Abstract
Manipulation of host protein post-translational modifications (PTMs) is used by various pathogens to interfere with host cell functions. Among these modifications, ubiquitin (UBI) and ubiquitin-like proteins (UBLs) constitute key targets because they are regulators of pathways essential for the host cell. In particular, these PTM modifiers control pathways that have been described as crucial for infection such as pathogen entry, replication, propagation, or detection by the host. Although bacterial pathogens lack eucaryotic-like UBI or UBL systems, many of them produce proteins that specifically interfere with these host PTMs during infection. In this review we discuss the different mechanisms used by bacteria to interfere with host UBI and the two UBLs, SUMO and NEDD8.
UBI and UBLs Constitute Essential Modifiers of Host Proteins
Protein PTMs are involved in the regulation of protein stability, activity, localization, and interactions with other cellular components. PTMs encompass a wide range of chemical processes. They include the cleavage of peptide bonds (proteolysis), the modification of specific amino acid sidechains by deamidation (the removal of an NH2 group) or eliminylation (the irreversible removal of a phosphate group), and the covalent addition of chemical moieties ranging from simple groups (such as phosphate, acetyl, or methyl groups) to more complex groups such as sugar, lipids, or even small polypeptides [1,2].
The human genome encodes several polypeptides that can be covalently linked, via their C-terminal glycine residues, to target proteins. The best-studied of these polypeptides, UBI, is a small polypeptide of 76 amino acids that was discovered more than 40 years ago [3–5]. In addition to UBI, other polypeptides such as SUMO (small ubiquitin-like modifier) [6], NEDD8 (neural precursor cell expressed developmentally downregulated protein 8) [7], ISG15 (interferon-stimulated gene 15) [8], and FAT10 (HLA-F-adjacent transcript 10) [9] can be similarly conjugated to target proteins. These polypeptides are grouped in the so-called UBL family and share high structural homology with UBI [10].
The consequences of UBI/UBL conjugation on the fate of the modified proteins are very diverse. UBI/UBL can alter the half-life of the modified proteins, for example by targeting them for proteasomal degradation [4–7,9]. They can change the structure of their target, thereby affecting their catalytic activity [4,6–8]. They can add new surfaces for interactions or mask internal binding domains and change the target interactome [4,6–8]. In particular, the cell encodes many ‘receptors’ containing UBI-binding domains (UBDs) or UBL-binding domains (such as SUMO interacting motifs, SIMs), that interact with proteins once conjugated to UBI/UBL and ‘decode’ these modifications into biochemical cascades in the cell [6,11]. Except for the well-known example of K48-UBI chain conjugation (see below), that addresses modified proteins to proteasomal degradation, it is usually difficult to anticipate the consequences of UBI or UBL conjugation to a given protein.
UBI/UBL are essential regulators of fundamental pathways in cell biology. Some of these pathways are crucial for the outcome of infection by pathogens. For example, UBI is a major regulator of the NF-κB pathway that triggers the expression of proinflammatory cytokines in response to pathogen detection [12]. SUMO is a central player in the regulation of type I interferon and in antiviral gene expression programs [13]. ISG15 plays several independent roles in antiviral defense and can restrict intracellular bacteria replication in vitro and in vivo [8,14,15]. FAT10 has been reported to be involved in xenophagy and in antimicrobial defense [9,16]. It is thus not surprising that pathogens have evolved strategies to target UBI/UBL and interfere with these different cellular processes.
In this review we present how pathogens interfere with host UBI/UBL systems. The UBI and UBL systems have been shown to be targeted by diverse pathogens such as viruses, bacteria, and parasites, including Plasmodium falciparum and Toxoplasma gondii [17–24]. We will focus on pathogenic bacteria because they display the widest variety of UBI/UBL-interfering strategies known to date. Although bacteria do not have eukaryotic-like UBI/UBL systems (Box 1), numerous species encode virulence factors that actually manipulate host UBI/UBL systems (Table 1). These factors can be toxins that are secreted into the extracellular space in the vicinity of the host cell, or effectors that are delivered directly into host cells via specialized systems such as type III secretion systems (T3SS) [25,26]. We discuss how bacterial pathogens (i) target UBI/UBLs conjugation machineries, (ii) increase or decrease the UBI/UBL conjugation of specific host factors, (iii) directly target UBI/UBL polypeptides, or (iv) use host UBI/UBL to modify their own proteins. We shed light on how these mechanisms allow bacterial pathogens to manipulate specific host cellular pathways to promote infection. Understanding how pathogens manipulate host UBI/UBL pathways is essential because it represents a prominent facet of host–pathogen interactions and may lead to the identification of new therapeutic targets.
Box 1. Prokaryotic UBLs.
Even if bacteria do not encode UBI, two types of bacterial polypeptides have been reported that can be conjugated to target proteins via covalent bonds [110,111]. The first type of bacterial modifier is called Pup (for prokaryotic ubiquitin-like protein) and is restricted to specific bacterial phyla such as Actinobacteria. Pupylation, namely the covalent fixation of Pup to target proteins, is a fascinating example of convergent evolution with respect to eukaryotic ubiquitination. It involves an enzymatic machinery distinct from ubiquitination, that deamidates Pup C-terminal Gln before linking it to target lysine residues [110,111]. This covalent modification targets bacterial proteins to proteasomal degradation. Several hundred pupylated targets have been identified to date, and these are involved in a variety of pathways. In Mycobacterium tuberculosis, pupylation is essential for the survival of bacteria in the host, and may thus constitute an interesting drug target [110–112]. The second type of bacterial modifier is constituted by polypeptides that differ from UBI in sequence but share a common structural β-grasp fold [110]. These UBI-fold proteins include the archeal SAMPs (small archeal modifier proteins) and Thermus TtuB (tRNA-two-thiouridine B) that function both as protein modifiers and sulfur carriers in the synthesis of sulfur-containing biomolecules [110]. Bacterial UBI-fold proteins linkage to lysine residues in target proteins involves UBI E1 homologs but not canonical E2 or E3 ubiquitin-like enzymes [110].
In addition to these modifiers that are analogous to UBI, some human commensal bacteria may even usurp eukaryotic UBI for their own purpose. Indeed, an UBI gene has been identified in the genome of Bacteroides fragilis strains [113,114]. Interestingly, this eukaryotic-like UBI, which was probably acquired via horizontal gene transfer, does not seem to be involved in bacterial protein modification since it lacks the crucial terminal glycine residue. This protein is instead secreted and acts as a bacterial toxin that targets and kills other intestinal bacteria [114]. Many other surprises such as this are probably still awaiting to be discovered and, even though the first report of a bacterium post-translationally modifying a host protein was almost 50 years ago [115], the field of pathogen and host PTMs is, without a doubt, still very promising.
Table 1. Examples of Bacterial Proteins Interfering with UBI/UBL Conjugation to Host Proteinsa.
| UBI/UBL target | Bacteria | Extra/intracellular bacteria | Effector | Enzymatic activity | Effect | Refs |
|---|---|---|---|---|---|---|
| UBI | Salmonella Typhimurium | Intracellular | SopA | E3 UBI ligase (HECT) | Regulation of host inflammation | [67] |
| UBI | EPEC, EHEC | Extracellular | NleL | E3 UBI ligase (HECT) | Regulation of actin pedestal formation | [101] |
| UBI | EPEC, EHEC | Extracellular | NleG | E3 UBI ligase (RING) | ND | [102] |
| UBI | Pseudomonas syringae | Extracellular | AvrPtoB | E3 UBI ligase (U-box) | Inhibition of plant pattern-triggered immunity | [103,104] |
| UBI | Shigella flexneri | Intracellular | OspI | Gln deamidase | Inactivation of UBE2N/UBC13 (E2 UBI enzyme (NF-κB pathway) | [34] |
| UBI | Shigella flexneri | Intracellular | IpaH1.4 | E3 UBI ligase (NEL) | Ubiquitination of LUBAC (NF-κB pathway) | [60] |
| UBI | Shigella flexneri | Intracellular | IpaH2.5 | E3 UBI ligase (NEL) | Ubiquitination of LUBAC (NF-κB pathway) | [60] |
| UBI | Shigella flexneri | Intracellular | IpaH0722 | E3 UBI ligase (NEL) | Ubiquitination of TRAF2 (NF-κB pathway) | [62] |
| UBI | Shigella flexneri | Intracellular | IpaH9.8 | E3 UBI ligase (NEL) | Ubiquitination of NEMO (NF-κB pathway) | [61] |
| UBI | Salmonella Typhimurium | Intracellular | Ssph1 | E3 UBI ligase (NEL) | Ubiquitination of PKN1 (NF-κB pathway) | [105] |
| UBI | Legionella pneumophila | Intracellular | SdeA | Non-eukaryotic UBI ligase | E1/E2-independent ubiquitination of RAB GTPases and RTN4 | [46–48] |
| UBI | Shigella flexneri | Intracellular | OspG | Kinase | Inhibition of UBCH5 (E2 UBI enzyme; NF-κB pathway) | [63] |
| UBI | EPEC, EHEC | Extracellular | NleB | Glycosyltransferase | Inhibition of TRAF2 ubiquitination (NF-κB pathway) | [106] |
| UBI | EPEC | Extracellular | ND | ND | Downregulation of UBE1 and UBA6 (E1 UBI enzymes) | [35] |
| UBI | EPEC | Extracellular | NleE | Cys methyltransferase | Inactivation of TAB2 and TAB3 (NF-κB pathway) | [67,107] |
| UBI | Legionella pneumophila | Intracellular | SidJ | Deubiquitylase | ND | [50] |
| UBI | Shigella flexneri | Intracellular | ShiCE | Deubiquitylase | ND | [54] |
| UBI | Chlamydia trachomatis | Intracellular | ChlaDUB1 | Deubiquitylase | Inhibition of NF-κB pathway activation | [108,109] |
| UBI | Burkholderia pseudomallei | Extracellular | CHBP | Gln deamidase | Deamidation of UBI | [57] |
| SUMO | Listeria monocytogenes | Intracellular | LLO | Pore-forming toxin | Downregulation of UBE2I/UBC9 (E2 SUMO enzyme) | [25] |
| SUMO | Clostridium perfringens | Extracellular | PFO | Pore-forming toxin | Downregulation of UBE2I/UBC9 (E2 SUMO enzyme) | [25] |
| SUMO | Streptococcus pneumoniae | Extracellular | PLY | Pore-forming toxin | Downregulation of UBE2I/UBC9 (E2 SUMO enzyme) | [25] |
| SUMO | Shigella flexneri | Intracellular | ND/Ca2+ influx | ND | Proteolytic cleavage of UBA2/SAE2 (E1 SUMO enzyme) | [39] |
| SUMO | Salmonella Typhimurium | Intracellular | ND/miRNAs | ND | Downregulation of UBE2I/UBC9 (E2 SUMO enzyme) | [38] |
| SUMO | Xanthomonas euvesicatoria | Extracellular | XopD | DeSUMOylase | DeSUMOylation of SIERF4 (plant immune response) | [51,52] |
| NEDD8 | EPEC | Extracellular | CIF | Gln deamidase | Deamidation of NEDD8 | [57,58] |
| NEDD8 | Chlamydia trachomatis | Intracellular | ChlaDUB1 | DeNEDDylase | Inhibition of NF-κB pathway activation | [108,109] |
Abbreviations: EPEC, enteropathogenic Escherichia coli; EHEC, enterohemorrhagic Escherichia coli; HECT, homologous to the E6-AP C terminus; ND, not determined; NEL, novel E3 ligase; RING, really interesting new gene.
UBI/UBL Conjugation Mechanisms in Eukaryotes
Ubiquitination, namely the conjugation of UBI, usually occurs on lysine residues of target proteins, although conjugation to other amino acids such as threonine, serine, or cysteine may also occur [5]. UBI itself contains seven lysines (K6, K11, K27, K29, K33, K48, and K63) that can serve as sites for additional cycles of UBI attachment, resulting in the formation of UBI chains. The topology of these chains is diverse, ranging from ‘homotypic’ K48- or K63-linked chains, composed of only one type of UBI linkage, to ‘mixed’ chains containing for example both K11 and K63 linkages [4,27]. An additional type of chain, called a ‘linear’ chain, is generated when UBI is attached to the N terminus of a second UBI [28]. Targeting of a given protein by UBI may thus result in mono-ubiquitination, multi-mono-ubiquitination (i.e., several mono-ubiquitinations on different amino acids), or poly-ubiquitination. UBI is attached to substrates by a three-step enzymatic cascade involving E1 (UBI-activating enzyme), E2 (UBI-conjugating enzyme), and E3 (UBI ligase) enzymes [4]. UBI is generally first activated in an ATP-dependent manner by E1, which links the C-terminal glycine residue of UBI via a thioester bond to a cysteine residue within the E1 active site. This activated UBI is then transferred to the catalytic cysteine residue of an E2 enzyme. E3 ligases then finally mediate the transfer of UBI from the E2 enzyme to specific substrates. There are two major classes of E3s: the HECT (homologous to the E6-AP C terminus) type and the RING (really interesting new gene)/U-box type. HECT-type E3 UBI ligases form a reactive intermediate with UBI before its transfer to the substrate protein, whereas RING/U-box-type E3 ligases mediate transfer of UBI from the E2 directly to the substrate protein, without forming an E3–UBI intermediate [29]. Of note, U-box protein E3 ligases display unique preferences for E2 and UBI chain formation compared to RING E3 ligases, and may be classified as an independent type of E3 [30]. In addition to HECT and RING/U-box E3 ligases, other classes of host E3 ligases have been described such as RBR (RING-between-RING) and RCR (RING-Cys-relay) ligases [31,32]. RBR ligases, such as parkin and HOIP, combine mechanistic features of RING and HECT-type E3 ligases [31], whereas RCR exhibits esterification activity and intrinsic selectivity for non-lysine residues [32]. Conjugation of UBI is a reversible process because several cellular isopeptidases (called deubiquitinases, DUBs) can cleave the covalent bond between UBI and its targets, and thereby remove UBI [33].
The mechanisms of UBL conjugation on target substrates is similar to that observed for ubiquitination. The enzymes required for all these modifications (i.e., E1 UBL-activating enzymes, E2 UBL-conjugating enzymes, and E3 UBL ligases) share highly conserved domain structures [10]. Of note, the number of UBL specific E1, E2, and E3 enzymes is usually smaller than for UBI. For example, SUMO conjugation to thousands of cellular targets seems to rely only on a single SUMO E1 enzyme (SAE1/UBA2), a single SUMO E2 enzyme (UBC9), and a dozen SUMO E3 ligases [6]. Similarly to UBI, the formation of UBL chains (where UBLs are conjugated to internal lysines of other UBLs) has been reported for SUMO and NEDD8 [6,7]. Finally, as for UBI, the host cell encodes several ULPs (UBL-specific proteases) that guarantee the reversibility of UBL conjugation [6–9].
Harnessing of Host UBI and UBL Conjugation by Bacterial Pathogens
Targeting of Host UBI and UBL Conjugation Machinery Enzymes
Targeting of host E1, E2, or E3 UBI enzymes constitutes a first strategy used by pathogens to dampen ubiquitination (Figure 1, Key Figure). This strategy is used for example by Shigella flexneri, the etiological agent of bacillary dysentery. This bacterium secretes through its T3SS an effector, named OspI, that deamidates the glutamine residue at position 100 in the human E2 UBI enzyme UBC13 [34]. This deamidation inactivates the UBC13 UBI-conjugating activity, leading to dampening of the UBI-dependent TRAF6-mediated signaling pathway which is involved in the activation of the NF-κB pathway (see below). This finally results in the inhibition of host inflammatory responses during infection [34]. Extracellular pathogens such as enteropathogenic Escherichia coli (EPEC) also target the host UBI conjugation machinery. Adhesion of these bacteria to human cells leads to the degradation of UBE1 and UBA6, the two E1 UBI enzymes [35]. This degradation involves aspartyl protease-dependent and proteasome-independent mechanisms, and triggers a global decrease of host protein ubiquitination [35].
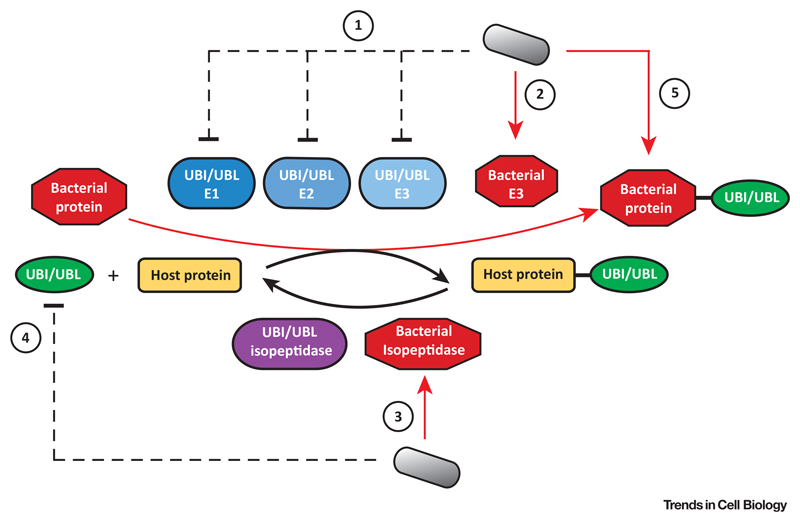
Figure 1. Key Figure: Main Strategies Used by Bacterial Pathogens to Interfere with Host Ubiquitin (UBI) or Ubiquitin-Like Protein (UBL) Modifications.
During infection, bacteria may inhibit or trigger the degradation of UBI/UBL enzymes such as E1 activating enzymes, E2 conjugating enzymes, or E3 ligases (1). These effects can be directly triggered by bacterial effectors or indirectly via the induction of host signaling cascades (dashed lines). Bacteria may also secrete effectors possessing either E3 ligase activity, which conjugate UBI/UBL to host targets, or isopeptidase activity, which remove UBI/UBL from their targets (3). Bacteria may also directly modify UBI or UBLs to block their conjugation to host targets (4). Finally, bacteria may hijack host UBI/UBL systems to modify their own proteins during infection (5). Red arrows, proteins secreted by bacteria.
The SUMO conjugation machinery constitutes another target for bacterial pathogens. Listeria monocytogenes, the bacterium responsible for human listeriosis, dampens SUMOylation of specific host factors by triggering the degradation of UBC9, the unique host E2 SUMO enzyme [25,36,37]. The degradation of UBC9 is triggered by the formation of pores in the host plasma membrane by the bacterial toxin listeriolysin O (LLO) [25]. Because LLO pores are not reported to affect the activity of host deSUMOylases, UBC9 degradation ultimately results in a shift in the SUMOylation/deSUMOylation equilibrium in the cell and the deSUMOylation of host proteins [25]. The deSUMOylation events triggered by LLO were shown to promote Listeria infection. Indeed, removal of SUMO from host factors that are essential for infection, such as specific transcription factors, may modulate their activity and favor bacterial replication or survival in host cells [25,36]. Of note, other toxins of the same family as LLO, and secreted by extracellular pathogens, were shown to downregulate UBC9, indicating that interference with host SUMOylation is a strategy conserved between different classes of pathogenic bacteria [25]. Inhibition of the SUMOylation machinery has also been observed during infection with Salmonella Typhimurium, a bacterium that is responsible for gastroenteritis in humans, and with Shigella flexneri, but the underlying mechanisms involved here do not rely on the production of bacterial toxins. In the case of Salmonella Typhimurium, infection leads to upregulation in the host cell of two small noncoding RNAs (miR30c and miR30e) that downregulate UBC9 levels [38]. In the case of Shigella flexneri, infection is associated with an influx of calcium into the host cell. This ion flux activates the host calpain proteases which cleave UBA2, one of the two components of the E1 SUMO enzyme [39]. The resulting inhibition of SUMOylation is associated with increased Shigella entry [39,40].
Interestingly, the Listeria LLO toxin was recently shown to decrease the level of various E2 conjugases, in addition to UBC9, such as UBI E2s and the NEDD8 E2 UBC12/UBE2M [41]. This suggests that several host UBI/UBL systems may be simultaneously altered in response to Listeria infection.
Secretion of Bacterial Effectors Mimicking Host UBI and UBL Enzymes
In addition to interfering with UBI- or UBL-conjugation machineries, bacterial pathogens produce proteins that can replace or act as components of these machineries (Figure 1). In particular, several bacterial effectors possess UBI E3-like activity (Table 1). Some of these bacterial effectors share structural homologies with the two major types of eukaryotic E3 ligases, namely the HECT type and the RING/U-box type E3 ligases [20–22]. These effectors may have been acquired by bacteria via horizontal transfer from diverse eukaryotic sources [42]. In addition to these types, three other classes of bacterial effectors display structures that are completely distinct from those of eukaryotic E3 ligases: NELs (novel E3 ligases) [42], XL-box-containing E3 ligases [43], and SidC ligase [44]. These ligases may represent structures evolved by pathogens to mimic the functions of these essential host enzymes. The NEL family encompasses effectors from different bacterial genera, including Salmonella (SlrP, SspH1, and SspH2 effectors) and Shigella (IpaH family effectors) [20,42]. These enzymes possess an E2-interacting domain (which hijacks host E2 charged with UBI), a cysteine residue for UBI transfer, and leucine-rich repeats (LRRs) that allow recognition of a wide array of targets [20,42]. These different classes of E3 ligases enable bacteria to conjugate UBI to specific host factors, thereby altering their stability or function, subcellular localization, or interaction with other cellular proteins [20,22,42]. Bacterial E3 ligases may in particular conjugate K48-UBI chains to host proteins, thereby triggering their proteasome-dependent degradation. By rerouting host factors to one of the most efficient proteolytic systems of the infected cell, bacteria can eliminate key host components that normally interfere with their replication and propagation [20,22]. Finally, bacterial E3 ligases can also target other bacterial effectors that are codelivered during infection, allowing tight restriction of their activity during a specific timeframe [21,22,45] (see below).
In contrast to bacterial effectors mimicking host UBI enzymes, a family of proteins secreted by the bacterial pathogen Legionella pneumophila, the causative agent of Legionnaires’ disease, were recently shown to catalyze the ubiquitination of host proteins without the need for E1 and E2 UBI enzymes [46–48]. The Legionella SdeA effector belongs to this family of enzymes: it conjugates UBI to endoplasmic reticulum (ER)-associated RAB GTPases and participates in bacterial virulence [46,47,49]. By acting independently of E1- and E2-UBI enzymes, SdeA extends the repertoire of proteins potentially modified by UBI. Conjugation of UBI to host targets by SdeA does not rely on ATP and does not occur on lysines. SdeA instead uses NAD+ to mediate phosphoribosyl-linked ubiquitination of serine residues in host proteins [46,47,49]. In addition to ER-associated RAB GTPases, the Legionella effector SdeA and other members of the Sde family ubiquitinate the host protein reticulon 4 (RTN4), leading to ER reorganization and the formation of Legionella-containing vacuoles, which are intracellular factories supporting bacteria replication [48]. Unconventional UBI conjugation by Sde effectors is reversible because L. pneumophila codes for a specific deubiquitinase, SidJ, which removes phosphoribosylated UBI from its substrate [50]. Whether functional homologs of SdeA exist in eukaryotes and what roles they may play remain to be determined.
Deconjugation of UBI and UBL from Host Targets by Bacterial Effectors
Another strategy used by bacteria to interfere with UBI or UBL conjugation involves secretion into host cells of effectors with isopeptidase activity, which removes UBI or UBL from their targets (Figure 1 and Table 1). XopD, for example, is a T3SS effector secreted by the plant pathogen Xanthomonas euvesicatoria, which possesses SUMO-specific isopeptidase activity [51]. Upon infection of tomato cells, it deconjugates SUMO from the SIERF4 transcription factor to suppress host ethylene production, which constitutes an important pathway for plant antibacterial immunity [52]. Many other bacterial proteases targeting UBI or UBLs have been identified in bacterial pathogens, including Salmonella, Shigella, Chlamydia, and Legionella, and some are specific for a single UBL while others display crossreactivity between different UBLs [53,54]. Interestingly, several bacterial effectors possessing deubiquitinase activity display a strong preference for K63-linked chains over K48 or K11 chains [54]. This may reveal a significant selection pressure for bacteria to interfere with this specific UBI modification to promote infection.
Direct targeting of UBI and UBL Polypeptides
UBI itself, as well as other UBLs, can be directly targeted and inactivated by bacterial effectors (Figure 1). Phosphoribosylation of UBI, for example, that is catalyzed by the Legionella SdeA effector, was reported to interfere with multiple steps in the ubiquitination cascade [47]. The presence of phosphoribosylated UBI in chains further confers resistance to various deubiquitinases [55]. SdeA, by both triggering E1 and E2-independent ubiquitination of specific host targets, and by inhibiting ubiquitination of others, thus efficiently controls the host ubiquitinome [47].
UBI and NEDD8 are also targeted by a family of bacterial T3SS effectors, called Cifs (for cycle-inhibiting factors), that are produced by diverse pathogenic bacteria such as some EPEC or Burkholderia pseudomallei [56]. Cifs directly target NEDD8 and UBI, and catalyze the deamidation of the Gln40 residue of these polypeptides [57]. Deamidation of UBI interferes with UBI chain formation, whereas deamidation of NEDD8 blocks the activity of NEDDylated cullin-RING E3 UBI ligases (CRLs) and impairs ubiquitination of several CRL substrates in EPEC-infected cells [57,58]. Cifs inhibit in particular the ubiquitination of perforin-2/MPEG1 (macrophage-expressed gene 1), an antimicrobial host protein that forms pores on bacterial cells, thereby blocking its intracellular trafficking and bactericidal activity [59].
Main Host Pathways Targeted by Bacteria and Regulated by UBI or UBLs
During infection, bacterial pathogens alter the conjugation of UBI or UBLs to many different host proteins. These proteins belong to different pathways that are often essential for bacteria to efficiently enter host cells and replicate therein, or to dampen host antibacterial responses. We detail here some of the pathways that are tightly regulated by UBI/UBL modifications and are frequently targeted by bacterial pathogens.
The NF-κB Pathway
The NF-κB pathway is an essential pillar of innate immunity and inflammation. Cytoplasmic NF-κB transcription factors are translocated into the nucleus within minutes after exposure to bacteria-derived molecules, and induce the transcription of a wide range of proinflammatory chemokines and cytokines [12]. Not surprisingly, many bacterial effectors target the NF-κB pathway to dampen the host innate immune response. A given pathogen may produce several independent effectors targeting this pathway [12]. This apparent redundancy of effectors, that all target the same signaling cascade, reflects the diversity of danger signals sensed by the host that trigger this pathway.
One common strategy used by bacterial pathogens to dampen the NF-κB signaling cascade consists of conjugating K48-UBI chains to essential components of this pathway, thereby triggering their proteasome-dependent degradation [22]. Shigella flexneri, for example, uses at least five different effectors that trigger UBI-dependent degradation of diverse components involved in essential branches of the NF-κB pathway: IpaH1.4 and IpaH2.5 ubiquitinate the catalytic component of the linear UBI chain assembly complex (LUBAC) and trigger its proteasomal degradation. Degradation of this component decreases LUBAC-mediated ubiquitination of the NF-κB modulator NEMO, which suppresses NF-κB activation [60]; NEMO may also be directly ubiquitinated by IpaH9.8 [61]; IpaH0722 ubiquitinates TRAF2, a factor involved in NF-κB pathway activation following the detection of intracytosolic bacteria [62].
In addition to triggering proteasome-dependent degradation of components of the NF-κB pathway, bacterial pathogens also interfere with the endogenous ubiquitination of key NF-κB regulators: as mentioned above, the Shigella OspI effector inhibits the host E2 enzyme UBC13, thereby blocking TRAF6-mediated activation of the NF-κB pathway [34]; OspG, another Shigella effector, binds to and inhibits the host E2 UBI enzyme UBCH5 that is involved in IκBα ubiquitination and subsequent degradation, which is a prerequisite for NF-κB nuclear translocation and NF-κB-dependent gene transcription [63]; the NleB effector, encoded by EPEC, possesses an N-acetylglucosamine transferase activity that modifies death domains in several proteins such as FADD and TRADD, and disrupts TNF signaling pathways including NF-κB signaling [64,65]. Interference with the NF-κB pathway may finally rely on inhibition of UBI-binding proteins that decode UBI signals into biochemical cascades [11]. TAB2 and TAB3 are two UBI chain binding proteins that are involved in NF-κB signaling. TAB2/3 bind to K63-linked polyubiquitin chains on target proteins and activate the IκBα kinase, leading to IκBα phosphorylation, ubiquitination, and degradation [66]. The NleE effector, secreted by EPEC, is an S-adenosyl-L-methionine-dependent methyltransferase that specifically modifies a zinc-finger cysteine in TAB2/3. This abolishes the binding of these proteins to UBI chains and disrupts NF-κB signaling [67].
The NF-κB pathway thereby constitutes a good example of the diverse mechanisms that bacteria have evolved to promote or inhibit ubiquitination of a large number of components in a coordinated fashion, resulting in dampening of an essential arm of the host antibacterial response. Of course, these interfering strategies are not restricted to the NF-κB pathway, and other important signaling cascades of the innate immune response, such as the IFN response or the activation of inflammasome, can be similarly targeted [21,68]. The Shigella effector IpaH9.8, for example, was reported to induce the degradation of targets other than NEMO, such as GBPs (guanylate binding proteins) [69,70]. Following infection, GBPs are normally recruited to bacteria-containing vacuoles or vacuole-escaped bacteria, and participate in bacterial clearance. IpaH9.8 ubiquitinates GBPs with K48-linked chains and targets these proteins for proteasomal degradation, thereby counteracting GBP-mediated inhibition of bacterial growth [69,70].
Autophagy
Autophagy is a cellular process by which intracellular cytosolic material is degraded by lysosomes. Specific substrates, such as intracellular pathogens, can be tagged for targeting to the autophagy pathway. They become encapsulated in de novo-generated double-membrane vesicles, called autophagosomes, that eventually fuse with lysosomes, leading to degradation of their contents [71,72]. This selective autophagy is essential for cell-autonomous defense against bacteria invading the cytosol. Tagging of invading bacteria involves deposition of an UBI coat consisting of multiple polyubiquitin chains. These UBI chains are synthesized by several host E3 ligases such as LRSAM1, parkin, SMURF1, RNF166, and LUBAC [73–77]. These chains allow the recruitment of host cargo receptors that induce autophagosome formation. The host E3 ligase LUBAC generates in particular linear UBI chains around intracytosolic bacteria that transform the bacterial surface into antibacterial and proinflammatory signaling platforms [77]. Indeed, these LUBAC-synthesized UBI chains recruit host adaptors that activate antibacterial immunity pathways such as the NF-κB pathway.
Professional cytosol-dwelling bacteria have evolved evasion strategies to overcome restriction by autophagy. Some of these strategies rely on interference with host UBI. Shigella flexneri, for example, remodels UBI chains normally deposited by host ligases. It antagonizes the deposition of linear UBI chains by targeting LUBAC via the IpaH1.4 effector, thereby interfering with NF-κB pathway activation [60,77]. Intracellular bacteria may also evade autophagy by targeting ATG8, which regulates autophagosome biogenesis and recruitment of specific cargos during selective autophagy [78]. ATG8 actually belongs to the UBL family along with SUMO, NEDD8, and ISG15. Interestingly, the RavZ effector from L. pneumophila was shown to target ATG8 and inhibit autophagy [79]. This effector cleaves the amide bond between the C-terminal glycine residue and an adjacent aromatic residue in ATG8. This produces an irreversibly inactivated form of ATG8 that cannot be reconjugated [79]. This example suggests that UBLs other than SUMO, NEDD8, and ISG15 may constitute pivotal targets for pathogens to promote infection.
Host Cytoskeleton
Remodeling of the host cytoskeleton is frequently used by intracellular bacterial pathogens to enter the targeted cells, create a niche where they can efficiently replicate, and disseminate to neighboring cells [80]. Several components of the host cytoskeleton are regulated by UBI. RHO GTPases, for example, which control actin cytoskeleton dynamics, are degraded by the proteasome following UBI conjugation [81]. Interestingly, the ubiquitination level of RHO GTPases can be modulated during Salmonella infection, suggesting that this bacterium may modulate RHO GTPase turnover [82]. SUMO can be conjugated to different components of the host cytoskeleton as well, including actin itself and actin regulatory proteins, septins, or intermediate filaments such as keratins and lamins [83,84]. The role of UBI and UBL modifications in the regulation of the cytoskeleton is only in its infancy, but one can anticipate that it may represent an important target enabling bacterial pathogens to manipulate the architecture of the cell.
Transcription Factors
To exploit host functions, bacterial pathogens remodel the proteome of infected cells. This remodeling may result from deregulation of gene transcription by injection of bacterial proteins, such as nucleomodulins, that act directly on the host nucleus [85], or by interference with host transcription factors, some of them being regulated by UBI or UBLs. L. monocytogenes, for example, dampens the SUMOylation of numerous transcription factors during infection [36]. Because SUMO conjugation either increases or decreases the activity of transcription factors, this decrease in SUMOylation may modulate the expression of a specific subset of genes and lead to reprogramming of host gene expression. As mentioned above, decreasing the SUMOylation of host transcription factors is a strategy also used by the plant pathogen Xanthomonas euvesicatoria that specifically targets SUMO–SIERF4 to dampen the host ethylene-mediated antibacterial response [52]. Finally, the colibactin toxin, produced by some Escherichia coli strains in the intestine, induces downregulation of the SUMO isopeptidase SENP1 and an increase in the SUMOylation of the transcription factor TP53. This ultimately results in the emergence of senescent cells secreting growth factors that may promote colorectal carcinogenesis [86].
Promyelocytic Leukemia Protein (PML) Nuclear Bodies
PML is a protein that polymerizes in discrete nuclear assemblies known as PML nuclear bodies (NBs) and plays essential roles in many different cellular processes [87]. Key to its function, PML can be post-translationally modified by SUMO [87]. In addition to its role in antiviral host defense [18], PML was recently identified as a sensor of bacteria producing pore-forming toxins [37]. Indeed, treatment of human cells with listeriolysin O toxin, that is secreted by L. monocytogenes, triggers massive deSUMOylation of PML. This deSUMOylation of PML, coupled to an oxidative stress-dependent multimerization of PML, initiates host-cell antibacterial responses leading to a decrease in Listeria intracellular replication [37]. The example of PML highlights how SUMO alterations of some specific host proteins can constitute danger signals for the cells and initiate antibacterial responses. The putative role of PML in other bacterial infections targeting host SUMOylation, such as Shigella or Salmonella, remains unknown but deserves further investigation.
PTMs of Bacterial Proteins during Infection
In addition to interfering with host protein PTMs, bacteria can hijack host UBI- or UBL-conjugation machineries to modify their own components (Figure 1). Exactly as for eukaryotic proteins, conjugation of UBI or UBL has diverse effects on bacterial effectors and may change their intracellular localization, stability, or interaction with other bacterial or host factors [88]. PTM of bacterial proteins couples their activity to their arrival in the host cell cytoplasm. Interestingly, PTM of bacterial proteins can also be used by the host to tag exogenous proteins and target them for degradation [88].
Ubiquitination of Salmonella proteins constitute good examples that illustrate the versatility of this PTM for modulating the activity of bacterial proteins. SopE and SptP are two Salmonella effectors that contribute to the transient remodeling of the host-cell cytoskeleton [89]. These two effectors, which are delivered simultaneously by Salmonella, exhibit different half-lives. SopE, which is involved in actin cytoskeleton rearrangement, membrane ruffling, and bacterial uptake, is rapidly polyubiquitinated and degraded by the host proteasome [89]. SptP, which deactivates the RHO GTPases turned on by SopE, exhibits much slower degradation kinetics, allowing recovery of normal actin cytoskeleton architecture a few hours after infection [89]. Conjugation of UBI to SopB, a phosphoinositide phosphatase secreted by Salmonella via T3SS, modifies its cellular localization [90]. Upon delivery, SopB associates with the host plasma membrane where it participates in actin-mediated bacterial entry. Later on, ubiquitination of SopB by TRAF6 leads to its translocation to Salmonella-containing vacuoles, where it modulates vesicle trafficking and interferes with the delivery of these vacuoles to lysosomes [90,91]. Mass spectrometry-based large-scale analysis of the ubiquitinome of cells infected by Salmonella recently provided additional examples of bacterial proteins modified by UBI [82]. In addition to SopE and SopB, several effectors were identified as being ubiquitinated during infection [82]. Interestingly, integral outer-membrane proteins were reported to be conjugated to UBI, and these may represent the targets that are modified to generate the UBI coat that surrounds intracytosolic bacteria and that is involved in host antibacterial autophagy [71,82].
In contrast to ubiquitination, only a few bacterial proteins have been reported to be modified by SUMO, and the biological consequences of these modifications during infection remain elusive [92,93]. These SUMO-modified bacterial proteins include two effectors, Trp120 and AmpA, that are secreted by two intracellular pathogens, Ehrlichia chaffeensis and Anaplasma phagocytophilum respectively [92,93]. OspF, an effector secreted by Shigella flexneri, constitutes another example of where SUMO conjugation is necessary for translocation of this effector into the host nucleus, where it modulates the expression of proinflammatory cytokines [94].
One can anticipate that recently developed techniques for large-scale proteomic studies of UBL conjugation will increase the list of bacterial proteins modified by SUMO or other UBLs, and thus provide new insights into the role of these modifications during infection.
Concluding Remarks and Future Directions
UBI and UBL are essential post-translational modifiers of eukaryotic cells. Thousands of UBI/UBL targets have been identified during recent years, suggesting that most proteins are modified by this type of PTM at some point in their cellular lifetime. It is thus not surprising that pathogens have evolved so many strategies to interfere with these particular PTMs so as to manipulate host cell physiology. Interfering with host UBI/UBL modifications is observed both for intracellular pathogens, that tightly interact with host-cell cytoplasmic components to create, for example, a protective niche where they can acquire nutrients from the host, and for extracellular pathogens, that manipulate host cells to favor their maintenance at the surface of the cells or to dampen host immune responses.
Two types of interfering strategies are used by bacterial pathogens: they may either globally dampen UBI/UBL systems, by targeting the conjugation machineries or by modifying the equilibrium between conjugation/deconjugation reactions, or they may alter the level of UBI/UBL-conjugation of specific host proteins involved in bacterial proliferation and antibacterial responses. In the case of global dampening of UBI/UBL systems, a wide range of host proteins display altered levels of UBI/UBL conjugation. Discrimination, in this pool of proteins, of host factors directly involved in infection from other factors that only represent ‘collateral damage’ may constitute a real challenge in studying the exact role of these PTMs during infection.
Owing to continuous improvement in proteomic analyses, the list of proteins known to be modified by UBI or UBLs has greatly expanded over the past few years. For example, it is now feasible to compare the variations of the ubiquitinome (or other ‘UBL-ome’) of cells during infection by a pathogen or after exposure to a bacterial toxin [36,82]. Some of these techniques are furthermore compatible with in vivo analysis, and comparison of the content of proteins modified by UBI/UBL in organs from infected versus control animals is now possible [95,96]. Interestingly, current proteomic-based approaches have not only revealed the identity of the proteins modified by UBI/UBL but also the modification sites. These data are crucial for further analysis of the role of these PTMs in the function of the identified protein, and hence for deciphering the consequences of bacterial alterations of these PTMs. Several recent studies on UBI conjugation have revealed that ubiquitination establishes a much more complex code than was originally thought. Indeed, in addition to ‘mixed’ UBI chains involving different types of linkages between UBI monomers, chains that mix UBI with other UBLs such as SUMO have also been reported [4,27,97]. In addition, UBI itself has recently been found to be post-translationally modified by acetylation or phosphorylation, which further expands the repertoire of ubiquitination [4,27,97]. We are only beginning to understand the tremendous diversity of UBI modifications and their roles in cell biology, but it is very likely that bacterial pathogens have long learned how to break this so-called ‘ubiquitin code’ and efficiently use it for their own profit (see Outstanding Questions).
Outstanding Questions.
Are the recently described non-canonical ubiquitination mechanisms (i.e., conjugation involving non-RING/non-HECT E3 ligases or E1/E2-independent ubiquitin conjugation) strictly confined to bacteria – or are functional homologs of these bacterial enzymes encoded by human cells?
Recent improvements in proteomic analyses now allow thorough monitoring of the changes in the host ubiquitinome/‘UBL-ome’ in response to infection. These approaches usually provide thousands of putative candidate proteins showing altered UBI/UBL-conjugation levels in response to infection. Which strategies should researchers use to cope with this complex set of data and identify key players affecting the outcome of infection?
Are there mutations in the human population that affect UBI/UBL systems, and thereby confer higher susceptibility to bacterial pathogens?
Would drugs targeting bacteria-specific enzymes that interfere with host UBI/UBL conjugation be an efficient means to treat infectious diseases?
Finally, although this review has focused on pathogenic bacteria, some non-pathogenic bacteria such as commensals of the intestinal microbiota have also been reported to interfere with host UBI/UBL systems [98]. For example, production of butyrate by commensal bacteria leads to the inactivation of the E2 NEDD8 enzyme in intestinal epithelial cells, and this was proposed to participate in the inflammatory tolerance of gut bacteria [99,100].
Manipulation of UBI/UBL conjugation by pathogenic bacteria constitutes a key facet of host–pathogen interactions. Studying how bacteria interfere with these PTMs is essential to complete our understanding of the infection process. In particular, identification of bacterial effectors harboring non-eukaryotic enzymatic activities and manipulating host UBI/UBL may provide potential new drug targets, which will be crucial in this age of bacterial resistance to antibiotics.
Highlights.
UBI and UBLs regulate essential pathways of the host cell involved in crucial steps of bacterial infections. Not surprisingly, bacterial pathogens have evolved multiple strategies to interfere with these host PTMs.
In addition to ubiquitin, UBLs such as SUMO and NEDD8 have recently emerged as prominent targets of bacterial pathogens.
Strategies used by bacteria to interfere with host UBI/UBL encompass the targeting of UBI/UBL conjugation machineries, the modulation of the UBI/UBL conjugation level of specific host factors, and the direct targeting of UBI/UBL proteins.
Host proteins modified by UBI/UBL and targeted by bacteria cluster into specific host-cell functions such as gene regulation, cytoskeleton dynamics, and cell-autonomous immunity.
Bacteria hijack the host UBI/UBL systems to modify their own proteins, thereby allowing regulation of their intracellular localization, stability, or interaction abilities.
Acknowledgments
We apologize to all colleagues whose work we were unable to include owing to space constraints. We thank L. Radoshevich for critical reading of this manuscript. P.C. received support from the Institut Pasteur, INSERM, INRA, the French National Research Agency (ANR) (ERANET Infect-ERA PROANTILIS ANR-13-IFEC-0004-02), the French Government Investissement d’Avenir program, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (ANR-10-LABX-62-IBEID), the European Research Council (ERC) (H2020-ERC-2014-ADG 670823-BacCellEpi), the Fondation le Roch les Mousquetaires, the Fondation Louis-Jeantet, and the International Balzan Prize Fondation. P.C. is a Senior International Research Scholar of the Howard Hughes Medical Institute. D.R. received support from INSERM, Rouen University, and the iXcore Foundation for Research.
References
- 1.Ribet D, Cossart P. Post-translational modifications in host cells during bacterial infection. FEBS Lett. 2010;584:2748–2758. doi: 10.1016/j.febslet.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Deribe YL, et al. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein G, et al. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon YT, Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. 2017;42:873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 7.Enchev RI, et al. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villarroya-Beltri C, et al. ISGylation – a key to lock the cell gates for preventing the spread of threats. J Cell Sci. 2017;130:2961–2969. doi: 10.1242/jcs.205468. [DOI] [PubMed] [Google Scholar]
- 9.Basler M, et al. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Mol Immunol. 2015;68:129–132. doi: 10.1016/j.molimm.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Streich FC, Jr, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 12.Rahman MM, McFadden G. Modulation of NF-kappaB signalling by microbial pathogens. Nat Rev Microbiol. 2011;9:291–306. doi: 10.1038/nrmicro2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decque A, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol. 2016;17:140–149. doi: 10.1038/ni.3342. [DOI] [PubMed] [Google Scholar]
- 14.Bogunovic D, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radoshevich L, et al. ISG15 counteracts Listeria mono-cytogenes infection. eLife. 2015;4:e06848. doi: 10.7554/eLife.06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spinnenhirn V, et al. The ubiquitin-like modifier FAT10 decorates autophagy-targeted Salmonella and contributes to Salmonella resistance in mice. J Cell Sci. 2014;127:4883–4893. doi: 10.1242/jcs.152371. [DOI] [PubMed] [Google Scholar]
- 17.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett RD, et al. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol. 2013;11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 19.Wimmer P, Schreiner S. Viral mimicry to usurp ubiquitin and SUMO host pathways. Viruses. 2015;7:4854–4872. doi: 10.3390/v7092849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maculins T, et al. Bacteria–host relationship: ubiquitin ligases as weapons of invasion. Cell Res. 2016;26:499–510. doi: 10.1038/cr.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashida H, Sasakawa C. Bacterial E3 ligase effectors exploit host ubiquitin systems. Curr Opin Microbiol. 2017;35:16–22. doi: 10.1016/j.mib.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Lin YH, Machner MP. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J Cell Sci. 2017;130:1985–1996. doi: 10.1242/jcs.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson VG. Viral interplay with the host sumoylation system. Adv Exp Med Biol. 2017;963:359–388. doi: 10.1007/978-3-319-50044-7_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruthi M, et al. Modulation of host cell SUMOylation facilitates efficient development of Plasmodium berghei and Toxoplasma gondii. Cell Microbiol. 2017;19:e12723. doi: 10.1111/cmi.12723. [DOI] [PubMed] [Google Scholar]
- 25.Ribet D, et al. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 2010;464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galan JE, Waksman G. Protein-injection machines (and not protein-unjection machines) Cell. 2018;172:1306–1318. doi: 10.1016/j.cell.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 28.Hrdinka M, Gyrd-Hansen M. The Met1-linked ubiquitin machinery: emerging themes of (de)regulation. Mol Cell. 2017;68:265–280. doi: 10.1016/j.molcel.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 30.Hatakeyama S, et al. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 31.Walden H, Rittinger K. RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns. Nat Struct Mol Biol. 2018;25:440–445. doi: 10.1038/s41594-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 32.Pao KC, et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature. 2018;556:381–385. doi: 10.1038/s41586-018-0026-1. [DOI] [PubMed] [Google Scholar]
- 33.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 34.Sanada T, et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- 35.Lin AE, Guttman JA. The Escherichia coli adherence factor plasmid of enteropathogenic Escherichia coli causes a global decrease in ubiquitylated host cell proteins by decreasing ubiquitin E1 enzyme expression through host aspartyl proteases. Int J Biochem Cell Biol. 2012;44:2223–2232. doi: 10.1016/j.biocel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Impens F, et al. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc Natl Acad Sci U S A. 2014;111:12432–12437. doi: 10.1073/pnas.1413825111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribet D, et al. Promyelocytic leukemia protein (PML) controls Listeria monocytogenes Infection. mBio. 2017;8:e02179–16. doi: 10.1128/mBio.02179-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma S, et al. Salmonella engages host MicroRNAs to modulate SUMOylation: a new arsenal for intracellular survival. Mol Cell Biol. 2015;35:2932–2946. doi: 10.1128/MCB.00397-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lapaquette P, et al. Shigella entry unveils a calcium/calpain-dependent mechanism for inhibiting sumoylation. eLife. 2017;6:e27444. doi: 10.7554/eLife.27444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fritah S, et al. Sumoylation controls host anti-bacterial response to the gut invasive pathogen Shigella flexneri. EMBO Rep. 2014;15:965–972. doi: 10.15252/embr.201338386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malet JK, et al. Rapid remodeling of the host epithelial cell proteome by the listeriolysin O (LLO) pore-forming toxin. Mol Cell Proteom. 2018;17:1627–1636. doi: 10.1074/mcp.RA118.000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hicks SW, Galan JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer AU, et al. A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 2013;9:e1003121. doi: 10.1371/journal.ppat.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu F, et al. The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. Proc Natl Acad Sci U S A. 2014;111:10538–10543. doi: 10.1073/pnas.1402605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubori T, et al. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu J, et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhogaraju S, et al. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell. 2016;167:1636–1649. doi: 10.1016/j.cell.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Kotewicz KM, et al. A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe. 2017;21:169–181. doi: 10.1016/j.chom.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalayil S, et al. Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature. 2018;557:734–738. doi: 10.1038/s41586-018-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu J, et al. A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res. 2017;27:865–881. doi: 10.1038/cr.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hotson A, et al. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim JG, et al. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe. 2013;13:143–154. doi: 10.1016/j.chom.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheedlo MJ, et al. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc Natl Acad Sci U S A. 2015;112:15090–15095. doi: 10.1073/pnas.1514568112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pruneda JN, et al. The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol Cell. 2016;63:261–276. doi: 10.1016/j.molcel.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puvar K, et al. Ubiquitin chains modified by the bacterial ligase SdeA are protected from deubiquitinase hydrolysis. Biochemistry. 2017;56:4762–4766. doi: 10.1021/acs.biochem.7b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taieb F, et al. Cycle inhibiting factors (Cifs): cyclomodulins that usurp the ubiquitin-dependent degradation pathway of host cells. Toxins. 2011;3:356–368. doi: 10.3390/toxins3040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui J, et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1318. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu C, et al. Gln40 deamidation blocks structural reconfiguration and activation of SCF ubiquitin ligase complex by Nedd8. Nat Commun. 2015;6 doi: 10.1038/ncomms10053. 10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCormack RM, et al. Enteric pathogens deploy cell cycle inhibiting factors to block the bactericidal activity of Perforin-2. eLife. 2015;4:e06505. doi: 10.7554/eLife.06505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Jong MF, et al. Shigella flexneri suppresses NF-kappaB activation by inhibiting linear ubiquitin chain ligation. Nat Microbiol. 2016;1:16084. doi: 10.1038/nmicrobiol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashida H, et al. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashida H, et al. Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to inhibit PKC–NF-kappaB activity in invaded epithelial cells. PLoS Pathog. 2013;9:e1003409. doi: 10.1371/journal.ppat.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim DW, et al. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li S, et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501:242–246. doi: 10.1038/nature12436. [DOI] [PubMed] [Google Scholar]
- 65.Pearson JS, et al. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature. 2013;501:247–251. doi: 10.1038/nature12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 67.Zhang L, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-kappaB activation. Nature. 2011;481:204–208. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki S, et al. Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc Natl Acad Sci U S A. 2014;111:E4254–E4263. doi: 10.1073/pnas.1324021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wandel MP, et al. GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe. 2017;22:507–518. doi: 10.1016/j.chom.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li P, et al. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature. 2017;551:378–383. doi: 10.1038/nature24467. [DOI] [PubMed] [Google Scholar]
- 71.Veiga E, Cossart P. Ubiquitination of intracellular bacteria: a new bacteria-sensing system? Trends Cell Biol. 2005;15:2–5. doi: 10.1016/j.tcb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 72.Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. 2013;16:339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 73.Huett A, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Franco LH, et al. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and antituberculous host defense. Cell Host Microbe. 2017;21:59–72. doi: 10.1016/j.chom.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heath RJ, et al. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 2016;17:2183–2194. doi: 10.1016/j.celrep.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noad J, et al. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat Microbiol. 2017;2:17063. doi: 10.1038/nmicrobiol.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choy A, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Souza Santos M, Orth K. Subversion of the cytoskeleton by intracellular bacteria: lessons from Listeria, Salmonella and Vibrio. Cell Microbiol. 2015;17:164–173. doi: 10.1111/cmi.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nethe M, Hordijk PL. The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci. 2010;123:4011–4018. doi: 10.1242/jcs.078360. [DOI] [PubMed] [Google Scholar]
- 82.Fiskin E, et al. Global analysis of host and bacterial ubiquitinome in response to Salmonella Typhimurium Infection. Mol Cell. 2016;62:967–981. doi: 10.1016/j.molcel.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 83.Alonso A, et al. Emerging roles of sumoylation in the regulation of actin, microtubules, intermediate filaments, and septins. Cytoskeleton. 2015;72:305–339. doi: 10.1002/cm.21226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ribet D, et al. SUMOylation of human septins is critical for septin filament bundling and cytokinesis. J Cell Biol. 2017;216:4041–4052. doi: 10.1083/jcb.201703096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bierne H, Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell Microbiol. 2012;14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- 86.Cougnoux A, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 87.Lallemand-Breitenbach V, de The H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154–161. doi: 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 88.Popa CM, et al. Modification of bacterial effector proteins inside eukaryotic host cells. Front Cell Infect Microbiol. 2016;6:73. doi: 10.3389/fcimb.2016.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kubori T, Galan JE. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 90.Patel JC, et al. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Knodler LA, et al. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol. 2009;11:1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dunphy PS, et al. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun. 2014;82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beyer AR, et al. The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol. 2015;17:504–519. doi: 10.1111/cmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jo K, et al. Host cell nuclear localization of Shigella flexneri effector OspF is facilitated by SUMOylation. J Microbiol Biotechnol. 2017;27:610–615. doi: 10.4014/jmb.1611.11066. [DOI] [PubMed] [Google Scholar]
- 95.Xu G, et al. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Becker J, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- 97.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–1083. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Collier-Hyams LS, et al. Bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J Immunol. 2005;175:4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- 99.Kumar A, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumar A, et al. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. J Immunol. 2009;182:538–546. doi: 10.4049/jimmunol.182.1.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Piscatelli H, et al. The EHEC type III effector NleL is an E3 ubiquitin ligase that modulates pedestal formation. PLoS One. 2011;6:e19331. doi: 10.1371/journal.pone.0019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu B, et al. NleG type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases. PLoS Pathog. 2010;6:e1000960. doi: 10.1371/journal.ppat.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Janjusevic R, et al. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 104.Abramovitch RB, et al. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rohde JR, et al. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 106.Gao X, et al. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe. 2013;13:87–99. doi: 10.1016/j.chom.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nadler C, et al. The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog. 2010;6:e1000743. doi: 10.1371/journal.ppat.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Misaghi S, et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 109.Le Negrate G, et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 110.Maupin-Furlow JA. Prokaryotic ubiquitin-like protein modification. Annu Rev Microbiol. 2014;68:155–175. doi: 10.1146/annurev-micro-091313-103447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Delley CL, et al. Prokaryotic ubiquitin-like protein and its ligase/deligase enyzmes. J Mol Biol. 2017;429:3486–3499. doi: 10.1016/j.jmb.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 112.Gandotra S, et al. The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide. PLoS Pathog. 2010;6:e1001040. doi: 10.1371/journal.ppat.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Patrick S, et al. A unique homologue of the eukaryotic protein-modifier ubiquitin present in the bacterium Bacteroides fragilis, a predominant resident of the human gastrointestinal tract. Microbiology. 2011;157:3071–3078. doi: 10.1099/mic.0.049940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chatzidaki-Livanis M, et al. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio. 2017;8:e01902–17. doi: 10.1128/mBio.01902-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Collier RJ, Cole HA. Diphtheria toxin subunit active in vitro. Science. 1969;164:1179–1181. doi: 10.1126/science.164.3884.1179. [DOI] [PubMed] [Google Scholar]