SUMMARY:
Texture analysis is a continuously evolving, noninvasive radiomics technique to quantify macroscopic tissue heterogeneity indirectly linked to microscopic tissue heterogeneity beyond human visual perception. In recent years, systemic oncologic applications of texture analysis have been increasingly explored. Here we discuss the basic concepts and methodologies of texture analysis, along with a review of various MR imaging texture analysis applications in glioma imaging. We also discuss MR imaging texture analysis limitations and the technical challenges that impede its widespread clinical implementation. With continued advancement in computational processing, MR imaging texture analysis could potentially develop into a valuable clinical tool in routine oncologic imaging.
Gliomas are central nervous system tumors of glial origin, with glioblastoma being the most common and aggressive subtype, having a median survival of 14.5 months and 10% survival at 5 years.1 Despite advanced imaging, accurate noninvasive prediction of glioma grade, survival, molecular status, and treatment response remains challenging. Brain biopsy remains the reference standard for histologic and genetic classification, but it is invasive and costly.2 Additionally, the inherently high molecular heterogeneity in gliomas may decrease the accuracy and prognostic value of stereotactic biopsy diagnosis. Moreover, despite stereotactic biopsy, the pathologic diagnosis may remain inconclusive in about 7%–15% of patients.3,4 This scenario necessitates preoperative identification of imaging surrogates to accurately assess global tumor heterogeneity and predict glioma grade, genetic milieu, and survival.5
Even though multiparametric MR imaging features show significant agreement in terms of morphologic features, some of which are strongly associated with poor survival, the accuracy of these imaging variables to predict genetic heterogeneity and prognosis is rather limited.6,7 Similarly, advanced MR imaging techniques such as diffusion, perfusion, and MR spectroscopy have also been beneficial, but with modest success.8 Texture analysis (TA) is a noninvasive method to quantify macroscopic tissue heterogeneity indirectly linked to microscopic tissue heterogeneity. Recently, MR imaging texture analysis (MRTA)-based studies have shown promise in predicting glioma grade, survival, molecular status, and response assessment. However, despite the continued work, consensus on the clinical role of MRTA remains elusive. Here we review the basic concepts behind MRTA, its applications in glioma imaging, its limitations, current challenges, and potential future directions.
MRTA: Concepts and Methodology
Texture, according to Merriam-Webster.com, is defined as “something composed of closely interwoven elements,” just as the structure formed by threads of a fabric identifies its character.9 Similarly, an image texture is a representation of pixel intensities, their distribution, and their interrelationships, which may or may not be discernible to the human eye. TA noninvasively measures tumor heterogeneity (through parameters like kurtosis, entropy, and pixel distribution that potentially correlate with cellular density, angiogenesis, and necrosis) and may better predict tumor biology.10,11
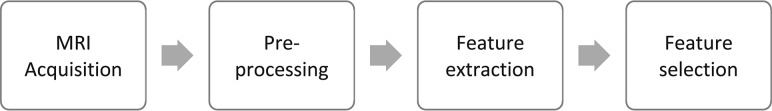
The workflow of MRTA is represented in the Figure.
FIGURE.
The workflow of MRTA.
Acquisition parameters such as magnet strength, spatial resolution, signal-to-noise ratio, and different pulse sequences may influence MRTA features.12 Most interesting, however, variations in these parameters can provide supplementary texture information, an added advantage of MR imaging over other imaging modalities.13 Postacquisition, an image undergoes preprocessing, which generally involves segmentation, image interpolation, intensity normalization, gray-level reduction, magnetic field inhomogeneity correction, and filtration. Performance of all these steps except segmentation is not a requirement for TA but helps enhance texture features and maintains uniformity and standardization.
Preprocessing steps can be performed on both open-source and commercially available software. The first step, segmentation, involves drawing an ROI manually or automatically either on a single 2D slice (or multiple slices) or a 3D-VOI.14 Next, to improve matrix resolution, interpolation is applied for which images are remapped to isotropic spacing to standardize the TA in all 3 directions. Furthermore, interpolation transforms the image into a higher matrix size and improves texture classification.15
Different MR imaging sequences have various ranges of intensities for the same image. This feature is addressed through intensity normalization, which extends the gray-level distribution of each MR image to the whole value range (0–255). It enhances the contrast between the tumor and background tissues and is achieved by either remapping the brightness to minimum or maximum value in the histogram, using mean ± 3 SD, or by using the histogram range between the first and the 99th percentile of the gray-scale image.
The undesirable effect of magnetic susceptibility on image texture can also be modified by use of the filtration process.16 Filtration can also be applied to derive new maps that individually extract and enhance subtle features otherwise lost while analyzing the original conventional image—that is, it converts an image into different anatomic scales varying from 2 mm (fine features), 3–5 mm (medium features), and 6 mm (coarse features). Furthermore, gray-level reduction is essential in the computation of gray-level matrices because TA can be computed on 16, 32, 64, 128, or 256 levels and actual MR imaging ranges up to 1024 levels.16 Because increasing the number of gray levels makes them computationally extensive without an added advantage, gray-level matrices are therefore computed at 5 or 6 bits per pixel.17
Feature extraction is the next step and includes agnostic and semantic features. Semantic features include shape, necrosis, vascularity, location, and speculation, and these can be quantified as well. Hundreds of features can be computed from available MRTA software.18 To overcome the issue of redundancy and overfitting that may be seen with multiple extracted features, several classifier models—Fischer coefficient, principal component analysis (PCA), linear/nonlinear discriminant analysis, regression models, support vector machine (SVM) with recursive feature elimination, artificial neural network, and random forest classifiers—are used as well as application of statistical methods to reduce the false discovery rate. These models extract the features that have the best discriminative power.19 Alternately, unsupervised deep learning models can also be used to agnostically generate discriminating texture features. This obviates generating thousands of random texture features and subsequent optimal feature selection as described above.
Types of TA
At present, statistical-, structural-, transform-, and spectral-based TAs are the most common agnostic methods used. Statistical-based TA depends on the pixel values, distribution, and spatial interrelationship in the defined ROI.20
First-order statistical TA is a histogram representation of image intensities in a predefined ROI and calculates mean, median, percentile, SD, skewness entropy, uniformity, and kurtosis. Mean is a measure of central tendency (average brightness), SD depicts dispersion from the mean, skewness reflects asymmetry of the histogram, kurtosis depicts the pointedness of the histogram (visual contrast), and entropy reflects the irregularity of the image-intensity distribution. The more heterogeneous the tumor, the higher the entropy is.20
Second-order or higher order statistical TA quantifies the image pattern on the basis of the spatial relationship or co-occurrence of the pixel value. It consists of several methods, including the 2 most common ones: gray-level co-occurrence matrix (GLCM) and gray-level run-length matrix (GLRLM). The GLCM measures the frequency of pixel pair distribution at a predefined distance,21 usually measured in 4 directions (0°, 45°, 90°, and 135°) for 2D and in 13 directions for 3D.14 GLCM features include homogeneity, inverse difference moment (IDM), dissimilarity, correlation, energy, and entropy. GLRLM observes the run of a specific pixel value over a chosen direction and consists of gray-level nonuniformity, run-length nonuniformity, short-run emphasis, and long-run emphasis. Both GLCM and GLRLM are calculated in different directions and averaged to make them rotationally invariant. GLCM may be measured over different pixel distances (for example, from 1 to 5), and similarly, GLRLM is computed over different run lengths to compute different texture features from the same ROI. GLCM and GLRLM over short distance and run provide fine texture, and over longer distance and run provide coarse texture. Different texture features can also be calculated in statistical methods by application of filters such as bandpass or nonorthogonal wavelet transform, which allow extraction of fine (≤2 mm), medium (3–5 mm), and coarse texture (>6 mm) using different filter values.22
Local binary patterns have high discriminative power and calculate the pixel value by comparing it with neighboring pixels and then assigning a binary value. Other higher order statistics include busyness, coarseness, and contrast, which calculate the spatial relationship among ≥3 gray-level pixel values.16
Structural (model-based) methods such as fractal analysis provide information about the self-symmetry of the objects. These are computationally extensive and less preferred. Spectral methods include wavelet, Gabor, and Fourier transforms and are based on transforming the spatial information of the image into spatial frequencies.22
In general, the first- and second-order statistical methods are used most commonly. First-order statistical methods provide global information, and second-order statistical methods provide additional information regarding the transition among pixel values. An important consideration is that 2 different tumors may have similar distribution of intensities but may differ in their spatial interrelationship; thus, histogram TA may be limited in such a setting. Second-order statistical TA may be preferable, especially for markedly heterogeneous tumors.23,24
MRTA Applications in Glioma Imaging
MRTA applications in gliomas are an active area of research, and multiple reports have shown promising results (On-line Tables 1–4). For the sake of simplicity, we have condensed various studies into 4 broad categories: MRTA for glioma grading, predicting survival, glioma radiogenomics, and a miscellaneous category of studies differentiating gliomas from other CNS tumors and assessing treatment changes.
Glioma Grading
The World Health Organization classifies gliomas as low grade (I and II) and high grade (III and IV).25 Pretherapy determination of glioma grade can help optimize treatment strategy, predicting therapeutic response, prognosis, and survival.10,26 On-line Table 1 summarizes prior studies evaluating MRTA for glioma grading.2,10,27–32 Some of these are briefly discussed below. In general, most studies used either ADC maps, T1-contrast-enhanced (CE) MR imaging, or a multiparametric technique along with a transform statistical (filtration-histogram) technique or purely statistical (first- or second-order) TA. Despite variabilities in TA software, entropy values of the ADC maps consistently showed promising results for differentiating low-grade gliomas (LGGs) from high-grade gliomas (HGGs). Skogen et al29 performed histogram-based TA in 95 patients using CE-MR images and found SD parameters at a fine texture highly significant (area under the curve [AUC], 0.910) in distinguishing LGGs from HGGs. Tian et al30 performed multiparametric TA in 153 patients (grades II–IV) using an SVM classifier model. They reported 98% accuracy of MRTA features for glioma grading. They also observed that while multiparametric TA performed better in comparison with single-sequence TA, T1-CE was the best single sequence. Xie et al31 observed that entropy (AUC = 0.885) and IDM (AUC = 0.901) of model-free and a dynamic contrast-enhanced MR imaging–based model were able to differentiate grade III from grade IV and grade II from grade III gliomas.
Glioma Survival Analysis
Prior studies have used features such as age, extent of resection, degree of necrosis, Karnofsky scores, and enhancing tumor size as prognostic predictors.33 On-line Table 2 summarizes MRTA studies predicting survival in gliomas.34–44 As mentioned above, these studies also had considerable heterogeneity in terms of methodologies and classifier models. Most interesting, most studies found CE-MR imaging sequences to be the most useful for predicting survival. Yang et al,36 for example, noted that even though several texture parameters predicted 12-month survival, CE-MR imaging sequences were the most accurate. They also mentioned that single-image features or MR images may not suffice because different combinations of image features and sequences are predictive for different tasks. Another multiparametric study by Kickingereder et al40 in 119 patients using supervised PCA predicted progression-free and overall survival after extracting 11 second-order texture features. The MRTA features outperformed clinical and radiologic risk models in predicting prognosis. Another multiparametric MRTA study by Upadhaya et al35 in 40 patients extracted the top 5 texture features from CE-MR images with an accuracy of 83% in predicting 15-month survival. Liu et al44 (n = 119) also noted the best survival prediction on CE-MR imaging sequences (AUC, 0.791; accuracy, 80.7%). They also discovered that texture features derived from CE-MR imaging were comparable with features derived from a combination of multiple sequences.
Glioma Radiogenomics
The 2016 World Health Organization classification update of gliomas incorporates genetic information for diagnosis. Radiogenomics refers to the relationship between imaging phenotypes and genomics that might allow improved decision-making and consequently improved patient outcomes.5 Established glioma biomarkers include isocitrate dehydrogenase (IDH), 1p/19q-codeletion, and methylguanine methyltransferase status. Immunohistochemistry combined with genome sequencing is a standard method for identifying glioma mutations.45,46 Many studies have correlated multiparametric imaging features with glioma mutations and, to date, have shown greater success for IDH status compared with other mutations. Currently, standard glioblastoma therapy does not include mutation-specific treatment based on molecular status.47
Multiple ongoing clinical trials are, however, evaluating targeted treatments in gliomas.1 On-line Table 3 summarizes the recently published MRTA studies on glioma radiogenomics.48–55 Using SVM-recursive feature elimination, Zhang et al48 (n = 152) extracted the top 15 texture features from CE-MR imaging and T2WI with 82% accuracy for predicting IDH status. Hsieh et al49 (n = 39) also reported similar results in predicting IDH status by extracting 14 GLCM textural features on CE-MR imaging. Han et al55 also showed that the joint variable derived from T1WI, T2WI, and CE-MR imaging histograms and GLCM features can be used for precise detection of IDH1-mutated gliomas. TA using B0 and fractional anisotropy maps has also shown a high accuracy of 95% in predicting IDH status.50 Bahrami et al51 reported greater FLAIR tissue heterogeneity and lower edge contrast in IDH wild-type compared with IDH mutants. Jakola et al53 also reported greater accuracy for predicting IDH mutation using 3D-FLAIR. IDH-mutant 1p/19q-codeleted gliomas also have shown similar results compared with an 1p/19q-intact group and an unmethylated group. Shofty et al52 used retrospective data from various MR imaging scanners with variable parameters. Despite the considerable data heterogeneity, they successfully predicted 1p/19q codeletion and discriminated LGGs on the basis of 1p/19q-codeletion status with an accuracy of 87% by extracting the top 39 texture features, mostly from CE-MR imaging and T2WI. Li et al56–58 accurately predicted alpha thalassemia mental-retardation syndrome, epidermal growth factor receptor, and p53 status in patients with LGG on T2WI. In general, the second-order TA on CE-MR imaging and FLAIR images mostly contributed to the high accuracy for predicting genomic status.
Miscellaneous Applications
Glioblastoma imaging features may overlap primary central nervous system lymphomas (PCNSLs) and metastases, rendering a noninvasive distinction truly challenging.59 Recent MRTA studies, however, have shown promising results in differentiating glioblastomas from PCNSLs and metastases (On-line Table 4).59–63,66 Kunimatsu et al60,61 differentiated glioblastomas from PCNSLs with 75% accuracy by selecting the top 4 best-performing texture features from CE-MR images. Xiao et al62 found skewness and kurtosis to be the best first-order features on CE-MR imaging in a similar population. Suh et al59 reported 90% accuracy of radiomics-based machine learning algorithms compared with visual analysis by 3 readers in differentiating PCNSLs from glioblastomas. Similarly, Alcaide-Leon et al63 showed superiority of the SVM classifier over human evaluation. Overall, better results were found using CE-MR imaging and machine-classification models. Dynamic histogram analysis is a novel technique using histogram-based texture parameter analysis on a time-series of dynamic susceptibility contrast MR imaging. Dynamic texture parameter analysis is a further extension of dynamic histogram analysis that analyzes a larger set of time-dependent texture maps from dynamic susceptibility contrast-enhanced series.64,65 By using dynamic texture parameter analysis, Verma et al66 extracted texture features from the earliest contrast phase of dynamic susceptibility contrast-enhanced perfusion maps and differentiated glioblastomas from PCNSLs. Skogen et al67 used MRTA on DTI-derived fractional anisotropy and ADC maps and reported significantly higher heterogeneity in peritumoral edema of glioblastomas compared with metastases.
Assessment of the therapeutic response based solely on Response Assessment in Neuro-Oncology criteria, which are based solely on the 2D size and enhancement, may be challenging.43 Recently, Ismail et al68 (n = 105) extracted the 2 most discriminative 3D shape features of the enhancing tumor on CE-MR imaging, FLAIR, and T2WI and noted that 3D shape features could distinguish pseudoprogression from true progression. TA may provide useful prognostic information regarding progression and survival in such a patient population. Grossmann et al41 found that “information correlation,” a GLCM parameter, had a significantly higher score in patients on bevacizumab surviving beyond 3 months. Bahrami et al43 reported that lower edge contrast of the FLAIR signal of gliomas correlated with poor survival after bevacizumab.
Despite the heterogeneity of the data and software, most studies demonstrate the robustness of the MRTA and its clinical transferability for diagnostic use. Second-order statistical TA showed promising results in most studies. Entropy also appears to be a key feature. Quite possibly, multisequence-based MRTA may have higher accuracy. However, it may be time-consuming, and not all advanced sequences are widely available. Performing MRTA on commonly available CE-MR imaging as well as T2-weighted/FLAIR sequences may be optimal for standardization, given the wide availability and promise shown in early studies. In studies involving LGGs, it may be better to perform MRTA on T2-weighted/FLAIR sequences because they better identify the tumor. On the other hand, CE-MR imaging appears to be the single best sequence in glioblastoma, as mentioned in a study by Liu et al.44
Challenges and Future Directions
Despite the advantages, widespread clinical implementation of MRTA is still limited, mostly due to nonuniformity and lack of standardization and quantification processes. The real challenge lies in the reproducibility and repeatability of these studies. Multiple studies used indigenous MRTA software, likely with varying algorithms. Thus, studies differ not only in image acquisition but also MRTA methodologies.
The other important aspect is use of a cancer imaging data base, which may suffice for conventional multiparametric assessment but nevertheless has considerable heterogeneity in sequences, protocols, and vendors. This is not confined just to the cancer data base but is a practical consideration for any multicenter study.
The impact of acquisition parameters on MRTA has been addressed in multiple studies. Ford et al,69 using a digital 3D phantom, concluded that multiple texture features vary considerably between T1-weighted images (spin-echo, gradient echo, gradient recalled-echo, and inversion recovery) and T1 maps. They also noted that TR/TE variations on T1WI and T2WI affect texture features. Another phantom-based study by Buch et al12 assessed the effect of magnet strength, flip angles, number of excitations, and different scanner platforms and concluded that some texture features are more robust (for example, except for histogram-related median, entropy, and GLCM contrast, all other histogram, GLCM, GLRLM, gray-level gradient matrix, and Law features did not show a significant difference from flip angles) and some are more susceptible to acquisition parameters (all Law features were significantly different for different magnetic strengths). Yang et al70 found that different reconstruction algorithms, noise levels, and parallel imaging acceleration factors can influence texture parameters. Texture features are also affected by a number of coil elements, coil arrangement, and k-space sampling.71 Rapid k-space sampling techniques can reduce SNR, thus affecting TA, especially histogram intensity-based features.12,71,72 The inclusion of preprocessing steps may also affect texture features. Mayerhoefer et al15 found zero-filling interpolation to be the most optimal with an interpolation factor of 4 to improve texture performance. Both Waugh et al72 and Mayerhoefer et al13 found spatial resolution to be the most important factor affecting MRTA and that variability in TR/TE, sampling bandwidth, and number of excitations is not significant at higher resolution. However, Molina et al73 found that several GLCM and GLRLM texture features computed on 3D segmentation of brain gliomas were not robust over different spatial resolution/matrix size and gray-level ranges. They found only entropy to be the most robust feature. For intensity normalization, Collewet et al74 found mean ± 3 SDs to be the most optimal strategy. Partial volume artifacts can be corrected by iterative optimal thresholding algorithms.12
In terms of analysis, the choice between analyzing multiple-versus-few sequences for MRTA also needs to be addressed. Spin-echo sequences are often acquired routinely in suspected brain tumor while advanced imaging may not be routinely performed, especially on the index scan. TA-based conventional sequences seem more practical in terms of generalizability, with T1-CE–based TA being the most optimal.
3D-TA appears more accurate than 2D, given the high spatial resolution of the acquired data. Similarly, results based on a VOI analysis appear more reliable than those based on a single slice (also a prominent limitation of multiple prior studies).75 However, more studies are needed to further establish better accuracy of 3D-MRTA and justify the additional time and effort.14
All these factors re-emphasize the need for standardization of MR imaging protocols, including uniform postprocessing techniques, to allow a more valid, multiple-institution comparison of MRTA results.
Challenges in processing include the inhomogeneity of MRTA software, which may be commercial, open-source, or developed in-house. The superiority of one over the other remains speculative at best.76 Future studies should assess the comparability and accuracy of results across multiple types of software, especially in terms of clinical outcomes, survival, and radiomic parameters, to help with standardization. Finally, adequate training of radiologists is also required for consistent evaluation and implementation in routine workflow.
Another factor is the problem of the “huge data” that need sorting to prevent redundancy. Several classifier models exist to accurately predict the optimal texture feature. However, there is no consensus as to whether one is superior to the others. Artificial intelligence may be helpful in this case, both in feature selection and building prediction models.
Additionally, even though MRTA has shown potential in neuroimaging, certain valid criticisms of this technique should also be acknowledged. One major criticism of MRTA is that it is not hypothesis-driven. In some ways, MRTA is essentially correlating different mathematic computations with various imaging and clinical parameters to see what is statistically significant. This is, however, problematic for 2 main reasons: First, there is no intuitive reason why mathematic variables would make physiologic sense. Whether these significant relationships are merely chance findings secondary to overfitting (see next paragraph) or reflect as-yet unexplored physiologic correlates currently remains unclear. Most interesting, some prior studies have shown correlations between CT texture parameters and histologic markers such as CD34 and Ki-67, findings that may support some tissue-level basis for texture parameters.11 These, however, remain to be fully determined and validated.
The other major limitation is the problem of overfitting, which can occur when the number of independent parameters being analyzed is larger than the number of data points/sample size. Generally, it is recommended that the sample size be 5–10 times the number of analyzed variables, which is often not the case, especially with studies using a smaller sample size. This issue could be addressed through either larger datasets or analysis of only a few preselected robust variables. Another way to avoid overfitting is to split the data into 3 mutually exclusive sets, one each for training, testing, and finally validation.
Finally, the role of MRTA should also be evaluated in the context of deep learning and neural networks. Even though unsupervised deep learning can self-identify features for itself and does not need manual input (thereby reducing interobserver bias in ROI selection) and feature selection,76 deep learning methods require higher processing powers and considerable high-quality ground truth data. The insatiable appetite of deep learning for large quantities of labeled training data (which are both expensive and difficult to produce) is another limitation of the deep learning approach.77 MRTA, on the other hand, is less data-hungry. Additionally, the internal algorithm feature vectors in unsupervised deep learning may not always be apparent (black box), while TA features can be explained more easily. However, ROI selection bias among observers can influence MRTA results and should be addressed prospectively.75 However, the 2 techniques may be complementary in terms of optimal feature selection (in deep learning) and ease of use for wider applicability (for MRTA), thereby providing optimal output without substantial changes to the clinical workflow.
Conclusions
MRTA has shown promising results in various glioma-related applications. The inclusion of tumor heterogeneity as a radiology-reporting variable appears to break with the notion of radiology being only diagnostic or qualitative and brings the shift toward prognostic value as an imaging biomarker for precision/personalized medicine. However, before widespread clinical applicability, prospective validation of accuracy, selection of robust sequences, interinstitutional congruity of results, and selection of the best possible technique need to be addressed. Last, development of automated segmentation tools with incorporation of machine learning is essential to expedite feature extraction and analysis, thus saving time and additional burden on the radiologist.
ABBREVIATIONS:
- AUC
area under the curve
- CE
contrast-enhanced
- GLCM
gray-level co-occurrence matrix
- GLRLM
gray-level run-length matrix
- HGG
high-grade glioma
- IDH
isocitrate dehydrogenase
- IDM
inverse difference moment
- LGG
low-grade glioma
- MRTA
MR imaging texture analysis
- PCNSL
primary central nervous system lymphoma
- PCA
principal component analysis
- SVM
support vector machine
- TA
texture analysis
REFERENCES
- 1. Auffinger B, Thaci B, Nigam P, et al. . New therapeutic approaches for malignant glioma: in search of the Rosetta stone. F1000 Med Rep 2012;4:18 10.3410/M4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kang Y, Choi SH, Kim YJ, et al. . Gliomas: histogram analysis of apparent diffusion coefficient maps with standard- or high-b-value diffusion-weighted MR imaging–correlation with tumor grade. Radiology 2011;261:882–90 10.1148/radiol.11110686 [DOI] [PubMed] [Google Scholar]
- 3. Gulsen S. Achieving higher diagnostic results in stereotactic brain biopsy by simple and novel technique. Open Access Maced J Med Sci 2015;3:99–104 10.3889/oamjms.2015.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akay A, Rüksen M, Islekel S. Magnetic resonance imaging-guided stereotactic biopsy: a review of 83 cases with outcomes. Asian J Neurosurg 2019;14:90–95 10.4103/ajns.AJNS_81_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol 2015;12:862–66 10.1016/j.jacr.2015.04.019 [DOI] [PubMed] [Google Scholar]
- 6. Law M, Yang S, Wang H, et al. . Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 2003;24:1989–98 [PMC free article] [PubMed] [Google Scholar]
- 7. Gutman DA, Cooper LA, Hwang SN, et al. . MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013;267:560–69 10.1148/radiol.13120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naveed MA, Goyal P, Malhotra A, et al. . Grading of oligodendroglial tumors of the brain with apparent diffusion coefficient, magnetic resonance spectroscopy, and dynamic susceptibility contrast imaging. Neuroradiol J 2018;31:379–85 10.1177/1971400918757217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Texture. Merriam-Webster.com. https://www.merriam-webster.com/dictionary/texture. Accessed January 10, 2019.
- 10. Raja R, Sinha N, Saini J, et al. . Assessment of tissue heterogeneity using diffusion tensor and diffusion kurtosis imaging for grading gliomas. Neuroradiology 2016;58:1217–31 10.1007/s00234-016-1758-y [DOI] [PubMed] [Google Scholar]
- 11. Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging 2013;13:400–06 10.1102/1470-7330.2013.9045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buch K, Kuno H, Qureshi MM, et al. . Quantitative variations in texture analysis features dependent on MRI scanning parameters: a phantom model. J Appl Clin Med Phys 2018;19:253–64 10.1002/acm2.12482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mayerhoefer ME, Szomolanyi P, Jirak D, et al. . Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Med Phys 2009;36:1236–43 10.1118/1.3081408 [DOI] [PubMed] [Google Scholar]
- 14. Depeursinge A, Foncubierta-Rodriguez A, Van De Ville D, et al. . Three-dimensional solid texture analysis in biomedical imaging: review and opportunities. Med Image Anal 2014;18:176–96 10.1016/j.media.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 15. Mayerhoefer ME, Szomolanyi P, Jirak D, et al. . Effects of magnetic resonance image interpolation on the results of texture-based pattern classification: a phantom study. Invest Radiol 2009;44:405–11 10.1097/RLI.0b013e3181a50a66 [DOI] [PubMed] [Google Scholar]
- 16. Larroza A, Bodí V, Moratal D. Texture analysis in magnetic resonance imaging: review and considerations for future applications. In: Constantinides C, ed. Assessment of Cellular and Organ Function and Dysfunction using Direct and Derived MRI Methodologies. Rijeka: IntechOpen; 2016 [Google Scholar]
- 17. Szczypiński PM, Klepaczko A. MaZda: a framework for biomedical image texture analysis and data exploration. In: Depeursinge A, Al-Kadi OS, Mitchell JR, eds. Biomedical Texture Analysis. Amsterdam: Elsevier; 2017:315–47 [Google Scholar]
- 18. Schaer R, Cid YD, Alkim E, et al. . Web-Based Tools for Exploring the Potential of Quantitative Imaging Biomarkers on Radiology. Waltham: Academic Press; 2017 [Google Scholar]
- 19. Di Cataldo S, Ficarra E. Mining textural knowledge in biological images: applications, methods and trends. Comput Struct Biotechnol J 2017;15:56–67 10.1016/j.csbj.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolossváry M, Kellermayer M, Merkely B, et al. . Cardiac computed tomography radiomics: a comprehensive review on radiomic techniques. J Thorac Imaging 2018;33:26–34 10.1097/RTI.0000000000000268 [DOI] [PubMed] [Google Scholar]
- 21. Bashir U, Siddique MM, McLean E, et al. . Imaging heterogeneity in lung cancer: techniques, applications, and challenges. AJR Am J Roentgenol 2016;207:534–43 10.2214/AJR.15.15864 [DOI] [PubMed] [Google Scholar]
- 22. Davnall F, Yip CS, Ljungqvist G, et al. . Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012;3:573–89 10.1007/s13244-012-0196-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lubner MG, Smith AD, Sandrasegaran K, et al. . CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 2017;37:1483–503 10.1148/rg.2017170056 [DOI] [PubMed] [Google Scholar]
- 24. Depeursinge A, Fageot J, Al-Kadi OS. Fundamentals of texture processing for biomedical image analysis: a general definition and problem formulation. Biomedical Texture Analysis. Amsterdam: Elsevier; 2017:1–27 [Google Scholar]
- 25. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803–20 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 26. Chand P, Amit S, Gupta R, et al. . Errors, limitations, and pitfalls in the diagnosis of central and peripheral nervous system lesions in intraoperative cytology and frozen sections. J Cytol 2016;33:93–97 10.4103/0970-9371.182530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ryu YJ, Choi SH, Park SJ, et al. . Glioma: application of whole-tumor texture analysis of diffusion-weighted imaging for the evaluation of tumor heterogeneity. PLoS One 2014;9:e108335 10.1371/journal.pone.0108335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kinoshita M, Sakai M, Arita H, et al. . Introduction of high throughput magnetic resonance T2-weighted image texture analysis for WHO grade 2 and 3 gliomas. PLoS One 2016;11:e0164268 10.1371/journal.pone.0164268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skogen K, Schulz A, Dormagen JB, et al. . Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 2016;85:824–29 10.1016/j.ejrad.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 30. Tian Q, Yan LF, Zhang X, et al. . Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging 2018;48:1518–28 10.1002/jmri.26010 [DOI] [PubMed] [Google Scholar]
- 31. Xie T, Chen X, Fang J, et al. . Textural features of dynamic contrast-enhanced MRI derived model-free and model-based parameter maps in glioma grading. J Magn Reson Imaging 2018;47:1099–111 10.1002/jmri.25835 [DOI] [PubMed] [Google Scholar]
- 32. Qi XX, Shi DF, Ren SX, et al. . Histogram analysis of diffusion kurtosis imaging derived maps may distinguish between low and high grade gliomas before surgery. Eur Radiol 2018;28:1748–55 10.1007/s00330-017-5108-1 [DOI] [PubMed] [Google Scholar]
- 33. Mazurowski MA, Zhang J, Peters KB, et al. . Computer-extracted MR imaging features are associated with survival in glioblastoma patients. J Neurooncol 2014;120:483–88 10.1007/s11060-014-1580-5 [DOI] [PubMed] [Google Scholar]
- 34. Brynolfsson P, Nilsson D, Henriksson R, et al. . ADC texture: an imaging biomarker for high-grade glioma? Med Phys 2014;41:101903 10.1118/1.4894812 [DOI] [PubMed] [Google Scholar]
- 35. Upadhaya T, Morvan Y, Stindel E, et al. . A framework for multimodal imaging-based prognostic model building: preliminary study on multimodal MRI in glioblastoma multiforme. IRBM 2015;36:345–50 10.1016/j.irbm.2015.08.001 [DOI] [Google Scholar]
- 36. Yang D, Rao G, Martinez J, et al. . Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys 2015;42:6725–35 10.1118/1.4934373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaddad A, Tanougast C. Extracted magnetic resonance texture features discriminate between phenotypes and are associated with overall survival in glioblastoma multiforme patients. Med Biol Eng Comput 2016;54:1707–18 10.1007/s11517-016-1461-5 [DOI] [PubMed] [Google Scholar]
- 38. Lee J, Jain R, Khalil K, et al. . Texture feature ratios from relative CBV maps of perfusion MRI are associated with patient survival in glioblastoma. AJNR Am J Neuroradiol 2016;37:37–43 10.3174/ajnr.A4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Molina D, Pérez-Beteta J, Luque B, et al. . Tumour heterogeneity in glioblastoma assessed by MRI texture analysis: a potential marker of survival. Br J Radiol 2016;89:20160242 10.1259/bjr.20160242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kickingereder P, Burth S, Wick A, et al. . Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016;280:880–89 10.1148/radiol.2016160845 [DOI] [PubMed] [Google Scholar]
- 41. Grossmann P, Narayan V, Chang K, et al. . Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol 2017;19:1688–97 10.1093/neuonc/nox092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaddad A, Daniel P, Desrosiers C, et al. . Novel radiomic features based on joint intensity matrices for predicting glioblastoma patient survival time. IEEE J Biomed Health Inform 2019;23:795–804 10.1109/JBHI.2018.2825027 [DOI] [PubMed] [Google Scholar]
- 43. Bahrami N, Piccioni D, Karunamuni R, et al. . Edge contrast of the FLAIR hyperintense region predicts survival in patients with high-grade gliomas following treatment with bevacizumab. AJNR Am J Neuroradiol 2018;39:1017–24 10.3174/ajnr.A5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Y, Zhang X, Feng N, et al. . The effect of glioblastoma heterogeneity on survival stratification: a multimodal MR imaging texture analysis. Acta Radiol 2018;59:1239–46 10.1177/0284185118756951 [DOI] [PubMed] [Google Scholar]
- 45. Combs SE, Rieken S, Wick W, et al. . Prognostic significance of IDH-1 and MGMT in patients with glioblastoma: one step forward, and one step back? Radiat Oncol 2011;6:115 10.1186/1748-717X-6-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia L, Wu B, Fu Z, et al. . Prognostic role of IDH mutations in gliomas: a meta-analysis of 55 observational studies. Oncotarget 2015;6:17354–65 10.18632/oncotarget.4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chow D, Chang P, Weinberg BD, et al. . Imaging genetic heterogeneity in glioblastoma and other glial tumors: review of current methods and future directions. AJR Am J Roentgenol 2018;210:30–38 10.2214/AJR.17.18754 [DOI] [PubMed] [Google Scholar]
- 48. Zhang X, Tian Q, Wu YX, et al. . IDH mutation assessment of glioma using texture features of multimodal MR images. Proceedings of SPIE 2017;10134 10.1117/12.2254212 [DOI] [Google Scholar]
- 49. Hsieh KL, Chen CY, Lo CM. Radiomic model for predicting mutations in the isocitrate dehydrogenase gene in glioblastomas. Oncotarget 2017;8:45888–97 10.18632/oncotarget.17585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eichinger P, Alberts E, Delbridge C, et al. . Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci Rep 2017;7:13396 10.1038/s41598-017-13679-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bahrami N, Hartman SJ, Chang YH, et al. . Molecular classification of patients with grade II/III glioma using quantitative MRI characteristics. J Neurooncol 2018;139:633–42 10.1007/s11060-018-2908-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shofty B, Artzi M, Ben Bashat D, et al. . MRI radiomics analysis of molecular alterations in low-grade gliomas. Int J Comput Assist Radiol Surg 2018;13:563–71 10.1007/s11548-017-1691-5 [DOI] [PubMed] [Google Scholar]
- 53. Jakola AS, Zhang YH, Skjulsvik AJ, et al. . Quantitative texture analysis in the prediction of IDH status in low-grade gliomas. Clin Neurol Neurosurg 2018;164:114–20 10.1016/j.clineuro.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 54. Rui W, Ren Y, Wang Y, et al. . MR textural analysis on T2 FLAIR images for the prediction of true oligodendroglioma by the 2016 WHO genetic classification. J Magn Reson Imaging 2018;48:74–83 10.1002/jmri.25896 [DOI] [PubMed] [Google Scholar]
- 55. Han L, Wang S, Miao Y, et al. . MRI texture analysis based on 3D tumor measurement reflects the IDH1 mutations in gliomas: a preliminary study. Eur J Radiol 2019;112:169–79 10.1016/j.ejrad.2019.01.025 [DOI] [PubMed] [Google Scholar]
- 56. Li Y, Liu X, Qian Z, et al. . Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol 2018;28:2960–68 10.1007/s00330-017-5267-0 [DOI] [PubMed] [Google Scholar]
- 57. Li Y, Liu X, Xu K, et al. . MRI features can predict EGFR expression in lower grade gliomas: a voxel-based radiomic analysis. Eur Radiol 2018;28:356–62 10.1007/s00330-017-4964-z [DOI] [PubMed] [Google Scholar]
- 58. Li Y, Qian Z, Xu K, et al. . MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin 2017;17:306–11 10.1016/j.nicl.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suh HB, Choi YS, Bae S, et al. . Primary central nervous system lymphoma and atypical glioblastoma: differentiation using radiomics approach. Eur Radiol 2018;28:3832–39 10.1007/s00330-018-5368-4 [DOI] [PubMed] [Google Scholar]
- 60. Kunimatsu A, Kunimatsu N, Yasaka K, et al. . Machine learning-based texture analysis of contrast-enhanced MR imaging to differentiate between glioblastoma and primary central nervous system lymphoma. Magn Reson Med Sci 2019;18:44–52 10.2463/mrms.mp.2017-0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kunimatsu A, Kunimatsu N, Kamiya K, et al. . Comparison between glioblastoma and primary central nervous system lymphoma using MR image-based texture analysis. Magn Reson Med Sci 2018;17:50–57 10.2463/mrms.mp.2017-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xiao DD, Yan PF, Wang YX, et al. . Glioblastoma and primary central nervous system lymphoma: preoperative differentiation by using MRI-based 3D texture analysis. Clin Neurol Neurosurg 2018;173:84–90 10.1016/j.clineuro.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 63. Alcaide-Leon P, Dufort P, Geraldo AF, et al. . Differentiation of enhancing glioma and primary central nervous system lymphoma by texture-based machine learning. AJNR Am J Neuroradiol 2017;38:1145–50 10.3174/ajnr.A5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Slotboom J, Schaer R, Ozdoba C, et al. . A novel method for analyzing DSCE-images with an application to tumor grading. Invest Radiol 2008;43:843–53 10.1097/RLI.0b013e3181893605 [DOI] [PubMed] [Google Scholar]
- 65. Verma RK, Slotboom J, Heldner MR, et al. . Characterization of microcirculation in multiple sclerosis lesions by dynamic texture parameter analysis (DTPA). PLoS One 2013;8:e67610 10.1371/journal.pone.0067610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Verma RK, Wiest R, Locher C, et al. . Differentiating enhancing multiple sclerosis lesions, glioblastoma, and lymphoma with dynamic texture parameters analysis (DTPA): a feasibility study. Med Phys 2017;44:4000–08 10.1002/mp.12356 [DOI] [PubMed] [Google Scholar]
- 67. Skogen K, Schulz A, Helseth E, et al. . Texture analysis on diffusion tensor imaging: discriminating glioblastoma from single brain metastasis. Acta Radiol 2019;60:356–66 10.1177/0284185118780889 [DOI] [PubMed] [Google Scholar]
- 68. Ismail M, Hill V, Statsevych V, et al. . Shape features of the lesion habitat to differentiate brain tumor progression from pseudoprogression on routine multiparametric MRI: a multisite study. AJNR Am J Neuroradiol 2018;39:2187–93 10.3174/ajnr.A5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ford J, Dogan N, Young L, et al. . Quantitative radiomics: impact of pulse sequence parameter selection on MRI-based textural features of the brain. Contrast Media Mol Imaging 2018;2018:1729071 10.1155/2018/1729071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang F, Dogan N, Stoyanova R, et al. . Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: a simulation study utilizing ground truth. Phys Med 2018;50:26–36 10.1016/j.ejmp.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 71. Schad LR. Problems in texture analysis with magnetic resonance imaging. Dialogues Clin Neurosci 2004;6:235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Waugh SA, Lerski RA, Bidaut L, et al. . The influence of field strength and different clinical breast MRI protocols on the outcome of texture analysis using foam phantoms. Med Phys 2011;38:5058–66 10.1118/1.3622605 [DOI] [PubMed] [Google Scholar]
- 73. Molina D, Pérez-Beteta J, Martínez-González A, et al. . Lack of robustness of textural measures obtained from 3D brain tumor MRIs impose a need for standardization. PLoS One 2017;12:e0178843 10.1371/journal.pone.0178843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 2004;22:81–91 10.1016/j.mri.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 75. Hainc N, Stippich C, Stieltjes B, et al. . Experimental texture analysis in glioblastoma: a methodological study. Invest Radiol 2017;52:367–73 10.1097/RLI.0000000000000354 [DOI] [PubMed] [Google Scholar]
- 76. Varghese BA, Cen SY, Hwang DH, et al. . Texture analysis of imaging: what radiologists need to know. AJR Am J Roentgenol 2019;212:520–28 10.2214/AJR.18.20624 [DOI] [PubMed] [Google Scholar]
- 77. Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017;10:257–73 10.1007/s12194-017-0406-5 [DOI] [PubMed] [Google Scholar]