Abstract
Bromodomain and extra terminal inhibitors (BETi) delay tumor growth, in part, through tumor cell intrinsic alterations and initiation of anti-tumor CD8+ T cell responses. By contrast, BETi effects on pro-tumoral immune responses remain unclear. Here, we show that the next generation BETi, PLX51107, delayed tumor growth to differing degrees in Braf V600E melanoma syngeneic mouse models. These differential responses were associated with the influx of tumor-associated macrophages during BETi treatment. Tumors that were poorly responsive to PLX51107 showed increased influx of colony stimulating factor-1 receptor (CSF-1R)-positive tumor-associated macrophages. We depleted CSF-1R+ tumor-associated macrophages with the CSF-1R inhibitor, PLX3397, in combination with PLX51107. Treatment with PLX3397 enhanced the efficacy of PLX51107 in poorly responsive Braf V600E syngeneic melanomas in vivo. These findings suggest that tumor-associated macrophage accumulation limits BETi efficacy and that co-treatment with PLX3397 can improve response to PLX51107, offering a potential novel combination therapy for metastatic melanoma patients.
Keywords: Melanoma, BET inhibitor, CSF-1R inhibitor, tumor-associated macrophages
Melanoma is the most lethal form of skin cancer. Despite recent therapeutic success with targeted inhibitors and immunotherapies, there is a need for new systemic therapies that target both the tumor and other cells/factors within the tumor microenvironment. Epigenetic modifiers, such as bromodomain and extra terminal inhibitors (BETi), induce cell death and cell cycle arrest in non-solid malignancies and solid tumors (Filippakopoulos & Knapp, 2014; Gallagher et al., 2015; Jung et al., 2015; Paoluzzi et al., 2016; Segura et al., 2013). BETi also have profound effects on both pro and anti-inflammatory responses to infections and autoimmunity (Chen et al., 2016; Klein et al., 2016; Sun et al., 2015). Recently, BETi have been shown to block tumor growth by inducing anti-tumor immune responses. Specifically, we and others have shown that BETi lowered tumoral PD-L1 expression, improving the function of tumor-infiltrating CD8+ T cells and inducing CD8-mediated tumor growth delay (Erkes et al., 2019; Hogg et al., 2017; Nikbakht et al., 2019; Zhu et al., 2016). Despite the clear effect of BETi on CD8+ T cells, their action on other tumor-infiltrating immune populations remains unclear.
The tumor immune microenvironment is a complex assortment of immune cells that act in an anti- or pro-tumoral capacity; in particular, tumor-associated macrophages (TAM) can play dual roles (Gabrilovich et al., 2012). During tumor initiation, TAM tend to be pro-inflammatory, stimulating CD8+ T cell responses by presenting antigen and releasing anti-tumor cytokines. As tumors grow, TAM shift to an immune-suppressive, pro-tumoral phenotype. These TAM release immune suppressive cytokines such as IL-10, induce T-regulatory influx via CCL22 release, and support angiogenesis by producing VEGF (Gabrilovich et al., 2012). Thus, targeting TAM can have beneficial therapeutic effects for cancer patients.
Melanoma has overexpressed BRD/BET proteins (Echevarría‐Vargas et al., 2018; Segura et al., 2013) and targeting these proteins with BETi alters tumor growth mechanisms and T cell responses in melanoma (Dar et al., 2015; Erkes et al., 2019; Gallagher et al., 2014a; Gallagher et al., 2014b; Gallagher et al., 2015; Nikbakht et al., 2019; Paoluzzi et al., 2016). BETi act by blocking the function of bromodomain proteins (BRDs), thereby altering a subset of genes driven from super-enhancers (Loven et al., 2013; Shi & Vakoc, 2014). Macrophages rely on BRDs for NF-κB directed production of nitric oxide and IL-6 (Filippakopoulos & Knapp, 2014). Additionally, BETi alter the differentiation, function, and recruitment of macrophages (Bandukwala et al., 2012; Belkina et al., 2013; Filippakopoulos & Knapp, 2014; Klein et al., 2016; Qiao & Ivashkiv, 2015; Wienerroither et al., 2014). Despite the critical role for BRDs in macrophage function, the impact of BETi on TAM is currently understudied (Groot & Pienta, 2018).
Here, we tested the next generation BETi, PLX51107, in immune-competent Braf V600E melanoma models and analyzed its effects on TAM. PLX51107 differentially altered the growth of mouse Braf V600E melanoma tumors; effects that were associated with the influx of TAM. PLX3397, a colony stimulating factor-1 receptor (CSF-1R) inhibitor, reduced TAM accumulation and improved the efficacy of BETi in poorly responsive tumors. Together, this work demonstrates that PLX51107 efficacy in melanoma can be improved by PLX3397 treatment.
Braf mutant mouse melanoma cell lines were utilized in this study. D4M3.A cells were donated by Dr. Constance E. Brinckerhoff (Dartmouth University, 2016) and grown in DMEM/F12 containing 5% FBS, 1% PenStrep, and 1% L-Glutamine. YUMM1.7 cells were donated by Dr. Marcus Bosenberg (Yale University, 2014) were maintained in DMEM F-12 50/50 supplemented with 10% FBS, 1% PenStrep, and 1% non-essential amino acids. To authenticate cell lines and determine that they were pathogen-free, cells were confirmed for Braf V600E mutation and IMPACT III PCR pathogen tested (IDEXX).
Animal experiments were performed at Thomas Jefferson University in a facility accredited by AAALAC after IACUC approval. Male C57BL/6 mice (Jackson, 6–10 weeks) were used for all studies. Tumors were implanted intradermally in 100 μL HBSS. Tumor volumes were tracked with caliper measurements: volume = (length × width2)*0.52. When the volume reached ~50 mm3, animals were fed either the appropriate drug-laced chow. For all chow experiments, mice were supplemented with ClearH2O DietGel® Recovery or intraperitoneal PBS injections to combat weight loss and dehydration. PLX51107 (80 and 90 ppm) and PLX3397 (for macrophage depletion, 275 ppm) were provided by Plexxikon Inc. Chow was purchased from Research Diets. Mice were switched from 90 ppm of PLX51107 + 275 ppm of PLX3397 to 80 ppm of PLX51107 + 275 ppm of PLX3397 if they lost more than 15% weight. The structures for PLX51107 and PLX3397 have previously been published (Ao et al., 2017; Ozer et al., 2018).
Tumor growth statistics were analyzed using the fixed effects in the linear mixed effects (LME) model included the treatment group as well as the interaction between treatment groups. Time-dependent trends were modeled as cubic polynomials. The random animal effects included only intercept, linear and quadratic coefficients. The residuals were evaluated to validate the assumptions of the models. Based on the fitted LME model, the overall comparisons of treatment groups were performed in terms of the growth rates (null hypotheses = controls are equal to the treatment groups).
To analyze tumor-infiltrating lymphocytes by flow cytometry, tumors were dissected and processed into single cell suspensions. Cells were stained with a fixable live/dead stain followed by surface antibody staining, as previously described (Erkes et al., 2019). Cells were surface stained with the following antibodies clones from Biolegend: CD45.2 (104), CD11c (N418), CD11b (M1/70), CD103 (2E7), F4/80 (BM8), CD3 (17A2), CD19 (6D5), CD206 (C068C2) I-A/I-E (M5/114.15.1), CD8α (53–6.7) and CSF-1R (AFS98). The BD Fortessa platform was used to analyze all samples. FlowJo was used to quantify flow cytometry data.
To measure blood plasma levels of PLX51107 and PLX3397, mice were bled retro-orbitally and plasma separated by centrifugation at 1000 RCF for 10 minutes and stored at −80°C. Analyses of PLX51107 and/or PLX3397 levels in plasma samples were carried out by Integrated Analytical Solutions (Berkeley, CA) using LC-MS/MS.
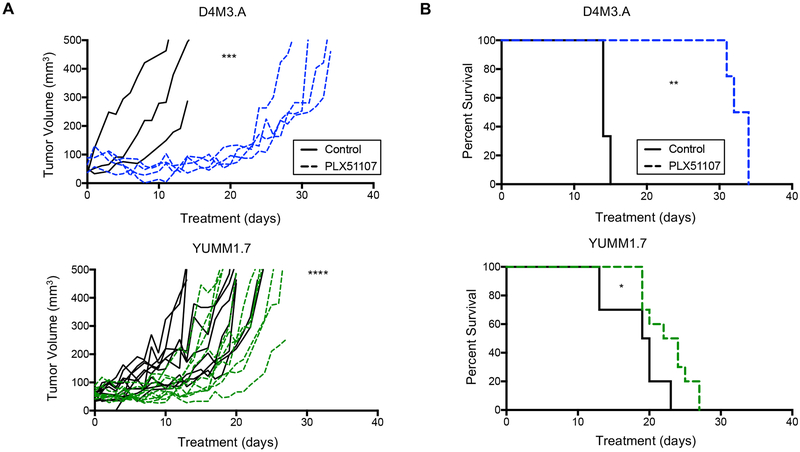
The impact of PLX51107 on cutaneous melanoma tumor growth and tumor infiltrating leukocytes was established in two syngeneic Braf V600E mouse melanoma models - D4M3.A and YUMM1.7 cells (Jenkins et al., 2014; Meeth et al., 2016). After intradermal cell implantation, tumors were grown to ~50 mm3 and then mice were fed either control or PLX51107-laced chow. D4M3.A tumors displayed robust tumor growth delay with PLX51107 treatment, as previously described (Erkes et al., 2019) but in contrast YUMM1.7 tumors exhibited only a modest growth delay (Fig. 1A). Accordingly, PLX51107 prolonged animal survival by ~20 days, but only ~5 days in mice bearing D4M3.A and YUMM1.7 tumors, respectively (Fig. 1B). These data imply that BETi differentially slows the growth of cutaneous melanoma tumors in immune competent models.
Figure 1: BETi treatment differentially delays the growth of Braf V600E melanoma.
Male C57BL/6 mice were intradermally implanted with one of two different mouse Braf V600E melanoma cell lines, D4M3.A (3 × 105 cells) or YUMM1.7 (2.5 × 105 cells). These tumors were grown to ~50 mm3 after which animals were given control (AIN-76A) or PLX51107 (90 ppm PLX51107 AIN-76A)-laced chow. D4M3.A: control n=3, PLX51107 n=4. YUMM1.7: control n=9, PLX51107 n=9. A) Tumor growth is represented as change of volume (mm3) over time from the start treatment. Significance ***p<0.001, ****p>0.0001. B) Kaplan Meier survival curves of mice bearing D4M3.A or YUMM1.7. Mice were considered ‘dead’ when tumors progressed beyond 400 mm3. Significance was determined using a log-rank test, *p<0.05, **p<0.01.
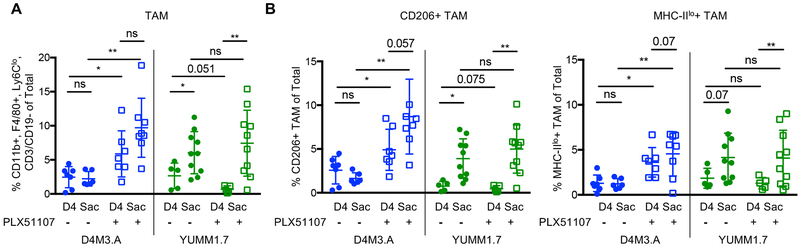
To determine possible causes of the differential BETi responses, we tested the impact of PLX51107 on phenotypically pro-tumoral TAM. Comparing day four on PLX51107 treatment to sacrifice, TAM significantly increased in poorly responsive YUMM1.7 tumors but not in highly responsive D4M3.A tumors (Fig. 2A and Supp. Fig. 1). Specifically, pro-tumoral TAM (CD206+ or MHC-IIlo) significantly accumulated in poorly responsive YUMM1.7 tumors during PLX51107 treatment; however, there was no significant difference in accumulation between day four and sacrifice in D4M3.A tumors (Fig. 2B and Supp. Fig. 1). Of note, TAM increased in the D4M3.A model after the addition of PLX51107, both in day four and at sacrifice harvested tumors. By contrast, TAMs increased over time with tumor growth in YUMM1.7 tumors in a PLX51107-independent manner (Fig. 2A and B). These data imply that the growth-associated influx of pro-tumoral TAM into YUMM1.7 tumors could be linked to their poor response to the BETi, PLX51107.
Figure 2: Accumulation of TAM during BETi.
Mice were implanted with 3 × 105 D4M3.A cells or 2.5 × 105 YUMM1.7 cells as in Figure 1 and TAM were phenotyped during PLX51107 treatment. A) Percent TAM in tumors at day 4 and sacrifice (Sac) on- and off-treatment. B) Pro-tumoral phenotype (CD206+ or MHC-IIlo) of TAM in tumors. Significance was assessed by unpaired t-test *p<0.05, **p<0.01.
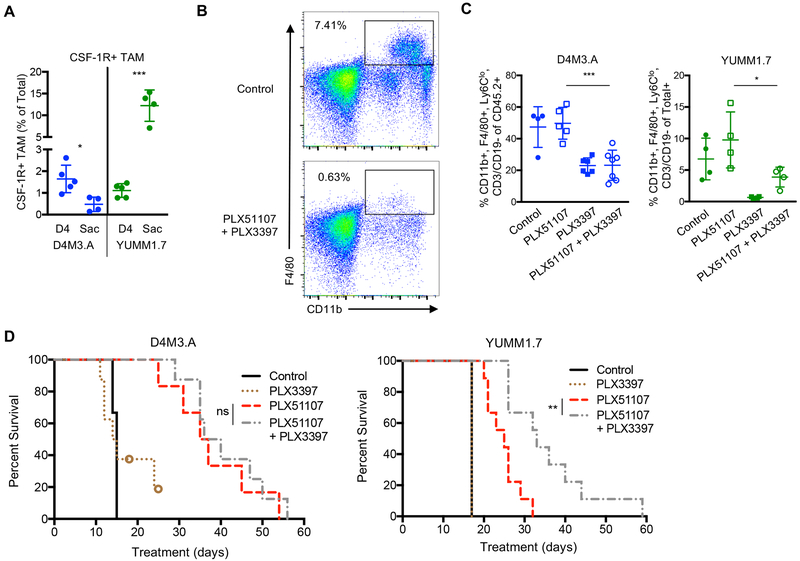
BETi efficacy is CD8+ T cell-dependent (Erkes et al., 2019; Hogg et al., 2017; Zhu et al., 2016), and in melanoma, T cells induce CSF-1 expression and subsequent TAM infiltration and resistance to therapy (Neubert et al., 2018). Thus, in an attempt to improve PLX51107 responsiveness, we focused on targeting CSF-1R+ TAM accumulation. CSF-1R expression on macrophages promotes their differentiation and accumulation into tissue and targeting CSF-1R+ TAM has demonstrated therapeutic efficacy in mouse melanoma models (Lenzo et al., 2012; Mok et al., 2014; Sluijter et al., 2014). YUMM1.7 cell lines expressed higher mRNA levels of molecules involved in macrophage infiltration into tissue (Erkes et al., 2017; Mantovani et al., 2017; Xuan et al., 2015) than D4M3.A cell lines (Supp. Fig. 2), suggesting that YUMM1.7 tumors may be more prone to macrophage infiltration. Indeed, we observed that poorly responsive YUMM1.7 tumors accumulated CSF-1R+ TAM (comparing sacrifice to day four), whereas highly responsive D4M3.A tumors did not (Fig. 3A).
Figure 3: CSF-1R inhibition improves BETi efficacy in poorly responsive tumors.
Mice were implanted with 3 × 105 D4M3.A cells or 2.5 × 105 YUMM1.7 cells as in Figure 1 and TAM were phenotyped. A) CSF-1R+ TAM in untreated tumors at day 4 and sacrifice. Significance was assessed by unpaired t-test *p<0.05, ***p<0.001. B-D) Mice were implanted with D4M3.A or YUMM1.7 cells and when tumors reached ~50 mm3, mice were given AIN-76A control, PLX51107 (90 ppm), PLX3397 (275 ppm), or PLX51107 (90 ppm) plus PLX3397 (275 ppm)-laced chow. Animals with >15% weight loss on PLX51107 plus PLX3397 were switched to 80 ppm of PLX51107 plus 275 ppm of PLX3397. B) Representative FACS plot of macrophages (CD11b+, F4/80+) in the blood of YUMM1.7 tumor-bearing animals 10 days after beginning treatment. C) TAM accumulation levels in D4M3.A or YUMM1.7 tumors at sacrifice. Significance was assessed by unpaired t-test comparing PLX51107 to PLX51107 plus PLX3397 *p<0.05, ***p<0.001. D) Kaplan Meier survival curves of mice bearing D4M3.A (control n=4, PLX3397 n=8, PLX51107 n=6, and PLX3397 plus PLX51107 n=8) or YUMM1.7 (control n=4, PLX3397 n=4, PLX51107 n=9, and PLX3397 plus PLX51107 n=9) tumors. Mice were considered ‘dead’ when tumors progressed beyond 400 mm3. Significance was determined using a log-rank test, **p<0.01.
To target this CSF-1R+ TAM accumulation, we used a CSF-1R inhibitor, PLX3397, in combination with PLX51107 (Mantovani et al., 2017; Sluijter et al., 2014). Time course studies established that PLX51107 was present at ~1–2 μg/mL and PLX3397 was present at ~20–60 μg/mL in the blood plasma of treated mice (Supp. Fig. 3A). PLX51107 significantly increased CD8+ T cells in D4M3.A tumors during treatment, as previously demonstrated (Erkes et al., 2019), but did not alter the influx of CD8+ T cells in YUMM1.7 tumors (Supp. Fig.3B). We previously showed that expression of PD-L1, a therapeutic target of BETi (Zhu et al., 2016), was decreased by PLX51107 treatment in D4M3.A tumors (Erkes et al., 2019), suggesting that BET inhibition is achieved in vivo. To ensure PLX3397 acted as intended, we tested for the depletion of macrophages. PLX3397 alone or in combination with PLX51107 decreased CSF-1R+ macrophages in the blood and tumors (Fig. 3B–C and Supp. Fig. 3C–D). As before, D4M3.A tumor-bearing animals were highly responsive to PLX51107, but co-treatment with PLX3397 showed no added growth or survival advantage (Fig. 3D and Supp. Fig. 3F), indicating the CSF-1R+ TAMs are not modulating the response to PLX51107 in this tumor model. In contrast, the addition of PLX3397 significantly improved survival and delayed tumor growth in poorly responsive YUMM1.7 tumor-bearing mice (Fig. 3D and Supp. Fig. 3F). Polymorphonuclear (PMN)-MDSCs have previously been shown to limit CSF-1R efficacy (Kumar et al., 2017) and effects of PLX3397 co-treatment were associated with a decrease in PMN-MDSCs in YUMM1.7, but did not have significant effects in D4M3.A tumors (Supp. Fig. 3F). Tumor-bearing mice experienced only minor weight loss during treatments (Supp. Fig. 3G). Collectively, these data demonstrate that PLX3397 improved the efficacy PLX51107 in poorly responsive YUMM1.7 tumors.
Receptor tyrosine kinase expression, such as CSF-1R, on melanoma cells can contribute to growth (Easty et al., 2011). To determine whether PLX3397 has a direct effect on tumor cells, we evaluated CSF-1R expression in untreated tumors. We observed low to moderate expression of CSF-1R on non-immune cells (Supp. Fig. 4A). Furthermore, in vitro treatment of D4M3.A and YUMM1.7 cells lines did not significantly alter cell growth at multiple concentrations tested (Supp. Fig. 4B). Together, these data suggest that the effects of PLX3397 were unlikely to be mediated by direct effects on the tumor cells.
Our study demonstrates that PLX3397 enhances the efficacy of PLX51107 in poorly responsive tumors. We established this novel combination by identifying the accumulation of immune-inhibitory and CSF-1R+ TAM in tumors poorly responsive to PLX51107 and attempting to deplete them with PLX3397. Our data demonstrated that PLX3397 depleted CSF-1R+ TAM and improved BETi efficacy in poorly responsive tumors. Thus, tumors that recruit CSF-1R+ TAM during BETi may be a target for PLX3397 combination treatment. We note that PLX3397 inhibits other tyrosine kinases besides CSF-1R, such as cKIT, FLT3 and PDGFR-ß, that may also contribute to the regulation melanoma growth (Cannarile et al., 2017). PLX51107 can inhibit cancer cell growth in vitro; however, in vivo we observe additional effects on the tumor immune microenvironment (Erkes et al., 2019). Further investigation is required to thoroughly understand the effects of PLX51107 on diverse immune cell types within the tumor. PLX3397 improved the efficacy of PLX51107 poorly responsive tumors and has demonstrated clinical efficacy (Mantovani et al., 2017), highlighting an opportunity for further studies combining BETi with PLX3397 in a clinical setting.
Supplementary Material
Significance.
Tumor-associated macrophages affect melanoma tumor progression by supporting tumor growth and the dampening of anti-tumor immune responses; thus, understanding the role of macrophages in the context of anti-tumor therapies is important. Previous studies by our group and others have shown that BET inhibitors are a potential therapy for melanoma; yet it is still unclear if macrophages are modulated during treatment. Here, we determined that macrophage influx is associated with BET inhibitor responsiveness and that depleting macrophages with PLX3397 improves BET inhibitor efficacy. Our study describes a new clinically relevant drug combination that could offer a new treatment strategy for metastatic melanoma.
Acknowledgments and Grant Support:
We thank Plexxikon Inc. (Berkeley, CA) for kindly providing us with PLX51107 and PLX3397. Work in the Villanueva and Aplin laboratories is partially supported by NCI P01 CA114046. These studies were also supported by grants from National Institutes of Health (NIH; R01 CA196278, R01 CA160495), and the Department of Defense (W81XWH-18-1-0224) to A.E. Aplin. D. A. Erkes is supported by an American Cancer Society - CEOs Against Cancer - PA Chapter Postdoctoral Fellowship (PF-18-096-01-LIB). The Sidney Kimmel Cancer Center Flow Cytometry and Translational Pathology core facilities are supported by NIH/NCI Cancer Center support grant (P30 CA056036).
Footnotes
Conflict of interest: A.E. Aplin reports receiving a commercial research grant from Pfizer Inc. (2013–2017), has ownership interest in patent number 9880150 and has consulted for SpringWorks Therapeutics and Fortress Biotech within the last 3 years. No potential conflicts of interest were disclosed by the other authors.
References
- Ao JY, Zhu XD, Chai ZT, Cai H, Zhang YY, Zhang KZ, … Sun HC (2017). Colony-Stimulating Factor 1 Receptor Blockade Inhibits Tumor Growth by Altering the Polarization of Tumor-Associated Macrophages in Hepatocellular Carcinoma. Mol Cancer Ther, 16, 1544–1554. doi: 10.1158/1535-7163.MCT-16-0866 [DOI] [PubMed] [Google Scholar]
- Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, … Rao A (2012). Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci USA, 103, 14532–14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Nikolajczyk BS, & Denis GV (2013). BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol, 190, 3670–3678. doi: 10.4049/jimmunol.1202838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarile MA, Weisser M, Jacob W, Jegg AM, Ries CH, & Ruttinger D (2017). Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer, 5, 53. doi: 10.1186/s40425-017-0257-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Campfield BT, Wenzel SE, McAleer JP, Kreindler JL, Kurland G, … Kolls JK (2016). Antiinflammatory effects of bromodomain and extraterminal domain inhibition in cystic fibrosis lung inflammation. JCI Insight, 1, 1–10. doi: 10.1172/jci.insight.87168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AA, Nosrati M, Bezrookove V, de Semir D, Majid S, Thummala S, … Kashani-Sabet M (2015). The role of BPTF in melanoma progression and in response to BRAF-targeted therapy. J Natl Cancer Inst, 107, 1–9. doi: 10.1093/jnci/djv034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easty DJ, Gray SG, O’Byrne KJ, O’Donnell D, & Bennett DC (2011). Receptor tyrosine kinases and their activation in melanoma. Pigment Cell Melanoma Res, 24, 446–461. doi: 10.1111/j.1755-148X.2011.00836.x [DOI] [PubMed] [Google Scholar]
- Echevarría‐Vargas IM, Reyes‐Uribe PI, Guterres AN, Yin X, Kossenkov AV, Liu Q, … Villanueva J, (2018). Co‐targeting BET and MEK as salvage therapy for MAPK and checkpoint inhibitor‐resistant melanoma. EMBO Mol Med, 10, e8446. doi: 10.15252/emmm.201708446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes DA, Field CO, Capparelli C, Tiago M, Purwin TJ, Chervoneva I, … Aplin AE (2019). The next-generation BET inhibitor, PLX51107, delays melanoma growth in a CD8-mediated manner. Pigment Cell Melanoma Res, 32, 687–696. doi: 10.1111/pcmr.12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes DA, Wilski NA, & Snyder CM (2017). Intratumoral infection by CMV may change the tumor environment by directly interacting with tumor-associated macrophages to promote cancer immunity. Hum Vaccin Immunother, 13, 1778–1785. doi: 10.1080/21645515.2017.1331795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, & Knapp S (2014). Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov, 13, 337–356. doi: 10.1038/nrd4286 [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Ostrand-Rosenberg S, & Bronte V (2012). Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol, 12, 253–268. doi: 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SJ, Mijatov B, Gunatilake D, Gowrishankar K, Tiffen J, James W, … Hersey P (2014a). Control of NF-kB activity in human melanoma by bromodomain and extra-terminal protein inhibitor I-BET151. Pigment Cell Melanoma Res, 27, 1126–1137. doi: 10.1111/pcmr.12282 [DOI] [PubMed] [Google Scholar]
- Gallagher SJ, Mijatov B, Gunatilake D, Tiffen JC, Gowrishankar K, Jin L, … Hersey P (2014b). The epigenetic regulator I-BET151 induces BIM-dependent apoptosis and cell cycle arrest of human melanoma cells. J Invest Dermatol, 134, 2795–2805. doi: 10.1038/jid.2014.243 [DOI] [PubMed] [Google Scholar]
- Gallagher SJ, Tiffen JC, & Hersey P (2015). Histone Modifications, Modifiers and Readers in Melanoma Resistance to Targeted and Immune Therapy. Cancers 7, 1959–1982. doi: 10.3390/cancers7040870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot AE, & Pienta KJ (2018). Epigenetic control of macrophage polarization: implications for targeting tumor-associated macrophages. Oncotarget, 9, 20908–20927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg SJ, Vervoort SJ, Deswall S, Ott CJ, Cluse LA, Beavis PA, … Johnstone RW (2017). BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep, 18, 2162–2174. doi: 10.1016/j.celrep.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MH, Steinberg SM, Alexander MP, Fisher JL, Ernstoff MS, Turk MJ, … Brinckerhoff CE (2014). Multiple murine BRAF melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res, 27, 495–501. doi: 10.1111/pcmr.12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, & Haendler B (2015). Targeting BET bromodomains for cancer treatment. Epigenomics, 7, 487–501. doi: 10.2217/epi.14.91 [DOI] [PubMed] [Google Scholar]
- Klein K, Kabala PA, Grabiec AM, Gay RE, Kolling C, Lin LL, … Reedquist KA (2016). The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann Rheum Dis, 75, 422–429. doi: 10.1136/annrheumdis-2014-205809 [DOI] [PubMed] [Google Scholar]
- Kumar V, Donthireddy L, Marvel D, Condamine T, Wang F, Lavilla-Alonso S, … Gabrilovich DI (2017). Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell, 32, 654–668.e655. doi: 10.1016/j.ccell.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzo JC, Turner AL, Cook AD, Vlahos R, Anderson GP, Reynolds EC, & Hamilton JA (2012). Control of macrophage lineage populations by CSF-1 receptor and GM-CSF in homeostasis and inflammation. Immunol Cell Biol, 90, 429–440. doi: 10.1038/icb.2011.58 [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, … Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell, 153, 320–334. doi: 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Marchesi F, Malesci A, Laghi L, & Allavena P (2017). Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol, 14, 399–416. doi: 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeth K, Wang JX, Micevic G, Damsky W, & Bosenberg MW (2016). The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res, 29, 590–597. doi: 10.1111/pcmr.12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S, Koya RC, Tsui C, Xu J, Robert L, Wu L, … Ribas A (2014). Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer Res, 74, 153–161. doi: 10.1158/0008-5472.CAN-13-1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert NJ, Schmittnaegel M, Bordry N, Nassiri S, Wald N, Martignier C, … Speiser DE (2018). T cell-induced CSF1 promotes melanoma resistance to PD1 blockade. Sci Transl Med, 10. doi: 10.1126/scitranslmed.aan3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikbakht N, Tiago M, Erkes DA, Chernova I, & Aplin AE (2019). BET inhibition modifies melanoma infiltrating T cells and enhances response to PD-L1 blockade. J Invest Dermatol, 139, 1612–1615. doi:doi: 10.1016/j.jid.2018.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer HG, El-Gamal D, Powell B, Hing ZA, Blachly JS, Harrington B, … Lapalombella R (2018). BRD4 profiling identifies critical Chronic Lymphocytic Leukemia oncogenic circuits and reveals sensitivity to PLX51107, a novel structurally distinct BET inhibitor. Cancer Discov, 8, 458–477. doi: 10.1158/2159-8290.CD-17-0902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoluzzi L, Hanniford D, Sokolova E, Osman I, Darvishian F, Wang J, … Hernando E (2016). BET and BRAF inhibitors act synergistically against BRAF-mutant melanoma. Cancer Med, 5, 1183–1193. doi: 10.1002/cam4.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, & Ivashkiv LB (2015). Effect and mechanism of BET bromodomain inhibition in macrophage transcriptional programming. Inflam Cell Signal, 2, 1–5. doi: 10.14800/ics.600 [DOI] [Google Scholar]
- Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, … Hernando E (2013). BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res, 73, 6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, & Vakoc Christopher R. (2014). The Mechanisms behind the Therapeutic Activity of BET Bromodomain Inhibition. Mol Cell, 54, 728–736. doi: 10.1016/j.molcel.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluijter M, van der Sluis TC, van der Velden PA, Versluis M, West BL, van der Burg SH, & van Hall T (2014). Inhibition of CSF-1R supports T-cell mediated melanoma therapy. PLoS One, 9, e104230–104236. doi: 10.1371/journal.pone.0104230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wang Y, Toubai T, Oravecz-Wilson K, Liu C, Methweson N, … Reddy P (2015). BET bromodomain inhibition suppresses graft-versus-host disease after allogeneic bone marrow transplantation in mice. Blood, 125, 2725–2728. doi: 10.1182/blood-2014-08-598037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienerroither S, Rauch I, Rosebrock F, Jamieson AM, Bradner JE, Muhar M, … Decker T (2014). Regulation of NO synthesis, local inflammation, and innate immunity to pathogens by BET family proteins. Mol Cell Biol, 34, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Qu Q, Zheng B, Xiong S, & Fan GH (2015). The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. J Leukoc Biol, 97, 61–69. doi: 10.1189/jlb.1A0314-170R [DOI] [PubMed] [Google Scholar]
- Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, … Zhang R (2016). BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep, 16, 2829–2837. doi: 10.1016/j.celrep.2016.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.