Abstract
The epidermis is the first line of defense against ultraviolet (UV) light from the sun. Keratinocytes and melanocytes respond to UV exposure by eliciting a tanning response dependent in part on paracrine signaling, but how keratinocyte:melanocyte communication is regulated during this response remains understudied. Here, we uncover a surprising new function for the keratinocyte-specific cell–cell adhesion molecule desmoglein 1 (Dsg1) in regulating keratinocyte:melanocyte paracrine signaling to promote the tanning response in the absence of UV exposure. Melanocytes within Dsg1-silenced human skin equivalents exhibited increased pigmentation and altered dendrite morphology, phenotypes which were confirmed in 2D culture using conditioned media from Dsg1-silenced keratinocytes. Dsg1-silenced keratinocytes increased melanocyte-stimulating hormone precursor (Pomc) and cytokine mRNA. Melanocytes cultured in media conditioned by Dsg1-silenced keratinocytes increased Mitf and Tyrp1 mRNA, TYRP1 protein, and melanin production and secretion. Melanocytes in Dsg1-silenced skin equivalents mislocalized suprabasally, reminiscent of early melanoma pagetoid behavior. Together with our previous report that UV reduces Dsg1 expression, these data support a role for Dsg1 in controlling keratinocyte:melanocyte paracrine communication and raise the possibility that a Dsg1-deficient niche contributes to pagetoid behavior, such as occurs in early melanoma development.
Keywords: desmosomes, environment, keratinocytes, melanocytes, melanoma, skin, sunlight
1 |. INTRODUCTION
The epidermis is a multi-layered organ composed of multiple cell types that work together to form a barrier against water loss and against environmental, pathogenic, chemical, and physical assaults. Melanocytes (MCs), pigment-producing cells, reside within the basal layer of the epidermis, where they interact through dendritic extensions with more abundant keratinocytes (KC) in a ratio of roughly 36 KCs: 1 MC to form the KC:MC pigmentary unit (Nguyen & Fisher, 2018; Weiner, Fu, Chirico, & Brissette, 2014). Melanin production and transfer from MCs to KCs is a critical protective response to ultraviolet (UV) light from the sun that helps prevent UV-induced DNA damage and subsequent mutagenesis leading to cancer (Brenner & Hearing, 2008; Yamaguchi et al., 2006). KCs and MCs directly associate through cadherin-based adhesive structures and indirectly communicate via paracrine signaling (Lee & Herlyn, 2007; Mescher et al., 2017; Serre, Busuttil, & Botto, 2018). The release of secreted factors (cytokines, chemokines, and growth factors) from KCs and other resident skin cells results in modulation of MC proliferation, differentiation, signaling, pigment production and secretion, and dendricity (Yuan & Jin, 2018).
Activation of the MC microphthalmia-associated transcription factor (MITF) in response to secretion of KC-derived factors (e.g., alpha MC-stimulating hormone [α–MSH], kit ligand [KITL], endothelin 1 [END-1]) following UV exposure results in upregulation of melanin-producing enzymes (including TYRP1, tyrosinase-related protein 1), and melanin production and secretion (Nguyen & Fisher, 2018; Serre et al., 2018). UV exposure also initiates signaling cascades in both KCs and MCs, resulting in secretion of cytokines and chemokines including interleukins (IL1, IL3, IL6, and IL8), interferon (IFN-γ), granulocyte-colony stimulating factor (G-CSF), and tumor necrosis factor α (TNF-α) (Schwarz & Luger, 1989; Terazawa & Imokawa, 2018; Yoshizumi et al., 2008), which control processes involving cross talk between cells such as the tanning response within the KC:MC unit. Lengthened dendrites and increased dendrite branching in response to UV light increase MC interactions with KCs to facilitate melanin transfer (López et al., 2015; Weiner et al., 2014). Further, secreted factors from KCs function to increase DNA damage repair pathways in MCs, reducing the risk of oncogenic mutations (Swope & Abdel-Malek, 2016).
While UV light can modulate the expression of cell adhesion proteins (Jamal & Schneider, 2002; Johnson et al., 2014), the extent to which adhesion-dependent versus signaling roles of cadherins modulate the KC:MC niche is unknown. We previously showed that in addition to its adhesive function, the KC-specific desmosomal cadherin desmoglein 1 (Dsg1) attenuates MAPK/ERK signaling as cells exit the basal proliferating layer of the epidermis to promote stratification and a tightly controlled program of differentiation (Getsios et al., 2009; Harmon et al., 2013; Nekrasova et al., 2018). We also showed that Dsg1 expression is selectively reduced in response to UV exposure in association with activation of the MAPK/ERK pathway, previously shown to increase epidermal inflammation following UV exposure (El-Abaseri, Hammiller, Repertinger, & Hansen, 2013; Johnson et al., 2014). In addition, loss-of-function mutations in Dsg1 lead to severe dermatitis, multiple allergies, and metabolic wasting (SAM) syndrome, accompanied by a KC autonomous increase in proallergy cytokines, and several pathological conditions resulting in downregulation of Dsg1 at the cell surface can induce cytokine production (Hammers & Stanley, 2013; Polivka et al., 2018; Samuelov et al., 2013).
Based on these previous studies, we addressed whether reduction in Dsg1, such as occurs following UV exposure (Johnson et al., 2014), serves to mediate paracrine communication between KCs and MCs to promote the tanning response. Our data show that KC-specific Dsg1 can modulate the epidermal microenvironment and MC signaling, pigment production and secretion, and dendricity through regulation of KC cytokines, chemokines, and other secreted factors in the absence of UV exposure. In addition to altering dendricity and increasing pigmentation, MCs within Dsg1-deficient 3D human skin equivalents mislocalized away from the basement membrane in a manner reminiscent of pagetoid behavior that occurs in early melanoma (Mo, Preston, & Zaidi, 2019). This study elucidates a new role for Dsg1 in regulating KC:MC cell–cell communication and suggests a mechanism by which the epidermis can sense and propagate signals in response to environmental stimuli such as UV light.
2 |. MATERIALS AND METHODS
2.1 |. Cell culture
KCs and MCs were isolated from neonatal foreskin provided by the Northwestern University Skin Disease Research Center (NUSDRC) as described in Halbert, Demers, and Galloway (1992). Collection of human tissue was reviewed and approved by the Northwestern University Institutional Review Board (STU00009443). Media conditions for cell culture are given in Appendix S1.
2.2 |. DNA constructs for viral transduction and transduction methods
LZRS-miR Dsg1 (Dsg1shRNA, shDsg1) and LZRS-Flag Dsg1 (FL) were generated as described (Getsios et al., 2009). LZRS–NTshRNA (shCTL) was generated with the following sequences inserted: NTshRNA-fwd 5′–GTATCTCTTCATAGCCTTAAA–3′ and NTshRNA-rev 5′–TTTAAGGCTATGAAGAGATAC–3′. KCs were transduced with retroviral supernatants produced from Phoenix cells (provided by G. Nolan, Stanford University, Stanford, CA) as previously described (Simpson, Kojima, & Getsios, 2010). Lentivirus (pLVX myristoylated tdTomato, Clontech) transduction of MCs utilized virus obtained from the NUSDRC. Briefly, MCs were plated at 30% confluence and the next day treated with 1 μg/ml polybrene plus lentivirus and incubated at 37°C overnight followed by washing.
2.3 |. siRNA transfection
KCs at approximately 30% confluency were transfected with siRNA oligonucleotides (siDsg3 [Dharmacon siGENOME SMARTpool, D-011646:5′-GCAAAUGACUGCUUGUUGA-3′, 5′-GAAUGGAAAUG ACCACUAA-3′, 5′-UGAAAGAUGUCAACGAUAA-3′, 5′-GAACCGAG AUUCUACUUUC-3′]) and scramble/non-targeting siRNA ([Dharmacon, D-001206–14–20]) at a final concentration of 20 nM via DharmaFECT.
2.4 |. Organotypic skin cultures
Organotypic cultures were grown as described previously (Arnette, Koetsier, Hoover, Getsios, & Green, 2016; Getsios et al., 2009) with the addition of the following steps: The collagen plug was coated with extracellular matrix using 804G supernatant (Langhofer, Hopkinson, & Jones, 1993). MCs were seeded onto the collagen plug overnight. KCs were then seeded at a 5 KCs:1 MC ratio. Details of fixation and processing of organotypic cultures for imaging are given in Appendix S1.
2.5 |. Preparation of conditioned media from KCs
KCs infected at 20% confluence with shCTL, shDsg1, or shDsg1 together with silencing-resistant Dsg1-Flag (shDsg1 + FL) were grown to 80% confluence, expanded to 3, 10-cm dishes, and grown to confluence. At confluence, cells were switched to high calcium medium and cultured for 3 days before replacing the medium. On day 3, the medium was replaced with new high calcium medium and harvested on day 5 (day 3–5 conditioned medium, a 48 hr of time period). All conditioned media was stored at −20°C in aliquots until use in experiments.
2.6 |. Quantitative real-time PCR and immunoblot analysis of proteins
Methods utilized for quantitative real-time PCR and immunoblot analysis of proteins, including gene-specific primers (Table S1), and antibodies used are given in Appendix S1.
2.7 |. Pigment analysis in 3D organotypic cultures
Pigment levels within the 3D organotypic skin cultures were measured using two independent methods. In whole-mount samples, five bright-field images were taken at random from each sample of 3D organotypic cultures containing shCTL- or shDsg1-infected KCs seeded together with MCs as described above (n = 3 pairs). Samples were unstained, allowing visualization of only pigmented cells and released pigment. Pigmented areas were identified by their low pixel intensity in the bright-field images after setting a threshold to exclude background using the shCTL sample from each pair. The percent area below the threshold was then calculated. Areas adjacent to MCs was analyzed, but pigmented MCs themselves were excluded.
In the second method, formalin-fixed tissue sections were stained according to the manufacturer’s instructions using the Fontana Masson Stain Kit (Abcam ab150669). Five random bright-field images were taken of each sample, and melanin-positive areas were identified by their low pixel intensity after setting a threshold to exclude other areas using ImageJ software. The percent area below the threshold was then calculated by setting the region of interest to the epidermal layer of the organotypic culture and calculating the area appearing positive for melanin.
2.8 |. Dendrite branch point measurements in organotypic cultures
Primary human MCs were infected with lentivirus expressing a myristoylated tdTomato construct to allow visualization of the MCs when incorporated into 3D organotypic cultures as described above. Five fields containing MCs were selected randomly from each sample and imaged. MC dendrites were modeled using the Simple Neurite Tracer plugin in Fiji (Longair, Baker, & Armstrong, 2011; Schindelin et al., 2012). Briefly, a point was selected within the cell body and another point along a dendrite was selected. The two points were connected by tracing within the plugin, and then, a new point was selected within the Z-stacked images until the entire dendrite and all branches were traced. These dendrite models were then used to determine the number of dendrite branch points per cell body.
2.9 |. Image tracing for visualization
Dendrite models generated using the Simple Neurite Tracer plugin as described above and in (Longair et al., 2011) were used to fill in the entire shape of the MCs from the raw images for the purpose of eliminating background fluorescence and allowing precise visualization of MCs for use in figures. Importantly, these filled images were only used to aid in visualization; all measurement and analyses were performed on raw image files.
2.10 |. Quantification of intracellular melanin and melanin secretion from monolayer-cultured MCs
MCs were grown to 50% confluence in MC media and subsequently switched to a 1:1 ratio of KC-conditioned media: MC media with fresh 1:1 media mixture added to existing media every 2 days up to 7 days. After 7 days, media was collected and subjected to centrifugation at 10,000 x g for 5 min. The resulting melanosome pellet was resuspended in 100 μl 1N NaOH plus 10% DMSO. Intracellular melanin content was quantified by trypsinizing the MCs on day 7 of culture in conditioned media. Cells were pelleted at 1500 x g and resuspended in 100 μl 1N NaOH plus 10% DMSO. Both media and intracellular suspensions were boiled for 30 min at 100°C prior to being transferred to the appropriate wells of a 96-well plate (BD Biosciences). The absorbance at 405 nm of each well was measured with an ELx800 Microplate Reader (Bio-Tek Instruments, Inc) (Laskin, Piccinini, Engelhardt, & Weinstein, 1982). When applicable, 100 nM recombinant human beta-defensin 3 (MilliporeSigma) was used as an MC1R inhibitor (Swope et al., 2012).
2.11 |. Dot blot analysis of cytokines
Conditioned media from KCs was concentrated 5× by centrifugation through a Centricon Plus-70 Centrifugal Filter with a cutoff of 3 kDa (MilliporeSigma), according to the manufacturer’s instructions. Protein concentration was normalized between paired samples, and a 1 ml sample was incubated, according to the manufacturer’s instructions, on a Human Cytokine C3 Array (RayBiotech). Dot blot images were digitized using an HP OfficeJet 5,610 All-in-One office scanner at 300 dpi and saved as grayscale mode TIFF documents. Densitometry was performed as described in Johnson et al. (2016) Densitometry values for each analyte were normalized to within-membrane-positive and within-membrane-negative control dots, and Dsg1 membrane dot values were normalized to the macrophage-derived chemokine (MDC) dots on the paired control membrane since this secreted factor was detectable on every membrane and had a low level of experiment to experiment variability.
2.12 |. Quantification of dendrite length in 2D culture
MCs were grown to 75% confluence in MC media and subsequently switched to a 1:1 ratio of KC-conditioned media:MC media. Cells were incubated overnight or for 7 days with fresh 1:1 media mixture added to existing media every 2 days before being fixed in 4% paraformaldehyde (MilliporeSigma) in PBS for 10 min at room temperature. Wide-field images were used to visualize MC dendrites. Dendrite length was quantified with ImageJ software and measured (in μm) from the edge of the cell body to the tip of the single longest dendrite for each cell. Dendrites were defined as extensions originating from the cell body that did not exhibit a lamellar morphology. When applicable, reparixin (Moriconi et al., 2007) (Cayman Chemical) was added to media at a final concentration of 5 μg/ml media. Recombinant IL6 and IL8 (Abcam) were added to media at final concentrations of 0.2 and 25 ng/ml, respectively.
2.13 |. Microscope imaging
Wide-field images were acquired on an Upright Leica Microscope (model DMR) fitted with an Orca-100 digital camera (model C4742-95; Hamamatsu Photonics) and a 40× 1.0 numerical aperture (NA) oil Plan Fluotar objective. Apotome images were acquired using an epifluorescence microscope system (AxioVision Z1; Carl Zeiss) fitted with an Apotome slide module, AxioCam MRm digital camera, and a 40× 0.5 EC Plan-Neofluar or 100× 1.4 NA oil Plan-Apochromat objective (Carl Zeiss). Confocal z-stacks (z-step size of 0.5 μm) of whole-mount samples were acquired using a Nikon A1R confocal laser microscope equipped with GaAsP detectors and a 60× Plan-Apochromat objective lambda with a NA of 1.4 and run by NIS-Elements software (Nikon). NIS-Elements (version 5.02) was used to generate 3D reconstructions of z-stacks using the Volume Viewer tool with z-depth coding blending (rainbow contrast look up table) and to determine the z position of the MC cell body centroid. For each experiment, images were acquired using the same imaging conditions.
2.14 |. Sample size and statistical analysis
All experiments were performed independently ≥3 times. The precise number of times the experiments were performed is provided in each figure legend. For each independent/biological replicate, multiple experimental/control arms were processed and analyzed in parallel. Graphs are displayed as either spaghetti plots, graphs showing all data points with mean and standard deviation (SD), or graphs showing mean with standard error of the mean (SEM) as stated in each figure legend. Power analyses to determine sample sizes were determined in consultation with the Northwestern University Quantitative Data Sciences Core. Two group comparisons were performed using two-tailed, two-sample equal variance Student’s t test. For comparisons of more than two groups, one-way ANOVA was used, followed by Tukey’s or Holm-Siddak’s multiple comparison tests. All analyses were performed using GraphPad Prism version 8.0 for Mac (GraphPad Software). Statistical significance was defined as p < .05.
3 |. RESULTS
3.1 |. MCs incorporated into KC Dsg1-deficient 3D organotypic cultures exhibit increased pigment distribution and dendrite branching compared with controls
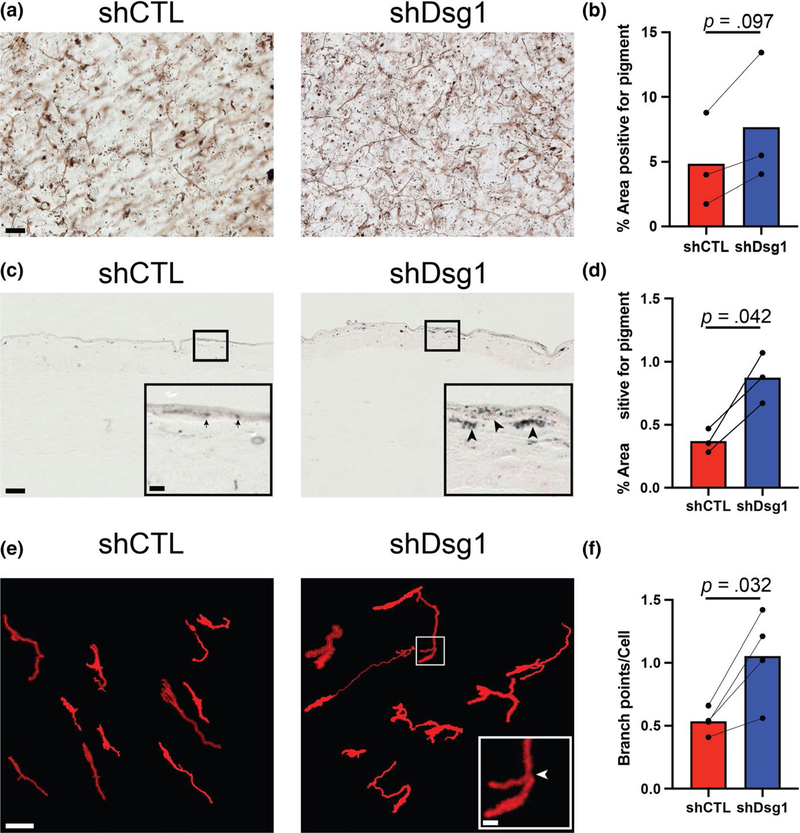
MC pigment production and secretion is a crucial protective response in the epidermis that can be regulated by paracrine signaling between KCs and MCs (Serre et al., 2018; Yuan & Jin, 2018). Since Dsg1 is known to be reduced after UV exposure of KCs (Johnson et al., 2014) and its suppression or mutation results in altered cytokine/chemokine profiles (Polivka et al., 2018; Samuelov et al., 2013), we hypothesized that Dsg1 may play a role in coordinating the skin’s paracrine-mediated tanning response. The 3D architecture of the skin propagates signals throughout the tissue through integration of multiple signaling and adhesive cues that differ from when cells are grown in 2D culture (Li, Fukunaga-Kalabis, & Herlyn, 2011). Therefore, we assessed whether reduction in KC Dsg1 in 3D organotypic cultures containing both KCs and MCs would result in changes in MC behavior. First, we addressed whether alterations in pigment deposition occurred in KC Dsg1-depleted organotypic cultures. Using both bright-field microscopy (Figure 1a,b) and Fontana Masson staining (Figure 1c,d), pigment deposition was greater throughout the epithelial tissue of Dsg1-depleted cultures compared with controls. Second, we addressed whether KC Dsg1 loss promoted changes in MC dendrite morphology. To address this question, MCs were infected with lentivirus expressing myristoylated tdTomato to aid cell visualization and tracking within the 3D skin structure. Increased dendrite branching was observed in the KC Dsg1-depleted cultures compared with controls (Figure 1e). The total number of dendrite branches arising from each cell body was quantified using the Simple Neurite Tracer (Longair et al., 2011) open-source software (Figure 1f). Figure S1 shows percent occurrence of each discrete number of dendrite branches (0–8 branch points), while Figure 1f reports the average number of branch points per MC. From these data, we conclude that reduction in KC Dsg1 alters MC behavior.
FIGURE 1.
MCs incorporated into Dsg1-deficient 3D organotypic cultures exhibit increased pigment distribution and dendrite branching compared with controls. (a) Silencing keratinocyte (KC) Dsg1 (shDsg1) resulted in increased pigment deposition throughout the organotypic cultures compared with controls (shCTL). (b) Quantification of the area positive for pigment in the images from 3D organotypic cultures prepared using the whole-mount method trends toward an increase in area positive for pigment in Dsg1-silenced cultures compared with controls. Areas selected for analysis excluded melanocytes (MCs) to focus on released pigment. (c) Fontana Masson staining of sections from shCTL and shDsg1 cultures shows increased pigment staining in the shDsg1 compared with shCTL cultures. Insets are provided to allow more detailed visualization of the darker stained, larger aggregates of pigment in the shDsg1 (arrowheads) compared with shCTL (arrows) cultures, scale bar for insets = 10 μm. (d) Quantification of the area positive for pigment from C. (e) Dsg1 knockdown in KCs increased the average number of branch points per MC in 3D organotypic cultures compared with controls. The open-source (FIJI) software Simple Neurite Tracer was used to trace tdTomato-expressing MCs in 3D as described in the Materials and Methods to aid visualization and analysis of MC dendrite branching and to exclude background fluorescence. Scale bar for insets = 10 μm. (f) Quantification of the average number of branch points per MC shows increased branch points in Dsg1-silenced cultures compared with controls. A histogram of the relative occurrence of 0 to 8 branch points per cell is shown in Figure S1. Data were statistically analyzed using the two-tailed paired t test. All scale bars except insets = 50 μm
3.2 |. Exposure to Dsg1-deficient KC-conditioned media increases MC pigmentation and pigment secretion
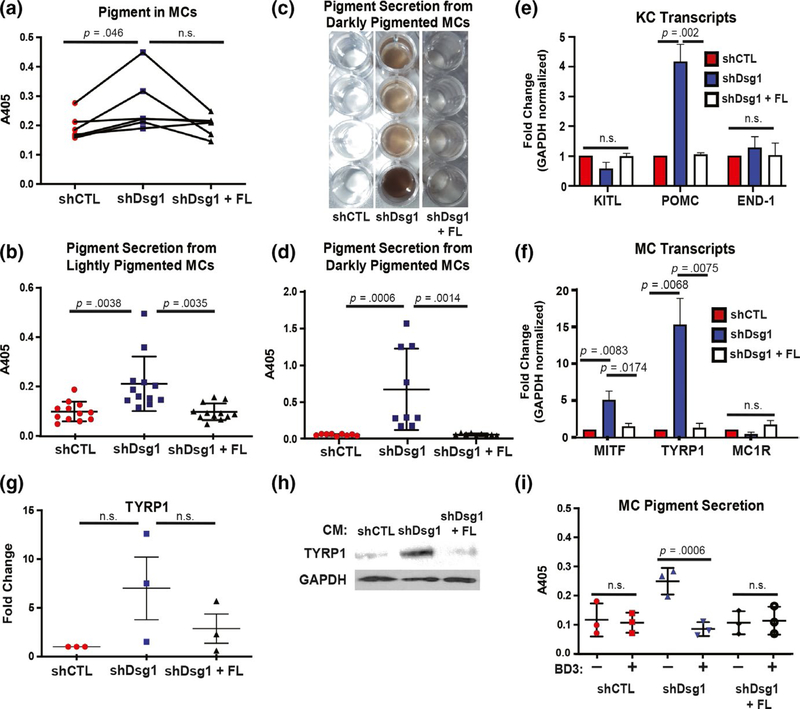
Since we cannot discriminate between paracrine and adhesive cues within the 3D skin structure, we turned to 2D culture for the remainder of the experiments to test the effects of paracrine signaling downstream of KC Dsg1 loss on MC behavior. After observing increased pigment deposition throughout KC Dsg1-depleted organotypic cultures containing MCs (Figure 1), we next measured MC pigment content and secretion following incubation with conditioned media from KCs with normal (shCTL), suppressed Dsg1 (shDsg1), or shDsg1 plus rescue by silencing-resistant (Dsg1 + FL) expression. The MTT assay was utilized to determine that the three conditions had similar numbers of viable cells on day 1 after treatment with conditioned media and on the day pigment was assessed (day 7), (Figure S2a). Growth curves of cells under the three conditions were also assessed through cell counting (Figure S2b). Intracellular pigment significantly increased in MCs treated with conditioned media from Dsg1-deficient KCs (Figure 2a). Conditioned media from Dsg1-deficient KCs increased MC pigment secretion into the media compared with media from shCTL-infected KCs or from shDsg1-infected KCs reconstituted with Dsg1 + FL(Figure 2b–d). MCs isolated from both lightly (Figure 2b) and darkly (Figure 2c,d) pigmented donors exhibited increased pigment secretion when grown in conditioned media from Dsg1deficient KCs.
FIGURE 2.
Exposure to conditioned media from Dsg1-deficient KCs increases MC pigmentation and pigment secretion. (a) Seven days of exposure to conditioned media from Dsg1-deficient (shDsg1) KCs increased MC pigment content (read as absorbance at 405 nm following lysis with NaOH) compared with conditioned media from control (CTL) KCs or media from KCs infected with shDsg1 plus silencing-resistant Dsg1-Flag (shDsg1 + FL) (N = 5 MC:KC-conditioned media pairs, plotted as spaghetti plots to represent each individual experiment). (b) Quantification of pigment secretion into the media from MCs from lightly pigmented donors (Caucasian, Asian, Hispanic, unknown) treated for 7 days with conditioned media from KCs infected with shCTL, shDsg1, shDsg1 + FL constructs (N = 13 MC:KC-conditioned media pairs). (c) Example of visual appearance of pigment secretion into the media (concentrated by centrifugation) from MCs from 4 individual darkly pigmented donors (African American) treated for 7 days with the indicated conditioned media. (d) Quantification of pigment secretion into the media from MCs from darkly pigmented donors (African American) treated for 7 days with the indicated conditioned media (N = 9 MC:KC-conditioned media pairs). Treatment with conditioned media from KCs depleted of Dsg1 significantly increased pigment secretion into the media compared with conditioned media from shCTL or shDsg1 + FL-infected KCs. For panels A, B, and D, data were analyzed using one-way ANOVA followed by Tukey’s post hoc analysis. (e) Pro-opiomelanocortin (POMC) mRNA was increased in shDsg1-infected KCs compared with shCTL and shDsg1 + FL-infected KCs, while KITL and END-1 mRNA levels were not significantly changed between groups (N = 3 independent experiments, KCs were harvested on day 3 after being induced to differentiate in high calcium-containing medium). (f) Transcripts involved in pigment production in MCs (microphthalmia-associated transcription factor [MITF] and tyrosinase-related protein 1 [TYRP1] were upregulated while melanocortin 1 receptor [MC1R]) were not significantly changed in MCs treated with conditioned media from shDsg1-infected KCs compared to MCs treated with conditioned media from shCTL or shDsg1 + FL-infected KCs (N = 3 independent experiments). For panels E and F, gene expression was normalized to GAPDH across samples and then reported as fold change compared with shCTL. Reported p values are results of one-way ANOVA of the fold change data followed by Tukey’s post hoc analysis. (g) Quantification of immunoblots performed after 3 MC isolates were treated with conditioned media from shCTL, shDsg1, or shDsg1 + FL-infected KCs. TYRP1 protein increased in all 3 MC isolates treated with conditioned media from shDsg1-infected KCs (did not reach the level of significance). (h) An example TYRP1 immunoblot is shown. GAPDH serves as a protein loading control. (i) Recombinant human beta-defensin 3 (BD3, 100 nM) in conditioned media (7 days) from KCs depleted of Dsg1 inhibits MC pigment secretion (N = 3, data were analyzed using one-way ANOVA followed by Holm-Sidak’s post hoc analysis for comparison between drug-treated and drug-untreated samples for each condition). Graphical data in B, D, G, and I are represented as mean and standard deviation since each individual data point represents a single experimental replicate. qPCR data in E and F are represented as mean and SEM
To determine whether the observed increase in pigment was due to classical pigment inducers upon KC Dsg1 silencing, we tested KCs for mRNA expression levels of KITL, Pro-opiomelanocortin (POMC—precursor for the melanocortin 1 receptor [MC1R] ligand αMSH), and END-1. POMC mRNA was increased upon Dsg1 suppression in KCs, while KITL and END-1 were not, compared with controls (Figure 2e). We next tested for induction of melanogenesis transcripts Mitf, Tyrp1, and MC1R in MCs treated with conditioned media. Mitf and Tyrp1, but not MC1R, were elevated in MCs incubated with conditioned media from KCs deficient in Dsg1 (Figure 2f). TYRP1 protein tended to be increased to various extents in 3 tested MC isolates treated with conditioned media from KCs deficient in Dsg1 compared with controls (Figure 2g,h). When MCs were incubated with human beta-defensin 3 (BD3), an inhibitor of MC1R (Swope et al., 2012), pigment induction was inhibited downstream of treatment with conditioned media from Dsg1-deficient KCs (Figure 2i). Together, these results indicate that reduction in Dsg1 in KCs induces paracrine signaling from KCs to increase pigment production and secretion in MCs, likely through activation of the MC1R pathway leading to activation of MITF and TYRP1.
3.3 |. Suppression of KC Dsg1 alters cytokine mRNA and secretion from keratinocytes
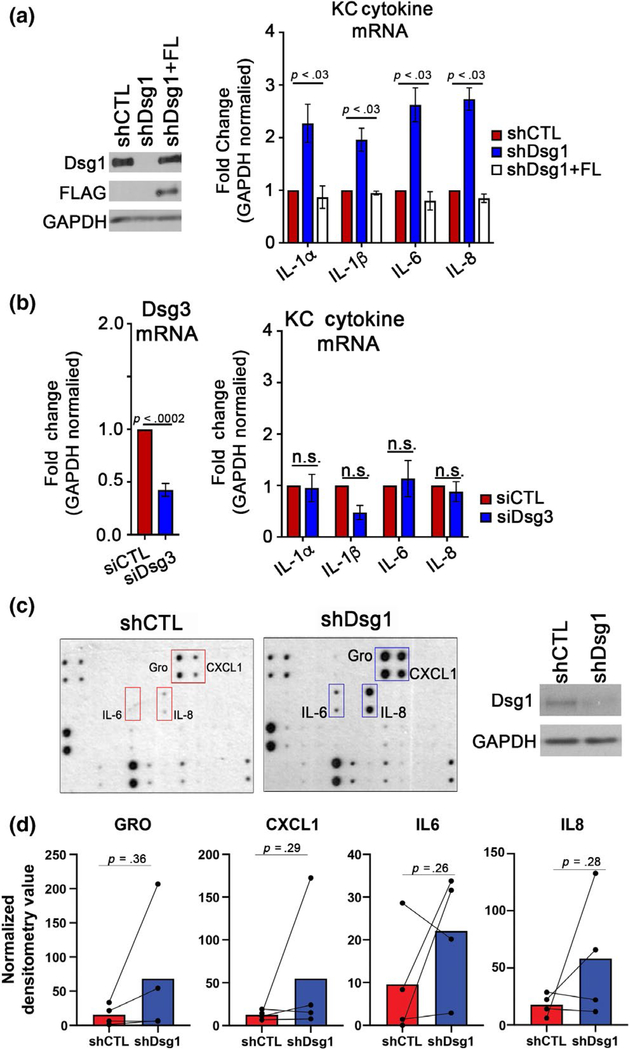
KCs exposed to UV light have been reported to increase secretion of IL6 and IL8 among other cytokines/chemokines (Schwarz & Luger, 1989; Yoshizumi et al., 2008). We sought to determine whether cytokines were altered upon Dsg1 suppression in KCs. KCs infected with shCTL, shDsg1, or shDsg1 + FL were grown to confluence, switched to high calcium medium to induce differentiation, and assessed by qRT-PCR after 3 days. Cytokine transcripts including IL1α, IL1β, IL6, IL8, and CXCL1 were significantly upregulated; IL2, IL4, and IL10 were significantly downregulated; and IL19, IL23, TNF-α, and IFN-γ were not significantly altered by suppression of Dsg1 (Figure 3a and Table 1). When a subset of these targets were tested, we found them to be returned to control levels upon restoration of Dsg1 + FL (Figure 3a). Depletion of another desmosomal cadherin in KCs, Dsg3, did not result in significant changes in tested cytokine mRNA levels (Figure 3b). To determine which of these altered cytokines were being secreted from KCs, conditioned media was collected from KCs and dot blot analysis of 42 targets was performed. Though IL1α and IL1β were increased at the mRNA level, we did not detect these cytokines in the secretome using the dot blot. However, though variable between the 4 KC isolates tested from 4 donors and not significantly changed between conditions, we detected IL6, IL8, GRO, and CXCL1 in the secretome (Figure 3c and quantified in Figure 3d).
FIGURE 3.
Suppression of KC Dsg1 alters cytokine mRNA and secretion from keratinocytes. (a) shCTL, shDsg1, and shDsg1 plus silencing-resistant Dsg1-Flag (shDsg1 + FL)-infected KCs were differentiated in high calcium-containing medium for 72 hr. The shDsg1-infected KCs exhibited increased mRNA levels of several cytokines compared with shCTL, while shDsg1 + FL restored cytokine levels to those of shCTL (see Table 1 for additional tested cytokines) (N = 5, analyzed by one-way ANOVA with Tukey’s post hoc test). (b) Cells transfected with siRNA targeting Dsg3 were found not to have significantly changed levels of cytokines compared with controls (N = 3; n.s. = not significant, analyzed by two-tailed Student’s t test). (c) Media conditioned by shCTL- and shDsg1-infected KCs were incubated with a Human Cytokine C3 Array (Raybiotech) to detect secretion of up to 42 cytokines, chemokines, and growth factors (one dot blot is shown together with the immunoblot of Dsg1 suppression in that specific KC isolate) (d) Densitometry was performed on dot blots from 4 shCTL and shDsg1-conditioned media pairs (from 4 KC isolations). No significant differences were observed between conditions (analyzed by two-tailed paired t tests). Graphical representations are spaghetti plots with bars representing the means of the four experiments
TABLE 1.
Average fold change in KC cytokine transcripts upon depletion of Dsg1 (shCTL versus shDsg1) to accompany Figure 3a
| Target | Avg. fold change | SEM | p-value |
|---|---|---|---|
| IL2 | 0.27 | 0.143 | .0314* |
| IL4 | 0.34 | 0.058 | .0105* |
| IL10 | 0.48 | 0.053 | .0090* |
| IL19 | 58.62 | 41.6 | .1191 |
| IL23 | 40.75 | 22.9 | .0791 |
| CXCL1 | 2.3 | 0.137 | .0149* |
| TNFα | 22.95 | 10.1 | .0816 |
| IFNγ | 43.06 | 22.7 | .0686 |
Note: Tested by qRT-PCR (N = 3 *p < .05).
Abbreviations: CXCL1, chemokine ligand 1; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor.
3.4 |. Dsg1-deficient KCs change MC dendricity, partially dependent upon cytokine/chemokine signaling
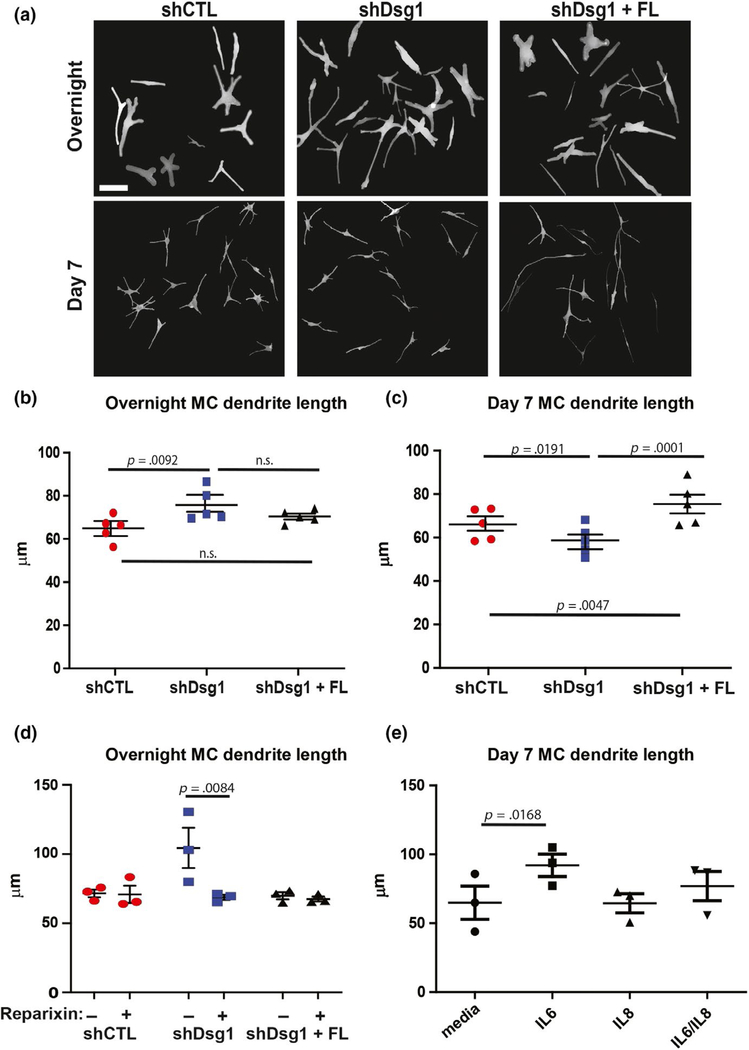
MC dendrites lengthen and branch in response to UV light, increasing MC interactions with KCs to facilitate melanin transfer (López et al., 2015; Weiner et al., 2014). To determine contributions of KC Dsg1 reduction to MC morphological changes, MCs were incubated with conditioned media from shCTL or shDsg1-infected KCs either overnight or for 7 days. The overnight incubation of MCs with Dsg1-deficient KC-conditioned media resulted in increased dendrite length when compared with MCs treated with shCTL KC-conditioned media (Figure 4a, upper panels and quantified in Figure 4b and Figure S3a). Increased dendrite length is an important determinant of the number of KCs that a MC can interact within normal skin (López et al., 2015; Weiner et al., 2014). However, 7 days of exposure to Dsg1-deficient KC-conditioned media resulted in shortening of MC dendrites compared with shCTL or shDsg1 + FL-infected KCs (Figure 4a, lower and quantified in Figure 4c and Figure S3b). Though we did not explicitly measure dendrite thickness or cell body appearance, dendrites in all conditions consistently appeared thinner and the total cell body appeared smaller at the day 7 time point compared with the day 1 time point. These morphological differences are likely due to normal changes that occur over time during spreading and differentiation (including dendritogenesis) in KC-conditioned media as described in Hirobe (2005). We tested whether individual cytokines played any role in the observed changes in MC dendricity. We inhibited the CXCL1/IL8 receptor (CXCR2) using reparixin, which inhibited the increase in MC dendrite length at the overnight time point when cultured in conditioned media from Dsg1-deficient KCs (Figure 4d and Figure S3c). We also queried whether recombinant IL6, IL8, or a combination of the 2 added into base MC media was sufficient to impact dendrite length. While neither individual cytokine nor their combination was sufficient to alter dendrite length after overnight incubation (data not shown), MC dendrites significantly lengthened in the presence of IL6, but not IL8 or their combination after incubation for 7 days (Figure 4e and Figure S3d). Together, these observations are consistent with the possibility that individual cytokines contribute to MC dendrite length changes, but are not sufficient to recapitulate the time course and/or signal propagation stimulated by the complete conditioned media from Dsg1-deficient KCs. Therefore, multiple paracrine factors may be required to elicit the MC dendrite changes observed upon the addition of conditioned media from Dsg1-deficient KCs. Together, these data indicate that paracrine signaling is at least partially responsible for altering MC dendrite length downstream of Dsg1 suppression.
FIGURE 4.
Dsg1-deficient KCs change MC dendricity, partially dependent upon cytokine/chemokine signaling. (a) MCs acutely exposed (overnight, top panels) or for 7 days (bottom panels) to conditioned media from KCs deficient in Dsg1 exhibit altered dendrite length compared to those exposed to conditioned media from shCTL- or shDsg1 + FL-infected KCs. Scale bar = 50 μm. The longest single dendrite was measured from each cell body, with at least 5 dendrites per field analyzed and over 50 dendrites measured for each condition. The experiment was performed with a total of 5 MC:KC-conditioned media pairs (N = 5). Images shown are traced using the Simple Neurite Tracer (FIJI software) to facilitate visualization. Not all cells in a given raw image were traced. Cells were excluded if dendrites extended beyond the field of view or if overlap between cells did not allow visualization of the full dendrite path. (b) Graphical representation of dendrite length in MCs exposed overnight to conditioned media from shCTL-, shDsg1-, or shDsg1 + FL-infected KCs. Acute (overnight) exposure to shDsg1-infected KC-conditioned media lengthened dendrites compared to MCs treated with shCTL KC-conditioned media. (c) Long-term exposure to shDsg1-infected KC-conditioned media (7 days) resulted in dendrite shortening, while rescue with FL Dsg1 resulted in dendrite lengthening compared with controls (N = 5 experimental replicates for both B and C. Graphical representations are mean and SEM. Experimental replicates [means] were analyzed using one-way ANOVA followed by Tukey’s post hoc test). (d) Incubation of MCs with 5 μg/ml reparixin, an inhibitor of the IL8/CXCL1 receptor CXCR2, abrogated the increase in MC dendrite length associated with the acute exposure (overnight) to conditioned media from shDsg1-infected KCs (N = 3 experimental replicates, means of experimental replicates were analyzed using one-way ANOVA followed by Holm-Sidak’s post hoc analysis for comparison between drug-treated and drug-untreated samples for each condition). (e) Incubation of MCs with IL6, IL8, or IL6 + IL8 in base MC media for 7 days. IL6 significantly increased MC dendrite length, while IL8 or the combination of the two cytokines did not have a significant impact on dendrite length (N = 3 experimental replicates, means analyzed using one-way ANOVA and Tukey’s post hoc test). Graphical representations are mean and SEM. Violin plots of the spread of lengths of the longest dendrite per cell for each condition and time point are shown in Figure S3
3.5 |. MCs are mislocalized within the 3D skin structure when KC Dsg1 is reduced
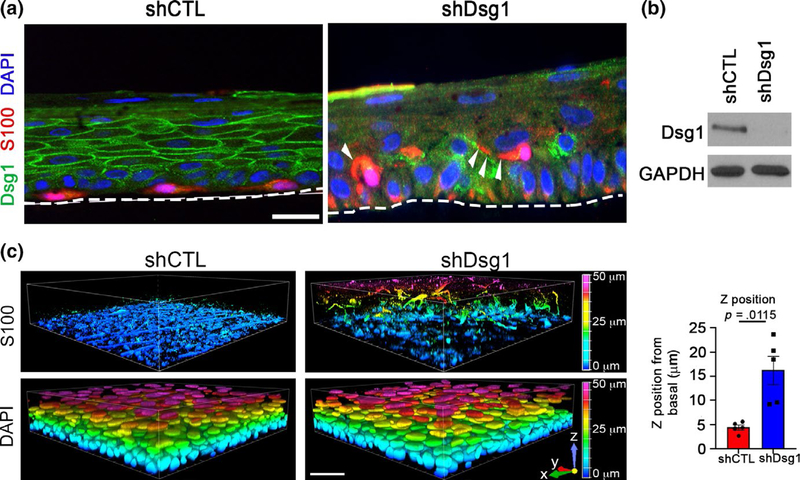
While analyzing melanocyte morphology in 3D organotypic cultures for Figure 1, we observed that MCs were mislocalized in KC Dsg1-deficient organotypic cultures (Figure 5a). Imaging of organotypic cultures (shCTL, shDsg1; representative immunoblot Figure 5b) prepared using the whole-mount method revealed that MCs moved into the upper epidermal layers throughout the entire shDsg1 organotypic culture while they remained in the basal layer in controls (Figure 5c, quantified to the right). Mislocalization suprabasally (Figure 5) was reminiscent of pagetoid behavior in MCs that have undergone transformation (Colebatch & Scolyer, 2018; Dhawan & Richmond, 2002; Mo et al., 2019).
FIGURE 5.
MCs are mislocalized within the 3D skin structure when KC Dsg1 is reduced. (a) Silencing Dsg1 (shDsg1) in KCs cocultured with MCs in 3D organotypic cultures resulted in mislocalization of MCs from basal to suprabasal layers compared with controls (shCTL). Dsg1 marks KCs and is mostly absent in the shDsg1 culture, S100 marks MCs, and DAPI marks nuclei. White arrowheads indicate MC dendrites. Scale bar = 10 μm. (b) Immunoblot of Dsg1 protein expression for organotypic cultures pictured in C. GAPDH is a loading control. (c) Images of 3D organotypic cultures of KCs and MCs prepared using the whole-mount method. S100 marks MCs, showing their movement into the suprabasal layers upon Dsg1 reduction (N = 5). Horizontal scale bar = 50 μm, vertical color scale bar = Z position. The graph represents the mean and SEM of movement in the Z position (μm) away from the basement membrane of at least 20 cells in each of 5 organotypic cultures
4 |. DISCUSSION
In addition to its importance in epidermal cell-cell adhesion and in regulating KC differentiation (Getsios et al., 2009; Harmon et al., 2013), our data support a model whereby Dsg1 also functions to regulate paracrine signaling. Our data indicate that loss of Dsg1, such as occurs following acute UV exposure (Johnson et al., 2014), helps trigger epithelial wide responses like the tanning response— that is, increased pigment production and release from MCs as well as alterations in MC dendrite morphology (Figure 6). The regulation of MCs via secretion of paracrine factors by KCs is a well-established phenomenon (Serre et al., 2018; Yuan & Jin, 2018). Our study places Dsg1 as a key regulator of the microenvironment within the KC:MC unit.
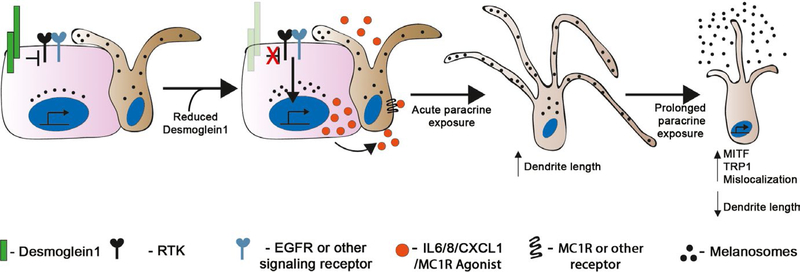
FIGURE 6.
Model: KC Dsg1 regulates the epidermal microenvironment and MC behavior. KCs (pink) and MCs (brown) in the skin communicate with each other through both direct contact and paracrine signaling. Reduction in the expression of KC Dsg1 initiates a signaling cascade (may include any receptor tyrosine kinase, [RTK], EGFR, NFkB, Stat3) that alters the production of paracrine factors including IL6, IL8, CXCL1, and a melanogenesis-stimulating factor that affects the signaling, morphology, pigment secretion, and localization of neighboring MCs
Protection against damage from environmental exposure, including UV light from the sun, is one of the key barrier functions of the epidermis. The classic KC:MC tanning response protects against mutagenesis (Brenner & Hearing, 2008; Swope & Abdel-Malek, 2016; Yamaguchi et al., 2006) and involves increase in POMC levels in KCs, production, and secretion of α MSH and other secreted factors, subsequent activation of the MC receptor MC1R, downstream activation of MITF, and pigment production through TYRP1 and TYRP2 (Serre et al., 2018). MCs have previously been shown to distribute melanin to KCs in part through release of shedding vesicles containing pigment (Ando et al., 2012; Scott, 2012), so increased pigment secretion may indicate increased ability to transfer melanin to KCs, though this was not tested. The POMC derivative αMSH also regulates the shedding of pigment granules from MCs (Ma et al., 2014). All human skin, regardless of skin type or melanin type and quantity (Wakamatsu et al., 2006), undergoes reaction to UV exposure including increased pigmentation (Alaluf et al., 2001). A recent study reported that MCs from darkly pigmented individuals more effectively increased melanogenesis after UVB exposure in the presence of KC-conditioned media compared with MCs from lightly pigmented individuals, suggesting an enhanced photoprotective mechanism in darker skin types (López et al., 2015). We show that reduction in KC Dsg1 increases pigment secretion in MCs from donors with multiple skin types with enhanced pigment release from darkly pigmented MCs, consistent with the previous report.
MC dendrites are crucial for mediating the tanning response following UV exposure, since they are the conduit through which melanosomes are transferred to multiple KCs (Gilchrest, Park, Eller, & Yaar, 1996). We showed that factors present in conditioned media from Dsg1-deficient KCs can signal to MCs to alter dendrite length. UV-exposed KCs increase secretion of IL6 and IL8 (Schwarz & Luger, 1989; Yoshizumi et al., 2008), similar to our findings of cytokine secretion downstream of Dsg1 suppression even in the absence of UV exposure. We showed that IL6 and the IL8/CXCL1 receptor (CXCR2) both impact MC dendricity downstream of Dsg1. However, the addition of IL6 and inhibition of CXCR2 were both insufficient to recapitulate the precise effects and timeline of complete media from Dsg1-deficient KCs. Dsg1 regulates KC differentiation (Getsios et al., 2009; Harmon et al., 2013), and differentiated KCs have been shown to regulate MC dendrite length (Kippenberger, Bernd, Bereiter-Hahn, Ramirez-Bosca, & Kaufmann, 1998). Thus, it may be that factors secreted from less differentiated KCs in the context of Dsg1 depletion played a role in both the initial dendrite extension (overnight) and eventual dendrite shortening (7 days). The dendrite shortening may have been due to pigment secretion or to tension and sheer after the initial extension, as reported in Kippenberger et al. (1998) and Wu et al. (2012). Further studies are needed to elucidate the roles of cytokines, chemokines, and differentiation-associated secreted factors in regulating the dendrite morphology of MCs downstream of Dsg1 loss.
While KC Dsg1 loss initially promotes a protective response within the KC:MC unit, several hallmarks of early melanoma were also identified downstream of Dsg1 loss. Normal KCs lacking Dsg1 upregulate IL6, IL8, and CXCL1 mRNA, which have been implicated in melanoma development and maintenance (Jobe et al., 2016; Payne & Cornelius, 2002). In fact, CXCL1 was one of the most highly modified markers allowing discrimination between common melanocytic and dysplastic nevi (Eliades & Tsao, 2016; Mitsui et al., 2016). An altered inflammatory microenvironment contributes to the growth and invasion of transformed MCs and melanoma cells (Richmond, Yang, & Su, 2009). Increased MITF levels have also been associated with MC transformation and melanoma progression (Hartman & Czyz, 2015). Suprabasal mislocalization, as observed in 3D epidermal skin equivalents where the KCs were depleted of Dsg1, is reminiscent of the pagetoid appearance of MCs in dysplastic nevi or in melanoma in situ (Colebatch & Scolyer, 2018; Dhawan & Richmond, 2002; Mo et al., 2019). It is possible that repeated cycles of acute UV exposure could prolong reduction in Dsg1 expression in KCs, leading to alterations in other KC:MC adhesive molecules (Mescher et al., 2017) or an altered microenvironment within the KC:MC unit to promote MC dysplasia. Transformed MCs themselves could secrete factors resulting in sustained loss of KC Dsg1.
Modulating KC Dsg1 levels through topical formulations could alter the epidermal microenvironment and cross talk within the KC:MC unit with potential ramifications for clinically managing pigmentation. Histone deacetylase (HDAC) inhibition using TSA was shown to moderately increase KC Dsg1 levels after UVB exposure (Johnson et al., 2014). Other desmogleins are being regulated through the use of glucocorticoids, rapamycin, and Stat3 inhibition in the treatment of pemphigus (Mao, Cho, Ellebrecht, Mukherjee, & Payne, 2017).
In conclusion, this study demonstrates a surprising new role for KC Dsg1 in regulating paracrine cross talk within the epidermal KC:MC unit. Reduction in Dsg1 results in KCs increasing both ligand-dependent and cytokine/chemokine-dependent signaling to MCs to stimulate MC pigment secretion and alter dendricity. Within the 3D epidermis, the presence of KC Dsg1 helps properly position MCs in the basal layer, as Dsg1 reduction results in suprabasal mislocalization of MCs, reminiscent of pagetoid behavior in early melanoma.
Supplementary Material
Significance.
Readers of our work will gain new insight into how epidermal cells in the skin coordinate the tissue-wide tanning response, a key protective reaction to environmental insult. This study places an adhesion molecule expressed in keratinocytes of the skin as a central player in regulating normal homeostasis within the keratinocyte:melanocyte unit.
ACKNOWLEDGEMENTS
This work was supported by NIH/NIAMS R01 AR041836 and NIH/NCI R01 CA122151 to KJG and by the Liz and Eric Lefkofsky Family Foundation Innovation Research Award to KJG and JLJ. Additional support was provided by the JL Mayberry endowment to KJG. CRA and QRR-C were supported through a NIH/NCI Ruth L. Kirschstein Training Grant through Northwestern University’s Robert H. Lurie Comprehensive Cancer Center (T32 CA070085) “Signal Transduction in Cancer.” CRA was also supported through a NIH/NCI Ruth L. Kirschstein National Research Service Award 1F32CA210498-01. HEB was supported through a NIH/NCI Ruth L. Kirschstein Training Grant through Northwestern University’s Robert H. Lurie Comprehensive Cancer Center (T32 CA080621-14) “Oncogenesis and Developmental Biology”. JAB was supported by NIH/NIAMS K01 AR075087. We thank Dr. Zalfa Abdel-Malek (University of Cincinnati) for critical reading of the manuscript. We acknowledge support and materials from the Northwestern University Skin Disease Research Center supported by 5P30AR057216. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. This work was presented in part in October 2018 at the joint Montagna Symposium on the Biology of Skin/Pan American Society for Pigment Cell Research Meeting. Funding for this conference was in part through R13AR009431-53 and R13AR07429-01 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), and co-funding was provided by the National Institute on Aging (NIA) and the National Institute of Environmental Sciences (NIEHS).
Abbreviations:
- Dsg1
desmoglein 1
- E-cad
epithelial cadherin
- KC
keratinocyte
- MC
melanocyte
- SAM
severe dermatitis, multiple allergies, and metabolic wasting syndrome
- UV
ultraviolet light
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
REFERENCES
- Alaluf S, Heath A, Carter N, Atkins D, Mahalingam H, Barrett K, … Smit N. (2001). Variation in melanin content and composition in type V and VI photoexposed and photoprotected human skin: The dominant role of DHI. Pigment Cell Research, 14, 337–347. 10.1034/j.1600-0749.2001.140505.x [DOI] [PubMed] [Google Scholar]
- Ando H, Niki Y, Ito M, Akiyama K, Matsui MS, Yarosh DB, & Ichihashi M. (2012). Melanosomes are transferred from melanocytes to keratinocytes through the processes of packaging, release, uptake, and dispersion. The Journal of Investigative Dermatology, 132, 1222–1229. 10.1038/jid.2011.413 [DOI] [PubMed] [Google Scholar]
- Arnette C, Koetsier JL, Hoover P, Getsios S, & Green KJ (2016). In vitro model of the epidermis: Connecting protein function to 3D structure. Methods in Enzymology, 569, 287–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M, & Hearing VJ (2008). The protective role of melanin against UV damage in human skin. Photochemistry and Photobiology, 84, 539–549. 10.1111/j.1751-1097.2007.00226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch AJ, & Scolyer RA (2018). Trajectories of premalignancy during the journey from melanocyte to melanoma. Pathology, 50, 16–23. 10.1016/j.pathol.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Dhawan P, & Richmond A. (2002). Role of CXCL1 in tumorigenesis of melanoma. Journal of Leukocyte Biology, 72, 9–18 [PMC free article] [PubMed] [Google Scholar]
- El-Abaseri TB, Hammiller B, Repertinger SK, & Hansen LA (2013). The epidermal growth factor receptor increases cytokine production and cutaneous inflammation in response to ultraviolet irradiation. ISRN Dermatol, 2013, 848705 10.1155/2013/848705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliades P, & Tsao H. (2016). New insights into the molecular distinction of dysplastic nevi and common melanocytic nevi-highlighting the keratinocyte-melanocyte relationship. The Journal of Investigative Dermatology, 136, 1933–1935. 10.1016/j.jid.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Getsios S, Simpson CL, Kojima S-I, Harmon R, Sheu LJ, Dusek RL, … Green KJ (2009). Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. Journal of Cell Biology, 185, 1243–1258. 10.1083/jcb.200809044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrest BA, Park HY, Eller MS, & Yaar M. (1996). Mechanisms of ultraviolet light-induced pigmentation. Photochemistry and Photobiology, 63, 1–10. 10.1111/j.1751-1097.1996-tb02988.x [DOI] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, & Galloway DA (1992). The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. Journal of Virology, 66, 2125–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers CM, & Stanley JR (2013). Desmoglein-1, differentiation, and disease. Journal of Clinical Investigation, 123, 1419–1422. 10.1172/JCI69071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon RM, Simpson CL, Johnson JL, Koetsier JL, Dubash AD, Najor NA, … Green KJ (2013). Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. Journal of Clinical Investigation, 123, 1556–1570. 10.1172/JCI65220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman ML, & Czyz M. (2015). MITF in melanoma: Mechanisms behind its expression and activity. Cellular and Molecular Life Sciences, 72, 1249–1260. 10.1007/s00018-014-1791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirobe T. (2005). Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Research, 18, 2–12. 10.1111/j.1600-0749.2004.00198.x [DOI] [PubMed] [Google Scholar]
- Jamal S, & Schneider RJ (2002). UV-induction of keratinocyte endothelin-1 downregulates E-cadherin in melanocytes and melanoma cells. Journal of Clinical Investigation, 110, 443–452. 10.1172/JCI0213729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe NP, Rösel D, Dvořánková B, Kodet O, Lacina L, Mateu R, … Brábek J. (2016). Simultaneous blocking of IL-6 and IL-8 is sufficient to fully inhibit CAF-induced human melanoma cell invasiveness. Histochemistry and Cell Biology, 146, 205–217. 10.1007/s00418-016-1433-8 [DOI] [PubMed] [Google Scholar]
- Johnson JL, Hoover P, Jovanovic BD, Green KJ, Friedewald JJ, & Robinson JK (2016). Epidermal desmoglein 1 expression is reduced in kidney transplant recipients compared with immunocompetent patients. The Journal of Investigative Dermatology, 136, 1908–1912 10.1016/j.jid.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Koetsier JL, Sirico A, Agidi AT, Antonini D, Missero C, & Green KJ (2014). The desmosomal protein desmoglein 1 aids recovery of epidermal differentiation after acute UV light exposure. The Journal of Investigative Dermatology, 134, 2154–2162. 10.1038/jid.2014.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippenberger S, Bernd A, Bereiter-Hahn J, Ramirez-Bosca A, & Kaufmann R. (1998). The mechanism of melanocyte dendrite formation: The impact of differentiating keratinocytes. Pigment Cell Research, 11, 34–37. 10.1111/j.1600-0749.1998.tb00708.x [DOI] [PubMed] [Google Scholar]
- Langhofer M, Hopkinson SB, & Jones JC (1993). The matrix secreted by 804G cells contains laminin-related components that participate in hemidesmosome assembly in vitro. Journal of Cell Science, 105(Pt 3), 753–764 [DOI] [PubMed] [Google Scholar]
- Laskin JD, Piccinini L, Engelhardt DL, & Weinstein IB (1982). Control of melanin synthesis and secretion by B16/C3 melanoma cells. Journal of Cellular Physiology, 113, 481–486. 10.1002/jcp.1041130318 [DOI] [PubMed] [Google Scholar]
- Lee JT, & Herlyn M. (2007). Microenvironmental influences in melanoma progression. Journal of Cellular Biochemistry, 101, 862–872. 10.1002/jcb.21204 [DOI] [PubMed] [Google Scholar]
- Li L, Fukunaga-Kalabis M, & Herlyn M. (2011). The three-dimensional human skin reconstruct model: A tool to study normal skin and melanoma progression. Journal of Visualized Experiments, 54, e2937 10.3791/2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, & Armstrong JD (2011). Simple Neurite Tracer: Open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics, 27, 2453–2454. 10.1093/bioinformatics/btr390 [DOI] [PubMed] [Google Scholar]
- López S, Alonso S, García de Galdeano A, & Smith-Zubiaga I. (2015). Garcia de Galdeano A, Smith-Zubiaga I. Melanocytes from dark and light skin respond differently after ultraviolet B irradiation: Effect of keratinocyte-conditioned medium. Photodermatology, Photoimmunology and Photomedicine, 31, 149–158. 10.1111/phpp.12169 [DOI] [PubMed] [Google Scholar]
- Ma HJ, Ma HY, Yang Y, Li PC, Zi SX, Jia CY, & Chen R. (2014). alpha-Melanocyte stimulating hormone (MSH) and prostaglandin E2 (PGE2) drive melanosome transfer by promoting filopodia delivery and shedding spheroid granules: Evidences from atomic force microscopy observation. Journal of Dermatological Science, 76, 222–230 [DOI] [PubMed] [Google Scholar]
- Mao X, Cho MJT, Ellebrecht CT, Mukherjee EM, & Payne AS (2017). Stat3 regulates desmoglein 3 transcription in epithelial keratinocytes. JCI Insight, 2, e92253 10.1172/jci.insight.92253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher M, Jeong P, Knapp SK, Rübsam M, Saynisch M, Kranen M, … Iden S. (2017). The epidermal polarity protein Par3 is a non-cell autonomous suppressor of malignant melanoma. Journal of Experimental Medicine, 214, 339–358. 10.1084/jem.20160596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Kiecker F, Shemer A, Cannizzaro MV, Wang CQ, Gulati N, … Krueger JG (2016). Discrimination of dysplastic nevi from common melanocytic nevi by cellular and molecular criteria. The Journal of Investigative Dermatology, 136, 2030–2040 [DOI] [PubMed] [Google Scholar]
- Mo X, Preston S, & Zaidi MR (2019). Macroenvironment-gene-microenvironment interactions in ultraviolet radiation-induced melanomagenesis. Advances in Cancer Research, 144, 1–54 [DOI] [PubMed] [Google Scholar]
- Moriconi A, Cesta MC, Cervellera MN, Aramini A, Coniglio S, Colagioia S, … Allegretti M. (2007). Design of noncompetitive interleukin-8 inhibitors acting on CXCR1 and CXCR2. Journal of Medicinal Chemistry, 50, 3984–4002. 10.1021/jm061469t [DOI] [PubMed] [Google Scholar]
- Nekrasova O, Harmon RM, Broussard JA, Koetsier JL, Godsel LM, Fitz GN, … Green KJ (2018). Desmosomal cadherin association with Tctex-1 and cortactin-Arp2/3 drives perijunctional actin polymerization to promote keratinocyte delamination. Nature Communications, 9, 1053 10.1038/s41467-018-03414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, & Fisher DE (2018). MITF and UV responses in skin: From pigmentation to addiction. Pigment Cell Melanoma Research, 32, 224–236. 10.1111/pcmr.12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AS, & Cornelius LA (2002). The role of chemokines in melanoma tumor growth and metastasis. The Journal of Investigative Dermatology, 118, 915–922. 10.1046/j.1523-1747.2002.01725.x [DOI] [PubMed] [Google Scholar]
- Polivka L, Hadj-Rabia S, Bal E, Leclerc-Mercier S, Madrange M, Hamel Y, … Smahi A. (2018). Epithelial barrier dysfunction in desmoglein-1 deficiency. The Journal of Allergy and Clinical Immunology, 142(702–6), e7 10.1016/j.jaci.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond A, Yang J, & Su Y. (2009). The good and the bad of chemokines/chemokine receptors in melanoma. Pigment Cell & Melanoma Research, 22, 175–186. 10.1111/j.1755-148X.2009.00554.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, … Sprecher E. (2013). Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nature Genetics, 45, 1244–1248. 10.1038/ng.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, … Cardona A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9, 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz T, & Luger TA (1989). Effect of UV irradiation on epidermal cell cytokine production. Journal of Photochemistry and Photobiology B: Biology, 4, 1–13 [DOI] [PubMed] [Google Scholar]
- Scott G. (2012). Demonstration of melanosome transfer by a shedding microvesicle mechanism. The Journal of Investigative Dermatology, 132, 1073–1074. 10.1038/jid.2012.20 [DOI] [PubMed] [Google Scholar]
- Serre C, Busuttil V, & Botto JM (2018). Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. International Journal of Cosmetic Science, 40(4), 328–347. 10.1111/ics.12466 [DOI] [PubMed] [Google Scholar]
- Simpson CL, Kojima S, & Getsios S. (2010). RNA interference in keratinocytes and an organotypic model of human epidermis. Methods in Molecular Biology, 585, 127–146 [DOI] [PubMed] [Google Scholar]
- Swope VB, & Abdel-Malek ZA (2016). Significance of the melanocortin 1 and endothelin B receptors in melanocyte homeostasis and prevention of sun-induced genotoxicity. Frontiers in Genetics, 7, 146 10.3389/fgene.2016.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swope VB, Jameson JA, McFarland KL, Supp DM, Miller WE, McGraw DW, et al. (2012). Defining MC1R regulation in human melanocytes by its agonist alpha-melanocortin and antagonists agouti signaling protein and beta-defensin 3. The Journal of Investigative Dermatology, 132, 2255–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terazawa S, & Imokawa G. (2018). Signaling cascades activated by UVB in human melanocytes lead to the increased expression of melanocyte receptors, endothelin B receptor and c-KIT. Photochemistry and Photobiology, 94, 421–431. 10.1111/php.12848 [DOI] [PubMed] [Google Scholar]
- Wakamatsu K, Kavanagh R, Kadekaro AL, Terzieva S, Sturm RA, Leachman S, … Ito S. (2006). Diversity of pigmentation in cultured human melanocytes is due to differences in the type as well as quantity of melanin. Pigment Cell Research, 19, 154–162. 10.1111/j.1600-0749.2006.00293.x [DOI] [PubMed] [Google Scholar]
- Weiner L, Fu W, Chirico WJ, & Brissette JL (2014). Skin as a living coloring book: How epithelial cells create patterns of pigmentation. Pigment Cell & Melanoma Research, 27, 1014–1031. 10.1111/pcmr.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XS, Masedunskas A, Weigert R, Copeland NG, Jenkins NA, & Hammer JA (2012). Melanoregulin regulates a shedding mechanism that drives melanosome transfer from melanocytes to keratinocytes. Proceedings of the National Academy of Sciences of the United States of America, 109, E2101–E2109. 10.1073/pnas.1209397109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takahashi K, Zmudzka BZ, Kornhauser A, Miller SA, Tadokoro T, … Hearing VJ (2006). Human skin responses to UV radiation: Pigment in the upper epidermis protects against DNA damage in the lower epidermis and facilitates apoptosis. The FASEB Journal, 20, 1486–1488. 10.1096/fj.06-5725fje [DOI] [PubMed] [Google Scholar]
- Yoshizumi M, Nakamura T, Kato M, Ishioka T, Kozawa K, Wakamatsu K, & Kimura H. (2008). Release of cytokines/chemokines and cell death in UVB-irradiated human keratinocytes, HaCaT. Cell Biology International, 32, 1405–1411. 10.1016/j.cellbi.2008.08.011 [DOI] [PubMed] [Google Scholar]
- Yuan XH, & Jin ZH (2018). Paracrine regulation of melanogenesis. British Journal of Dermatology, 178, 632–639. 10.1111/bjd.15651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.