Forty patients with MS were prospectively included and scanned (3T) to acquire synthetic MR imaging and conventional FLAIR images. Synthetic FLAIR images were created with the SyMRI software. Acquired data were divided into 30 training and 10 test datasets. A conditional generative adversarial network was trained to generate improved FLAIR images from raw synthetic MR imaging data using conventional FLAIR images as targets. The peak signal-to-noise ratio, normalized root mean square error, and the Dice index of MS lesion maps were calculated for synthetic and deep learning FLAIR images against conventional FLAIR images, respectively. Lesion conspicuity and the existence of artifacts were visually assessed. The peak signal-to-noise ratio and normalized root mean square error were significantly higher and lower, respectively, in generated-versus-synthetic FLAIR images in aggregate intracranial tissues and all tissue segments. The Dice index of lesion maps and visual lesion conspicuity were comparable between generated and synthetic FLAIR images. Using deep learning, the authors conclude that they improved the synthetic FLAIR image quality by generating FLAIR images that have contrast closer to that of conventional FLAIR images and fewer granular and swelling artifacts, while preserving the lesion contrast.
Abstract
BACKGROUND AND PURPOSE:
Synthetic FLAIR images are of lower quality than conventional FLAIR images. Here, we aimed to improve the synthetic FLAIR image quality using deep learning with pixel-by-pixel translation through conditional generative adversarial network training.
MATERIALS AND METHODS:
Forty patients with MS were prospectively included and scanned (3T) to acquire synthetic MR imaging and conventional FLAIR images. Synthetic FLAIR images were created with the SyMRI software. Acquired data were divided into 30 training and 10 test datasets. A conditional generative adversarial network was trained to generate improved FLAIR images from raw synthetic MR imaging data using conventional FLAIR images as targets. The peak signal-to-noise ratio, normalized root mean square error, and the Dice index of MS lesion maps were calculated for synthetic and deep learning FLAIR images against conventional FLAIR images, respectively. Lesion conspicuity and the existence of artifacts were visually assessed.
RESULTS:
The peak signal-to-noise ratio and normalized root mean square error were significantly higher and lower, respectively, in generated-versus-synthetic FLAIR images in aggregate intracranial tissues and all tissue segments (all P < .001). The Dice index of lesion maps and visual lesion conspicuity were comparable between generated and synthetic FLAIR images (P = 1 and .59, respectively). Generated FLAIR images showed fewer granular artifacts (P = .003) and swelling artifacts (in all cases) than synthetic FLAIR images.
CONCLUSIONS:
Using deep learning, we improved the synthetic FLAIR image quality by generating FLAIR images that have contrast closer to that of conventional FLAIR images and fewer granular and swelling artifacts, while preserving the lesion contrast.
The synthetic MR imaging technique can be used to create any contrast-weighted image, including T1-weighted, T2-weighted, and FLAIR images, based on the R1 and R2 relaxation rates (inverse of T1 and T2 relaxation times) and proton density.1 Synthetic MR imaging has become clinically feasible due to the development of rapid and simultaneous relaxometric methods,2 which have high repeatability and reproducibility across different vendors.3 The synthetic MR imaging quality4–6 and its clinical utility for evaluating brain diseases7–10 have been widely investigated. Synthetic MR imaging has the potential to reduce scan times in clinical settings, where multiple contrast-weighted images are usually required. Although the synthetic T1-weighted and T2-weighted image quality is comparable with that of conventional images, the synthetic FLAIR image quality is generally inferior to that of conventional FLAIR images,4,6 thus hindering the introduction of synthetic MR imaging into routine clinical practice. Synthetic FLAIR images reportedly show more artifacts than other contrasts,6 including hyperintensity on the brain-CSF interface with apparently swollen brain parenchyma,7,11 and granular hyperintensities in the CSF, neither of which have been reported for conventional FLAIR.6 Hence, methods that can improve the synthetic FLAIR image quality are necessary.
One such method, deep learning (DL), has been applied to medical images for various purposes,12,13 particularly generative adversarial networks (GANs),14 which have been used for noise reduction15 and resolution improvement.16 The GAN technique uses an image generator, which generates a new image similar to the input target image, and a discriminator, which differentiates the target and generated images. The image generator and discriminator are simultaneously trained to transcend each other. A conditional GAN (cGAN) is a newly proposed technique that conditions the output on the input to render the same underlying information between the input and output, ultimately keeping the generated image realistic.17 However, a problem in applying DL to clinical image processing is its generative nature, which is driven by down/up sampling operations, including convolution and deconvolution. When DL is used to process nonmedical images, it often generates artificial objects in empty space or eliminates observed objects.17 While these are useful features when using DL to synthesize photos from label maps and colorize images,17 these features may be harmful when applied to clinical images.
To solve these issues, we propose using a pixel-by-pixel neural network function as a generator to improve the synthetic FLAIR image quality. This pixel-by-pixel neural network operation sets the kernel size and stride of all convolutional layers to 1, thus minimizing the possibility of generating artificial objects or erasing true information by ensuring that the 2 identical input pixel values become the same output pixel value. Because conventional FLAIR images, which were used as the target, were acquired separately from the input images, we trained the generator model with a cGAN-type learning system to avoid any adverse misregistration effects between the input and target images.
Materials and Methods
Study Participants
This prospective study recruited 40 patients with MS between December and May 2017. Patients were diagnosed according to standard criteria.18 The MR imaging data were screened for severe motion artifacts. The first 30 consecutive subjects (11 men; mean age, 43.37 ± 9.36 years; median Expanded Disability Status Scale score, 1; range, 0–6; mean disease duration, 10.6 ± 6.94 years) were used as a training set, and the last 10 subjects (2 men; mean age, 44.7 ± 12.27 years; median Expanded Disability Status Scale score, 2.5; range, 0–4.5; mean disease duration, 10.56 ± 8.62 years) were used as a test set. The institutional review board of Juntendo University Hospital, Japan, approved this Health Insurance Portability and Accountability–compliant study, and all participants provided written informed consent.
Image Acquisition
All MR imaging was performed on a 3T system (Discovery MR750w; GE Healthcare, Milwaukee, Wisconsin) with a 19-channel head coil. All patients underwent synthetic MR imaging and conventional FLAIR imaging. We performed MR relaxometry with a 2D axial pulse sequence. This is a multisection, multiecho, multisaturation delay saturation-recovery turbo spin-echo acquisition method in which images are collected with different TE and saturation delay time combinations.2 Typically, 2 TEs and 4 delay times are used to generate a matrix of 8 complex images, which are then used to quantify the longitudinal R1 relaxation and transverse R2 relaxation rates and proton density.19 The acquisition parameters for quantitative synthetic MR imaging were as follows: TE, 16.9 and 84.5 ms; delay times, 146, 546, 1879, and 3879 ms; TR, 4.0 seconds; FOV, 240 × 240 mm; matrix, 320 × 320; echo-train length, 10; bandwidth, 31.25 kHz; section thickness/gap, 4.0/1.0 mm; slices, 30; and acquisition time, 7 minutes 12 seconds. The SyMRI software (Version 8.0; SyntheticMR, Linköping, Sweden) was used to retrieve the R1, R2, and proton-density maps on the basis of the acquired data and to create synthetic FLAIR (with postprocessing TR, 15,000 ms; TE, 100 ms; TI, 3000 ms) and T1-weighted (with postprocessing TR, 500 ms; TE, 10 ms) images. Intracranial tissue masks were also created with the SyMRI software.20 Postprocessing time on the SyMRI software was around 30 seconds in total using a workstation (HP Z230 Tower Workstation; Hewlett-Packard Japan, Tokyo, Japan) comprising Windows 7 (64-bit version; 16 GB memory) and a central processing unit (Xeon Processor E3–1281 v3; Intel, Santa Clara, California).
The acquisition parameters for conventional FLAIR imaging were as follows: TR, 9000 ms; TE, 124 ms; TI, 2472 ms; FOV, 240 × 240 mm; matrix, 320 × 224; echo-train length, 16; section thickness/gap, 4.0/1.0 mm; number of slices, 30; and acquisition time, 2 minutes 33 seconds.
DL Framework
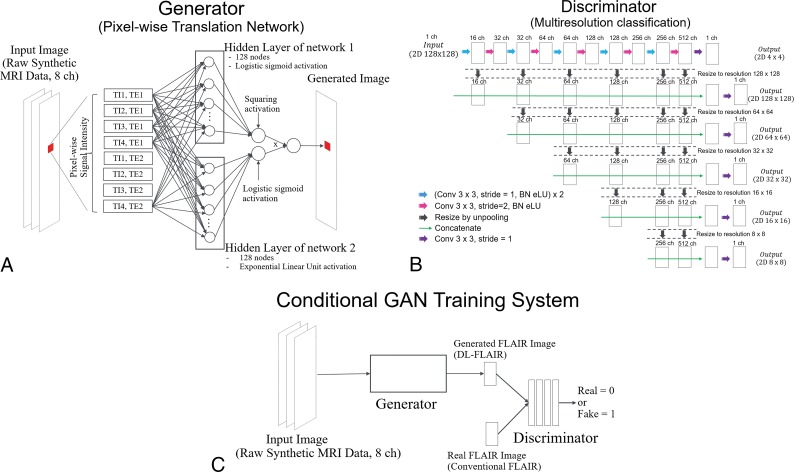
We designed a pixel-wise translation network that receives raw synthetic MR imaging data and outputs FLAIR images by translating the input signal intensities into FLAIR images pixel by pixel using the same weight function across all pixels. Because some of the raw data information may have been lost when creating the synthetic FLAIR images using previous Bloch simulation-based algorithms,2 we supposed that using the raw data as the DL input would improve the synthetic FLAIR image quality. Figure 1A shows the precise architecture of the generator model. The image generator used herein includes 2 parallel fully connected neural network streamlines that accept the same input from their common former layer. Each of the parallel networks outputs 1 value; then, these are multiplied by each other to rejoin the divided streamlines immediately before the final output. One of the 2 networks contains 1 hidden layer consisting of 128 nodes with logistic sigmoid activation and 1 output node with squaring activation to ensure that the output value is non-negative. Squaring activation was chosen here to simulate FLAIR contrast that usually takes the absolute signal values from the acquired data while keeping the differential coefficients continuous. However, this network generated T2-weighted-like images rather than FLAIR-like images. Hence, to suppress the signals of CSF, we applied 1 more network that contains 1 hidden layer consisting of 128 nodes with exponential linear unit activation and 1 output node with logistic sigmoid activation to ensure that the output value is between 0 and 1. We used the exponential linear unit here rather than the rectified linear unit because the exponential linear unit is less nonlinear and the learning process may become more stable.21
Fig 1.
A, Illustration describing the generator. B, Illustration describing the discriminator. C, The framework describing the training phase of our proposed cGAN model for improving the synthetic FLAIR image quality. BN indicates batch normalization; Conv, convolution; eLU, exponential linear unit; ch, channel.
The training routine is divided into 2 steps: In the first step, the generator produces coarse FLAIR images, which allow the next GAN optimization to start with a condition that is relatively close to the convergent state. The training model is also constructed in this step to minimize the mean absolute error compared with the corresponding synthetic FLAIR images. These synthetic FLAIR images were preprocessed before this step with additional signal suppression of some specific areas, namely part of the CSF, to avoid misleading the generator to create hyperintense signals on the brain surface, a known major artifact in synthetic FLAIR images. Specifically, we roughly estimated the apparent inversion recovery time and T2 signal intensity of each voxel and then manually modified the original synthetic FLAIR pixel value to zero where the inversion recovery time was >0.3 and the T2 signal intensity was >0.7. We estimated the apparent inversion recovery time by subtracting the signal intensity ratio of the fifth-to-seventh raw data images from 1. Similarly, the apparent T2 signal intensity was estimated by subtracting the signal intensity ratio of the eighth-to-seventh raw data images from 1. Although this estimation process was rough, it was sufficient to achieve the first step because miscalibration and suppression of signals in other areas similar to those of CSF, if any, are supposed to be corrected in the GAN training step 2. During the training step 1, fifteen patches (128 × 128 pixels) were randomly selected from the synthetic MR imaging raw data (512 × 512 pixels) as a batch, and the model was trained batch-wise for 100,000 iterations with updating of the generator by the Adam22 rule with α = 0.0005, β1 = 0.9, and β2 = 0.999. Simultaneously, for efficiency, the discriminator used in step 2 was pretrained by receiving generated and target images of the same size and updated by the Adam rule with α = 0.000001, β1 = 0.9, and β2 = 0.999. Figure 1B shows the precise architecture of the discriminator.
In the second phase of the 3 training steps, we set up a cGAN training strategy with patches. Figure 1C shows a conceptual description of the cGAN training system. We set the pretrained discriminator as the initial state and continued the training to allow it to discriminate generated images and conventional true FLAIR images while also updating the generator. From the synthetic MR imaging raw data, 15 patches were randomly selected as a batch and fed into generator to yield FLAIR images. The generated images were again fed into the discriminator, which received target images (conventional FLAIR patches) and minimized the classification loss. Classification loss was determined as the average of C(D) − C(1 −D), where C is the cross-entropy item and D is the logistic sigmoid output of the discriminator. This averaging operation was performed along both batches and pixels to normalize the loss. From the discriminator, the generator received the negative of the classification loss for its generated images. To stabilize the training, the discriminator was designed to return the classification results pixel by pixel in different resolutions. We used multiresolution classification that is similar to the structure of U-Net (https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/), which has been mainly used for semantic segmentation.23 With this segmentation task-like architecture of the discriminator, the mode collapse phenomenon that frequently occurs in GAN training24 was not observed in this study.
Although several techniques for stabilizing GAN training have been discussed in the field of machine learning,25–27 the discriminator architecture devised herein for stabilizing GAN training has not been reported previously. In this study, we used the same weight across different pixels and resolutions. For training, 15 patches (128 × 128 pixels) were randomly selected from the synthetic MR imaging raw data (512 × 512 pixels) as a batch and 100,000 iterations were performed with updating of the generator by the Adam rule in which α = 0.00001, β1 = 0.5, and β2 = 0.999 and updating of the discriminator by the Adam rule where α = 0.000001, β1 = 0.9, and β2 = 0.999. All model training was performed on a computer with 64 GB of CPU memory, Xeon E5–2670 v3 CPU (Intel), and a TITAN Xp graphics processing unit (NVIDIA, Santa Clara, California). The computer program was coded with Python 3.6 (https://www.python.org/downloads/release/python-360/) and the DL framework of Chainer 3.2.0 (http://chainer.org/). The FLAIR images generated by the proposed DL scheme are hereafter denoted as DL-FLAIR images.
Evaluation of the Model
Quantitative Evaluation.
To quantitatively compare the synthetic and DL-FLAIR image quality, we used the peak signal-to-noise ratio (PSNR), normalized root mean square error (NRMSE), and lesion maps derived from these images. Conventional FLAIR images were registered to synthetic FLAIR images using the FMRIB Software Library (FSL; http://www.fmrib.ox.ac.uk/fsl).28 First, mean square error maps were calculated for synthetic and DL-FLAIR images against registered conventional FLAIR images, and the PSNR was defined as:
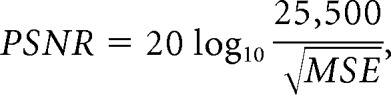 |
where 25,500 is the maximum range of the FLAIR signal intensity. Next the squared root of the mean square errors was calculated and scaled by dividing by the mean signal intensity of synthetic and DL-FLAIR images, respectively, after applying the intracranial masks, to produce NRMSE maps. Notably, images with a higher PSNR and lower NRMSE theoretically exhibit higher image quality.29 Synthetic T1-weighted images, which were inherently aligned to synthetic FLAIR images, were skull-stripped by the intracranial masks and segmented into GM, WM, and CSF using the FMRIB Automated Segmentation Tool (FAST; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/fast). No gross error was visually observed in this segmentation step. Each tissue map and intracranial mask were used to extract the PSNR and NRMSE metrics in the GM, WM, CSF, and aggregate intracranial tissues.
Conventional FLAIR images were linearly coregistered to synthetic FLAIR images using SPM 12 software (http://www.fil.ion.ucl.ac.uk/spm). The WM lesions on these images were automatically segmented using a lesion-prediction algorithm30 in the Lesion Segmentation Toolbox, Version 2.0.15 (http://www.applied-statistics.de/lst.html),31 running under SPM 12. Lesion maps were manually corrected by an experienced neuroradiologist (A.H.) with 7 years of experience. The Dice index, which represents the percentage of overlap between 2 image sets, of lesion maps was calculated between synthetic or DL-FLAIR images and conventional FLAIR images.
Qualitative Evaluation.
We screened the training and test DL-FLAIR image sets for artificial object creation or large signal dropouts and confirmed that no DL-FLAIR image had these artifacts. Image quality was visually assessed by an experienced neuroradiologist (C.A.) with 8 years of experience. Synthetic and DL-FLAIR images were assessed in random order. Lesion conspicuity in each patient was rated on a 5-point scale (1, very bad; 2, bad; 3, acceptable; 4, good; and 5, excellent). The existence of artifacts (surface hyperintensity, granular artifacts, and other artifacts that substantially degrade image quality) was also rated as follows: 1, none; 2, minimal; 3, moderate; 4, remarkable; and 5, highly remarkable. Because parenchymal swelling artifacts were difficult to evaluate on separate images, synthetic and DL-FLAIR images for each patient were simultaneously shown to the neuroradiologist (C.A.) for further evaluation using the overlay function of the FSLeyes viewer (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLeyes).
Statistical Analysis
Statistics were computed using R (version 3.2.1; http://www.r-project.org/). Because not all the datasets were normally distributed when analyzed by the Shapiro-Wilk test, we used the nonparametric Wilcoxon signed rank test to compare the quantitative and qualitative scores between synthetic and DL-FLAIR images. The PSNR and NRMSE values were also compared among the GM, WM, and CSF on synthetic or DL-FLAIR images. Significance was set at P < .05 (2-sided). Multiple comparison correction was not performed.
Results
Figure 2 shows images from a representative patient. The Table shows the PSNR and NRMSE of synthetic and DL-FLAIR images calculated against conventional FLAIR images. The PSNR was significantly higher for DL-FLAIR than synthetic FLAIR images in GM, WM, CSF, and aggregate intracranial tissues (all P < .001). The PSNR of synthetic and DL-FLAIR images was the lowest in GM, with WM showing lower values than CSF (all P < .001). The NRMSE was significantly lower for DL-FLAIR than synthetic FLAIR images in GM, WM, CSF, and aggregate intracranial tissues (all P < .001). The NRMSE of synthetic and DL-FLAIR images was the highest in GM, with WM showing higher values than CSF (all P < .001). The mean Dice index of lesion maps against conventional FLAIR images was comparable between DL-FLAIR (0.57 ± 0.17) and synthetic FLAIR images (0.55 ± 0.14) (P = 1).
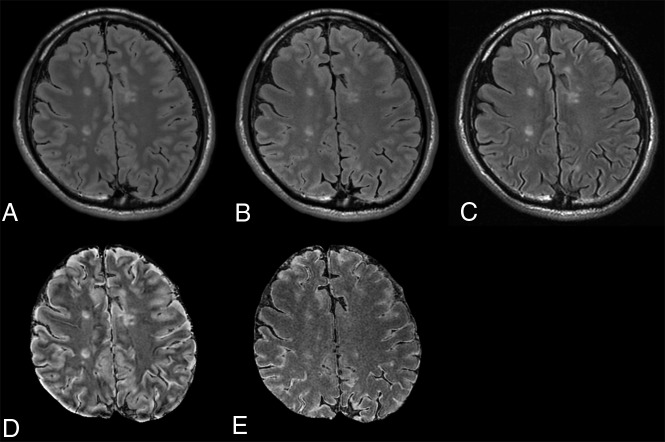
Fig 2.
Synthetic FLAIR (A), DL-FLAIR (B), and conventional FLAIR (C) images of a representative patient. The overall image contrast of the DL-FLAIR image is more similar to that of the conventional FLAIR image than the contrast of the synthetic FLAIR image, while preserving the lesion contrast. The NRMSE maps of synthetic FLAIR (D) and DL-FLAIR (E) images against conventional FLAIR images are also shown. The NRMSE in the intracranial tissues is much larger in the synthetic FLAIR image than in the DL-FLAIR image. Note that the parenchymal surface shows patchy high NRMSE values on the synthetic FLAIR image, which are reduced but still visible in the DL-FLAIR image.
The PSNR and NRMSE of synthetic FLAIR and DL-FLAIR images against conventional FLAIR images in various regionsa
| Synthetic FLAIR Images | DL-FLAIR Images | |
|---|---|---|
| GM | ||
| PSNR | 27.16 ± 0.54b,c | 34.03 ± 1.33b,c (+25.31%) |
| NRMSE | 0.47 ± 0.039b,c | 0.33 ± 0.028b,c (−30.44%) |
| WM | ||
| PSNR | 29.51 ± 0.44b,c | 35.45 ± 1.48b,c (+20.14%) |
| NRMSE | 0.37 ± 0.025b,c | 0.27 ± 0.024b,c (−27.32%) |
| CSF | ||
| PSNR | 34.83 ± 1.37b,c | 40.47 ± 1.37b,c (+16.21%) |
| NRMSE | 0.29 ± 0.027b,c | 0.21 ± 0.017b,c (−27.36%) |
| Aggregate intracranial tissues | ||
| PSNR | 29.79 ± 0.71b | 35.90 ± 1.21b (+20.54%) |
| NRMSE | 0.38 ± 0.024b | 0.27 ± 0.016b (−28.66%) |
Values are means ± SD. Percentage changes in the PSNR and NRMSE for DL-FLAIR vs synthetic FLAIR images are in parentheses.
P < .001 for synthetic FLAIR vs DL-FLAIR images.
P < .001 for GM vs WM, GM vs CSF, and WM vs CSF.
No significant differences in lesion conspicuity, existence of surface hyperintensity artifacts, or the presence of other artifacts were identified between synthetic FLAIR and DL-FLAIR images (3.8 ± 0.40 versus 3.7 ± 0.46, P = .59; 3 ± 0.77 versus 3.1 ± 1.04, P = .78; and 1 ± 0 versus 1 ± 0; P = 1, respectively). However, fewer granular artifacts were present in the CSF of DL-FLAIR (2.3 ± 0.90) than synthetic FLAIR (4 ± 1.1) images (P = .003; Fig 3). During the simultaneous evaluation of synthetic and DL-FLAIR images, the neuroradiologist agreed that though hyperintensity artifacts were still visible in some parts of the brain surface on DL-FLAIR images, the brain parenchyma looked grossly swollen on all synthetic FLAIR images compared with the DL-FLAIR images (Fig 4).
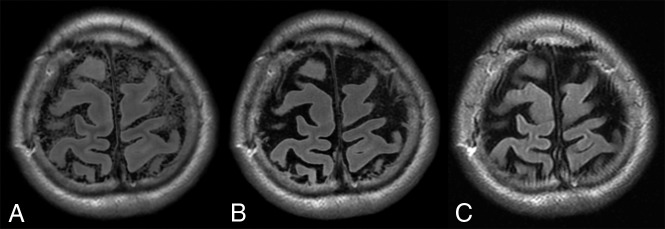
Fig 3.
A, Granular artifacts in the CSF on a synthetic FLAIR image. B, The artifact is almost invisible (successfully deleted) on the DL-FLAIR image. C, Conventional FLAIR image is shown for comparison.
Fig 4.
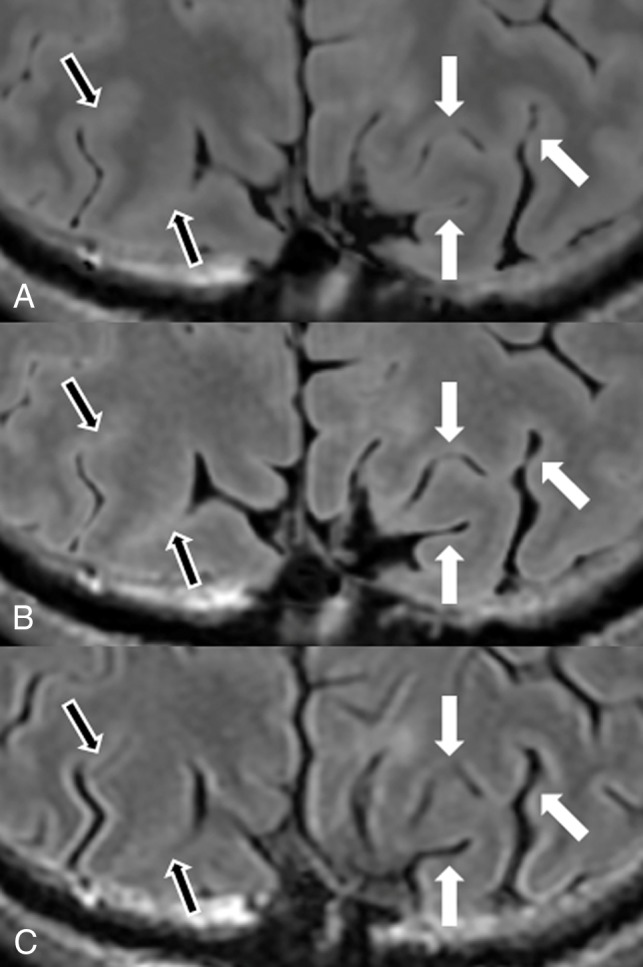
Magnified images from Fig 2. Synthetic FLAIR (A), DL-FLAIR (B), and conventional FLAIR (C) images are shown. Sulci are wider in B and C in some areas than they are in A (white arrows). However, for areas with tight sulci on the conventional FLAIR image (C), the sulci are tighter and more hyperintense on both synthetic FLAIR (A) and DL-FLAIR (B) images than they are on the conventional FLAIR image (black arrows).
Discussion
To our knowledge, our study is the first to use a DL algorithm to improve the synthetic FLAIR image quality. Here, the PSNR and NRMSE were higher and lower, respectively, in all tissue segments in DL-FLAIR than in synthetic FLAIR images, meaning that DL-FLAIR images more accurately replicated conventional true FLAIR images. Although synthetic FLAIR images rely on the well-established Bloch equation,2,32 this approach may not be the best for replicating true FLAIR images.
In our study, lesion delineation was comparable between synthetic and DL-FLAIR images in patients with MS. Hence, the proposed DL scheme might enable reliable delineation of parenchymal lesions. However, this should be confirmed by a future study with a systematic reading session for various diseases including brain tumors or encephalitis.
Herein, while some perceived surface hyperintensity remained, brain parenchymal swelling artifacts were improved in all patients. Hagiwara et al7 reported that 3 false-positives and 157 true MS plaques were detected by an experienced neuroradiologist on synthetic MR images. Because these false-positives were located in the sulci, our proposed model may decrease the number of false-positives in clinical practice. Although it has not been previously discussed, these artifacts may derive from the unavoidable partial volume effects at tissue boundaries, considering that the NRMSE on synthetic FLAIR images in our study was the highest in the GM compared with the WM and CSF. However, the current synthetic MR imaging technique adopted a monoexponential decay model assuming a homogeneous voxel2 and may not appropriately produce the FLAIR signal in a voxel with >1 tissue compartment. While our proposed DL model may have succeeded in partially solving the multiexponential signal decay at tissue boundaries, applying the single function to all voxels may not be ideal, especially for voxels with different tissue mixtures. Future studies should focus on using different weight functions for different tissue mixtures in a voxel using DL.
In the current study, granular artifacts in the CSF were minimized by the DL model. Although the exact origin of such artifacts remains unknown, Tanenbaum et al6 reported that they were observed in up to 59.2% of synthetic FLAIR images and typically disappeared on rescanning. The DL model proposed herein can reduce the number of rescans to avoid these artifacts. However, when the possibility of CSF pathology is minimal on the basis of clinical history, one may just ignore these granular artifacts. While prior studies confirmed the high repeatability of quantitative values derived from quantitative synthetic MR imaging,3,33 the evaluated value ranges mainly focused on brain tissues; hence, quantitative values in the CSF range may not be precisely measured by quantitative synthetic MR imaging, resulting in granular artifacts in the CSF on synthetic FLAIR images.
Our GAN training approach using a pixel-wise simple neural network as a generator was successful in this study. Our model did not generate obvious artificial objects because its architecture ensured that the 2 identical pixel values in the input became the same output pixel values. This property is essential for medical imaging because it will not mislead clinicians. An important contribution of our study is the implementation of such a structurally restricted generator trained by a GAN. Recently, a pixel-wise neural network translation model was used to approximate the dictionary in MR fingerprinting,34 and the training method used the same acquired data for the input and target, allowing it to learn the time-consuming conventional reconstruction method of MR fingerprinting by DL. Because we aimed to train the generator to create FLAIR images resembling conventional FLAIR images from raw synthetic MR imaging data, positional differences between the input and target data were unavoidable. The 2 approaches for overcoming positional difference were to use a GAN based on patches and to calibrate the positional deviation and take the absolute error as a loss.
In the first approach based on patches, adversarial loss represents the complex reality expressions of generated patches by comparing them with patches of target images (conventional true FLAIR images herein) rather than pixel-wise similarity between generated images and target images. Thus, the GAN trains a generator to create visually realistic images while ignoring the effects of positional differences between input and target images. Conversely, the latter method to take error as a loss reportedly leads a generator to create unrealistic blurry images.35 Therefore, we adopted a GAN approach based on patches. Because there was an imbalance of power in the competing generator and discriminator owing to fewer parameters in the generator without a convolutional architecture, the training required various stabilization measures, including learning-rate adjustments, guidance to the initial state by a mean absolute error loss function with synthetic FLAIR images, and segmentation-like discriminator architecture. To our knowledge, this study is the first to train a generator using a GAN without down/up sampling operations.
The present study has some limitations. First, we did not explicitly compare the qualities of DL-FLAIR and conventional FLAIR images. Additional studies are required to investigate whether DL-FLAIR images with further image-quality improvement can serve as substitutes for conventional FLAIR images in clinical settings. Second, data from only 1 scanner were used in this study. Hence, future studies should assess the generalizability of our results using more patients and several scanners. Third, we did not perform a reading study to count the number of MS plaques. This was due to broad diffusely abnormal white matter and/or fused plaques in some of our patient data. Fourth, only 30 cases were used for training the DL algorithm used in this study. Considering the large number of pixels per case, we thought that 30 scans could substantially cover the possible signal distributions of the synthetic MR imaging raw data. However, this assumption should be confirmed by checking the signal distributions of large datasets. In the future, the products of the DL algorithm in our study should also be applied to and validated with disease processes spanning various appearances and tissue compartments.
Conclusions
We successfully improved the synthetic FLAIR image quality using DL, thus creating FLAIR images that have contrast similar to that of conventional FLAIR images, with fewer swelling artifacts and minimal granular artifacts in the CSF, while preserving lesion contrast. The proposed DL algorithm may facilitate the introduction of synthetic MR imaging into clinical practice.
ABBREVIATIONS:
- cGAN
conditional generative adversarial network
- DL
deep learning
- GAN
generative adversarial network
- NRMSE
normalized root mean square error
- PSNR
peak signal-to-noise ratio
Footnotes
Disclosures: Akifumi Hagiwara—RELATED: Grant: Japan Society for the Promotion of Science KAKENHI, ABiS, Japanese Society for Magnetic Resonance in Medicine, Impulsing Paradigm Change through Disruptive Technologies, Agency for Medical Research and Development, UNRELATED: Payment for Lectures Including Service on Speakers Bureaus: GE Healthcare, Comments: luncheon seminar. Yujiro Otsuka—UNRELATED: Employment: Milliman Inc, Comments: I am receiving a salary as an employee. Yasuhiko Tachibana—UNRELATED: Employment: National Institute of Radiological Sciences, QST; Grants/Grants Pending: Grant-in-Aid for Scientific Research, from the Japan Society for the Promotion of Science, Comments: KAKENHI No. 17K10385. This grant is for a research in the same field as this article but is unrelated to the current study. Koji Kamagata—UNRELATED: Grants/Grants Pending: Japan Society for the Promotion of Science KAKENHI (JP16K19854). Shigeki Aoki—RELATED: Grant: Japan Society for the Promotion of Science KAKENHI, ABiS, Japanese Society for Magnetic Resonance in Medicine, Impulsing Paradigm Change through Disruptive Technologies, Agency for Medical Research and Development, UNRELATED: Grants/Grants Pending: Nihon Medi-Physics, Toshiba, Bayer Yakuhin, Daiichi Sankyo, Eisai, Fujiyakuhin, FUJIFILM RI Pharma, Toshiba, Canon, Japan Society for the Promotion of Science KAKENHI, ABiS, Japanese Society for Magnetic Resonance in Medicine, Impulsing Paradigm Change through Disruptive Technologies, Agency for Medical Research and Development, Payment for Lectures Including Service on Speakers Bureaus: honorarium for lectures/chair from GE Healthcare, Toshiba, Hitachi, Siemens, Bayer Yakuhin, Daiichi Sankyo, Eisai, Fujiyakuhin, FUJIFILM RI Pharma, Nihon Medi-Physics, Meiji Seika Pharma, Canon, Guerbet. Mariko Takemura—RELATED: Grant: Japan Society for the Promotion of Science KAKENHI, Comments: grant No. 16K10327.
This work was supported by Japan Society for the Promotion of Science KAKENHI grant No. 16K19852; grant No. 16K10327; grant No. JP16H06280, Grant-in-Aid for Scientific Research on Innovative Areas, resource and technical support platforms for promoting research “Advanced Bioimaging Support”; the Japanese Society for Magnetic Resonance in Medicine; the Impulsing Paradigm Change through Disruptive Technologies (ImPACT) Program of the Council for Science, Technology and Innovation (cabinet office, Government of Japan); the program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from the Japan Agency for Medical Research and Development (AMED); and AMED under grant number JP18lk1010025.
References
- 1. Bobman SA, Riederer SJ, Lee JN, et al. Cerebral magnetic resonance image synthesis. AJNR Am J Neuroradiol 1985;6:265–69 [PMC free article] [PubMed] [Google Scholar]
- 2. Warntjes JB, Leinhard OD, West J, et al. Rapid magnetic resonance quantification on the brain: optimization for clinical usage. Magn Reson Med 2008;60:320–29 10.1002/mrm.21635 [DOI] [PubMed] [Google Scholar]
- 3. Hagiwara A, Hori M, Cohen-Adad J, et al. Linearity, bias, intrascanner repeatability, and interscanner reproducibility of quantitative multidynamic multiecho sequence for rapid simultaneous relaxometry at 3 T: a validation study with a standardized phantom and healthy controls. Invest Radiol 2019;54:39–47 10.1097/RLI.0000000000000510 [DOI] [PubMed] [Google Scholar]
- 4. Blystad I, Warntjes JB, Smedby O, et al. Synthetic MRI of the brain in a clinical setting. Acta Radiol 2012;53:1158–63 10.1258/ar.2012.120195 [DOI] [PubMed] [Google Scholar]
- 5. Granberg T, Uppman M, Hashim F, et al. Clinical feasibility of synthetic MRI in multiple sclerosis: a diagnostic and volumetric validation study. AJNR Am J Neuroradiol 2016;37:1023–29 10.3174/ajnr.A4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tanenbaum LN, Tsiouris AJ, Johnson AN, et al. Synthetic MRI for clinical neuroimaging: results of the Magnetic Resonance Image Compilation (MAGiC) prospective, multicenter, multireader trial. AJNR Am J Neuroradiol 2017;38:1103–10 10.3174/ajnr.A5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hagiwara A, Hori M, Yokoyama K, et al. Synthetic MRI in the detection of multiple sclerosis plaques. AJNR Am J Neuroradiol 2017;38:257–63 10.3174/ajnr.A5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagiwara A, Hori M, Yokoyama K, et al. Analysis of white matter damage in patients with multiple sclerosis via a novel in vivo magnetic resonance method for measuring myelin, axons, and g-ratio. AJNR Am J Neuroradiol 2017;38:1934–40 10.3174/ajnr.A5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andica C, Hagiwara A, Nakazawa M, et al. Synthetic MR imaging in the diagnosis of bacterial meningitis. Magn Reson Med Sci 2017;16:91–92 10.2463/mrms.ci.2016-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wallaert L, Hagiwara A, Andica C, et al. The advantage of synthetic MRI for the visualization of anterior temporal pole lesions on double inversion recovery (DIR), phase-sensitive inversion recovery (PSIR), and myelin images in a patient with CADASIL. Magn Reson Med Sci 2018;17:275–76 10.2463/mrms.ci.2017-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helmersson T. Evaluation of Synthetic MRI for Clinical Use [master's thesis]. Department of Medical and Health SciencesAccessed August 30, 2018:71 Linköping: Linköping University; 2010. http://liu.diva-portal.org/smash/record.jsf?pid=diva2%3A389732&dswid=9594. Accessed August 30, 2018:71. [Google Scholar]
- 12. Yasaka K, Akai H, Kunimatsu A, et al. Deep learning with convolutional neural network in radiology. Jpn J Radiol 2018;36:257–72 10.1007/s11604-018-0726-3 [DOI] [PubMed] [Google Scholar]
- 13. Nakao T, Hanaoka S, Nomura Y, et al. Deep neural network-based computer-assisted detection of cerebral aneurysms in MR angiography. J Magn Reson Imaging 2018;47:948–53 10.1002/jmri.25842 [DOI] [PubMed] [Google Scholar]
- 14. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: Proceedings of Advances in Neural Information Processing Systems; 2014:2672–80 [Google Scholar]
- 15. Wolterink JM, Leiner T, Viergever MA, et al. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging 2017;36:2536–45 10.1109/TMI.2017.2708987 [DOI] [PubMed] [Google Scholar]
- 16. Kim KH, Do WJ, Park SH. Improving resolution of MR images with an adversarial network incorporating images with different contrast. Med Phys 2018;45:3120–31 10.1002/mp.12945 [DOI] [PubMed] [Google Scholar]
- 17. Isola P, Zhu J, Zhou T, et al. Image-to-image translation with conditional adversarial networks. arXiv:161107004Accessed August 13, 2018 2017. http://adsabs.harvard.edu/abs/2016arXiv161107004I. Accessed August 13, 2018.
- 18. Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302 10.1002/ana.22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagiwara A, Warntjes M, Hori M, et al. SyMRI of the brain: rapid quantification of relaxation rates and proton density, with synthetic MRI, automatic brain segmentation, and myelin measurement. Invest Radiol 2017;52:647–57 10.1097/RLI.0000000000000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ambarki K, Lindqvist T, Wählin A, et al. Evaluation of automatic measurement of the intracranial volume based on quantitative MR imaging. AJNR Am J Neuroradiol 2012;33:1951–56 10.3174/ajnr.A3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clevert D, Unterthiner T, Hochreiter S. Fast and accurate deep network learning by exponential linear units (ELUs). arXiv:151107289Accessed October 5, 2018 2015. http://adsabs.harvard.edu/abs/2015arXiv151107289C. Accessed October 5, 2018.
- 22. Kingma D, Ba J. Adam: a method for stochastic optimization. arXiv:14126980Accessed August 22, 2018 2014. https://arxiv.org/abs/1412.6980. Accessed August 22, 2018.
- 23. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. arXiv:150504597Accessed October 6, 2018 2015. http://adsabs.harvard.edu/abs/2015arXiv150504597R. Accessed October 6, 2018.
- 24. Lin G, Khetan G, Fanti G, et al. PacGAN: the power of two samples in generative adversarial networks. arXiv:171204086Accessed August 23, 2018 2017. https://arxiv.org/abs/1712.04086. Accessed August 23, 2018.
- 25. Salimans T, Goodfellow I, Zaremba W, et al. Improved techniques for training GANs. arXiv:160603498Accessed August 28, 2018 2016. http://adsabs.harvard.edu/abs/2016arXiv160603498S. Accessed August 28, 2018.
- 26. Huszár F. How (not) to train your generative model: scheduled sampling, likelihood, adversary? arXiv:151105101Accessed August 28, 2018 2015. http://adsabs.harvard.edu/abs/2015arXiv151105101H. Accessed August 28, 2018.
- 27. Arjovsky M, Bottou L. Towards principled methods for training generative adversarial networks. arXiv:170104862Accessed August 28, 2018 2017. http://adsabs.harvard.edu/abs/2017arXiv170104862A. Accessed August 28, 2018.
- 28. Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. Neuroimage 2012;62:782–90 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y, Yu B, Wang L, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage 2018;174:550–62 10.1016/j.neuroimage.2018.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Egger C, Opfer R, Wang C, et al. MRI FLAIR lesion segmentation in multiple sclerosis: does automated segmentation hold up with manual annotation? Neuroimage Clin 2017;13:264–70 10.1016/j.nicl.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt P, Gaser C, Arsic M, et al. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 2012;59:3774–83 10.1016/j.neuroimage.2011.11.032 [DOI] [PubMed] [Google Scholar]
- 32. Maitra R, Riddles JJ. Synthetic magnetic resonance imaging revisited. IEEE Trans Med Imaging 2010;29:895–902 10.1109/TMI.2009.2039487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krauss W, Gunnarsson M, Andersson T, et al. Accuracy and reproducibility of a quantitative magnetic resonance imaging method for concurrent measurements of tissue relaxation times and proton density. Magn Reson Imaging 2015;33:584–91 10.1016/j.mri.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 34. Cohen O, Zhu B, Rosen MS. MR fingerprinting Deep RecOnstruction NEtwork (DRONE). Magn Reson Med 2018;80:885–94 10.1002/mrm.27198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Welander P, Karlsson S, Eklund A. Generative adversarial networks for image-to-image translation on multi-contrast MR images: a comparison of CycleGAN and UNIT. arXiv:180607777Accessed August 28, 2018 2018. https://arxiv.org/abs/1806.07777. Accessed August 28, 2018.