One hundred two IDH-mutant lower grade gliomas with preoperative MR imaging and known 1p/19q status from The Cancer Genome Atlas composed a training dataset. Two neuroradiologists in consensus analyzed the training dataset for various imaging features: tumor or cyst texture, margins, cortical infiltration, T2-FLAIR mismatch, tumor cyst, T2* susceptibility, hydrocephalus, midline shift, maximum dimension, primary lobe, necrosis, enhancement, edema, and gliomatosis. Statistical analysis of the training data produced a multivariate classification model for codeletion prediction based on a subset of MR imaging features and patient age. Training dataset analysis produced a 2-step classification algorithm with 86.3% codeletion prediction accuracy, based on the following: 1) the presence of the T2-FLAIR mismatch sign, which was 100% predictive of noncodeleted lowergrade gliomas; and 2)a logistic regression model based on texture, patient age, T2* susceptibility, primary lobe, and hydrocephalus. Independent validation ofthe classification algorithm rendered codeletion prediction accuracies of 81.1% and 79.2% in 2 independent readers.
Abstract
BACKGROUND AND PURPOSE:
Isocitrate dehydrogenase (IDH)-mutant lower grade gliomas are classified as oligodendrogliomas or diffuse astrocytomas based on 1p/19q-codeletion status. We aimed to test and validate neuroradiologists' performances in predicting the codeletion status of IDH-mutant lower grade gliomas based on simple neuroimaging metrics.
MATERIALS AND METHODS:
One hundred two IDH-mutant lower grade gliomas with preoperative MR imaging and known 1p/19q status from The Cancer Genome Atlas composed a training dataset. Two neuroradiologists in consensus analyzed the training dataset for various imaging features: tumor texture, margins, cortical infiltration, T2-FLAIR mismatch, tumor cyst, T2* susceptibility, hydrocephalus, midline shift, maximum dimension, primary lobe, necrosis, enhancement, edema, and gliomatosis. Statistical analysis of the training data produced a multivariate classification model for codeletion prediction based on a subset of MR imaging features and patient age. To validate the classification model, 2 different independent neuroradiologists analyzed a separate cohort of 106 institutional IDH-mutant lower grade gliomas.
RESULTS:
Training dataset analysis produced a 2-step classification algorithm with 86.3% codeletion prediction accuracy, based on the following: 1) the presence of the T2-FLAIR mismatch sign, which was 100% predictive of noncodeleted lower grade gliomas, (n = 21); and 2) a logistic regression model based on texture, patient age, T2* susceptibility, primary lobe, and hydrocephalus. Independent validation of the classification algorithm rendered codeletion prediction accuracies of 81.1% and 79.2% in 2 independent readers. The metrics used in the algorithm were associated with moderate-substantial interreader agreement (κ = 0.56–0.79).
CONCLUSIONS:
We have validated a classification algorithm based on simple, reproducible neuroimaging metrics and patient age that demonstrates a moderate prediction accuracy of 1p/19q-codeletion status among IDH-mutant lower grade gliomas.
The revised World Health Organization (WHO) 2016 classification of diffuse gliomas integrates isocitrate dehydrogenase (IDH) gene status and whole-arm codeletion of chromosome arms 1p and 19q with histologic findings to classify grades II and III diffuse lower grade gliomas (LGGs).1,2 More than 80% of LGGs are IDH-mutant; of those, 37%–50% carry the 1p/19q codeletion.3,4 The 1p/19q-codeleted IDH-mutant LGGs (oligodendrogliomas; IDHmut-Codel) show better overall survival compared with noncodeleted IDH-mutant LGGs (astrocytomas; IDHmut-Noncodel) and are more sensitive to adjuvant chemotherapy with procarbazine, lomustine, and vincristine.5–7
The integration of genomic data in the updated WHO classification of LGGs has accelerated efforts to noninvasively predict genetic signatures of diffuse gliomas using neuroimaging techniques. While numerous studies have identified neuroimaging features that correlate with 1p/19q-codeletion status in LGG subtypes,8–28 many were performed before the 2016 WHO update, affecting their patient-selection process (ie, no accounting for IDH status). Moreover, these studies have applied variable imaging metrics and neuroimaging analysis, rendering it difficult to assess the relative and combined diagnostic performance of the various neuroimaging metrics reported to be associated with 1p/19q-codeletion status. Finally, simple metrics extrinsic to the glioma (eg, hydrocephalus, midline shift) have not been tested. The purpose of our study was to test and validate the combined accuracy of simple neuroimaging features to predict 1p/19q-codeletion status among cohorts of IDH-mutant LGGs.
Materials and Methods
This was a Health Insurance Portability and Accountability Act–compliant retrospective study conducted with the University of Virginia Health System institutional review board approval.
The study consisted of 2 phases. First, analysis of a training dataset yielded a multivariate classification algorithm for predicting 1p/19q-codeletion status. Second, the classification algorithm was validated using a separate dataset of cases and separate neuroradiologist readers.
Training Dataset Analysis
The cases composing the training dataset were accrued from The Cancer Imaging Archive (TCIA), an LGG on-line data base.29 TCIA data base houses MR imaging data for 199 LGGs, with molecular data (including IDH and 1p/19q statuses) available through The Cancer Genome Atlas (TCGA). The inclusion criteria were the following: 1) LGG with histopathologic assessment and grade, 2) LGG with an IDH mutation, 3) LGG with known 1p/19q-codeletion status, and 4) preoperative MR imaging (or MR imaging after a small-needle biopsy) containing imaging sequences relevant to the below-described neuroimaging classification. IDH wild-type glioma (n = 42), cases with incomplete molecular/pathologic data (n = 2), and cases with insufficient MR imaging data (n = 53) were excluded from the study, rendering 102 IDH-mutant LGGs included in the training dataset.
Two neuroradiologists, with 5 and 13 years of experience, blinded to the 1p/19q-codeletion status, analyzed the MR images from the training dataset in consensus. They measured 14 neuroimaging metrics: 1) primary lobe: yes/no centered on frontal lobe; 2) texture: more or less than 75% of the tumor showing homogeneous signal intensity on T1WI/T2WI; 3) margins: more or less than 75% of the tumor showing sharp/circumscribed margins; 4) T2-FLAIR mismatch sign: the presence or absence of complete/near-complete hyperintense signal on T2WI and relatively hypointense signal on FLAIR except for a hyperintense peripheral rim19; 5) T2* susceptibility blooming: present or absent; 6) contrast enhancement: present or absent; 7) cysts: present or absent; 8) necrosis: present or absent; 9) maximum tumor diameter (centimeter); 10) cortical infiltration: present or absent; 11) peritumoral edema: present or absent; 12) gliomatosis: yes/no involvement of ≥3 lobes; 13) midline shift (centimeter); and 14) hydrocephalus: present or absent. Figures 1 and 2 show the characteristic imaging appearance of IDHmut-Noncodel and IDHmut-Codel LGGs, respectively, including a description of several of the above imaging metrics. Univariate and multivariate logistic regression analysis of the MR imaging characteristics and patient age for predicting the 1p/19q-codeletion status was undertaken. On the basis of these results, a classification algorithm for 1p/19q prediction was developed.
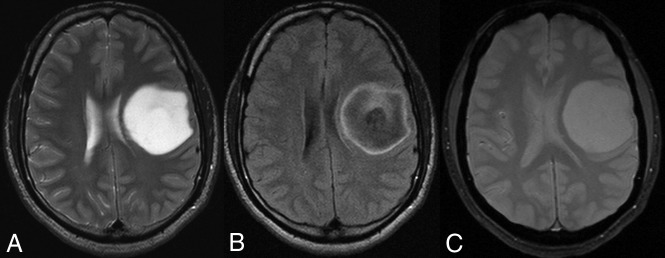
Fig 1.
A 38-year-old man with a left frontal lobe diffuse astrocytoma, IDH-mutant and 1p/19q-noncodeleted, showing characteristic imaging features. A, On T2WI, the mass is homogeneously hyperintense, sharply marginated, and without significant cortical infiltration. B, FLAIR sequence shows central suppression of signal compared with the T2WI, except for a peripheral rim (ie, T2-FLAIR mismatch sign). C, T2*WI shows lack of susceptibility blooming.
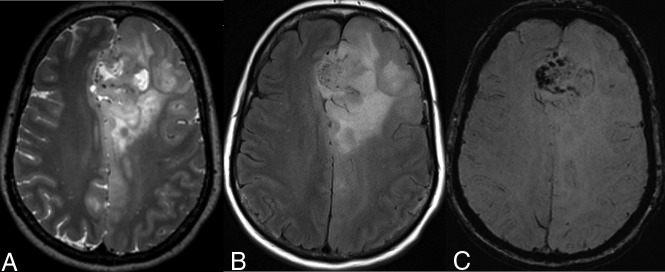
Fig 2.
A 54-year-old woman with a left frontal lobe oligodendroglioma, IDH-mutant and 1p/19q-codeleted, showing characteristic imaging features. A and B, T2WI and FLAIR demonstrate a heterogeneous and poorly marginated mass with significant cortical infiltration and no T2-FLAIR mismatch sign. C, T2*WI shows regions of striking susceptibility blooming.
Validation Analysis
To validate the classification algorithm developed with the training dataset, two new neuroradiologists analyzed a separate institutional cohort of IDH-mutant LGGs. The same selection criteria used for the training dataset were applied, and 106 IDH-mutant LGGs consecutively accrued from an institutional neuro-oncology/neuroradiology data base between 2010 and 2017 composed the validation cohort. The neuroradiologists (reader A with 3 years of experience, reader B with 19 years of experience), blinded to the 1p/19q-codeletion status, independently reviewed the MR images of these cases. The readers analyzed the MR imaging metrics relevant to the classification model with the same criteria used for the training dataset. Interreader agreement for the neuroimaging metrics and independent reader performance in predicting 1p/19q-codeletion status were determined.
Neuropathology
For TCIA/TCGA cohort, histopathologic assignment and molecular classification were derived from supplemental material in Ceccarelli et al,30 in 2016, and included somatic mutation analysis of IDH1 or IDH2 from whole-exome sequencing and codeletion of chromosome arms 1p and 19q from the SNP Array 6.0 (Affymetrix, Santa Clara, California).
For the validation cohort, the IDH and 1p/19q statuses were retrieved from the electronic medical record. Both markers were tested in the Clinical Laboratory Improvement Amendments–certified molecular pathology laboratory at our institution. IDH mutation status was first determined by immunohistochemistry using an IDH1 R132H mutant-specific clinically validated antibody (DIA-H09; Dianova GmbH, Hamburg, Germany).31,32 In immunohistochemistry cases negative for IDH1 R132H mutation, IDH mutation status was assessed by the clinically validated DNA pyrosequencing assay.33 1p/19q status was determined by fluorescence in situ hybridization on paraffin-embedded tissue, using human probes localizing 1p, 1q, 19p, and 19q (Locus Specific Identifier 1p36/1q25 and 19p13/19q13 Dual-Color Probes; Vysis, Downers Grove, Illinois).
Statistical Analysis
The following is an abbreviated description of the statistical methodology; a full description is included in the On-line Appendix.
Training Dataset.
Univariate logistic regression analysis of the 14 MR imaging characteristics and patient age for predicting 1p/19q-codeletion status was undertaken for the TCIA/TCGA–derived training dataset. Because the presence of the T2-FLAIR mismatch sign showed 100% positive predictive value (PPV) for the IDHmut-Noncodel molecular subtype (see Results below), these cases were segregated from the cohort. A multivariate logistic regression using the remaining aforementioned predictor variables was applied in a 2-step analytic process to the remaining cases in the cohort (either negative for the T2-FLAIR mismatch sign or had no T2-FLAIR match sign information available). First, a full model was constructed with the goal of identifying predictor variables that contribute unique information about 1p/19q-codeletion status based on a set of type III Wald χ2 tests at the α = .10 threshold. Second, a reduced model was constructed using only the unique predictors of 1p/19q-codeletion status. The regression equation of the reduced multivariate model was then used to compute the predicted probability for codeletion status, and these predicted probabilities were used to derive a classification algorithm rule. The predicted probability threshold for the classification rule of the algorithm was derived by identifying the predicted probability threshold that produced the largest Youden J statistic (J = Diagnostic Sensitivity + Diagnostic Specificity − 1).34
Validation Dataset.
Interreader agreement for readers A and B was evaluated via the unweighted κ statistic sign. Cases that were deemed by the independent readers to be positive for the T2-FLAIR mismatch sign were included as “true-negatives” (ie, 1p/19q noncodeleted), and the training set–derived reduced multivariate logistic regression model equation was applied to the reader data in cases negative for T2-FLAIR mismatch. Cases could be classified as either 1p/19q codeleted or noncodeleted based on whether the predicted probability was greater than, equal to, or less than the established classification algorithm predicted probability threshold, respectively. The overall diagnostic classification performance was assessed per reader.
Statistical Software.
The statistical software package Spotfire S+, Version 8.2 (TIBCO, Palo Alto, California) was used to conduct the multivariate logistic regression (MLR) analyses, and the pROC package of R (http://www.r-project.org/) was used to conduct the diagnostic classification performance analyses.35,36
Results
Training Dataset Analysis
Of the 102 patients with IDH-mutant LGGs in the training dataset, 51% were women (n = 52) and 49% were men (n = 50). The median age was 41 years (range, 20.0–75.0 years; interquartile range, 33.0–53.0 years). Of the 102 LGGs, 62.7% (n = 64) were IDHmut-Noncodel and 37.3% (n = 38) were IDHmut-Codel, 57.8% (n = 59) were WHO grade II, and 42.2% (n = 43) were WHO grade III.
Univariate Analyses.
Univariate logistic regression analyses showed that several metrics were significantly associated with 1p/19q-codeletion status, including texture (OR, 12.33; 95% CI, 4.66–31.58; P < .001), T2* susceptibility blooming (OR, 6.92; 95% CI, 2.04–23.49; P = .002), T2-FLAIR mismatch sign (OR, 22.50; 95% CI, 6.26-∞; P < .001), location (OR, 5.68; 95% CI, 2.08–15.44; P = .001), midline shift (OR, 4.27; 95% CI, 1.49–12.23; P = .027), and patient age (OR, 3.38; 95% CI, 1.71–6.71; P < .001) (Table 1). Notably, the T2-FLAIR mismatch sign showed PPV = 100% and negative predictive value = 44% for the IDHmut-Noncodel subtype. In 100% of cases (n = 21) in which the T2-FLAIR mismatch sign was present, the glioma was 1p/19q-noncodeleted. Therefore, the cases in which the T2-FLAIR mismatch sign was present were segregated from the cohort, and a multivariate logistic regression analysis was undertaken in the remaining cases (n = 81).
Table 1:
Univariate logistic regression analyses for predicting 1p/19q codeletion among the training dataset
| Predictor | Ratio | Odds Ratio (95% CI) | P Value |
|---|---|---|---|
| Maximum diameter (cm) | 1st Quartile: 3rd quartile | 1.33 (0.70–2.49) | .381 |
| Margins | Irregular: sharp | 1.04 (0.47–2.33) | .917 |
| Texture | Homogeneous (<75%): homogeneous (>75%) | 12.33 (4.66–31.58) | <.001a |
| Peritumoral edema | Yes: no | 1.42 (0.62–3.23) | .973 |
| Hydrocephalus | No: yes | 2.32 (0.88–6.11) | .089 |
| Midline shift (cm)b | 1st Quartile: 3rd quartile | 4.27 (1.49–12.23) | .027a |
| Enhancement | Yes: no | 1.28 (0.57–2.86) | .555 |
| Necrosis | Yes: no | 2.18 (0.61–7.69) | .228 |
| T2* blooming | Yes: no | 6.92 (2.04–23.49) | .002a |
| Cortical infiltration | Yes: no | 2.02 (0.67–6.10) | .212 |
| Cyst | No: yes | 1.18 (0.48–2.91) | .715 |
| T2 FLAIR mismatch signc | No: yes | 22.50 (6.26–∞) | <.001a |
| Gliomatosis | Yes: no | 1.13 (0.18–7.08) | .896 |
| Primary lobe | Frontal: nonfrontal | 5.68 (2.08–15.44) | .001a |
| Age | 3rd Quartile: 1st quartile | 3.38 (1.71–6.71) | <.001a |
| Sex | Female: male | 1.55 (0.69–3.50) | .283 |
Significant.
Analyzed as a restricted cubic spline function of the predictor.
Median unbiased estimate derived with exact logistic regression.
Multivariate Analyses.
On the basis of a full multivariate logistic regression model analysis, tumor texture (P < .001), patient age (P = .010), T2* susceptibility blooming (P = .022), primary lobe (P = .039), and hydrocephalus (P = .052) were determined to be uniquely associated with 1p/19q-codeletion status, and these metrics were used to create a reduced multivariate logistic regression model. A predicted probability threshold of 0.40 resulted in the largest Youden J statistic for the reduced multivariate logistic regression model. Finally, a 2-step classification algorithm was created on the basis of the following: 1) the presence of the T2-FLAIR mismatch sign; and 2) a reduced multivariate logistic regression model, with application of the Youden J statistic–derived predicted probability threshold of 0.40 (Fig 3). The 2-step classification algorithm demonstrated 86.3% accuracy in predicting the 1p/19q-codeletion status among the IDH-mutant LGGs in the training dataset (Table 2).
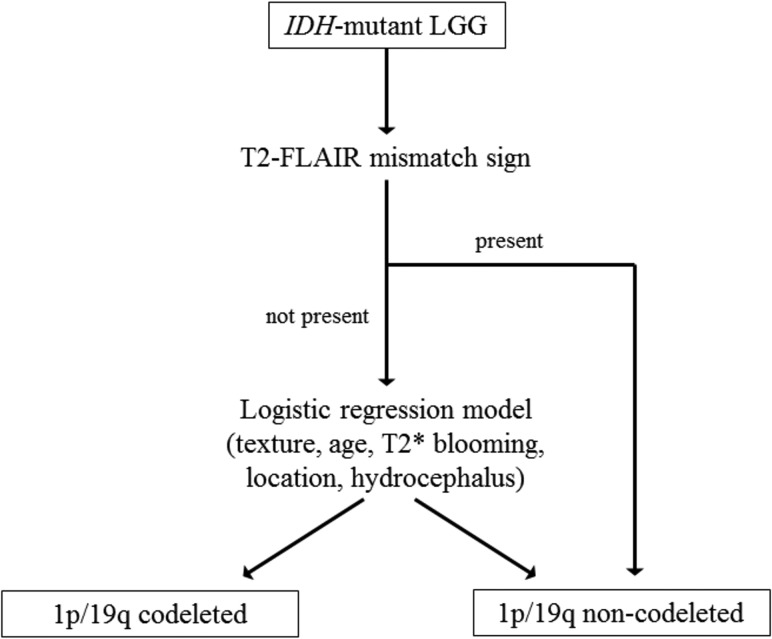
Fig 3.
Classification algorithm for 1p/19q-codeletion status in IDH-mutant LGGs, derived from an analysis of the TCGA/TCIA training cohort. The first step of the algorithm is an assessment of the T2-FLAIR mismatch sign; when present, it indicates, with high certainty, the IDH-mutant 1p/19q-noncodeleted subtype. When the T2-FLAIR mismatch sign is absent (or unavailable), a multivariate logistic regression model based on tumor texture, patient age, T2* blooming, tumor location, and hydrocephalus is applied. The equation for the logistic regression model is the following: ρ codeleted = exp[(Xβ) / (1+exp(Xβ)′], where Xβ = −4.8834 + 2.7842 × (texture <75% homogeneous) + 0.0587 × (patient age) + 3.1948 × (T2* susceptibility blooming present) + 1.6646 × (primary lobe = frontal) − 3.4496 × (hydrocephalus = present). A predicted probability threshold of 0.40 was established for the logistic regression model, based on the Youden J statistic. Cases were classified as either 1p/19q codeleted or non-codeleted based on whether their predicted probability was ≥ or < 0.40, respectively.
Table 2:
Training dataset classification summary for predicting 1p/19q codeletion based on the classification algorithm in Fig 3a
| Classification Algorithm Prediction | Molecular Status |
|
|---|---|---|
| 1p/19q Codeleted | 1p/19q Noncodeleted | |
| 1p/19q Codeleted | 34 | 10 |
| 1p/19q Noncodeleted | 4 | 54 |
Overall prediction accuracy was 86.3%.
Validation Analysis
The validation dataset accrued from our institution was composed of 106 patients with IDH-mutant LGGs, of which 50% were female (n = 53). The median age was 38.5 years (range, 17.0–70.0 years; interquartile range, 32.0–50.8 years). Of the 106 LGGs, 47.2% (n = 50) were IDHmut-Noncodel, 52.8% (n = 56) were IDHmut-Codel, 70.8% (n = 75) were WHO grade II, and 29.2% (n = 31) were WHO grade III.
Readers A and B demonstrated moderate interreader agreement with respect to the T2-FLAIR mismatch sign (κ = 0.56; 95% CI, 0.34–0.77) and substantial interreader agreement with respect to tumor texture (κ = 0.69; 95% CI, 0.56–0.83), T2* blooming (κ = 0.74; 95% CI, 0.58–0.90), primary lobe (κ = 0.79; 95% CI, 0.68–0.89), and hydrocephalus (κ = 0.72; 95% CI, 0.51–0.93).
The performance of the 2-step classification algorithm (Fig 3) was assessed using the independently collected data from readers A and B. Reader A identified the T2-FLAIR mismatch sign in 19 cases; thus, the remaining 87 cases were assessed by applying the reduced logistic regression model based on texture, patient age, T2* susceptibility blooming, primary lobe, and hydrocephalus. Reader B identified the T2-FLAIR mismatch sign in 16 cases, with the remaining 90 cases assessed by applying the reduced logistic regression model. The 2-step classification algorithm for predicting 1p/19q-codeletion status had 81.1% accuracy for reader A and 79.2% accuracy for reader B (Table 3). Notably, the T2-FLAIR mismatch sign demonstrated PPV = 100% for predicting the IDHmut-Noncodel subtype for both independent readers.
Table 3:
Validation dataset classification summary for predicting 1p/19q codeletion with 2 independent readers based on the classification algorithm in Fig 3a
| Reader A Prediction | Molecular Status |
Reader B Prediction | Molecular Status |
||
|---|---|---|---|---|---|
| 1p/19q Codeleted | 1p/19q Noncodeleted | 1p/19q Codeleted | 1p/19q Noncodeleted | ||
| 1p/19q Codeleted | 48 | 12 | 1p/19q Codeleted | 48 | 15 |
| 1p/19q Noncodeleted | 8 | 38 | 1p/19q Noncodeleted | 7 | 36 |
Overall prediction accuracy was 81.1% for reader A and 79.2% for reader B.
Discussion
Prior studies have reported multiple morphologic imaging features in LGGs that were associated with 1p/19q-codeletion status.8–28 IDHmut-Codel LGGs commonly localize to the frontal lobe and typically have indistinct borders, calcification, and tumor heterogeneity. IDHmut-Noncodel LGGs are more typically homogeneous, circumscribed, lack calcification, and more frequently localize to the insula and temporal lobe. Before the 2016 WHO classification update, studies assessing neuroimaging associations with 1p/19q-codeletion status frequently limited their analysis to histologically defined oligodendrogliomas or oligoastrocytomas.8–15 The impact of excluding diffuse astrocytomas from these earlier studies is unknown. Although recent studies have adopted the 2016 WHO classification scheme, many have limited their analyses to select MR imaging sequences, select morphologic imaging features, or single-institution datasets without training/validation methodology.20,23,26,28
Four recent studies have used training/validation methodology for analyzing imaging features associated with 1p/19q-codeletion status.19,24,25,27 Park et al,24 in 2018, analyzed the neuroimaging features of a discovery set of 175 LGGs and a validation set of 40 LGGs and reported that mixed restricted diffusion and pial invasion were associated with 1p/19q codeletion among IDH1-mutant LGGs. Limitations to their methodology included single-institution analysis, lack of T2-FLAIR mismatch sign assessment, and lack of IDH2 testing. Kanazawa et al,27 in 2018, analyzed a discovery cohort (n = 45) and validation cohort (n = 52) of LGGs and found that when at least 3 of the following imaging features were present—calcification, paramagnetic susceptibility, indistinct tumor border, and cystic component—there was >95% specificity for 1p/19q codeletion. Limitations to their methodology included a lack of interreader agreement determination, mostly nonradiologist readers, lack of T2-FLAIR mismatch sign assessment, and overlap between calcification and paramagnetic susceptibility. Patel et al,19 in 2017, assessed LGG MR imaging features in training (n = 125) and validation (n = 60) datasets and were the first to report 100% PPV of the T2-FLAIR mismatch sign for predicting the IDHmut-Noncodel molecular subtype. Limitations to their methodology included the small number of imaging metrics tested (n = 4).
Notably, Broen et al,28 in 2018, confirmed the 100% PPV for the T2-FLAIR mismatch sign in predicting IDH-mutant noncodeleted astrocytomas in a multi-institution cohort of LGGs (n = 154), though they did not use a training/validation methodology. Finally, Lasocki et al,25 in 2018, analyzed the MR imaging features of an LGG cohort comprising 69 patients (n = 10 in the training cohort, n = 59 in the validation cohort). They found 100% PPV of >50% T2-FLAIR mismatch for lack of 1p/19q codeletion and high specificity of calcification for underlying 1p/19q codeletion. Limitations to their methodology included low cohort size (only 10 patients used for training), inclusion of cases with unknown IDH status, and single-institution analysis. None of the above described studies used completely different readers for their training and validation analyses.
In our study, we strictly adhered to the molecular classification of diffuse LGGs defined in the 2016 WHO update and excluded cases without relevant molecular data. We excluded IDH wild-type LGGs because our aim was to determine imaging features associated with each of the 2 subgroups among IDH-mutant LGGs, as defined by their 1p/19q-codeletion status. The TCGA/TCIA data base was selected as our training dataset because it had the highest likelihood for generalizability: Cases were accrued from multiple institutions, MR imaging examinations were performed on a variety of scanners with marked variability in imaging quality, and the molecular data were reliable and comprehensive. To further explore the generalizability and reproducibility of our results, we used a large-validation cohort and completely different readers for the training and validation analyses. Our neuroimaging metrics are simple, mostly binary, and can be easily deduced from routine neuroimaging sequences. Unique to our study, we assessed simple extrinsic morphologic features such as hydrocephalus and midline shift, as well as patient age. Aside from Lasocki et al,25 no prior study assessed the utility of T2-FLAIR mismatch in a multivariate model for predicting 1p/19q codeletion.
Our classification algorithm achieved good accuracy (86.3%) for predicting the codeletion status among the TCGA/TCIA IDH-mutant LGGs, and the validation analysis showed comparable accuracy (81.1% and 79.2% for readers A and B). In addition, we revalidated the high PPV of the T2-FLAIR mismatch sign for predicting the IDHmut-Noncodel subtype (PPV = 100% in both training and validation analyses). We also report a novel association between hydrocephalus and midline shift with codeletion status. We found tumor heterogeneity, frontal lobe location, and T2* susceptibility blooming to be significant predictors of the presence of 1p/19q-codeletion status, concordant with prior studies. However, in contrast to prior studies, tumor margin was not a useful discriminatory feature for determining codeletion status in our study.10,17 This could be partly due to differences in the cohorts chosen for analysis or may reflect the subjective nature of this imaging metric.
Although we did not include IDH wild-type LGGs in our analysis, prediction of IDH status is critical to a neuroimaging-based classification of LGGs. This topic has been extensively studied in recent years, including with the use of conventional neuroimaging metrics,37,38 advanced methods such as MR spectroscopic detection of 2-hydroxyglutarate (an oncometabolite that accumulates in IDH-mutant gliomas),39 and machine learning techniques.40 Our work may complement neuroimaging-based methods for IDH prediction and contribute to a comprehensive prediction of molecular status in LGGs.
Our study has limitations. We followed a retrospective design, and prospective validation of our results would be desirable. The training and validation cohorts had differing frequencies of 1p/19q codeletions and WHO grades, which may have affected our results. The moderate accuracy achieved by our classification algorithm for predicting codeletion status underlines the fact that molecular testing of surgical specimens will remain the criterion standard for LGG classification in the foreseeable future. However, prediction by neuroimaging may be useful for patient counseling in the preoperative setting, in cases in which biopsy or resection is challenging or pathologic tissue is insufficient for accurate rendering of molecular results, and in cases of laboratory error or misinterpretation (eg, misinterpretation of fluorescence in situ hybridization–based 1p/19q-codeletion results in the setting of chromosomal polysomy or partial chromosome arm deletions).41–43
Conclusions
A 2-step classification algorithm based on the T2-FLAIR mismatch sign and a multivariate logistic regression model using tumor texture, patient age, T2* blooming, location, and hydrocephalus demonstrates an overall moderate prediction accuracy for 1p/19q-codeletion status in IDH-mutant LGGs. We validated the high PPV of the T2-FLAIR mismatch sign for predicting the IDHmut-Noncodel LGG subtype and report novel associations between midline shift/hydrocephalus and the IDHmut-Noncodel LGG subtype.
ABBREVIATIONS:
- IDH
isocitrate dehydrogenase
- IDHmut-Codel
1p/19q-codeleted IDH-mutant LGGs, oligodendrogliomas
- IDHmut-Noncodel
noncodeleted IDH-mutant LGGs, astrocytomas
- LGG
lower grade glioma
- MLR
multivariate logistic regression
- PPV
positive predictive value
- TCGA
The Cancer Genome Atlas
- TCIA
The Cancer Imaging Archive
- WHO
World Health Organization
Footnotes
Disclosures: Camilo E. Fadul—UNRELATED: Grants/Grants Pending: Novartis, Comments: funding for a clinical trial,* Maria-Beatriz Lopes—UNRELATED: Royalties: Elsevier, Comments: book editor; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: International Society of Neuropathology, Comments: partial travel expenses to present at the International Congress of Neuropathology, Tokyo, October 2018. Rajan Jain—UNRELATED: Consultancy: Cancer Panels; Royalties: Thieme. Sohil Patel—RELATED: Grant: Radiological Society of North America, Comments: I received support for my contributions to this article from a Radiological Society of North America Research Scholar Grant.* David Schiff—UNRELATED: Board Membership: Monteris Medical, Orbus, Comments: Data Safety and Monitoring Committees; Consultancy: Cavion, Comments: preclinical drug development*; Royalties: UpToDate. *Money paid to the institution.
S.H. Patel was supported by a Radiological Society of North America Research Scholar Grant (RSCH1819).
Preliminary data previously presented at: Annual Meeting of the American Society of Neuroradiology and the Foundation of the ASNR Symposium, June 2–7, 2018; Vancouver, British Columbia, Canada.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803–20 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 2. Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer J 2014;20:66–72 10.1097/PPO.0000000000000020 [DOI] [PubMed] [Google Scholar]
- 3. Yan H, Parsons DW, Jin G, et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brat DJ, Verhaak RG, Aldape KD, et al. ; Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372:2481–98 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van den Bent MJ, Brandes AA, Taphoorn MJ, et al. . Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 2013;31:344–50 10.1200/JCO.2012.43.2229 [DOI] [PubMed] [Google Scholar]
- 6. Chamberlain MC, Born D. Prognostic significance of relative 1p/19q codeletion in oligodendroglial tumors. J Neurooncol 2015;125:249–51 10.1007/s11060-015-1906-y [DOI] [PubMed] [Google Scholar]
- 7. van den Bent MJ. Chemotherapy for low-grade glioma: when, for whom, which regimen? Curr Opin Neurol 2015;28:633–938 10.1097/WCO.0000000000000257 [DOI] [PubMed] [Google Scholar]
- 8. Zlatescu MC, TehraniYazdi A, Sasaki H, et al. . Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res 2001;61:6713–15 [PubMed] [Google Scholar]
- 9. Megyesi JF, Kachur E, Lee DH, et al. . Imaging correlates of molecular signatures in oligodendrogliomas. Clin Cancer Res 2004;10:4303–06 10.1158/1078-0432.CCR-04-0209 [DOI] [PubMed] [Google Scholar]
- 10. Jenkinson MD, du Plessis DG, Smith TS, et al. . Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain 2006;129(Pt 7):1884–91 10.1093/brain/awl108 [DOI] [PubMed] [Google Scholar]
- 11. Brown R, Zlatescu M, Sijben A, et al. . The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res 2008;14:2357–62 10.1158/1078-0432.CCR-07-1964 [DOI] [PubMed] [Google Scholar]
- 12. Sherman JH, Prevedello DM, Shah L, et al. . MR imaging characteristics of oligodendroglial tumors with assessment of 1p/19q deletion status. Acta Neurochir (Wien) 2010;152:1827–34 10.1007/s00701-010-0743-1 [DOI] [PubMed] [Google Scholar]
- 13. Kim JW, Park CK, Park SH, et al. . Relationship between radiological characteristics and combined 1p and 19q deletion in World Health Organization grade III oligodendroglial tumours. J Neurol Neurosurg Psychiatry 2011;82:224–27 10.1136/jnnp.2009.178806 [DOI] [PubMed] [Google Scholar]
- 14. Fellah S, Caudal D, De Paula AM, et al. . Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? AJNR Am J Neuroradiol 2013;34:1326–33 10.3174/ajnr.A3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chawla S, Krejza J, Vossough A, et al. . Differentiation between oligodendroglioma genotypes using dynamic susceptibility contrast perfusion-weighted imaging and proton MR spectroscopy. AJNR Am J Neuroradiol 2013;34:1542–49 10.3174/ajnr.A3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smits M. Imaging of oligodendroglioma. Br J Radiol 2016;89:20150857 10.1259/bjr.20150857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson DR, Diehn FE, Giannini C, et al. . Genetically defined oligodendroglioma is characterized by indistinct tumor borders at MRI. AJNR Am J Neuroradiol 2017;38:678–84 10.3174/ajnr.A5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Darlix A, Deverdun J, Menjot de Champfleur N, et al. . IDH mutation and 1p19q codeletion distinguish two radiological patterns of diffuse low-grade gliomas. J Neurooncol 2017;133:37–45 10.1007/s11060-017-2421-0 [DOI] [PubMed] [Google Scholar]
- 19. Patel SH, Poisson LM, Brat DJ, et al. . T2-FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res 2017;23:6078–85 10.1158/1078-0432.CCR-17-0560 [DOI] [PubMed] [Google Scholar]
- 20. Yoon HJ, Ahn KJ, Lee S, et al. . Differential diagnosis of oligodendroglial and astrocytic tumors using imaging results: the added value of perfusion MR imaging. Neuroradiology 2017;59:665–75 10.1007/s00234-017-1851-x [DOI] [PubMed] [Google Scholar]
- 21. Akkus Z, Ali I, Sedlář J, et al. . Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging 2017;30:469–76 10.1007/s10278-017-9984-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smits M, van den Bent MJ. Imaging correlates of adult glioma genotypes. Radiology 2017;284:316–31 10.1148/radiol.2017151930 [DOI] [PubMed] [Google Scholar]
- 23. Delfanti RL, Piccioni DE, Handwerker J, et al. . Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol 2017;135:601–09 10.1007/s11060-017-2613-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park YW, Han K, Ahn SS, et al. . Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. AJNR Am J Neuroradiol 2018;39:37–42 10.3174/ajnr.A5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lasocki A, Gaillard F, Gorelik A, et al. . MRI features can predict 1p/19q status in intracranial gliomas. AJNR Am J Neuroradiol 2018;39:687–92 10.3174/ajnr.A5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park YW, Han K, Ahn SS, et al. . Whole-tumor histogram and texture analyses of DTI for evaluation of IDH1-mutation and 1p/19q-codeletion status in World Health Organization grade II gliomas. AJNR Am J Neuroradiol 2018;39:693–98 10.3174/ajnr.A5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanazawa T, Fujiwara H, Takahashi H, et al. . H. Imaging scoring systems for preoperative molecular diagnoses of lower-grade gliomas. Neurosurg Rev 2018. April 26. [Epub ahead of print] 10.1007/s10143-018-0981-x [DOI] [PubMed] [Google Scholar]
- 28. Broen MP, Smits M, Wijnenga MM, et al. . The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncol 2018;20:1393–99 10.1093/neuonc/noy048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedano N, Flanders AE, Scarpace L, et al. . Radiology Data from The Cancer Genome Atlas Low Grade (TCGA-LGG) collection. The Cancer Imaging Archive. 10.7937/K9/TCIA.2016.L4LTD3TK. Accessed January 10, 2019. [DOI]
- 30. Ceccarelli M, Barthel FP, Malta TM, et al. . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016;164:550–63 10.1016/j.cell.2015.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Capper D, Weissert S, Balss J, et al. . Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol 2010;20:245–54 10.1111/j.1750-3639.2009.00352.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Capper D, Zentgraf H, Balss J, et al. . Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol 2009;118:599–601 10.1007/s00401-009-0595-z [DOI] [PubMed] [Google Scholar]
- 33. Felsberg J, Wolter M, Seul H, et al. . Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol 2010;119:501–07 10.1007/s00401-010-0647-4 [DOI] [PubMed] [Google Scholar]
- 34. Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32–35 [DOI] [PubMed] [Google Scholar]
- 35. Robin X, Turck N, Hainard A, et al. . pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna; R Foundation for Statistical Computing; 2017 [Google Scholar]
- 37. Qi S, Yu L, Li H, et al. . Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 2014;7:1895–902 10.3892/ol.2014.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Metellus P, Coulibaly B, Colin C, et al. . Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 2010;120:719–29 10.1007/s00401-010-0777-8 [DOI] [PubMed] [Google Scholar]
- 39. Choi C, Ganji SK, DeBerardinis RJ, et al. . 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 2012;18:624–29 10.1038/nm.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang K, Bai HX, Zhou H, et al. . Residual convolutional neural network for determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res 2018;24:1073–81 10.1158/1078-0432.CCR-17-2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel KS, Carter BS, Chen CC. Role of biopsies in the management of intracranial gliomas. Prog Neurol Surg 2018;30:232–43 10.1159/000464439 [DOI] [PubMed] [Google Scholar]
- 42. Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol 2011;21:57–73 10.1111/j.1750-3639.2010.00453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Riemenschneider MJ, Jeuken JW, Wesseling P, et al. . Molecular diagnostics of gliomas: state of the art. Acta Neuropathol 2010;120:567–84 10.1007/s00401-010-0736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]