Lipids Inc. | 330001 |
| IgM isotype control | Invitrogen | MA110438 |
| PGC-1α antibody | Novus Biologicals | NBP104676 |
| PGC-1α antibody | Santa Cruz Biotechnology | SC-517380 |
| SOD2/MnSOD antibody | Abcam | Ab110300 |
| CD45 | Biolegend | 304017 |
| F4/80 antibody | Biolegend | 123110 |
| CD146 antibody | Biolegend | 134714 |
| CD11b | BD Biosciences | 563553 |
| Ly-6G antibody | Biolegend | 127616 |
| Ly-6C antibody | Biolegend | 128041 |
| CX3CR1 antibody | Biolegend | 149004 |
| Tim4 antibody | Biolegend | 130006 |
| KLH antibody | Rockland | 600-405-466 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM high glucose | Corning | 10-013-CV |
| FBS | Omega Biosciences | FB-12 |
| 100X Penicillin/Streptomycin | GIBCO | 10378-016 |
| Dynabeads Protein G | Thermo Fisher Scientific | 10004D |
| Speedbeads | GE Healthcare | 65152105050250 |
| Millipore Ultrafree MC column | Millipore | UFC30HVNB |
| Formaldehyde | Thermo Fisher Scientific | BP531-500 |
| Glycine | Thermo Fisher Scientific | BP3815 |
| DSG | Peirce | PI20593 |
| ATP | Roche | R0441 |
| GTP | Roche | R0461 |
| CTP | Roche | R0451 |
| Postscript II reagents | NEB | E6560S |
| T4 RNA ligase 2 truncated | NEB | M0242S |
| SUPERase | Ambion | AM2696 |
| RNA 5’-Pyrophosphohydrolase | NEB | M0356S |
| NEB Quick Ligation reagents | NEB | M2200S |
| Oligo dT primer | Invitrogen | 18418020 |
| Agentcout RNA Clean XP Beads | Beckman Coulter | A63987 |
| DNA polymerase I | DNA polymerase I | P7050L |
| Superscript III | Invitrogen | 18080092 |
| Blue Buffer | Enzymatics | P7050L |
| 2x Superscript III first-strand buffer | Invitrogen | 18080400 |
| Carbon tetrachloride | Sigma-Aldrich | 319961 |
| Corn oil | Sigma-Aldrich | C8267 |
| STREPTOZOTOCIN | ENZO Life Sciences | 89149-802 |
| Collagenase I | Worthington Biochemical Co. | LS004194 |
| HBSS (no calcium, no magnesium) | Life Technologies | 14185052 |
| HBSS (calcium, magnesium) | Life Technologies | 14065056 |
| TRIzol Reagent | Life Technologies | 15596018 |
| Percoll | Sigma-Aldrich | P1644 |
| Protease inhibitor tablet | Sigma-Aldrich | 11873580001 |
| HEPES buffer | Life Technologies | 15630080 |
| Trypsin inhibitor | Worthington Biochemical Co. | LS003570 |
| Liberase TM | Sigma-Aldrich | 5401127001 |
| Deoxyribonuclease I | Fisher Scientific | NC9709009 |
| Flavopiridol hydrochloride | Sigma-Aldrich | F3055 |
| Fast Green | Fisher Chemical | F9910 |
| Direct Red 80 | Sigma-Aldrich | 365548 |
| Oil Red O | Fisher Scientific | 501201162 |
| Sudan IV | Sigma-Aldrich | 198102 |
| Donkey serum | Fisher Scientific | 507533005 |
| Normal Serum Block | Biolegend | 927501 |
| EDTA | Fisher Scientific | NC9141704 |
| EGTA | BIOWORLD | 40520008-3 |
| PC-KLH | Biosearch Technologies | PC-1013-5 |
| Zombie NIR | Biolegend | 423106 |
| Recombinant SOD2 protein | Abcam | ab82656 |
| Mitotracker Red FM | Life Technologies | M22425 |
| Lumi-Phos 530 | Lumigen Inc. | P-5000 |
| Critical Commercial Assays | ||
| ChIP DNA Clean & Concentrator | Zymo Research | D5205 |
| NEXTflex® DNA Barcodes | Bioo Scientific | NOVA-514104 |
| Ultra-sensitive mouse insulin ELISA kit | Crystal Chem | 90080 |
| Triglycerides Quantification kit | BIOVISION | K622 |
| Total Cholesterol kit | BIOVISION | K603 |
| ApoBrdU DNA Fragmentation kit | BIOVISION | K401-60 |
| NEFA-HR(2) assay | Wako | 434-91795 |
| Total ROS detection kit | Enzo Life Sciences Inc. | ENZ-51011 |
| MitoSOX Mitochondrial Superoxide Indicator | Life Technologies | M36008 |
| Free glycerol determination kit | Sigma-Aldrich | F6428 |
| Fatty acid oxidation assay | Abcam | Ab217602 |
| Extracellular oxygen consumption assay | Abcam | ab197243 |
| Image-iT TMRM Reagent | Life Technologies | I34361 |
| NAD/NADH assay kit | Abcam | ab65348 |
| SIRT1 activity assay kit | Abcam | ab156065 |
| TBARS assay kit | Cayman Chemicals | 10009055 |
| Hydroxyproline assay kit | Abcam | ab222941 |
| Superoxide Dismutase Activity Assay Kit | Abcam | ab65354 |
| Dil-OxLDL | Life Technologies | L34358 |
| Dil-nLDL | Life Technologies | L3482 |
| BODIPY-fatty acids | Life Technologies | D3823 |
| PURELINK DNA MINI KIT | Life Technologies | K182002 |
| PURELINK RNA MINI KIT | Life Technologies | 12183025 |
| Deposited Data | ||
| Transcriptomic analysis | This paper | GEO: GSE138419 |
| Experimental Models: Cell Lines | ||
| HepG2 cells | ATCC | HB-8065 |
| hTERT-immortalized HSCs | Gift from David Brenner | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: E06sc-Fv | Bred at UCSD | |
| Mouse: Ldlr−/− | Bred at UCSD | |
| Mouse: C57BL/6J | Jackson Laboratory | 000664 |
| Oligonucleotides | ||
| Human Acta2 forward primer 5’-GTGTTGCCCCTGAAGAGCAT-3’ | This paper | N/A |
| Human Acta2 reverse primer 5’-GCTGGGACATTGAAAGTCTCA-3’ | This paper | N/A |
| Human Col1a1 forward primer 5’-GTGCGATGACGTGATCTGTGA-3’ | This paper | N/A |
| Human Col1a1 reverse primer 5’-CGGTGGTTTCTTGGTCGGT-3’ | This paper | N/A |
| Human Col3a1 forward primer 5’-GGAGCTGGCTACTTCTCGC-3’ | This paper | N/A |
| Human Col3a1 reverse primer 5’-GGGAACATCCTCCTTCAACAG-3’ | This paper | N/A |
| Human Tgfb1 forward primer 5’-GGCCAGATCCTGTCCAAGC-3’ | This paper | N/A |
| Human Tgfb1 reverse primer 5’-GTGGGTTTCCACCATTAGCAC-3’ | This paper | N/A |
| Human Ddr2 forward primer 5’-GCTATATGCCGCTATCCTCTGG-3’ | This paper | N/A |
| Human Ddr2 reverse primer 5’-ACTCTGACCACTGACTGGAAG-3’ | This paper | N/A |
| Human Timp1 forward primer 5’-CTTCTGCAATTCCGACCTCGT-3’ | This paper | N/A |
| Human Timp1 reverse primer 5’-ACGCTGGTATAAGGTGGTCTG-3’ | This paper | N/A |
| Mouse D-loop forward primer 5’-AATCTACCATCCTCCGTGAAACC-3’ | This paper | N/A |
| Mouse D-loop reverse primer 5’-TCAGTTTAGCTACCCCCAAGTTTAA-3′ | This paper | N/A |
| Mouse Tert forward primer 5’-CTAGCTCATGTGTCAAGACCCTCTT-3′ | This paper | N/A |
| Mouse Tert reverse primer 5’-GCCAGCACGTTTCTCTCGTT-3′ | This paper | N/A |
| Mouse B2m forward primer 5’-ATGGGAAGCCGAACATACTG-3′ | This paper | N/A |
| Mouse B2m reverse primer 5’-CAGTCTCAGTGGGGGTGAAT-3′ | This paper | N/A |
| Mouse non-NUMT forward primer 5’-CTAGAAACCCCGAAACCAAA-3′ | This paper | N/A |
| Mouse non-NUMT reverse primer 5’-CCAGCTATCACCAAGCTCGT-3′ | This paper | N/A |
| Software and Algorithms | ||
| UCSC genome browser | Kent et al., 2002 | https://genome.ucsc.edu/ |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| R package: DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R package: RcolorBrewer | Neuwirt, 2014 | https://cran.r-project.org/web/packages/RColorBrewer/index.html |
| R package: gplots | R main package | https://cran.r-project.org/web/packages/gplots/index.html |
| Metascape | Tripathi et al., 2015 | http://metascape.org |
| ImageJ | NIH | N/A |
| GraphPad Prism 8 | GraphPad Software | N/A |
| IBM SPSS Statistics | IBM | N/A |
| Other | ||
| HFD | Research Diets | D12492 |
| AMLN Diet (45% kcal% fat) | Research Diets | D09100301 |
QUANTIFICATION AND STATISTICAL ANALYSIS
All data of animal and human studies are shown as mean ± SEM. Replicates are indicated in figure legends. n represents the number of experimental replicates. F-test was performed to determine the equality of variance. When comparing two groups, statistical analysis was performed using a two-tailed Student’s t-test, except when the f-test suggested that variances were statistically different. For analysis of more than two groups, we used analysis of variance (ANOVA) to determine equality of variance. Comparisons between groups were performed with Tukey-Krammer post-hoc analysis. For all tests, P < 0.05 was considered statistically significant.
DATA and SOFTWARE AVAILABILITY
The accession number for the transcriptomic data reported in this paper is GEO: GSE138419.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Witztum lab and the Glass lab for helpful discussions. We thank Karen Bowden, Jennifer Pattison and Elena Alekseeva from the La Jolla Atherosclerosis Morphology Lab; Ying Jones in the Cellular and Molecular Medicine (CMM) Electron Microscopy Core; and the UCSD Histology Core for tissue sectioning, staining and imaging support. We thank the UCSD School of Medicine Microscopy Core for multiple microscopy services. We thank Marilyn Hardee and Animal Phenotyping Core for support of CLAMs indirect calorimetry and DEXA studies, and Zeng Liu, Qiongyu Chen in the Hematology Core for liver function measurements. We thank Dr. Walter Shaw of Avanti Polar Lipids for providing sterile oxidized PAPC and E06-IgM antibody. We thank Dr. Yury Miller for critical reading of the manuscript. This work was supported by NIH grants HL088093 (C.K.G, X.Q., X.S., R. E., J.L.W.); HL136275 (J.L.W.); HL135737 (J.L.W.); American Heart Association grants 18POST34060088 (X.S.); and NIH Award S10OD023527 in support of the CEM Electron Microscopy Core. P. Z. was supported by NIH grant K99 HL143277. T.D.T was supported by P30 DK063491, T32 DK007044, and NRSA T32CA009523.
Footnotes
DECLARATION OF INTEREST
J.L.W, S.T., X.Q. and X.S. are named inventors on patent applications or patents related to the use of oxidation-specific antibodies held by UCSD. J.L.W, S.T. and X.Q. are founding members of Oxitope, Inc, and J.L.W. and S.T. of Kleanthi Diagnostics. J.L.W. is a consultant for Ionis Pharmaceuticals and S.T. is employed by UCSD and by Ionis Pharmaceuticals. The other authors declare that they have no competing interests.
REFERENCES
- Aharoni-Simon M, Hann-Obercyger M, Pen S, Madar Z, and Tirosh O (2011). Fatty liver is associated with impaired activity of PPARgamma-coactivator 1alpha (PGC1alpha) and mitochondrial biogenesis in mice. Lab Invest 91, 1018–1028. [DOI] [PubMed] [Google Scholar]
- Alisi A, Carpino G, Oliveira FL, Panera N, Nobili V, and Gaudio E (2017). The Role of Tissue Macrophage-Mediated Inflammation on NAFLD Pathogenesis and Its Clinical Implications. Mediators Inflamm 2017, 8162421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes C, and Bhimji SS (2018). Fatty Liver In StatPearls (Treasure Island (FL)). [Google Scholar]
- Ayala A, Munoz MF, and Arguelles S (2014). Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev 2014, 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldan A, Gonen A, Choung C, Que X, Marquart TJ, Hernandez I, Bjorkhem I, Ford DA, Witztum JL, and Tarling EJ (2014). ABCG1 is required for pulmonary B-1 B cell and natural antibody homeostasis. J Immunol 193, 5637–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara M, Scott A, and Alkhouri N (2018). New insights into genetic predisposition and novel therapeutic targets for nonalcoholic fatty liver disease. Hepatobiliary Surg Nutr 7, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieghs V, Verheyen F, van Gorp PJ, Hendrikx T, Wouters K, Lutjohann D, Gijbels MJ, Febbraio M, Binder CJ, Hofker MH, et al. (2012). Internalization of modified lipids by CD36 and SR-A leads to hepatic inflammation and lysosomal cholesterol storage in Kupffer cells. PLoS One 7, e34378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder CJ, Chang MK, Shaw PX, Miller YI, Hartvigsen K, Dewan A, and Witztum JL (2002). Innate and acquired immunity in atherogenesis. Nat Med 8, 1218–1226. [DOI] [PubMed] [Google Scholar]
- Binder CJ, Papac-Milicevic N, and Witztum JL (2016). Innate sensing of oxidation-specific epitopes in health and disease. Nat Rev Immunol 16, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, and Auwerx J (2009). AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapper JR, Hendricks MD, Gu G, Wittmer C, Dolman CS, Herich J, Athanacio J, Villescaz C, Ghosh SS, Heilig JS, et al. (2013). Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am J Physiol Gastrointest Liver Physiol 305, G483–495. [DOI] [PubMed] [Google Scholar]
- Devisscher L, Scott CL, Lefere S, Raevens S, Bogaerts E, Paridaens A, Verhelst X, Geerts A, Guilliams M, and Van Vlierberghe H (2017). Non-alcoholic steatohepatitis induces transient changes within the liver macrophage pool. Cell Immunol 322, 74–83. [DOI] [PubMed] [Google Scholar]
- Dhibi M, Brahmi F, Mnari A, Houas Z, Chargui I, Bchir L, Gazzah N, Alsaif MA, and Hammami M (2011). The intake of high fat diet with different trans fatty acid levels differentially induces oxidative stress and non alcoholic fatty liver disease (NAFLD) in rats. Nutr Metab (Lond) 8, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes C, Razavi H, Loomba R, Younossi Z, and Sanyal AJ (2018). Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 67, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T (2012). Signal transduction by mitochondrial oxidants. J Biol Chem 287, 4434–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman P, Horkko S, Steinberg D, Witztum JL, and Dennis EA (2002). Correlation of antiphospholipid antibody recognition with the structure of synthetic oxidized phospholipids. Importance of Schiff base formation and aldol condensation. J Biol Chem 277, 7010–7020. [DOI] [PubMed] [Google Scholar]
- Friedman SL, Neuschwander-Tetri BA, Rinella M, and Sanyal AJ (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat Med 24, 908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Ikura Y, Arimoto J, Sugioka K, Iezzoni JC, Park SH, Naruko T, Itabe H, Kawada N, Caldwell SH, et al. (2009). Expression of perilipin and adipophilin in nonalcoholic fatty liver disease; relevance to oxidative injury and hepatocyte ballooning. J Atheroscler Thromb 16, 893–901. [DOI] [PubMed] [Google Scholar]
- Gorden DL, Myers DS, Ivanova PT, Fahy E, Maurya MR, Gupta S, Min J, Spann NJ, McDonald JG, Kelly SL, et al. (2015). Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res 56, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrikx T, Watzenbock ML, Walenbergh SM, Amir S, Gruber S, Kozma MO, Grabsch HI, Koek GH, Pierik MJ, Staufer K, et al. (2016). Low levels of IgM antibodies recognizing oxidation-specific epitopes are associated with human non-alcoholic fatty liver disease. BMC Med 14, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gea V, and Friedman SL (2011). Pathogenesis of liver fibrosis. Annu Rev Pathol 6, 425–456. [DOI] [PubMed] [Google Scholar]
- Huber J, Vales A, Mitulovic G, Blumer M, Schmid R, Witztum JL, Binder BR, and Leitinger N (2002). Oxidized membrane vesicles and blebs from apoptotic cells contain biologically active oxidized phospholipids that induce monocyte-endothelial interactions. Arterioscler Thromb Vasc Biol 22, 101–107. [DOI] [PubMed] [Google Scholar]
- Ikura Y, Ohsawa M, Suekane T, Fukushima H, Itabe H, Jomura H, Nishiguchi S, Inoue T, Naruko T, Ehara S, et al. (2006). Localization of oxidized phosphatidylcholine in nonalcoholic fatty liver disease: impact on disease progression. Hepatology 43, 506–514. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, et al. (2008). Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133, 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadl A, Sharma PR, Chen W, Agrawal R, Meher AK, Rudraiah S, Grubbs N, Sharma R, and Leitinger N (2011). Oxidized phospholipid-induced inflammation is mediated by Toll-like receptor 2. Free Radic Biol Med 51, 1903–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsyuba E, Mottis A, Zietak M, De Franco F, van der Velpen V, Gariani K, Ryu D, Cialabrini L, Matilainen O, Liscio P, et al. (2018). De novo NAD(+) synthesis enhances mitochondrial function and improves health. Nature 563, 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CW, Addy C, Kusunoki J, Anderson NN, Deja S, Fu X, Burgess SC, Li C, Ruddy M, Chakravarthy M, et al. (2017). Acetyl CoA Carboxylase Inhibition Reduces Hepatic Steatosis but Elevates Plasma Triglycerides in Mice and Humans: A Bedside to Bench Investigation. Cell Metab 26, 394–406 e396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, et al. (2005). Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321. [DOI] [PubMed] [Google Scholar]
- Koyama Y, and Brenner DA (2017). Liver inflammation and fibrosis. J Clin Invest 127, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Birukov KG, Romanoski CE, Springstead JR, Lusis AJ, and Berliner JA (2012). Role of phospholipid oxidation products in atherosclerosis. Circ Res 111, 778–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, and Bickel J (2011). Measuring reproducibility of high-throughput experiments. The Annals of Applied Statistics 5, 1752–1779. [Google Scholar]
- Li R, Chen W, Yanes R, Lee S, and Berliner JA (2007). OKL38 is an oxidative stress response gene stimulated by oxidized phospholipids. J Lipid Res 48, 709–715. [DOI] [PubMed] [Google Scholar]
- Link VM, Duttke SH, Chun HB, Holtman IR, Westin E, Hoeksema MA, Abe Y, Skola D, Romanoski CE, Tao J, et al. (2018). Analysis of Genetically Diverse Macrophages Reveals Local and Domain-wide Mechanisms that Control Transcription Factor Binding and Function. Cell 173, 1796–1809 e1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Czajka A, and Cunningham P (2016). Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion 29, 59–64. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Gattolliat CH, and Asselah T (2018). Mitochondrial Dysfunction and Signaling in Chronic Liver Diseases. Gastroenterology 155, 629–647. [DOI] [PubMed] [Google Scholar]
- Medeiros DM (2008). Assessing mitochondria biogenesis. Methods 46, 288–294. [DOI] [PubMed] [Google Scholar]
- Mederacke I, Dapito DH, Affo S, Uchinami H, and Schwabe RF (2015). High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc 10, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller YI, Choi SH, Wiesner P, Fang L, Harkewicz R, Hartvigsen K, Boullier A, Gonen A, Diehl CJ, Que X, et al. (2011). Oxidation-specific epitopes are danger-associated molecular patterns recognized by pattern recognition receptors of innate immunity. Circ Res 108, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, and Nagata S (2007). Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439. [DOI] [PubMed] [Google Scholar]
- Muriel PR-T, E.; Montes-Páez G; Buendía-Montaño LD (2017). Experimental Models of Liver Damage Mediated by Oxidative Stress In Liver Pathophysiology Therapies and Antioxidants (Elsevier; ), pp. 529–546. [Google Scholar]
- Muse ED, Yu S, Edillor CR, Tao J, Spann NJ, Troutman TD, Seidman JS, Henke A, Roland JT, Ozeki KA, et al. (2018). Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc Natl Acad Sci U S A 115, E4680–E4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassir F, and Ibdah JA (2014). Role of mitochondria in nonalcoholic fatty liver disease. Int J Mol Sci 15, 8713–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navab M, Ananthramaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fonarow GC, Vahabzadeh K, Hama S, Hough G, Kamranpour N, et al. (2004). The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J Lipid Res 45, 993–1007. [DOI] [PubMed] [Google Scholar]
- Oishi Y, Spann NJ, Link VM, Muse ED, Strid T, Edillor C, Kolar MJ, Matsuzaka T, Hayakawa S, Tao J, et al. (2017). SREBP1 Contributes to Resolution of Pro-inflammatory TLR4 Signaling by Reprogramming Fatty Acid Metabolism. Cell Metab 25, 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen BM, Ding X, Morgan DA, Coate KC, Bookout AL, Rahmouni K, Kliewer SA, and Mangelsdorf DJ (2014). FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metab 20, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Gugiu B, Fox PL, et al. (2002). Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J Biol Chem 277, 38503–38516. [DOI] [PubMed] [Google Scholar]
- Popat RJ, Hakki S, Thakker A, Coughlan AM, Watson J, Little MA, Spickett CM, Lavender P, Afzali B, Kemper C, et al. (2017). Anti-myeloperoxidase antibodies attenuate the monocyte response to LPS and shape macrophage development. JCI Insight 2, e87379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que X, Hung MY, Yeang C, Gonen A, Prohaska TA, Sun X, Diehl C, Maatta A, Gaddis DE, Bowden K, et al. (2018). Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi A, Boekholdt SM, Mallat Z, Talmud PJ, Kastelein JJ, Wareham NJ, Miller ER, Benessiano J, Tedgui A, Witztum JL, et al. (2011). Relationship of IgG and IgM autoantibodies and immune complexes to oxidized LDL with markers of oxidation and inflammation and cardiovascular events: results from the EPIC-Norfolk Study. J Lipid Res 52, 1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector RS, Thyfault JP, Uptergrove GM, Morris EM, Naples SP, Borengasser SJ, Mikus CR, Laye MJ, Laughlin MH, Booth FW, et al. (2010). Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. J Hepatol 52, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, and Kietzmann T (2016). Reactive oxygen species and fibrosis: further evidence of a significant liaison. Cell Tissue Res 365, 591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Konzack A, Pihlajaniemi T, Heljasvaara R, and Kietzmann T (2015). Redox-fibrosis: Impact of TGFbeta1 on ROS generators, mediators and functional consequences. Redox Biol 6, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanoski CE, Che N, Yin F, Mai N, Pouldar D, Civelek M, Pan C, Lee S, Vakili L, Yang WP, et al. (2011). Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ Res 109, e27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero F, Shah D, Duong M, Penn RB, Fessler MB, Madenspacher J, Stafstrom W, Kavuru M, Lu B, Kallen CB, et al. (2015). A pneumocyte-macrophage paracrine lipid axis drives the lung toward fibrosis. Am J Respir Cell Mol Biol 53, 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S, Cortes VA, Rashid S, Anderson NN, McDonald JG, Liang G, Moon YA, Hammer RE, and Horton JD (2017). Expression of SREBP-1c Requires SREBP-2-mediated Generation of a Sterol Ligand for LXR in Livers of Mice. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhanizadeh M, Hwang J, Clempus RE, Marcu L, Lassegue B, Sevanian A, and Hsiai TK (2005). Oxidized-1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphorylcholine induces vascular endothelial superoxide production: implication of NADPH oxidase. Free Radic Biol Med 39, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl B, Choi YH, Olsen JC, Hagedorn CH, and Brenner DA (2002). Immortal activated human hepatic stellate cells generated by ectopic telomerase expression. Lab Invest 82, 323–333. [DOI] [PubMed] [Google Scholar]
- Scott CL, T’Jonck W, Martens L, Todorov H, Sichien D, Soen B, Bonnardel J, De Prijck S, Vandamme N, Cannoodt R, et al. (2018). The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity 49, 312–325 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, et al. (2016). Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun 7, 10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimon TA, Nadolski MJ, Liao X, Magallon J, Nguyen M, Feric NT, Koschinsky ML, Harkewicz R, Witztum JL, Tsimikas S, et al. (2010). Atherogenic lipids and lipoproteins trigger CD36-TLR2-dependent apoptosis in macrophages undergoing endoplasmic reticulum stress. Cell Metab 12, 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, and Schwabe RF (2007). TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 13, 1324–1332. [DOI] [PubMed] [Google Scholar]
- Senders ML, Que X, Cho YS, Yeang C, Groenen H, Fay F, Calcagno C, Meerwaldt AE, Green S, Miu P, et al. (2018). PET/MR Imaging of Malondialdehyde-Acetaldehyde Epitopes With a Human Antibody Detects Clinically Relevant Atherothrombosis. J Am Coll Cardiol 71, 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbulea V, Upchurch CM, Ahern KW, Bories G, Voigt P, DeWeese DE, Meher AK, Harris TE, and Leitinger N (2018). Macrophages sensing oxidized DAMPs reprogram their metabolism to support redox homeostasis and inflammation through a TLR2-Syk-ceramide dependent mechanism. Mol Metab 7, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbulea V, Upchurch CM, Schappe MS, Voigt P, DeWeese DE, Desai BN, Meher AK, and Leitinger N (2018). Macrophage phenotype and bioenergetics are controlled by oxidized phospholipids identified in lean and obese adipose tissue. Proc Natl Acad Sci U S A 115, E6254–E6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, Vrbanac AF, Li W, Perkins A, Matsutani T, et al. (2017). Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 551, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PX, Horkko S, Chang MK, Curtiss LK, Palinski W, Silverman GJ, and Witztum JL (2000). Natural antibodies with the T15 idiotype may act in atherosclerosis, apoptotic clearance, and protective immunity. J Clin Invest 105, 1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha N, Chand L, Han MK, Lee SO, Kim CY, and Jeong YJ (2016). Glutamine inhibits CCl4 induced liver fibrosis in mice and TGF-beta1 mediated epithelial-mesenchymal transition in mouse hepatocytes. Food Chem Toxicol 93, 129–137. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. (2006). Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408. [DOI] [PubMed] [Google Scholar]
- Tiniakos DG, Vos MB, and Brunt EM (2010). Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol 5, 145–171. [DOI] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. (2015). Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop MH, Speakman JR, Arch JR, Auwerx J, Bruning JC, Chan L, Eckel RH, Farese RV Jr., Galgani JE, Hambly C, et al. (2011). A guide to analysis of mouse energy metabolism. Nat Methods 9, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantoulas D, Perkmann T, Afonyushkin T, Mangold A, Prohaska TA, Papac-Milicevic N, Millischer V, Bartel C, Horkko S, Boulanger CM, et al. (2015). Circulating microparticles carry oxidation-specific epitopes and are recognized by natural IgM antibodies. J Lipid Res 56, 440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimikas S, Brilakis ES, Lennon RJ, Miller ER, Witztum JL, McConnell JP, Kornman KS, and Berger PB (2007). Relationship of IgG and IgM autoantibodies to oxidized low density lipoprotein with coronary artery disease and cardiovascular events. J Lipid Res 48, 425–433. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, and Friedman SL (2017). Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 14, 397–411. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Lee YA, Fujiwara N, Ybanez M, Allen B, Martins S, Fiel MI, Goossens N, Chou HI, Hoshida Y, et al. (2018). A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol 69, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar F, Sezer S, Erdogan S, Akyol S, Armutcu F, and Akyol O (2013). The relationship between oxidative stress and nonalcoholic fatty liver disease: Its effects on the development of nonalcoholic steatohepatitis. Redox Rep 18, 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lenten BJ, Wagner AC, Navab M, and Fogelman AM (2001). Oxidized phospholipids induce changes in hepatic paraoxonase and ApoJ but not monocyte chemoattractant protein-1 via interleukin-6. J Biol Chem 276, 1923–1929. [DOI] [PubMed] [Google Scholar]
- Wang X, Zheng Z, Caviglia JM, Corey KE, Herfel TM, Cai B, Masia R, Chung RT, Lefkowitch JH, Schwabe RF, et al. (2016). Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab 24, 848–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win S, Than TA, Zhang J, Oo C, Min RWM, and Kaplowitz N (2018). New insights into the role and mechanism of c-Jun-N-terminal kinase signaling in the pathobiology of liver diseases. Hepatology 67, 2013–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, and Ahmed A (2015). Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 148, 547–555. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. (1999). Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124. [DOI] [PubMed] [Google Scholar]
- Wynn TA, and Barron L (2010). Macrophages: master regulators of inflammation and fibrosis. Semin Liver Dis 30, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MQ, Du Q, Goswami J, Varley PR, Chen B, Wang RH, Morelli AE, Stolz DB, Billiar TR, Li J, et al. (2018). Interferon regulatory factor 1-Rab27a regulated extracellular vesicles promote liver ischemia/reperfusion injury. Hepatology 67, 1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi ZM, Loomba R, Anstee QM, Rinella ME, Bugianesi E, Marchesini G, Neuschwander-Tetri BA, Serfaty L, Negro F, Caldwell SH, et al. (2018). Diagnostic modalities for nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, and associated fibrosis. Hepatology 68, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi ZM, Loomba R, Rinella ME, Bugianesi E, Marchesini G, Neuschwander-Tetri BA, Serfaty L, Negro F, Caldwell SH, Ratziu V, et al. (2018). Current and future therapeutic regimens for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 68, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Marsh S, Hu J, Feng W, and Wu C (2016). The Pathogenesis of Nonalcoholic Fatty Liver Disease: Interplay between Diet, Gut Microbiota, and Genetic Background. Gastroenterol Res Pract 2016, 2862173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Springstead JR, and Kagan JC (2017). By Capturing Inflammatory Lipids Released from Dying Cells, the Receptor CD14 Induces Inflammasome-Dependent Phagocyte Hyperactivation. Immunity 47, 697–709 e693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Wong KI, Sun X, Reilly SM, Uhm M, Liao Z, Skorobogatko Y, and Saltiel AR (2018). TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell 172, 731–743 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the transcriptomic data reported in this paper is GEO: GSE138419.