Abstract
Background:
Although early survival from sepsis has improved with timely resuscitation and source control, survivors frequently experience persistent inflammation and develop chronic critical illness (CCI). We examined whether increased copy number of endogenous alarmins, mitochondrial DNA (mtDNA), and nuclear DNA (nuDNA) are associated with the early ‘genomic storm’ in blood leukocytes and the development of CCI in hospitalized patients with surgical sepsis.
Methods:
A prospective, observational, cohort study of critically ill septic patients was performed at a United States tertiary health care center. Blood samples were obtained at multiple time points after the onset of sepsis. Droplet digital™ PCR was performed to quantify RHO (nuDNA) and MT-CO2 (mtDNA) copies in plasma. Leukocyte transcriptomic expression of 63 genes was also measured in whole blood.
Results:
We enrolled 112 patients with surgical sepsis. Two experienced early death, 69 rapidly recovered rapidly, and 41 developed CCI. Both mtDNA and nuDNA copy number were increased in all sepsis survivors, but early nuDNA, and not mtDNA, copy number was further increased in patients who developed CCI. Cell-free DNA (cfDNA) copy number was associated with in-hospital but not long-term (180 day and 365 day) mortality, and were only weakly correlated with leukocyte transcriptomics.
Conclusions:
Increased cfDNA copy number persists in survivors of sepsis but is not strongly associated with leukocyte transcriptomics. nuDNA but not mtDNA copy number is associated with adverse, short-term, clinical trajectories and outcomes.
Keywords: sepsis, cfDNA, nuclear, mitochondrial, transcriptomic
TOC Statement- 20190981
We found that cell-free nuclear, but not mitochondrial, DNA copy number was particularly increased in survivors of surgical sepsis who developed chronic critical illness. The importance of this finding is that, contrary to current dogma, mitochondrial DNA copy number was not an important predictor of either transcriptomic changes or clinical outcomes from surgical sepsis.
INTRODUCTION
Sepsis is a leading cause of death and long-term morbidity with annual costs over $24 billion in the United States.1 Despite advances in early recognition, source control, and critical care management, mortality from sepsis remains high at 18–28%.2 In addition, many survivors of sepsis develop chronic critical illness (CCI), which is characterized by prolonged stays in the Intensive Care Unit (ICU), long-term organ dysfunction, and persistent immunosuppression, all od which can lead to secondary infections and sepsis recidivism.3, 4 The origins of CCI are unknown, but early sepsis-related inflammation and organ injury and late secondary infections may promote the release of alarmins which perpetuate long-term persistent inflammation by activating innate immunity.
Cell-free, double-stranded DNA is an important source of alarmins, also called damage associated molecular patterns (or DAMPS); alarmins are comprised in part of both nuclear DNA (nuDNA) and mitochondrial DNA (mtDNA). mtDNA in particular has been proposed as an important alarmin given that it is not bound to nucleosomes in the circulation and is therefore, more likely to be internalized and recognized by pattern recognition receptors to initiate inflammation.5, 6 Both nuDNA and mtDNA can be recognized by these multiple pattern recognition receptors located primarily intracellularly, including TLR9, AIM2, cGAS, DAI, and IFI16.7 One example is their ability to signal through MyD88 or STING, thereby activating both type I interferon and NF-kB-dependent early gene expression.8
Previous work has demonstrated that plasma cell-free DNA (cfDNA) levels are increased during sepsis.9–11 Although some studies have utilized cfDNA levels as prognostic markers in early sepsis, there is little evidence regarding alterations in cfDNA levels over time during recovery form sepsis recovery and the association between cfDNA levels and transcriptomic changes. We hypothesized that if plasma cfDNA levels regulate systemic inflammatory responses during sepsis, cfDNA concentrations would correlate with leukocyte transcriptomic changes and clinical trajectories, reflecting alarmin-induced inflammation and organ injury.
MATERIALS AND METHODS
Enrollment
This prospective, observational, cohort study was approved by the institutional review board and performed between 2014 and 2017 at UF Health Shands Hospital, a 996-bed, academic, tertiary care center. The purpose of this study, conducted by the Sepsis and Critical Illness Research Center at UF under protocols described previously, was to define the epidemiology, dysregulated immunity, and long-term consequences of surgical ICU patients with newly diagnosed sepsis.12, 13 Patients with suspected sepsis were enrolled consecutively in the study. Inclusion criteria were the following: admission to the surgical ICU; age ≥18 years; clinical diagnosis of either sepsis, severe sepsis, or septic shock; and initiation of the sepsis clinical management protocol which includes antibiotics and source control with goal-directed resuscitation and frequent clinical reassessment. Exclusion criteria were the following: refractory shock with expected demise within the first 24 hours of initiation of the protocol; inability to achieve source control; pre-sepsis expected life-span less than three months; patient or family wishes not to pursue aggressive treatment; New York Heart Association Class IV heart disease; Child-Pugh Class C cirrhosis; known HIV with CD4+ count <200 cells/mm3; organ transplant recipient or chronic use of corticosteroids or immunosuppressants; pregnancy; institutionalized patients; chemotherapy or radiation within three days; severe traumatic brain injury with evidence on computed tomography of intracranial injury with Glasgow Coma Scale [GCS] <8); spinal cord injury resulting in permanent sensory or motor deficits; or inability to obtain informed consent from the patient or next of kin. These criteria excluded patients whose baseline conditions would be a primary determinant of their long-term outcomes and thus confound outcome assessment.
Signed informed consent was obtained from the individual subject or their legally-appointed representative within 96 hours of the onset of sepsis. This delayed consent was approved by the University of Florida IRB due to the vulnerable nature of the subject and their immediate family. Families were given time to understand the gravity of the clinical situation, the purpose, and the minimal risks of participating in the study. If consent could not be obtained within 96 hours, all collected data and blood samples were destroyed. This study was registered with clinicaltrials.gov ().
In addition, we recruited 19 control subjects who were age, sex, and race/ethnicity matched; informed consent was obtained, and a single venous blood sample collected. Only subject demographics were collected, and subjects were excluded if they had a history of advanced, recurrent, or metastatic cancer, autoimmune disease, or recent infection.
Definitions
Enrolled patients were classified as having sepsis, severe sepsis, or septic shock according to consensus definitions from the 2001 International Sepsis Definitions Conference.14 Survivors of sepsis were classified into rapid recovery and CCI groups; CCI was defined as an ICU duration of stay ≥14 days with persistent organ dysfunction after the start of the protocol(with Sequential Organ Failure Assessment [SOFA] cardiovascular score ≥1 or any other organ system score ≥2).4, 13 Patients with ICU durations of stay,14 days were also classified as having CCI if discharged to either another hospital, to a long-term acute care facility, or to hospice with evidence of ongoing organ dysfunction at discharge. Patients who died within 14 days of onset of the protocol were classified as experiencing early death. Patients not meeting criteria for CCI or early death were classified as experiencing rapid recovery. Clinical outcomes were adjudicated prospectively by study investigators prior to predictive modeling.
Sample Processing
Blood samples were collected in EDTA-anticoagulated tubes within 12 h of protocol onset, again at 24 h, then at 4, 7, 14, 21, and 28 days. For plasma cytokine studies, blood samples were centrifuged at 200 × g and stored at −80° C until processing using the Luminex Magpix™ (Austin, TX, USA) platform according to the manufacturer’s specification. For measurements of cfDNA, plasma was centrifuged at 5000 × g to remove microaggregates, microparticles, and circulating mitochondria. Fernando et al. have suggested that up to 93% of circulating cfDNA resides in the exosomes, which would be included in our preparations.15 Droplet digital™ PCR (ddPCR) was then performed to quantify the number of copies of a representative mtDNA and nuDNA sequence using the Bio-Rad QX 200 ddPCR™ System with EvaGreen™ fluorescent dye (Hercules, CA, USA). Human mitochondrial cytochrome C oxidase subunit III (MT-CO3) represented mtDNA, while rhodopsin (RHO) DNA sequences represented nuDNA; measurements were reported as copies/μL. For total blood leukocyte transcriptomic measurements, total blood leukocytes were processed and lysed as described previously.16 The NanoString Flex™ platform was then utilized to measure expression of 63 genes; this 63-gene metric (Supplementary Table 1) has been validated prospectively to predict outcomes following severe trauma.17
Statistical Analysis
Continuous data are presented as medians and quartiles and were compared using the Kruskal-Wallis test. Categorical variables are presented as frequencies and percentage and were compared using Fischer’s exact test. Univariate and multivariate analyses were performed with results presented with unadjusted and adjusted odds ratios with 95% confidence intervals. For multivariate logistic regression analysis, stepwise analysis with a threshold of statistical of p<0.10 was performed. For predictive models, discrimination and fit were reported with area under the receiver operating curve (AUC) values and Hosmer-Lemeshow goodness-of-fit tests, respectively. Spearman correlation coefficients were calculated to determine relationships between quantitative variables. All analyses were performed using SAS version 9.4 (Cary, NC, USA).
RESULTS
Enrollment and Patient Characteristics
We enrolled 112 patients with surgical sepsis in the study (Table 1). Of those, 2 experienced early death (< 14 days), 6 recovered 9 rapidly, and 41 developed CCI. Overall in-hospital and 28-day mortality were both 8% (n=9). By 180 days, mortality increased to 18% (n=20). The median age of enrolled patients was 62 years, and 54% of patients were male. There were no differences between groups who either recovered rapidly or developed CCI in terms of sex, age, race/ethnicity, or body mass index (BMI). CCI patients had a greater baseline Charlson Comorbidity Index than patients who recovered rapidly(5 vs. 4, p = 0.040). CCI patients also had a greater median Acute Physiology and Chronic Health Evaluation (APACHE II) score than patients who recovered rapidly (22 vs. 15, p = 0.0004).rec0vered
Table 1:
Patient demographics and baseline characteristics. RAP, rapid recovery; CCI, chronic critical illness; BMI, body mass index (kg/m2); APACHE II, Acute Physiology, Age, Chronic Health Evaluation II; mL, milliliters.
| Overall | RAP | CCI | Early Death | p-value | |
|---|---|---|---|---|---|
| (n=112) | (n=69) | (n=41) | (n=2) | (CCI vs RAP) | |
| Age, median (25th, 75th) | 62 (53, 69.5) | 61 (51, 69) | 64 (58, 71) | 64 (62, 66) | 0.0935 |
| Male sex, n (% when appr0priate) | 60 (53.6) | 34 (49) | 25 (61) | 1 | 0.2443 |
| Race, n (%) | 0.7797 | ||||
| White | 99 (88.4) | 60 (87) | 38 (93) | 1 | |
| African American | 9 (8) | 6 (9) | 2 (5) | 1 | |
| American Indian | 1 (0.9) | 1 (1.5) | 0 | 0 | |
| Asian | 1 (0.9) | 1 (1.5) | 0 | 0 | |
| Other | 1 (0.9) | 0 | 1 (2.4) | 0 | |
| Unknown | 1 (0.9) | 1 (2) | 0 | 0 | |
| Ethnicity (non-Hispanic), n (%) | 109 (97.3) | 66 (96) | 41 (100) | 2 (100) | 0.1188 |
| BMI, median (25th, 75th) | 29.1 (24.5, 35.9) | 29 (24.9, 35.4) | 29 (23.4, 35.6) | 37.4 (29.3, 45.5) | 0.7557 |
| Number of comorbidities, n (%) | 0.2348 | ||||
| 0 | 28 (25) | 20 (29) | 7 (17) | 1 | |
| 1 | 31 (27.7) | 20 (29) | 10 (25) | 1 | |
| 2 | 22 (19.6) | 14 (20) | 8 (20) | 0 | |
| ≥3 | 31 (27.7) | 15 (22) | 16 (39) | 0 | |
| Charlson Comorbidity Index, median (25th, 75th) | 4 (2, 6) | 4 (2, 6) | 5 (3, 8) | 3.5 (3, 4) | 0.0406 |
| APACHE II, median (25th, 75th) | 18 (12, 24) | 15 (10, 22) | 22 (18, 26) | 38 (33, 43) | 0.0004 |
| Total crystalloid within 24 h, median mL (25th, 75th) | 3400 (2400, 4775) | 3350 (2311, 4410) | 3550 (2440, 5175) | 3485 (2400, 4570) | 0.4538 |
| Worst base deficit within 24 h, median (25th, 75th) | 4.2 (1.9, 7.2) | 3.9 (1.8, 7.1) | 5.2 (1.2, 7.5) | 6 (3.6, 8.4) | 0.3912 |
| Highest lactate within 24 h, median (25th, 75th) | 1.7 (1.2, 2.7) | 1.6 (1.2, 2.5) | 1.7 (1.2, 3.6) | 3.7 (1.4, 6) | 0.4986 |
Clinical Outcomes
Clinical outcomes are described in Table 2. The leading source of sepsis for patients overall was intra-abdominal (36%), followed by pneumonia (22%) and surgical site infection (18%). There were no differences between outcome groups in terms of source of sepsis. Overall, patients had a hospital duration of stay at 18 days, which was expectedly greater among CCI patients than patients who recovered rapidly (28 vs. 11 days, p < 0.0001). Thirty-two percent of patients developed non-infectious complications during their hospitalization, which was particularly greater in the CCI group compared to the rapid recovery group (59% vs. 15%, p < 0.0001). Overall, 46% of patients had a “good” discharge disposition, defined as discharge to home, to home with home health care, or a rehabilitation facility. Fifty-five percent of patients had a “poor” disposition to either another hospital, skilled nursing facility (SNF), long-term acute care facility (LTAC), hospice, or in-hospital death.18 “Poor” disposition was markedly more likely among patients who experienced CCI (90% vs. 32%, p < 0.0001).
Table 2:
Patient clinical outcomes and discharge disposition. RAP, rapid recovery; CCI, chronic critical illness; LOS, length of stay; ICU, intensive care unit; MOF, multiple organ failure; SOFA, sequential organ failure assessment score; NSTI, necrotizing soft tissue infection; UTI, urinary tract infection; CLABSI, central-line associated bloodstream infection; SNF, skilled nursing facility; LTAC, long-term acute care facility.
| Overall | RAP | CCI | Early Death | p-value | |
|---|---|---|---|---|---|
| (n=112) | (n=69) | (n=41) | (n=2) | (CCI vs RAP) | |
| Hospital days, median (25th, 75th) | |||||
| Hospital Duration of stay | 18 (8, 29) | 11 (7, 20) | 28 (22, 43) | 5.5 (5, 6) | <.0001 |
| ICU days | 8 (4, 18) | 5 (3, 9) | 21 (16, 29) | 5.5 (5, 6) | <.0001 |
| ICU-free days (28-day) | 19 (7, 24) | 23 (19, 25) | 6 (0, 11) | 0 (0, 0) | <.0001 |
| Mechanically ventilated, n (%) | 80 (71.4) | 40 (58.0) | 38 (92.7) | 2 (100) | <.0001 |
| Ventilator days | 2 (0,6) | 0 (0, 2) | 6 (3, 15) | 5 (5, 5) | <.0001 |
| Ventilator-free days (28-day) | 26 (20, 28) | 28 (26, 28) | 19 (8, 24) | 0 (0, 0) | <.0001 |
| MOF, n (%) | 58 (51.8) | 24 (34.8) | 32 (78.1) | 2 (100) | <.0001 |
| Maximum SOFA score in first 24 h, median (25th, 75th) | 8 (4, 11) | 6 (3, 9) | 10 (8, 12) | 21.5 (21, 22) | <.0001 |
| Noninfectious complications, n (%) | 36 (32.1) | 10 (14.5) | 24 (58.5) | 2 (100) | <.0001 |
| Sourse of Infection, n (%) | 0.8206 | ||||
| Intra-abdominal sepsis | 40 (35.7) | 25 (36) | 14 (34) | 1 | |
| NSTI | 16 (14.3) | 11 (16) | 5 (12) | 0 | |
| Pneumonia | 25 (22.3) | 13 (19) | 11 (27) | 1 | |
| Surgical Site Infection | 20 (17.9) | 12 (17) | 8 (20) | 0 | |
| UTI | 7 (6.3) | 5 (7) | 2 (5) | 0 | |
| Other | 1 (0.9) | 1 (2) | 0 | 0 | |
| CLABSI | 2 (1.8) | 2 (3) | 0 | 0 | |
| Empyema | |||||
| Time to 1st nosocomial infection, median days (25th, 75th) | 8 (5, 17.5) | 6 (3, 14) | 9 (6, 20) | NA | 0.1356 |
| Number of nosocomial infections per patient, n (%) | <.0001 | ||||
| 0 | 80 (71.4) | 58 (84) | 20 (9) | 0 | |
| 1 | 16 (14.3) | 8 (12) | 8 (20) | 0 | |
| ≥2 | 16 (14.3) | 3 (4) | 13 (32) | 0 | |
| Discharge disposition (n, %) | <.0001 | ||||
| ”Good” Disposition | 51 (45.5) | 47 (68) | 4 (10) | 0 | <.0001 |
| Home | 22 (19.6) | 22 (32) | 0 | 0 | |
| Homecare | 25 (22.3) | 22 (32) | 3 (7) | 0 | |
| Rehab | 4 (3.6) | 3 (4) | 1 (2) | 0 | |
| “Poor” Disposition | 61 (54.5) | 22 (32) | 37 (90) | 2 | <.0001 |
| Another Hospital | 8 (7.1) | 0 | 8 (20) | 0 | |
| SNF | 27 (24.1) | 21 (30) | 6 (15) | 0 | |
| LTAC | 15 (13.4) | 1 (1) | 14 (34) | 0 | |
| Hospice | 2 (1.8) | 0 | 2 (5) | 0 | |
| Death (in-hospital) | 9 (8) | 0 | 7 (17) | 2 (100) | 0.0007 |
| Mortality within 28 days, n (%) | 9 (8) | 1 (1) | 6 (15) | 2 (100) | 0.0105 |
| Mortality within 180 days, n (%) | 20 (17.8) | 2 (3) | 16 (39) | 2 (100) | <.0001 |
| Mortality within 1 year, n (%) | 24 (21.4) | 4 (6) | 18 (44) | 2 (100) | <.0001 |
Cytokine and Cell-Free DNA Levels in Septic Patients
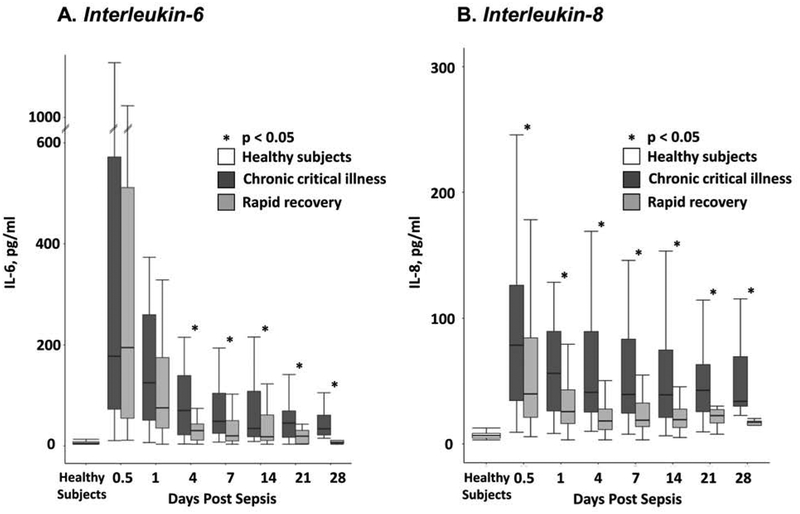
Plasma cytokine analyses revealed statistically significantly greater concentrations of IL-6 and IL-8 in CCI patients compared to patients who recovered rapidly, which persisted at later time points out to 28 days after onset of the protocol (Figure 1). Patients who recovered rapidly also had an early increase in IL-6 after onset of the protocol, but IL-6 levels returned to levels seen in healthy subjects by 21 days post sepsis.
Figure 1:
Plasma cytokine concentrations (Panel A, IL-6; Panel B, IL-8) over time in patients with surgical sepsis who developed CCI or who rapidly recovered. IL = interleukin; pg = picograms; ml = milliliter; CCI = chronic critical illness.
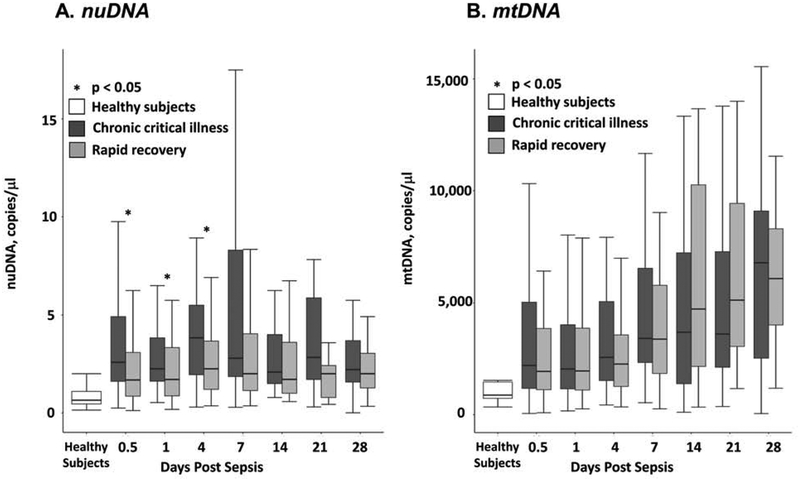
Cell-free DNA copy numbers over time are described in Figure 2. Enrolled septic patients had uniformly greater levels of nuDNA and mtDNA than healthy control subjects at all measured time points (p < 0.05). CCI patients had greater nuDNA copy number at 12 hours, 24 hours, 4 days, and 7 days after onset of the protocol compared to patients who recovered rapidly (p < 0.05). Surprisingly, mtDNA copy number was not different between CCI and patients who recovered rapidly patients at all measured time points after onset of surgical sepsis. Although the 9 patients who died within 28 days had greater mean nuDNA levels 12 hours after sepsis than survivors (12 vs. 2 copies/μL), this trend was driven by the group of patients who died early; median nuDNA and mtDNA levels were not different at this time point (median 3 vs. 2 copies/μL). When comparing cfDNA levels between long-term survivors and non-survivors, patients who survived by 180 days had lesser nuDNA levels 12 hours after sepsis compared to non-survivors (median 2 vs. 3 copies/μL, p = 0.0111). Non-survivors at one year also had lesser mtDNA levels 7 days after sepsis compared to survivors (median 1324 vs. 2218 copies/μL, p = 0.0467).
Figure 2:
Nuclear (Panel A) and mitochondrial DNA (Panel B) copy number changes over time in surgical sepsis patients who either experienced chronic critical illness (CCI) or rapidly recovered (RAP). Asterisks indicate statistically significant differences at those time points between patients who developed CCI or recovered rapidly (p<0.05).
Cell-free DNA Correlation with Clinical Parameters and Transcriptomics
Correlations between nuDNA and mtDNA copy number over time with select clinical parameters and transcriptomics are described in Table 3. We found that nuDNA and mtDNA copy number were positively correlated throughout the first seven days, and nuDNA, but not mtDNA, copy number 12 hours after the initiation of the sepsis protocol had a positive correlation with maximum SOFA score within 24 hours of onset of the protocol and their APACHE II score. Twenty-four hours after onset of the sepsis, nuDNA copy number was only weakly correlated with blood leukocyte expression of the 63 genes in the transcriptomic metric (Spearman ρ= 0.21, p = 0.041). The nuDNA copy number at 24 hours also correlated significantly with the maximum SOFA score within 24 hours of onset of the protocol. Surprisingly, mtDNA copy number 12 hours after protocol onset was negatively correlated with the transcriptomic metric (Spearman ρ = −0.20, p = 0.0429).
Table 3:
Spearman correlation coefficients comparing cell-free DNA levels, clinical scores, and the S63 DFR transcriptomic metric.
| nuDNA 12 hours | nuDNA 1 day | nuDNA 4 days | nuDNA 7 days | mtDNA 12 hours | mtDNA 1 day | mtDNA 4 days | mtDNA 7 days | Max SOFA first 24 hours | APACHE II | IL-6 12 hours | S63 DFR 12 hours | S63 DFR 1 day | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nuDNA 12 hours | 1 | 0.5804 | 0.4146 | 0.4547 | 0.4752 | 0.3583 | 0.2481 | 0.2630 | 0.2262 | 0.2560 | 0.0179 | 0.0115 | 0.0696 |
| p-value | <.0001 | <.0001 | <.0001 | <.0001 | 0.0002 | 0.0229 | 0.0267 | 0.0170 | 0.0067 | 0.8524 | 0.9083 | 0.5030 | |
| nuDNA 1 day | 1 | 0.5258 | 0.3246 | 0.2545 | 0.2931 | 0.1982 | 0.2078 | 0.2411 | 0.1611 | −0.0237 | −0.0305 | 0.2129 | |
| p-value | <.0001 | 0.0065 | 0.0098 | 0.0027 | 0.0742 | 0.0867 | 0.0141 | 0.1040 | 0.8119 | 0.7665 | 0.0405 | ||
| nuDNA 4 days | 1 | 0.6248 | 0.2720 | 0.3357 | 0.4097 | 0.3867 | −0.0178 | −0.0778 | −0.0811 | 0.0064 | 0.1323 | ||
| p-value | <.0001 | 0.0123 | 0.0020 | <.0001 | 0.0013 | 0.8713 | 0.4789 | 0.4606 | 0.9548 | 0.2613 | |||
| nuDNA 7 days | 1 | 0.4405 | 0.3648 | 0.5587 | 0.6266 | 0.0785 | −0.0583 | −0.0061 | 0.1236 | 0.1808 | |||
| p-value | 0.0001 | 0.0021 | <.0001 | <.0001 | 0.5152 | 0.6290 | 0.9595 | 0.3115 | 0.1562 | ||||
| mtDNA 12 hours | 1 | 0.7155 | 0.5989 | 0.4228 | 0.0280 | 0.0891 | −0.1446 | −0.2000 | −0.1700 | ||||
| p-value | <.0001 | <.0001 | 0.0002 | 0.7706 | 0.3527 | 0.1300 | 0.0429 | 0.0995 | |||||
| mtDNA 1 day | 1 | 0.7064 | 0.5948 | −0.1213 | −0.0945 | −0.1446 | −0.1875 | −0.1514 | |||||
| p-value | <.0001 | <.0001 | 0.2224 | 0.3424 | 0.1449 | 0.0660 | 0.1474 | ||||||
| mtDNA 4 days | 1 | 0.6442 | −0.1917 | −0.1567 | −0.2046 | −0.2430 | −0.3309 | ||||||
| p-value | <.0001 | 0.0788 | 0.1521 | 0.0603 | 0.0288 | 0.0040 | |||||||
| mtDNA 7 days | 1 | 0.0105 | −0.1707 | −0.1437 | −0.1805 | −0.0718 | |||||||
| p-value | 0.9305 | 0.1546 | 0.2319 | 0.1377 | 0.5762 | ||||||||
| Max SOFA first 24 hours | 1 | 0.6458 | 0.1958 | 0.1981 | 0.2265 | ||||||||
| p-value | <.0001 | 0.0385 | 0.0438 | 0.0273 | |||||||||
| APACHE II | 1 | 0.1020 | 0.2824 | 0.1096 | |||||||||
| p-value | 0.2846 | 0.0037 | 0.2903 | ||||||||||
| IL-6 12 hours | 1 | 0.6101 | 0.5600 | ||||||||||
| p-value | <.0001 | <.0001 | |||||||||||
| S63 DFR 12 hours | 1 | 0.6857 | |||||||||||
| p-value | <.0001 | ||||||||||||
| S63 DFR 1 day | 1 | ||||||||||||
| p-value |
Prediction Models
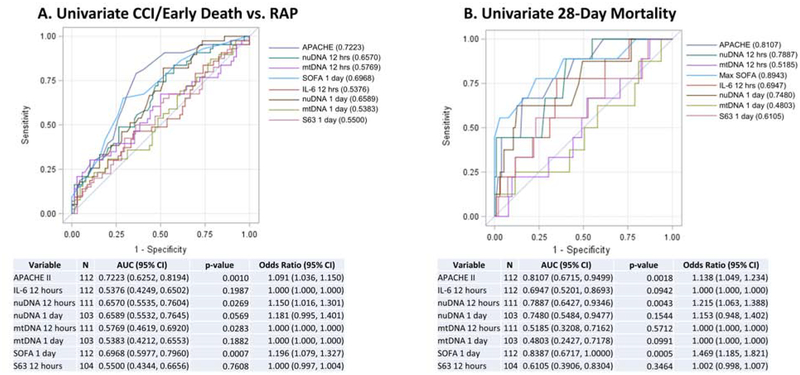
We developed univariate (Figure 3) and multivariate prediction models to predict the occurrence of CCI versus a rapid recovery, as well as 28-day and long-term mortality. Explanatory variables included in univariate analysis were nuDNA and mtDNA copy number at 12 hours and at 1 , 4 , and 7 days after protocol initiation, APACHE II score, IL-6 concentrations, the 63 gene transcriptomic metric at 12 and 24 hours after onset of the protocol onset16, 17, and the maximum SOFA score within 24 hours of onset of the protocol. Univariate analysis revealed that APACHE II score, IL-6 level, nuDNA and mtDNA levels at 12 hours, and SOFA score were predictors of CCI (p<0.05 each). For prediction of 28-day mortality, univariate analysis revealed that APACHE II and SOFA score, and nuDNA copy number at 12 hours after the onset of sepsis were predictors (p < 0.05 each). Controlling for potential confounders using multivariate regression, nuDNA copy number at 12 hours and SOFA score emerged as statistically significant independent predictors of 28-day mortality (AUC 0.8466). On multivariate prediction of long-term mortality (180-day and 1-year), only markers of organ dysfunction and severity of physiologic derangement remained independent predictors (SOFA score at the onset of the protocol for predicting 180-day mortality, APACHE II score for predicting 1-year mortality).
Figure 3:
Univariate analysis predicting chronic critical illness CCI) or rapid death versus rapid recovery (Panel A) and 28-Day Mortality (Panel B).
CCI = chronic critical illness; RAP = rapid recovery; APACHE II = Acute Physiology and Chronic Health Evaluation; SOFA = Sequential Organ Failure Assessment; S63 = Difference-From-Related S63 transcriptomic score.
DISCUSSION
Our work revealed that patients with surgical sepsis have an early increase in circulating nuDNA and mtDNA copy numbers, particularly in the first week after their infection. Interestingly, both the nuDNA and mtDNA copy numbers remained increased over these patients’ entire hospitalization period, and the mtDNA copy number actually increased over time while hospitalized. Surprisingly, early nuDNA but not mtDNA copy number was increased in patients who developed CCI versus those who recovered rapidly. nuDNA, but not mtDNA, copy number was also increased in the patients with sepsis who died during within 28 days; nuDNA also correlated with clinical parameters, including the APACHE II and SOFA scores. Additionally, early nuDNA copy number along with markers of organ dysfunction emerged as statistically significant independent predictors of 28-day mortality. Neither nuDNA nor mtDNA correlated strongly with the transcriptomic responses in the first 24 hours after sepsis.
Prior work has suggested that mtDNA copy number is increased after injury and sepsis and can be recognized as endogenous alarmins by mainly intracellular pattern-recognition receptors.19, 20 mtDNA is thought to activate innate immunity and inflammation via multiple intracellular signaling pathways.21 In particular, Harrington et al. described mtDNA as potent damage-associated molecular patterns (DAMPs) driving inflammation, and thereby influencing TLR9 receptors and inflammasomes.22 Here, although mtDNA copy number was increased after sepsis compared to healthy controls in our study, we were unable to demonstrate consistent association of mtDNA copy number with clinical outcomes; moreover, early nuDNA but not mtDNA copy numbers were only weakly associated with early transcriptomic changes in blood leukocytes. In fact, mtDNA copy number was negatively correlated with early leukocyte transcription. This finding is surprising, because we would have assumed that if cfDNA concentrations were driving inflammatory responses as endogenous alarmins, then there should have been a much stronger relationship between the two. The absence of any strong correlation suggests that other alarmins may be contributing to the early genomic storm which illustrates the redundancy in the signaling of the immediate inflammatory response.
Our laboratory is not the first to report this discrepancy. Jansen et al. studied mtDNA levels in patients with the systemic inflammatory response syndrome (SIRS), proposing that patients who developed acute kidney injury (AKI) would have increased renal cellular damage and increased mtDNA in the plasma and urine.23 Surprisingly, that study found no increase in mtDNA among AKI patients with SIRS.
Despite being primarily bound to nucleosomes in the circulation, prior studies have demonstrated an association between nuDNA levels and clinical outcomes after sepsis. Timmermans et al. also showed that nuDNA but not mtDNA, copy number was correlated with plasma levels of inflammatory cytokines, infusion rate of norepinephrine in septic shock, and evidence of end-organ injury, including total bilirubin and creatinine levels.24
Aswani et al. proposed previously that decreasing cfDNA concentrations might be an experimental approach to limit the incidence and severity of multiple organ dysfunction after severe trauma, with a nucleic acid scavenging polymer emerging as a potential therapeutic.25 Our current work emphasizes the importance of nuDNA as a predictor of outcomes in this cohort of surgical patients with sepsis. Further work is needed to evaluate prospectively whether cfDNA-neutralizing therapeutics improve outcomes both in patients with trauma and surgical sepsis.
Our study has several limitations that require discussion. nuDNA and mtDNA copy numbers were estimated quantitatively based on the concentration of a single representative sequence of each using ddPCR. Multiple representative sequences may assist with more accurate representation of cfDNA levels. Additionally, our use of the 63-gene metric as an estimate for transcriptomic changes is based on prior work validating those 63 genes to predict clinical outcomes and endotypes after severe blunt trauma.17 Sepsis is very different than trauma in terms of eliciting endogenous and exogenous alarmins, and results in more wide-ranging transcriptomic changes. The 63 genes selected for the metric are important in sepsis (Supplementary Table 1), but may not represent a global transcriptomic response. Our work was also limited to patients in surgical ICUs who developed sepsis, potentially causing selection bias and rendering the results perhaps less applicable to the overall cohort of patients with sepsis. Additionally, although our control group was matched as closely as possible to the sepsis cohort, the controls were healthy subjects and not those admitted to the hospital with non-sepsis conditions. It is possible that non-healthy subjects such as those with chronic conditions might have baseline increases in cfDNA or transcriptomic changes.
Another issue that needs to be recognized is that this study was initiated prior to the release of the Sepsis-3 guidelines and used the 2001 consensus definitions for sepsis, severe sepsis, and septic shock. When the 112 subjects with sepsis were re-evaluated using the Sepsis-3 criteria, only five patients would have been excluded. We believe the low mortality in this population is due to the exclusion of those individuals who would not survive 24 hours and the early recognition protocols used for these hospitalized patients developing sepsis.
Despite these limitations, we can conclude that cfDNA copy numbers are increased in patients after surgical sepsis. We cannot conclude, however, that mtDNA copy number has any relationship with the magnitude of the ‘genomic storm’ in blood leukocytes or clinical outcomes. We can infer, however, that nuDNA but not mtDNA copy number early in sepsis correlates with clinical trajectory and outcome. Our work suggests the possibility of using nuDNA rather than mtDNA as a useful biomarker in surgical sepsis. Future work should focus on tissue sources of cfDNA early in sepsis as well as further examination of mechanistic causes and effects of cfDNA on inflammatory processes, as well as the possibility of cfDNA-neutralizing agents as therapeutic options to decrease organ dysfunction and improve outcomes.
Supplementary Material
Acknowledgments
The authors would like to thank all clinicians, technicians, and support staff at UF Shands Health Hospital and the Sepsis and Critical Illness Research Center. Specifically we would like to thank Jennifer Lanz, Ricardo Ungaro, Marvin Dirain, Dina Nacionales, Jaimar Rincon, McKenzie Hollen, Jillianne Brakenridge, Ruth Davis, Ashley McCray, and Elizabeth McRee for their invaluable work and contributions to this project. The work was supported in part by grants, U54 GM062119, P50 GM111152, and R01 GM113945. RBH, JAS, MCC were all supported by a NIGMS training grant T32 GM008721. DCH was funded from an individual training grant for M.D./Ph.D. students, F30 GM119285.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The datasets generated during and analyzed for this study are available from the corresponding author on reasonable request.
COI/DISCLOSURES: The authors have no competing interests to disclose.
References
- 1.Paoli CJ, Reynolds MA, Sinha M, Gitlin M, Crouser E. Epidemiology and Costs of Sepsis in the United States-An Analysis Based on Timing of Diagnosis and Severity Level. Crit Care Med. 2018;46:1889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee C, Dantes R, Epstein L, Murphy DJ, Seymour CW, Iwashyna TJ, et al. Incidence and Trends of Sepsis in US Hospitals Using Clinical vs Claims Data, 2009–2014. JAMA. 2017;318:1241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guirgis FW, Brakenridge S, Sutchu S, Khadpe JD, Robinson T, Westenbarger R, et al. The long-term burden of severe sepsis and septic shock: Sepsis recidivism and organ dysfunction. J Trauma Acute Care Surg. 2016;81:525–32. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins RB, Raymond SL, Stortz JA, Horiguchi H, Brakenridge SC, Gardner A, et al. Chronic Critical Illness and the Persistent Inflammation, Immunosuppression, and Catabolism Syndrome. Front Immunol. 2018;9:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser CJ, Otterbein LE. Danger signals from mitochondrial DAMPS in trauma and post-injury sepsis. Eur J Trauma Emerg Surg. 2018;44:317–24. [DOI] [PubMed] [Google Scholar]
- 6.Thurairajah K, Briggs GD, Balogh ZJ. The source of cell-free mitochondrial DNA in trauma and potential therapeutic strategies. Eur J Trauma Emerg Surg. 2018;44:325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampson P, Dinsdale RJ, Wearn CM, Bamford AL, Bishop JRB, Hazeldine J, et al. Neutrophil Dysfunction, Immature Granulocytes, and Cell-free DNA are Early Biomarkers of Sepsis in Burn-injured Patients: A Prospective Observational Cohort Study. Ann Surg. 2017;265:1241–9. [DOI] [PubMed] [Google Scholar]
- 10.Huang T, Yang Z, Chen S, Chen J. [Predictive value of plasma cell-free DNA for prognosis of sepsis]. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2018;30:925–8. [DOI] [PubMed] [Google Scholar]
- 11.Long Y, Zhang Y, Gong Y, Sun R, Su L, Lin X, et al. Diagnosis of Sepsis with Cell-free DNA by Next-Generation Sequencing Technology in ICU Patients. Arch Med Res. 2016;47:365–71. [DOI] [PubMed] [Google Scholar]
- 12.Loftus TJ, Mira JC, Ozrazgat-Baslanti T, Ghita GL, Wang Z, Stortz JA, et al. Sepsis and Critical Illness Research Center investigators: protocols and standard operating procedures for a prospective cohort study of sepsis in critically ill surgical patients. BMJ Open. 2017;7:e015136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stortz JA, Mira JC, Raymond SL, Loftus TJ, Ozrazgat-Baslanti T, Wang Z, et al. Benchmarking clinical outcomes and the immunocatabolic phenotype of chronic critical illness after sepsis in surgical intensive care unit patients. J Trauma Acute Care Surg. 2018;84:342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–6. [DOI] [PubMed] [Google Scholar]
- 15.Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One. 2017;12:e0183915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuenca AG, Gentile LF, Lopez MC, Ungaro R, Liu H, Xiao W, et al. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013;41:1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raymond SL, Hawkins RB, Wang Z, Mira JC, Stortz JA, Han F, et al. Prospective Validation of a Transcriptomic Metric in Severe Trauma. Ann Surg. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prescott HC, Langa KM, Liu V, Escobar GJ, Iwashyna TJ. Increased 1-year healthcare use in survivors of severe sepsis. Am J Respir Crit Care Med. 2014;190:62–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIlroy DJ, Jarnicki AG, Au GG, Lott N, Smith DW, Hansbro PM, et al. Mitochondrial DNA neutrophil extracellular traps are formed after trauma and subsequent surgery. J Crit Care. 2014;29:1133 e1–5. [DOI] [PubMed] [Google Scholar]
- 20.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577; discussion e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins LV, Hajizadeh S, Holme E, Jonsson IM, Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol. 2004;75:995–1000. [DOI] [PubMed] [Google Scholar]
- 22.Harrington JS, Choi AMK, Nakahira K. Mitochondrial DNA in Sepsis. Curr Opin Crit Care. 2017;23:284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen MPB, Pulskens WP, Butter LM, Florquin S, Juffermans NP, Roelofs J, et al. Mitochondrial DNA is Released in Urine of SIRS Patients With Acute Kidney Injury and Correlates With Severity of Renal Dysfunction. Shock. 2018;49:301–10. [DOI] [PubMed] [Google Scholar]
- 24.Timmermans K, Kox M, Scheffer GJ, Pickkers P. Plasma Nuclear and Mitochondrial DNA Levels, and Markers of Inflammation, Shock, and Organ Damage in Patients with Septic Shock. Shock. 2016;45:607–12. [DOI] [PubMed] [Google Scholar]
- 25.Aswani A, Manson J, Itagaki K, Chiazza F, Collino M, Wupeng WL, et al. Scavenging Circulating Mitochondrial DNA as a Potential Therapeutic Option for Multiple Organ Dysfunction in Trauma Hemorrhage. Front Immunol. 2018;9:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.