Supplemental digital content is available in the text.
Key Words: EXERCISE TRAINING, GUT MICROBIOTA, METABOLIC ENDOTOXEMIA, INTESTINAL GLUCOSE UPTAKE, INTESTINAL FREE FATTY ACID UPTAKE, SPRINT INTERVAL TRAINING
ABSTRACT
Introduction
Intestinal metabolism and microbiota profiles are impaired in obesity and insulin resistance. Moreover, dysbiotic gut microbiota has been suggested to promote systemic low-grade inflammation and insulin resistance through the release of endotoxins particularly lipopolysaccharides. We have previously shown that exercise training improves intestinal metabolism in healthy men. To understand whether changes in intestinal metabolism interact with gut microbiota and its release of inflammatory markers, we studied the effects of sprint interval (SIT) and moderate-intensity continuous training (MICT) on intestinal metabolism and microbiota in subjects with insulin resistance.
Methods
Twenty-six, sedentary subjects (prediabetic, n = 9; type 2 diabetes, n = 17; age, 49 [SD, 4] yr; body mass index, 30.5 [SD, 3]) were randomized into SIT or MICT. Intestinal insulin-stimulated glucose uptake (GU) and fatty acid uptake (FAU) from circulation were measured using positron emission tomography. Gut microbiota composition was analyzed by 16S rRNA gene sequencing and serum inflammatory markers with multiplex assays and enzyme-linked immunoassay kit.
Results
V˙O2peak improved only after SIT (P = 0.01). Both training modes reduced systematic and intestinal inflammatory markers (tumor necrosis factor-α, lipopolysaccharide binding protein) (time P < 0.05). Training modified microbiota profile by increasing Bacteroidetes phylum (time P = 0.03) and decreasing Firmicutes/Bacteroidetes ratio (time P = 0.04). Moreover, there was a decrease in Clostridium genus (time P = 0.04) and Blautia (time P = 0.051). Only MICT decreased jejunal FAU (P = 0.02). Training had no significant effect on intestinal GU. Colonic GU associated positively with Bacteroidetes and inversely with Firmicutes phylum, ratio Firmicutes/Bacteroidetes and Blautia genus.
Conclusions
Intestinal substrate uptake associates with gut microbiota composition and whole-body insulin sensitivity. Exercise training improves gut microbiota profiles and reduces endotoxemia.
Gut microbiota has been recognized to play a key role in human health and well-being. Gut microbial profiles have been shown to differ in healthy subjects compared with subjects with obesity, metabolic syndrome, and inflammatory bowel disease (IBD) (1). It is suggested that the impairments in gut microbiota composition induce metabolic endotoxemia through the release of endotoxins, particularly lipopolysaccharide (LPS), which promote systemic low-grade inflammation and insulin resistance (2).
The effect of exercise training on gut microbiota remains elusive. It has been suggested that there is a link between physical fitness and health-associated gut microbial parameters, such as taxonomic diversity (3) and richness (4). Previously, Mika and Fleshner (5) have suggested that regular physical activity promotes the psychological and metabolic health through the development of diverse microbiota in childhood and adolescence. Exercise training has been shown to effect the gut microbiota first, through its effects on autonomic nervous system (vagal tone), which is also known as “brain–gut axis” (6), and second by its impact on improvement of immune function (7). It has been suggested that alterations in the vagal nerve influence the gut microbiota through their control of inflammatory alterations, and that exercise training improves this vagal tone by improving gut microbiota composition (8). Additionally, inflammation in the intestinal mucosa associated with the IBD has been shown to alter gut microbiota (9). Training has been demonstrated to be an effective treatment for multiple inflammatory conditions, including IBD (10). Even though there is lot of speculation about the beneficial effects of exercise training on gut microbiota (4), studies in humans are scarce.
Recently, we reported in healthy middle-age sedentary men, that short term moderate-intensity continuous training (MICT) improves intestinal insulin-stimulated glucose uptake (GU) (from circulation into intestine) and fasting free fatty acid uptake (FAU) more efficiently compared with sprint interval training (SIT) (11). We also demonstrated a positive correlation between intestinal insulin GU and whole-body GU (i.e., insulin sensitivity) (11). However, it is unclear whether the changes in the intestinal metabolism are associated with gut microbiota composition and activity and whether this interaction further reflects whole-body metabolism.
In the present study, we investigated the effects of short-term training on intestinal insulin-stimulated GU, fasting FAU, gut microbiota composition, and metabolic endotoxemia (LPS binding protein [LBP]) in prediabetic and type 2 diabetes (T2D) subjects. Based on our previous study, we hypothesized that training responses are more rapidly detectable in intestinal metabolism and gut microbiota after MICT than SIT training.
MATERIALS AND METHODS
Study Design
This subproject is part of a bigger study entitled “The effects of short-term high-intensity interval training on tissue glucose and fat metabolism in healthy subjects and in patients with type 2 diabetes” (NCT01344928). All studies were performed at Turku PET Centre, University of Turku, Turku University hospital, Åbo Akademi University (Turku, Finland) and Paavo Nurmi Centre (Turku, Finland). The various studies performed before and after the exercise intervention are illustrated in Figs. 1A and B. Participants were also asked to abstain from any caffeinated and alcoholic drinks, avoid strenuous exercise, and stop all oral hypoglycemic medication 48 h before these studies. The study was approved by the ethics committee of the Hospital district of South-Western Finland (decision 95/180/2010 §228) and carried out in compliance with the declaration of Helsinki. The purpose, nature, and potential risks involved with the study were explained in detail, and informed consent was obtained before any measurements were performed.
FIGURE 1.
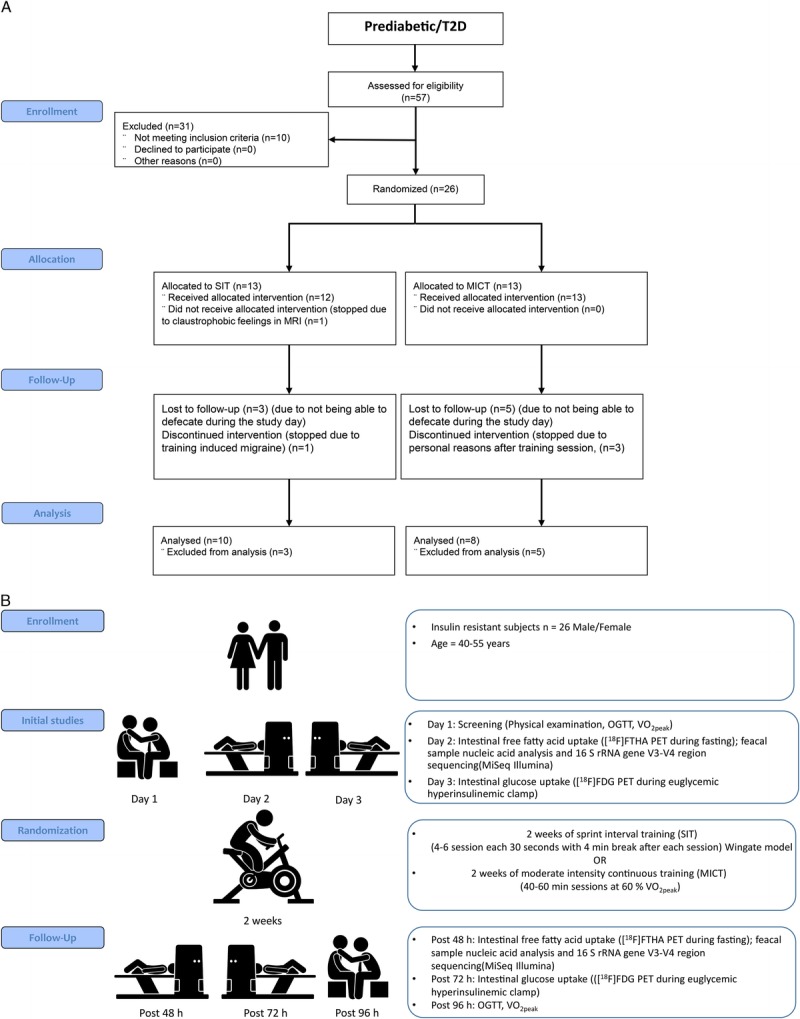
A, Consort flow diagram showing the total number of subjects recruited and analyzed. B, Study design: Subjects were studied on three separate days before and after the exercise intervention. OGTT, oral glucose tolerance test.
Subjects
Twenty-six sedentary middle-age insulin-resistant subjects (prediabetic, n = 9; T2D, n = 17; males/females, 16/10) were randomized either into SIT or MICT group. The randomization was done by permuted blocks of 1:1 ratio. The subjects (age, 40–55 yr; V˙O2peak, <40 mL·kg−1·min−1) had no previous background of exercise training. All 26 subjects met the criteria of defective glucose tolerance set by American Diabetes Association criteria (12) and had an glycosylated hemoglobin (HbA1c) less than 7.5 mmol·L−1. Of the 26 subjects, 17 (male n = 11, female n = 6) met the criteria of type 2 diabetes mellitus (T2DM) (median duration for T2D 4 yr), and 9 had prediabetes, having either impaired fasting glucose and/or impaired glucose tolerance (12). Of the 17 T2DM subjects, 13 were treated with oral hypoglycemic medication (11 metformin; 5 DPP-IV, and 1 sulfonylurea), whereas the remaining 4 were diagnosed at the screening and were not taking any medication for T2DM. All diabetes medication was discontinued before any studies were performed. The inclusion and exclusion criteria have been explained previously in detail (13).
Three of the subjects failed to complete the study during the intervention one due to training induced migraine and two due to personal reasons. The fecal samples were available from 18 of the 26 subjects, and therefore, the results presented here are from these 18 subjects.
Exercise Interventions
In both training interventions participants exercised six times (three per week) over 2 wk. All sessions were performed under supervision. The training protocol is described in detailed previously (14). During the screening phase, participants were familiarized with SIT (2 × 30 s bouts). Each SIT session consisted 30-s exercise bouts (4–6) of all out cycling efforts (Wingate protocol, load 10% of fat-free mass in kilograms, Monark Ergomedic 828E, Monark, Vansbro, Sweden) with 4 min of recovery in between the exercise bouts. The MICT training involved 40 to 60 min of moderate-intensity (60% of V˙O2peak intensity) cycling (Tunturi E85, Tunturi Fitness, Almere, Netherlands). Both training interventions were progressive; with number of exercise bouts increasing from four to five and finally to six in the SIT group while in the MICT group the training duration was increased from 40 to 50 min and then to 60 min after every second training session.
Primary Outcomes
Positron emission tomography scans
The positron emission tomography (PET) imaging method has been described in detail previously (11). Briefly, intestinal FAU and GU [18F]fluoro-6-thia-heptadecanoic acid ([18F]FTHA) PET and ([18F]FDG) PET imagining were performed on two different days using 14(R,S)-FTHA and 2-[18F]fluoro-2-deoxy-d-glucose (FDG) radiotracers. The [18F]FTHA PET was done under fasting conditions, whereas the [18F]FDG PET was done under euglycemic hyperinsulinemic clamp. The PET raw data were corrected for attenuation, dead time, and decay. The data were reconstructed with 3D-OSEM method and were analyzed with Carimas software (version 2.9, www.turkupetcentre.fi/carimas). Regions of interest were drawn manually on sections of intestine (duodenum, jejunum, and colon) as explained previously (11). From these tubular regions of interest, tissue time activity curves were obtained. Fractional uptake rate was calculated from the plasma and tissue time activity curves using graphical analysis (15). The regional (duodenal, jejunal, and colonic) FAU and GU were calculated by multiplying the regional fractional uptake rate with plasma-free fatty acid or glucose concentration, respectively, during the scan.
Fecal DNA extraction and specific quantitative real-time polymerase chain reaction
Stool samples were collected before and after the exercise intervention and kept frozen at −80°C until processed for analysis. Total DNA was isolated from the fecal samples using the MasterPure Complete DNA & RNA Purification Kit (Epicenter) according to the manufacturer’s instructions with some modifications as described previously (16).
Polymerase chain reaction (PCR) primers targeted to Bacteroides group, Bifidobacterium group, and also, Enterobacteriaceae family were used as previously described (17). Specific quantitative real-time PCR was performed in LightCycler® 480 Real-Time PCR System (Roche®) by use of SYBR® Green PCR Master Mix (Roche®). The fluorescent products were detected in the last step of each amplification cycle. A melting curve analysis was made at the end of the PCR to distinguish the untargeted PCR product. The bacterial concentration in each sample was calculated comparing the Ct values obtained from standard curves.
16S rRNA gene sequencing analysis
Total DNA concentrations were measured using a Qubit® 2.0 Fluorometer (Life Technology, Carlsbad, CA) and diluted to 5 ng·μL−1. 16S rDNA gene (V3-V4 region) was amplified by PCR using Illumina adapter overhang nucleotide sequences following Illumina protocols. After 16S rDNA gene amplification, the mutiplex step was performed using Nextera XT Index Kit (Illumina, San Diego, CA). PCR product was checked with a Bioanalyzer DNA 1000 chip (Agilent Technologies, Santa Clara, CA), and libraries were sequenced using a 2 × 300pb paired-end run (MiSeq Reagent kit v3) on a MiSeq-Illumina platform (FISABIO sequencing service, Valencia, Spain) according to manufacturer’s instructions (Illumina).
Data processing was performed using the QIIME pipeline (version 1.9.0) (18). The sequences were clustered into operational taxonomic units at 97% similarity and checked for chimeras. Representative sequences were obtained for each microbial phylotype, and taxonomy was assigned using Greengenes GG 13.8 database. Sequences that could not be classified to domain level, or were classified as Cyanobacteria and Chloroplasts, were removed from the data set because they likely represent ingested plant material.
Biomarkers in plasma and stool LBP, C-Reactive Protein, and Tumor Necrosis Factor α measurements
Concentrations of interleukins IL-1B, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12 P70, IL-13, interferon gamma (IFN-g), tumor necrosis factor (TNF)-α, and also, C-reactive protein (CRP) were measured by multiplex bead assay analysis (12-plex, Luminex® Performance Assay Multiplex Kit; Procarta® Immunoassay Affymetrix, Santa Clara US) according to the manufacturer’s instructions. Plasma samples were analyzed with LUMINEX® 200™ using the Luminex xPonent software (Luminex, USA). Concentrations of some of the interleukins were under the detection limit and were therefore not included in the final analysis. Human LBP was determined by ELISA Kit (FineTest® Ref.-EH1560, Wuhan Fine Biotech Co., Ltd.) in both plasma and fecal samples. Levels of human calprotectine (FineTest® Ref.-EH4140, Wuhan Fine Biotech Co., Ltd.) and zonulin (MyBiosource Ref. MBS749365-96) in fecal samples were also analyzed by ELISA.
Secondary Outcomes
Other measurements
Euglycemic hyperinsulinemic clamp technique was performed to calculate the whole-body insulin sensitivity (M value) (19). A cycle ergometer (Ergoline 800s; VIASYS Healthcare, Germany) was used to determine the V˙O2peak as previously described (14). Bioimpedence monitor (Inbody 720; Megaelectonics Ltd., Kuopio, Finland) was used to measure the body composition. Abdominal subcutaneous and visceral fat masses were measured using magnetic resonance imaging (MRI) and SliceOmatic software version 4.3 (http://www.tomovision.com/products/sliceomatic.html) as previously described (11).
Statistics
The sample size for the whole study (NCT01344928) was based on the primary outcome skeletal muscle GU (quadriceps femoris). To achieve >90% power of detecting a 20% unit change in insulin-stimulated GU in quardriceps femoris a total of 20 prediabetic/T2D subjects (SIT = 10 and MICT = 10) were required. To accommodate for the drops outs and technical problems during the study, an extra six subjects were recruited as described previously (20). No sample size calculations were done specifically on the outcomes measures of the current study. Descriptive statistics shown in the tables are presented by model-based means and 95% confidence intervals (CI), whereas the figures are based on model-based means and (95% CI). Association between anthropometrics, glucose profile, lipid profile and training groups, time points, training–time, interaction were performed with hierarchical linear mixed models, using the compound symmetry covariance structure for repeated measurements. The medication status (taking oral hypoglycemic medication/not taking oral hypoglycemic medication) and gender were used as additional factors for all the analyses. Subjects with one value, but another missing (drop outs, technical problems) are included in this model and therefore model-based mean (SAS least square means) values are reported for all the parameters. Transformations (logarithmic or square root) were done to (whole-body fat percentage, subcutaneous and visceral fat volume, glucose fasting, insulin fasting, FFA clamp, ratio Firmicutes/Bacteroidetes, and Clostridium genus) variables to achieve the normal distribution assumption. Multivariate redundancy analysis (RDA) plots were created with Calypso Multivariate tool, an online platform for mining, visualizing, and comparing multiple microbial community composition data (cgenome.net/calypso). In addition, Pearson correlation coefficient was calculated. All tests were performed as two-sided, with a significance level set at 0.05. The analyses were performed using SAS System, version 9.3 for Windows (SAS Institute Inc., Cary, NC).
RESULTS
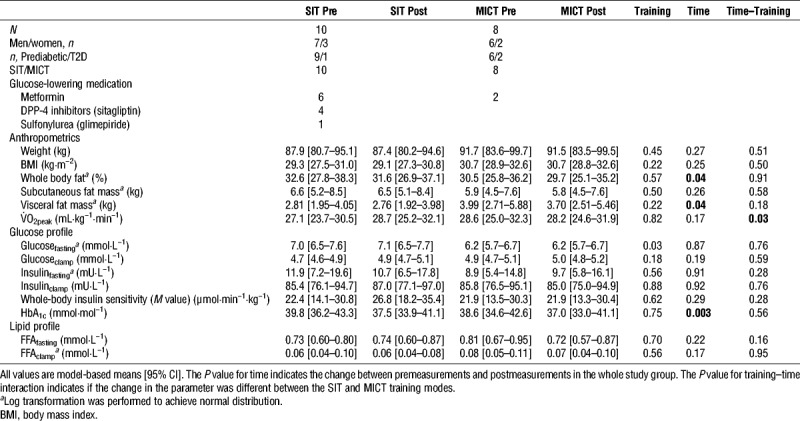
Aerobic capacity (V˙O2peak) improved only after SIT and not after MICT training (time–training = 0.03). However, both training modes reduced whole-body fat percentage and the abdominal visceral fat mass (both time P = 0.04) and improved HBA1c (time, P = 0.003) (Table 1).
TABLE 1.
Subject characteristics at baseline and the changes induced after the exercise intervention.
Both training modes significantly reduced systemic inflammatory marker TNF α (time, P = 0.03) and tended to reduce CRP (time, P = 0.08) and intestinal inflammatory marker LBP (time, P = 0.02) (Figs. 2A–C). There were no changes in other plasma cytokines and intestinal inflammatory markers (calprotectin and zonulin) measured from fecal samples (Supplemental Fig. 1, Supplemental Digital Content 1, Changes in the inflammatory markers, http://links.lww.com/MSS/B712). LBP correlated positively with HBA1c (r = 0.54; P = 0.02) (Supplemental Fig. 2, Supplemental Digital Content 2, correlations between different parameters, http://links.lww.com/MSS/B713).
FIGURE 2.
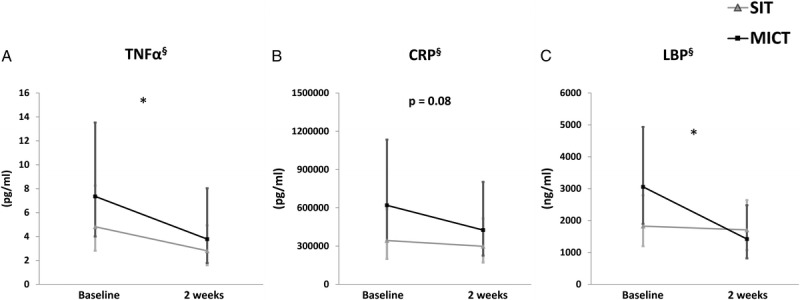
Impact of exercise intervention on specific inflammatory markers. A, TNF α, (B) CRP, and (C) LBP. All values are expressed as model-based means and bars are 95% CI. (§) Log transformation was performed to achieve normal distribution. *P value for time interaction (i.e., the groups (SIT + MICT) behaved similarly for the change in the parameter with a significant difference between them.
Gut microbiota composition was influenced by exercise training. Both training modes decreased the ratio of Firmicutes/Bacteroidetes (time, P = 0.04), mainly due to the increase in the relative abundance of Bacteroidetes phyla (time, P = 0.03) (Figs. 3A, B), whereas no change was found in the Firmicutes levels. At genus level, both training modes decreased the abundance of Blautia spp. (time P = 0.051) and Clostridium spp. (time P = 0.04) (Figs. 3C, D). Lachnospira genus was present in higher abundance after SIT compared with baseline (P = 0.025) and significant higher abundance of Veillonella genus (and also, Veillonella dispar) was observed after MICT compared to baseline (P = 0.036) and compared with SIT (P = 0.055). Interestingly, the abundance of Faecalibacterium genus (F. prausnitzii) was increased after MICT (P = 0.057) while no change after SIT was found (Supplemental Fig. 3, Supplemental Digital Content 3, changes in relative abundance of Feacalibacterium and Akkermansia, http://links.lww.com/MSS/B714). There were no differences in microbiome richness and diversity between the training modes (by Chao1, Shannon, observed OTU and PD whole tree) (Supplemental Fig. 4, Supplemental Digital Content 4, effects of exercise on microbial diversity, http://links.lww.com/MSS/B715). While at OTU level, MICT training increased significantly (P = 0.035) the relative abundance of Veillonella dispar (OTU 4034), whereas no change was seen after SIT. In addition, multivariate RDA test showed significant differences in the gut microbiota according to the training response (SIT vs MICT) (Supplemental Fig. 5, Supplemental Digital Content 5, multivariate redundancy analysis, http://links.lww.com/MSS/B716).
FIGURE 3.
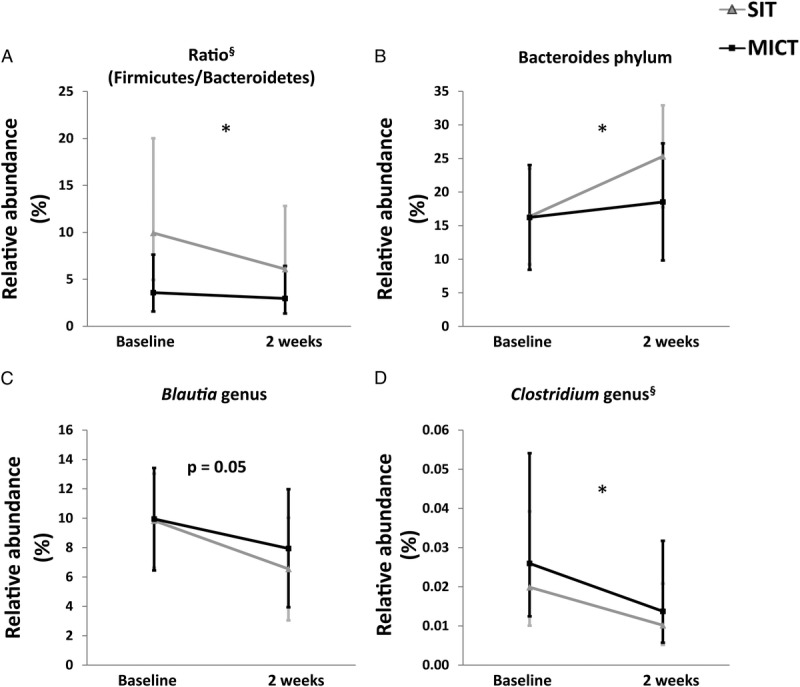
Impact of exercise intervention on the gut microbiota composition. A, Ratio (Firmicutes/Bacteroidetes), (B) Bacteroidetes, and the genus (C) Blautia and (D) Clostridium. All values are expressed as model-based means and bars are 95% CI. (§) Log transformation was performed to achieve normal distribution. *P value for time interaction (i.e., the groups (SIT + MICT) behaved similarly for the change in the parameter with a significant difference between them.
There was no change in insulin-stimulated intestinal GU in either training group (Fig. 4A). Although MICT reduced the fasting FAU in the jejunum, no changes were observed either in duodenum or colon (Fig. 4B) or after SIT. Intestinal FAU correlated negatively with whole-body insulin sensitivity (r = −0.81; P = 0.049) after MICT training only (Supplemental Fig. 2, Supplemental Digital Content 2, http://links.lww.com/MSS/B713). Interestingly, at baseline, insulin-stimulated colonic GU associated inversely with the abundance of Firmicutes (r = −0.59; P = 0.03), Firmicutes/Bacteroidetes ratio (r = −0.62; P = 0.024) and Blautia (r = −0.57; P = 0.049) and positively with the abundance of Bacteroidetes (r = 0.71; P = 0.007) (Figs. 5A–D). In addition, lower abundance of Blautia genus was associated with better whole-body insulin sensitivity (M value) (r = −0.53; P = 0.04) (Supplemental Fig. 2, Supplemental Digital Content 2, correlations between different parameters, http://links.lww.com/MSS/B713).
FIGURE 4.
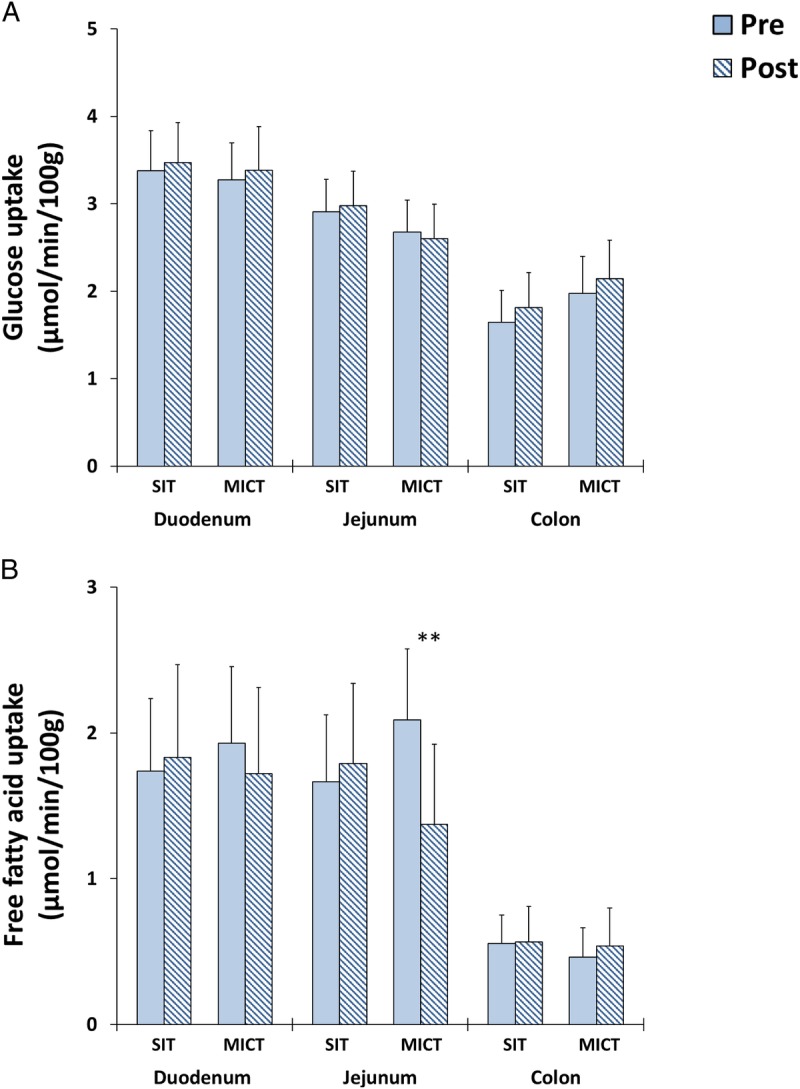
A, Insulin-stimulated GU and (B) fasting free FAU in different parts of intestine before and after 2 wk of either SIT or MICT. All values are expressed as model-based means and bars are 95% CI. (§) Log transformation was performed to achieve normal distribution. **P value for time–training interaction (i.e., the groups behaved differently for the change in the parameter with a significant difference between them).
FIGURE 5.
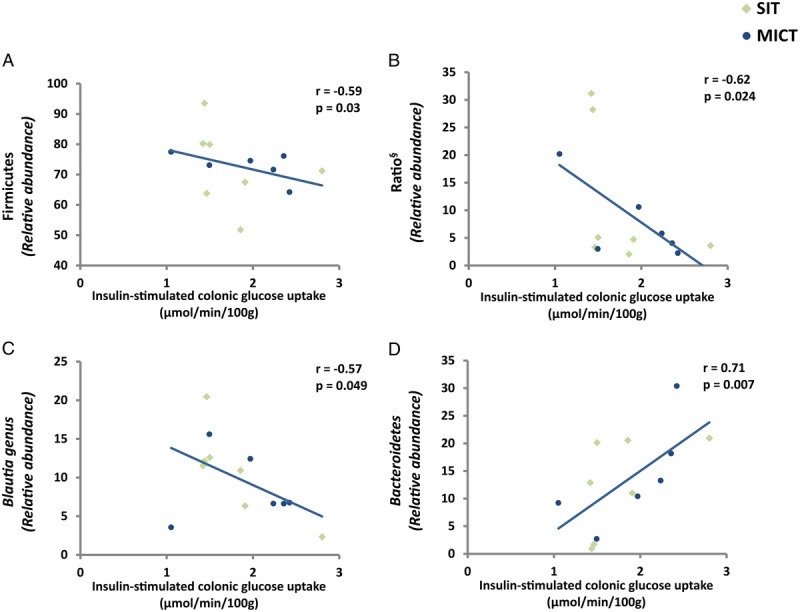
Correlation between insulin-stimulated colonic GU and (A) Firmicutes (B) Ratio (Firmicutes/Bacteroidetes), (C) Blautia, and (D) Bacteroides phylum in pooled analysis of both SIT and MICT subjects at baseline. (◊) SIT and (●) MICT. (§) Log transformation was performed to achieve normal distribution.
DISCUSSION
The present study shows for the first time that exercise training reduces intestinal inflammation and modulates gut microbiota profiles in insulin-resistant subjects. Both training modes reduced endotoxemia by decreasing the intestinal inflammatory marker (LBP). Training also decreased the ratio of Firmicutes/Bacteroidetes (obesity) (21), Clostridium genus (immune response) (1) and Blautia genus (inflammation) (22), and increased Bacteroidetes (protection against obesity) (23). Only MICT decreased jejunal FAU, whereas no training response was observed in intestinal insulin sensitivity.
Gut microbiota has been suggested to induce whole-body systematic low-grade inflammation through the release of inflammatory products (LPS, TNF α) (22,24). Healthy gut microbiota has been reported to release small amount of LPS in the blood, which is essential for the maintenance and development of the host immune system. However, when LPS is released in massive amounts, it is associated with pathophysiological reactions in various organs, in adipose tissue (induces inflammation and insulin resistance), in liver (damages hepatocytes leading to progression from simple fatty liver to steatohepatitis), in endothelium (contributes to plagues formation and rupture), and in some cases, irreversible shock (2). Due to the limitations in measuring LPS in biologic fluids (25), endogenous protein LBP has been used as an alternative clinical marker for measuring endotoxemia and immune response (26,27). LBP binds to LPS and enhances the binding of LPS to “Cluster of Differentiation 14” (CD14) (28). The CD14–LPS complex is transduced into the cell nucleus, and it initiates a cascade of inflammatory cytokines (29). Elevated LBP levels have been associated with obesity, T2D, and metabolic syndrome (30). In our study, both SIT and MICT significantly reduced LBP after 2 wk of training. This significant reduction in the LBP levels can be due to changes in gut microbiota. Cani et al. (24) showed that high fat diet significantly reduces the number of Bifidobacterium, Eubacterium rectale-Blautia coccoides, and Bacteroides genus. This reduction leads to an increase in the Gram-negative to Gram-positive ratio, leading to an increase in the levels of LPS in blood (24). Compared with the Cani et al. data after the high-fat diet, our findings indicate the opposite. We found a reduction in Firmicutes/Bacteroidetes ratio, mainly due to the increase in the relative abundance of Bacteroidetes and a decrease in the abundance of Blautia spp. and Clostridium spp. at genus level. These modulations can possibly explain the observed improvement in the LBP level in our study as the improvement in the relative abundance of Bacteroidetes can lead to a reduction in Gram-negative to Gram-positive ratio leading to a decrease in the LPS levels. Moreover, the improvement in Bacteroidetes levels can also lead to an improvement in the intestinal inflammation as Bacteroidetes induces regulatory T cells to produce IL-10 (anti-inflammatory cytokine) inside the gut (31). Furthermore, in the present study, Bacteroidetes at the species level correlated negatively with plasma inflammatory makers LBP, TNF α, and CRP levels (data not shown), highlighting the importance of Bacteroidetes in intestinal inflammation.
Exercise training also reduced the relative abundance of Clostridium and tended to reduce the Blautia genus. Clostridium bacteria have been suggested to play an important role in whole-body immune responses (22). Blautia genus has been shown to be one of the most abundant genus in prediabetes and T2D compared with healthy subjects (32) and has been suggested to increase the release of pro inflammatory cytokines (TNFα, cytokines) (22). Interestingly, in the present study, Blautia did decrease (P = 0.051), and we also observed a significant reduction in the TNFα after 2 wk of training (Fig. 2A). This reduction is important as TNFα plays a critical role in the inflammatory processes, such as IBD (33). Thus, our data suggest that exercise training reduces the inflammation in the gut and whole body, and thereby, another manner through which training reduces the risk of acquiring various diseases.
One of the most interesting results in our study is the reduction of Firmicutes/Bacteroidetes ratio. The ratio has been shown to have a significant relevance in the normal/healthy human gut microbiome (23). In obesity, the Firmicutes/Bacteroidetes ratio is elevated (34,35), it is reversible after dietary intervention and correlates with body weight loss (23). One of the reasons how Firmicutes contribute to obesity is the speculation that they are able to harvest more energy from food (34,36). In our study, 2 wk of training reduced the ratio between Firmicutes/Bacteroidetes, mainly through the significant increase in the relative abundance of Bacteroidetes at the phylum level (Fig. 3B). In addition, in measuring the Bacteroidetes relative abundance, we also measured its levels in the feces using quantitative real-time PCR. Both exercise modes also tended to increase the overall Bacteroidetes levels measured with quantitative real-time PCR (P = 0.07). The increase in Bacteroidetes is significant because it plays an essential role in the metabolic conversions of complex sugar polymers and degradation of proteins (37). Additionally, in obesity and irritable bowel syndrome, there is reduced relative abundance of Bacteroidetes (23,38).
In addition, to quantify bacteria at the phylum and genus level, we also performed multivariate RDA. Redundancy analysis identifies significant associations between microbial communities according to their composition at baseline and after the intervention (SIT and MICT). The RDA analysis showed that the training adaptation was different between SIT and MICT at the OTU level (Supplemental Fig. 3, Supplemental Digital Content 3, changes in relative abundance of Feacalibacterium and Akkermansia, http://links.lww.com/MSS/B714). OTU are a cluster of microorganisms which share similar sequences and represent more phylogenetically similar organisms. MICT increased Veillonella OTU significantly compared with baseline, whereas no OTU were different after the SIT intervention. However, when analyzed further, we did not find any differences between SIT and MICT at the genus and phylum level. This observation needs to be further studied with longer intervention duration and higher number of study subjects.
Previous cross-sectional studies in humans have shown that physically active subjects have higher microbial abundance Akkermansia muciniphila, Faecalibacterium prausnitzi, and Roseburia and higher microbial diversity compared with sedentary subjects (4,39). Many training intervention studies in animals have suggested that exercise indeed has a positive effect on the gut microbiome composition (increased Bacteroidetes and reduced Firmicutes phylum, as well as increased Actinobacteria phylum toward Bifidobacterium genus) (40,41). Moreover, intervention studies done with humans have shown an increase in the abundance of the six of the major bacterial phyla/genera (42); an increase in Akkermansia, Coriobacteriaceae, and Succinivibrionaceae; a decrease in Proteobacteria phylum (43,44); and positive impact on the butyrate producing bacteria (45,46). Exercise training has been suggested to modify the gut microbiota through its anti-inflammatory effects (7), mainly via the release of cytokines and peptides (also known as “myokines”) by the contracting skeletal muscles (36). Another way through which exercise modifies gut microbiota is through the digestive physiology. Moderate-intensity exercise has been shown to accelerate the digestive transit time (34), this is important because it has been suggested that with advancing age, the gut microbiome composition alters due to changes in the digestive time compared with younger adults (37). In our study, the digestive transit might have changed after both SIT and MICT, but unfortunately, we did not measure it in this study. However, this hypothesis warrants further studies.
Our data show that already short-term MICT decreases jejunal FAU in insulin-resistant subjects. This finding is consistent with our previous data regarding the training responses in healthy subjects (11). And also in line with recent study by Koffert et al. (47) showing increased jejunal FAU in morbidly obese compared with normal weight healthy subjects. One of the probable explanations for the change in intestinal FAU is the change in the plasma FFA supply to the intestine. In morbidly obese subjects, circulating FFA levels are increased; in contrast, training decreases FFA levels by reducing lipolysis (reduction in visceral mass and increase in insulin sensitivity). In the present study, plasma FFA level decreased after MICT by −11%, but the decrease did not reach statistical significance.
Opposite to our previous data in healthy humans (11) training did not improve insulin-stimulated GU in insulin-resistant subjects in the present study. It might be that 2-wk training period is too short to induce adaptations in insulin-resistant intestine.
In the present study, colonic FAU correlated negatively with whole-body insulin sensitivity (r = −0.81; P = 0.049) in the MICT group. Additionally, Firmicutes/Bacteroidetes ratio (increased in obesity) and Firmicutes correlated negatively and Bacteroidetes (reduced in obesity) positively with the insulin-stimulated colonic GU (Figs. 5A, B, and D). These results highlight the importance of intestinal substrate uptake on the whole body, and that the changes especially in the GU might have a positive effect on the gut microbiota as well. In this study, colonic GU correlated negatively with Blautia genus (Fig. 5C), which releases pro inflammatory markers (TNFα). As we know, exercise reduces the risk of IBD (10), the reduction in Blautia genus can be one of the mechanism through which exercise reduces the risk of acquiring IBD. Moreover, as we know, the risk of acquiring T2D, cardiovascular disease, and certain cancers is directly proportional to the degree of obesity (48). Exercise by altering the Firmicutes/Bacteroidetes ratio can reduce the comorbidities associated with obesity.
Limitations
Our study contains some limitations. First, we did not have dietary control in our study, but subjects were asked to maintain their dietary habit throughout the study period. However, the possible effects of diet on the intestinal metabolism cannot be ruled out. Furthermore, there were more men than women in the study and differences in medication between the subjects. Previous studies have shown contradictory findings regarding the effects of gender on microbiota (21,22). We found no differences in the microbiota composition between the sexes in our study at baseline. Moreover, metformin has been shown to have a therapeutic impact on gut microbiota by increasing the relative abundance of Butyrivibrio, Bifidobacterium bifidum, and Megasphaera (production of short chain fatty acids) and Akkermansia muciniphila (that use mucin, a complex glycosylated protein, as a carbon and nitrogen source) which plays an important role in the maintenance of intestinal mucosa (49,50). Metformin was taken into account as a covariate in the statistics and were found to have no effect on the results. Additionally, diabetic status (i.e., prediabetic and T2D) was also taken into account as a covariate in the statistics, and it was shown to have no effect on the results. The study assessments were done 48 to 96 h posttraining. As the training intervention was short, it is possible that due to the detraining effect some training adaptations were not detectable anymore 72 or 96 h posttraining. Also, it is highly likely that with longer training intervention period more changes would have been detected.
In conclusion, this study suggests that short-term exercise training improves the gut microbiota at the phylum and genus levels. Both training modes induced changes mostly in the major bacteria inhabiting the intestine and reduced the whole-body systemic and gut microbiota specific endotoxemia markers. The RDA analysis indicated differences in the gut microbiota after SIT and MICT, observation to be further studied. Moreover, baseline correlations between the microbiota, intestinal substrate uptake, and whole-body insulin sensitivity suggest another possible mechanism by which exercise can alter the gut and whole-body metabolism.
Supplementary Material
Acknowledgments
The authors would like to thank the staff at the Turku PET Centre and Paavo Nurmi Centre. University of Turku, Åbo Akademi University and Turku University Hospital for their excellent assistance in the study. The results of the study are presented clearly, honestly, and without fabrication, falsification, or inappropriate data manipulation, and statement that results of the present study do not constitute endorsement by ACSM.
K. K. K. and J. C. H. designed the study. J. J. E., K. A. V., K. K. K., and J.C. H. collected data. K. K. M. analyzed PET images. M. C. C. and S. E. performed fecal data analysis. K. K. M., M. C. C., and E. L. analyzed data; K. K.M., M. C. C., P. N., and J. C. H. interpreted data. K. K. M. and M. C. C. wrote the article and prepared the figures. All authors critically reviewed the manuscript and approved the final version. J. C. H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
This study was conducted within the Centre of Excellence in Cardiovascular and Metabolic Diseases and supported by the Academy of Finland, the University of Turku, Turku University Hospital, and Åbo Akademi University. The study was financially supported by the European Foundation for the Study of Diabetes, the Finnish Cultural Foundation, Varsinais-Suomi Regional Fund, Juho Vainio Foundation, Emil Aaltonen Foundation, Hospital District of Southwest Finland, Orion Research Foundation, Finnish Diabetes Foundation, Ministry of Education of the State of Finland, Academy of Finland (grants 251399 and 256470), Paavo Nurmi Foundation, Novo Nordisk Foundation and the Centre of Excellence funding.
The authors declare conflicts of interest.
Trial registration: The Effects of Short-time High-intensity Interval Training on Tissue Glucose and Fat Metabolism in Healthy Subjects and Patients With Type 2 Diabetes (HITPET), ClinicalTrials.gov, NCT01344928.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.acsm-msse.org).
REFERENCES
- 1.Cronin O, O’Sullivan O, Barton W, Cotter PD, Molloy MG, Shanahan F. Gut microbiota: implications for sports and exercise medicine. Br J Sports Med. 2017;51(9):700–1. [DOI] [PubMed] [Google Scholar]
- 2.Manco M, Putignani L, Bottazzo GF. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev. 2010;31(6):817–44. [DOI] [PubMed] [Google Scholar]
- 3.Estaki M, Pither J, Baumeister P, et al. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome. 2016;4(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913–20. [DOI] [PubMed] [Google Scholar]
- 5.Mika A, Fleshner M. Early-life exercise may promote lasting brain and metabolic health through gut bacterial metabolites. Immunol Cell Biol. 2016;94(2):151–7. [DOI] [PubMed] [Google Scholar]
- 6.O’Mahony SM, Clarke G, Dinan TG, Cryan JF. Early-life adversity and brain development: is the microbiome a missing piece of the puzzle? Neuroscience. 2017;342:37–54. [DOI] [PubMed] [Google Scholar]
- 7.Cook MD, Allen JM, Pence BD, et al. Exercise and gut immune function: evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol Cell Biol. 2016;94(2):158–63. [DOI] [PubMed] [Google Scholar]
- 8.O’Sullivan O, Cronin O, Clarke SF, et al. Exercise and the microbiota. Gut Microbes. 2015;6(2):131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ringel Y. The gut microbiome in irritable bowel syndrome and other functional bowel disorders. Gastroenterol Clin North Am. 2017;46(1):91–101. [DOI] [PubMed] [Google Scholar]
- 10.Khalili H, Ananthakrishnan AN, Konijeti GG, et al. Physical activity and risk of inflammatory bowel disease: prospective study from the nurses’ health study cohorts. BMJ. 2013;347:f6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motiani KK, Savolainen AM, Eskelinen JJ, et al. Two weeks of moderate-intensity continuous training, but not high-intensity interval training, increases insulin-stimulated intestinal glucose uptake. J Appl Physiol (1985). 2017;122(5):1188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Diabestes Association Classification and diagnosis of diabetes. Diabetes Care. 2015;38(Suppl):S8–16. [DOI] [PubMed] [Google Scholar]
- 13.Heiskanen MA, Sjoros TJ, Heinonen IHA, et al. Sprint interval training decreases left-ventricular glucose uptake compared to moderate-intensity continuous training in subjects with type 2 diabetes or prediabetes. Sci Rep. 2017;7(1):10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiviniemi AM, Tulppo MP, Eskelinen JJ, et al. Cardiac autonomic function and high-intensity interval training in middle-age men. Med Sci Sports Exerc. 2014;46(10):1960–7. [DOI] [PubMed] [Google Scholar]
- 15.Patlak CS, Blasberg RG. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations. J Cereb Blood Flow Metab. 1985;5(4):584–90. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Diaz J, Garcia-Mantrana I, Vila-Vicent S, et al. Relevance of secretor status genotype and microbiota composition in susceptibility to rotavirus and norovirus infections in humans. Sci Rep. 2017;7:45559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010;92(5):1023–30. [DOI] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237(3):E214–23. [DOI] [PubMed] [Google Scholar]
- 20.Sjoros TJ, Heiskanen MA, Motiani KK, et al. Increased insulin-stimulated glucose uptake in both leg and arm muscles after sprint interval and moderate-intensity training in subjects with type 2 diabetes or prediabetes. Scand J Med Sci Sports. 2018;28(1):77–87. [DOI] [PubMed] [Google Scholar]
- 21.Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuovinen E, Keto J, Nikkila J, Matto J, Lahteenmaki K. Cytokine response of human mononuclear cells induced by intestinal clostridium species. Anaerobe. 2013;19:70–6. [DOI] [PubMed] [Google Scholar]
- 23.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3. [DOI] [PubMed] [Google Scholar]
- 24.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–81. [DOI] [PubMed] [Google Scholar]
- 25.Cohen J. The detection and interpretation of endotoxaemia. Intensive Care Med. 2000;26(1 Suppl):S51–6. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Quintela A, Alonso M, Campos J, Vizcaino L, Loidi L, Gude F. Determinants of serum concentrations of lipopolysaccharide-binding protein (LBP) in the adult population: the role of obesity. PLoS One. 2013;8(1):e54600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albillos A, de la Hera A, González M, et al. Increased lipopolysaccharide binding protein in cirrhotic patients with marked immune and hemodynamic derangement. Hepatology. 2003;37(1):208–17. [DOI] [PubMed] [Google Scholar]
- 28.Tobias PS, Ulevitch RJ. Lipopolysaccharide binding protein and CD14 in LPS dependent macrophage activation. Immunobiology. 1993;187(3–5):227–32. [DOI] [PubMed] [Google Scholar]
- 29.Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002;23(6):301–4. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, Yu Z, Ye X, et al. A marker of endotoxemia is associated with obesity and related metabolic disorders in apparently healthy Chinese. Diabetes Care. 2010;33(9):1925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5. [DOI] [PubMed] [Google Scholar]
- 32.Egshatyan L, Kashtanova D, Popenko A, et al. Gut microbiota and diet in patients with different glucose tolerance. Endocr Connect. 2016;5(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pache I, Rogler G, Felley C. TNF-alpha blockers in inflammatory bowel diseases: practical consensus recommendations and a user’s guide. Swiss Med Wkly. 2009;139(19–20):278–87. [DOI] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. [DOI] [PubMed] [Google Scholar]
- 35.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1792–801. [DOI] [PubMed] [Google Scholar]
- 39.Bressa C, Bailen-Andrino M, Perez-Santiago J, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One. 2017;12(2):e0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambert JE, Myslicki JP, Bomhof MR, Belke DD, Shearer J, Reimer RA. Exercise training modifies gut microbiota in normal and diabetic mice. Appl Physiol Nutr Metab. 2015;40(7):749–52. [DOI] [PubMed] [Google Scholar]
- 41.Evans CC, LePard KJ, Kwak JW, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. 2014;9(3):e92193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla SK, Cook D, Meyer J, et al. Changes in gut and plasma microbiome following exercise challenge in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). PLoS One. 2015;10(12):e0145453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munukka E, Ahtiainen JP, Puigbó P, et al. Six-week endurance exercise alters gut Metagenome that is not reflected in systemic metabolism in over-weight women. Front Microbiol. 2018;9:2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X, Zhang Z, Hu B, Huang W, Yuan C, Zou L. Response of gut microbiota to metabolite changes induced by endurance exercise. Front Microbiol. 2018;9:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen JM, Mailing LJ, Niemiro GM, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc. 2018;50(4):747–57. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell CM, Davy BM, Hulver MW, Neilson AP, Bennett BJ, Davy KP. Does exercise alter gut microbial composition? A systematic review. Med Sci Sports Exerc. 2019;51(1):160–7. [DOI] [PubMed] [Google Scholar]
- 47.Koffert J, Stahle M, Karlsson H, et al. Morbid obesity and type 2 diabetes alter intestinal fatty acid uptake and blood flow. Diabetes Obes Metab. 2018;20(6):1384–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Hajifathalian K, Ezzati M, Woodward M, Rimm EB, Danaei G. Metabolic mediators of the effects of body-mass index, overweight, and obesity on coronary heart disease and stroke: a pooled analysis of 97 prospective cohorts with 1·8 million participants. Lancet. 2014;383(9921):970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40(1):54–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


