The authors applied an optimized TSE T2 sequence to washed postmortem brain samples to reveal exquisite and reproducible brain stem anatomic MR imaging contrast comparable with histologic atlases. Direct TSE MR imaging sequence discrimination of brain stem anatomy can help validate other MR imaging contrasts, such as diffusion tractography, or serve as a structural template for extracting quantitative MR imaging data in future postmortem investigations.
Abstract
BACKGROUND AND PURPOSE:
The brain stem is compactly organized with life-sustaining sensorimotor and autonomic structures that can be affected by numerous pathologies but can be difficult to resolve on conventional MR imaging.
MATERIALS AND METHODS:
We applied an optimized TSE T2 sequence to washed postmortem brain samples to reveal exquisite and reproducible brain stem anatomic MR imaging contrast comparable with histologic atlases. This resource-efficient approach can be performed across multiple whole-brain samples with relatively short acquisition times (2 hours per imaging plane) using clinical 3T MR imaging systems.
RESULTS:
We identified most brain stem structures at 7 canonical axial levels. Multiplanar or oblique planes illustrate the 3D course and spatial relationships of major brain stem white matter pathways. Measurements of the relative position, course, and cross-sectional area of these pathways across multiple samples allow estimation of pathway location in other samples or clinical subjects. Possible structure-function asymmetries in these pathways will require further study—that is, the cross-sectional area of the left corticospinal tract in the midpons appeared 20% larger (n = 13 brains, P < .10).
CONCLUSIONS:
Compared with traditional atlases, multiplanar MR imaging contrast has advantages for learning and retaining brain stem anatomy for clinicians and trainees. Direct TSE MR imaging sequence discrimination of brain stem anatomy can help validate other MR imaging contrasts, such as diffusion tractography, or serve as a structural template for extracting quantitative MR imaging data in future postmortem investigations.
The human brain stem is phylogenetically the oldest brain region, serving critical integrative functions and linking the spinal cord, cerebellum, basal ganglia, limbic system, and neocortex. The brain stem consists of numerous small fiber tracts and nuclei that regulate sensory, motor, and autonomic functions.1 Small lesions due to diverse disorders (eg, multiple sclerosis,2 neoplasm,3 infection,4 or neurodegeneration5) can cause devastating consequences due to the compact juxtaposition of vital structures. Furthermore, the brain stem also contains anatomic targets for functional neurosurgery.6,7 Conventional MR imaging does not provide adequate contrast or spatial resolution of many brain stem substructures to define their involvement in specific clinical cases or direct precise surgical targeting.
Because clinical 3T MR imaging cannot reliably discriminate many small brain stem structures, clinicians and researchers must infer brain stem anatomy relative to craniocaudal position, a few identifiable internal features, and surface topography. Susceptibility-weighted MR imaging8 demonstrates some additional internal features that can improve indirect localization, but images from this sequence are also vulnerable to distortion from the skull base. Ultra-high-field in vivo MR imaging9–12 and advanced diffusion methods13–15 also improve discrimination of more brain stem structures. Diffusion methods though are vulnerable to spatial distortions, require long acquisition times, and depend on modeling assumptions that are difficult to validate directly in human tissue.16–18 Ultra-high-field MR imaging is limited by increased geometric distortion and signal loss at the skull base and is only available at major academic centers with dedicated technical support staff.
MR imaging microscopy can help characterize dissected, isolated ex vivo human brain stem samples19,20 and can illustrate detailed anatomy for teaching and guiding image interpretation in living subjects. These acquisitions require long scan times (>12 hours) using small dedicated radiofrequency coils that cannot accommodate the whole brain and are limited to single or few specimens. Furthermore, image contrast may be altered relative to typical clinical MR imaging in living subjects by MR imaging water-relaxation changes associated with higher field strengths,21 the postmortem interval,22 and formaldehyde fixation.23–25 We recently developed a rapid 3T postmortem anatomic MR imaging protocol26 to screen postmortem whole brains in sudden unexplained death of childhood (SUDC).27 This protocol washes the brain thoroughly, then uses optimized-but-conventional MR imaging sequences, a 3T MR imaging system, and a head coil available at most institutions. The optimized 2D TSE sequence, in particular, produces exquisite anatomic contrast for subcortical structures in all 3 planes, comparable with neuroanatomic atlases with histologic stains.28–31 Here we demonstrate how the optimized TSE sequence can precisely delineate brain stem anatomy across multiple samples.
Materials and Methods
Sample Procurement and Preparation
Whole-brain samples were obtained from an institutional review board (New York University)–approved and Health Insurance Portability and Accountability Act–compliant multisite research study, the SUDC Registry and Research Collaborative,32 which used ex vivo MR imaging screening before gross pathologic assessment, brain cutting, and histopathology for forensic investigation. For each subject, the postmortem brain was removed intact by the local medical examiner; then, it was immersion-fixed in a 4% formaldehyde solution for at least 21 days to reach near equilibrium with presumed fixative-induced nervous tissue T2 changes.24 The brain was shipped to our institution and was then washed continuously in water for 48 hours to eliminate MR imaging relaxation changes from the free aldehyde fixative solution.23 Individual brains with MR imaging data included for the figures and tables in this study (n = 13) met the following criteria: 1) transected at or below the pyramidal decussation; 2) no MR imaging or pathological abnormality (outside the hippocampus) identified by a board-certified neuroradiologist and neuropathologist respectively; 3) no T1-hyperintense fixation bands in the brain stem or diencephalon structures due to variable fixation penetration25; and 4) a prerefrigeration postmortem interval of <24 hours.22
Whole-Brain MR Imaging Protocol
Each brain was immersed under water within a custom 3D-printed container specifically designed to conform to a 64-channel head and neck coil on a 3T Magnetom Prisma MR imaging scanner (Siemens, Erlangen, Germany). Sealed water-filled disposable powderless latex medical gloves were gently wedged between the container and brain to prevent motion and to optimize the coil-filling factor. Scout sequences identified the brain position; then, 2D high-resolution TSE MR imaging sequences of the whole brain were obtained in coronal, sagittal, and axial planes relative to the anterior/posterior commissure (ACPC) plane. In selected cases, additional images were obtained in oblique planes to illustrate specific anatomic relationships within the brain stem. T2-weighted TSE sequence parameters were the following: TR = 5380 ms, TE = 53 ms, echo-train length = 7, echo spacing = 10.8 ms, bandwidth = 415 Hz/pixel, slice thickness = 0.8 mm, 116 slices (no interslice gap), in-plane resolution = 0.35 × 0.35 mm, concatenations = 2, averages = 10, total time = 2 hours (full protocol available on request). Optimization of sequence parameters for contrast resolution within the brain stem, diencephalic structures, and cerebral hemispheres using TSE sequences with 3T MR imaging is reported separately.26
MR Imaging Data Anatomic Analysis
For each subject, we characterized brain stem detail at 7 canonical axial levels for anatomy and reproducibility parallel to the ACPC plane33: rostral and caudal midbrain; rostral, middle, and caudal pons; and rostral and caudal medulla (Fig 1). The MR images were labeled with standard nomenclature.20,29,30 Only tracts and nuclei identified in all samples by consensus between 2 board-certified neuroradiologists are reported. We measured 4 major brain stem white matter tracts (the corticospinal tract [CST], medial lemniscus [ML], medial longitudinal fasciculus [MLF], and the central tegmental tract [CTT]) for shape and cross-sectional area in the axial plane at the caudal midbrain, middle pons, and cranial medulla levels. The area was derived from the formula for an ellipse in all structures and reproducibility was assessed by repeating measurements of the left and right CST in the cranial medulla and midpons on 3 separate days for 3 randomly selected brains. We compared the right and left CST cross-sectional areas for possible asymmetries using an unpaired 2-tailed t test. The craniocaudal extent of these pathways was measured; then, the 3D course of the tracts was precisely defined. The cranial limit of the CST was defined as its superiormost extent in the cerebral peduncle before joining the posterior limb of the internal capsule. The cranial extent of the ML was its fibers before entering the thalamus. The inferior limit of the CST and ML was the inferior margins of their respective medullary decussations. The visualized MLF and CTT remained within the brain stem. The inferior half of the dentatorubrothalamic tract within the superior cerebellar peduncle also was analyzed at midbrain levels.
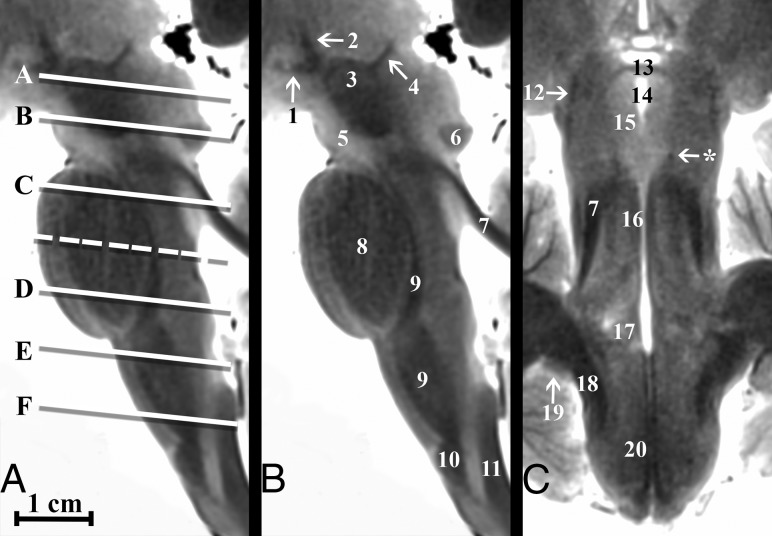
Fig 1.
Parasagittal and coronal T2-weighted MR images of the postmortem human brain stem. A, Canonical axial brain stem levels parallel to the anterior/posterior commissure plane that are found in Fig 2 are represented with the solid lines and On-line Fig 1 with the dashed line. Only selected brain stem substructures are labeled in coaligned sagittal and coronal images to orient the reader relative to the craniocaudal axial slice positions. The On-line Table provides a complete list of labeled anatomy for all figures, indicated by the numbers in parentheses in the legends. Note the trochlear nerve (asterisk, C) only seen in some brains.
Results
Axial images of the brain stem at 7 canonical anatomic levels are shown in Fig 2 and On-line Fig 1, with labeled substructures (see The On-line Table for the complete list of labeled substructures). While brain stem anatomy is typically shown in the axial plane, Fig 1 also demonstrates selected sagittal and coronal views with axial section positions on the sagittal image. On-line Fig 1 demonstrates the reproducibility of anatomic contrast for 4 selected brains at both the caudal midbrain and middle pons levels. Videos of the brain stem in 3 planes are provided in On-line Videos 1–3. All numbered structures could be directly identified for each subject by both board-certified neuroradiologists. The mean postnatal age for the subjects included in this study was 32.1 ± 6.1 months.
Fig 2.
Axial modified T2-weighted TSE images at 6 canonical levels of the postmortem brain stem orientated parallel to the anterior/posterior commissure plane. Upper row: A, cranial midbrain; B, caudal midbrain. Middle row: C, cranial pons; D, caudal pons. Lower row: E, cranial medulla, F, caudal medulla. Improved image contrast from the modified TSE sequence directly demonstrates even small structures like the medial longitudinal fasciculus (16). Note the sensory decussation of the medial lemniscus in the caudal medulla (asterisk, F). The motor decussation is demonstrated in Fig 3.
We briefly describe the course, orientation, and shape of 5 major white matter pathways based on the 13 whole-brain specimens (all measurements are reported as mean ± SD). The numbers in parentheses refer to the numbers of the brain structures listed in the On-line Table. The corticospinal tract (28) (Fig 3) is the major motor pathway controlling the voluntary movements of the limbs and trunk. The CST at the midbrain level is 1.3 ± 0.1 cm lateral to the midsagittal plane and descends within the cerebral peduncle (24) from the posterior limb of the internal capsule at a 31° ± 7° angle from superolateral to inferomedial in the coronal plane. The center of the tract is 0.6 ± 0.05 cm deep to the ventral surface and 0.4 ± 0.09 cm lateral to the midsagittal plane at the midpons, while maintaining a rounded shape before converging fibers descend to the medullary pyramids (34) at a less steep superolateral-to-inferomedial 14° ± 2° angle. The pyramids form the ventral surface of the medulla and are 0.26 ± 0.04 cm lateral to the midsagittal plane. The CST descends at a 5° ± 2° anterosuperior-to-inferoposterior angle relative to the long axis of the brain stem in the sagittal plane. CST signal intensity remains T2-hypointense even with tract dispersion in the pontine levels (Fig 2).
Fig 3.
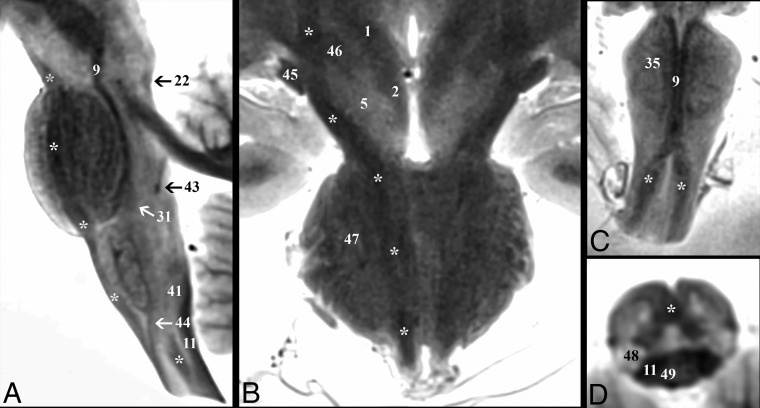
Demonstration of the corticospinal tract (asterisk) throughout the brain stem. A, Parasagittal image depicts the corticospinal tract descending within the brain stem from the cerebral peduncle to the upper cervical cord. B, Coronal image shows the course of the corticospinal tract from the posterior limb of the internal capsule to the most superior aspect of the medullary pyramids. Note in the diencephalic junction, the close relationship of the corticospinal tract to the optic tract (45) laterally and the subthalamic nucleus (46) medially. Oblique coronal (C) and oblique axial (D) images highlight the decussation of the corticospinal tracts at the cervicomedullary junction. C, The paramedian dark lines are the medial lemniscus (9), which is superficial to the corticospinal tract on this oblique axial image.
After the internal arcuate fibers (37) decussate, the medial lemniscus (9) (Fig 4) is in the central paramedian medulla with an elongated ovoid shape and its long axis oriented anterior to posterior on axial images. The ML is a sensory pathway conveying fine touch, vibration, and proprioception of the skin and joints. As the ML ascends, its long axis rotates at 56° ± 11°, 80° ± 9°, 115° ± 13°, and 130° ± 4° angles relative to the midsagittal plane at the caudal pons, middle pons, cranial pons, and caudal midbrain levels, respectively. The tract is located 0.3 ± 0.05 cm and 0.8 ± 0.06 cm lateral to the midsagittal plane at the pons and midbrain levels, respectively. In the sagittal plane, the ML ascends at a 4° ± 1° anteroinferior-to-posterosuperior angle relative to the long axis of the brain stem at the medulla but pivots posteriorly 18° ± 5° at the pontomedullary junction and pivots again posteriorly 17° ± 3° at the midbrain. The ML maintains uniform signal intensity until the fibers become less distinct just before terminating in the ventral posterolateral thalamic nucleus (50).
Fig 4.
Demonstration of the medial lemniscus (asterisk) throughout the brain stem. A, Coronal image shows the change in the long-axis orientation of the medial lemniscus from anteroposterior to transverse as it ascends the medulla to the pontomedullary junction. B, Parasagittal image highlights the terminations of the medial lemniscus in the ventral posterolateral thalamic nucleus (50). C, Axial image angled anteroinferior to posterosuperior 20° relative to the ACPC plane through the inferior colliculus (6) shows the relationship of the medial lemniscus to the spinothalamic tract (12) and lateral lemniscus (22) at the lateral tegmentum.
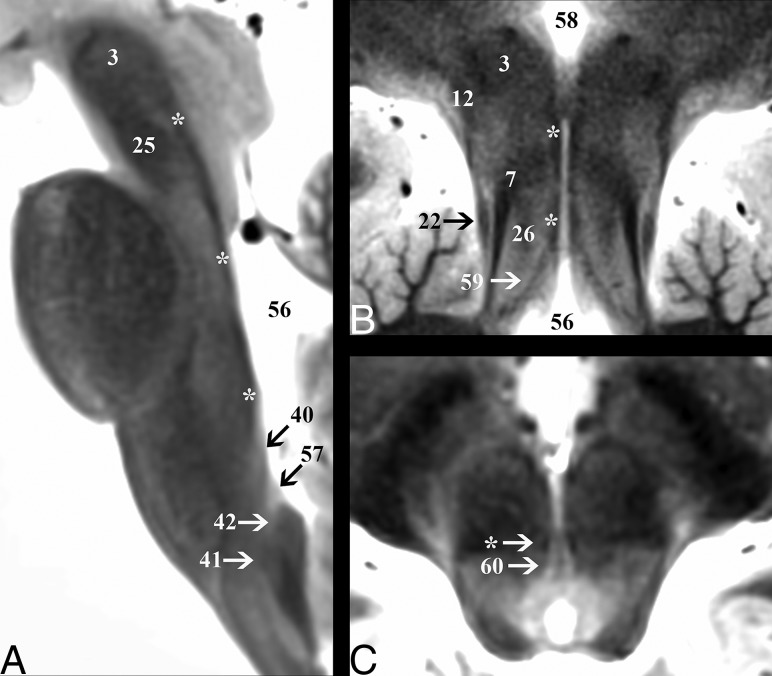
The medial longitudinal fasciculus (16) (Fig 5) is a small tear drop–shaped tract just deep to the rhomboid fossa, 0.05 ± 0.01 cm lateral to the midsagittal plane. The MLF coordinates connections among the oculomotor, trochlear, and abducens nuclei for control of conjugate eye movements. The tract ascends at a 5° ± 2° angle posteroinferior to anterosuperior relative to the long axis of the brain stem on sagittal images in the medullary and pontine levels. At the midbrain, the tract takes a 20° ± 6° ventral turn to terminate along the walls of the inferior third ventricle (58). At its cranial termination, the MLF signal becomes less conspicuous. On axial midbrain slices, the MLF is 0.16 ± 0.02 cm lateral to the midsagittal plane with the long axis oriented at 137° ± 7° anteromedial to posterolateral.
Fig 5.
Demonstration of the medial longitudinal fasciculus (asterisk) throughout the brain stem. A, Sagittal image depicts the dorsal course of the medial longitudinal fasciculus from its origin in the cranial medulla just superior to the hypoglossal nucleus (40) to the level of the red nucleus (3). B, Coronal oblique image that is perpendicular to the long axis of the hippocampus (structure not shown) at the level of the posterior commissure shows the terminations of the tract in the inferior walls of the third ventricle (58). This coronal image also highlights vertical columns of the central midbrain from lateral to medial: lateral lemniscus (22), superior cerebellar peduncle (7), central tegmental tract (26), and medial longitudinal fasciculus (asterisk). C, Axial cranial midbrain image angled anterosuperior to posteroinferior 20° relative to the ACPC plane highlights the close relationship of the medial longitudinal fasciculus to the oculomotor nucleus (60).
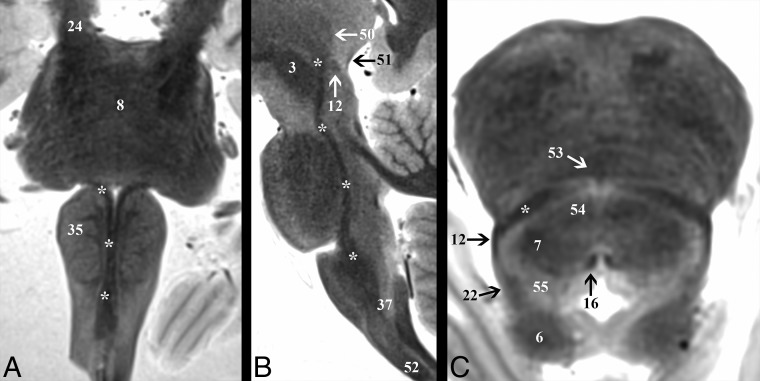
Fibers descending from the red nucleus (3) to the ipsilateral inferior medullary olive (35) are within the central tegmental tract (26) (On-line Fig 2), located 0.3 ± 0.04 cm lateral to the midsagittal plane. The tract is round and maintains a less distinct hypointense signal compared with the other major tracts described here. The CTT courses inferiorly and parallel to the long axis of the brain stem in the sagittal plane until a 10° ± 2° anterior bend to meet the inferior olive. The CTT contains ascending taste fibers from the solitary nucleus, whereas the descending fibers are part of a feedback circuit (dentatorubro-olivary) responsible for modulating motor activity.
The inferior half of the dentatorubrothalamic tract (On-line Fig 3) ascends to the superior cerebellar peduncle (7) at a 27° ± 3° inferoposterior to the anterosuperior angle in the sagittal plane. On axial images, the superior cerebellar peduncle has a parabolic configuration with an inner concave angle of 114° ± 8°. The apex is 0.6 ± 0.07 cm lateral to the midsagittal plane. The fibers become less distinct just before entering the superior cerebellar decussation (25) but then have very T2-hypointense signal in the decussation. The center point of the superior cerebellar decussation is 1.5 ± 0.1 cm inferior to the ACPC plane and 0.3 ± 0.06 cm deep to the interpeduncular fossa surface. The dentatorubrothalamic tract acts to coordinate the initiation, planning, and timing of movement.
The Table provides measurements of 5 major craniocaudally oriented white matter tracts at selected axial levels of the brain stem for all 13 brains. The MLF (16) was the smallest tract in the cross-sectional area at the midbrain, pons, and medulla levels (eg, transverse dimension, <0.8 mm). The long axis of the ML in the axial plane rotates as the tract ascends (9), but the ML showed the least variation in the cross-sectional area within the 3 levels. The CST (28) had the largest cross-sectional area and was largest in the midpons where the fibers intermix with the pontocerebellar fibers (47). The mean transaxial cross-sectional area of the left CST was 20% larger than the right CST in the medulla (1.5-mm2 difference, P = .099) and pons (5.7-mm2 difference, P = .063). The left and right CST cross-sectional areas were measured for 3 brains at 4 separate sessions at the same cranial medulla and midpons levels to assess repeatability; within-subject SDs were 0.8 and 1.6 mm2, respectively. There were no additional observable left-right asymmetries in the other measured major white matter tracts.
Selected measurements for 5 major brain stem white matter tracts at 3 canonical axial planesa
| Tract | Fig | CC | Cranial Medulla |
Mid Pons |
Caudal Midbrain |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AP | TV | Area | AP | TV | Area | AP | TV | Area | |||
| CST (L) | 3 | 51.7 ± 4.8 | 2.9 ± 0.6 | 3.7 ± 0.5 | 8.8 ± 2.6b | 6.6 ± 1.0 | 6.5 ± 0.7 | 33.9 ± 6.8b | 6.1 ± 0.6 | 3.5 ± 0.5 | 17.1 ± 3.0b |
| CST (R) | 3 | 51.7 ± 4.8 | 2.7 ± 0.3 | 3.3 ± 0.4 | 7.3 ± 1.4b | 5.7 ± 0.9 | 6.2 ± 0.8 | 28.2 ± 8.1b | 6.1 ± 0.8 | 3.2 ± 0.8 | 15.5 ± 5.2b |
| ML | 4 | 46.9 ± 3.5 | 5.6 ± 0.8 | 0.6 ± 0.1 | 2.9 ± 0.6 | 1.2 ± 0.4 | 4.6 ± 0.7 | 4.7 ± 1.9 | 2.0 ± 0.4 | 3.0 ± 0.4 | 4.9 ± 1.4 |
| MLF | 5 | 39.6 ± 3.4 | 1.0 ± 0.3 | 0.5 ± .06 | 0.5 ± 0.2 | 1.4 ± 0.3 | 0.8 ± 0.1 | 0.9 ± 0.2 | 3.2 ± 0.4 | 0.7 ± 0.1 | 1.9 ± 0.5 |
| CTT | 6 | 37.7 ± 4.0 | 2.6 ± 0.4 | 1.6 ± 0.3 | 3.4 ± 0.8 | 1.7 ± 0.2 | 2.1 ± 0.4 | 2.9 ± 0.8 | 3.2 ± 0.3 | 3.6 ± 0.4 | 9.1 ± 1.5 |
Note:—CC indicates craniocaudal; AP, anteroposterior; TV, transverse; Fig, figure; L, left; R, right.
Units are millimeters or square millimeters, and data are mean ± SD, with 13 SUDC samples.
All measurements of the right and left corticospinal tracts were compared separately. Cross-sectional areas trended toward small statistical differences in the medulla (P = .099) and pons (P = .063), but not the midbrain (P = .361).
Major brain stem nuclei were also consistently identified directly (On-line Fig 4). At the cranial midbrain level, the red nucleus (3), substantia nigra (5), and superior colliculus (62) were seen. The oculomotor nucleus (60) was best visualized with an oblique axial plane tipped 20° superiorly anterosuperior to posteroinferior relative to the ACPC plane (Fig 5), but the Edinger-Westphal nucleus was not identified. The trochlear nucleus could not be discriminated from the medial longitudinal fasciculus (16) in the caudal midbrain, but its fascicles were identifiable for some brains in the coronal plane (Fig 1). The mesencephalic trigeminal nucleus (23) was seen with an oblique axial image angled 10° superiorly anterosuperior to posteroinferior relative to the ACPC plane (On-line Fig 2). The interpeduncluar nucleus (53) was most clearly identified on an oblique axial image 20° superior relative to the ACPC plane through the level of the inferior colliculi (6) (Fig 4). At the cranial pons, the locus coeruleus was not identified in any subject.
The midpons contains the spinal sensory (48) and motor nuclei (66) of the trigeminal nerve. The trigeminal motor nucleus was best seen at the cranial-midpons level junction just medial to the superior cerebellar peduncles (7). The trigeminal sensory spinal nucleus was not reliably seen in the pons in all brains but could be identified at the cervicomedullary junction (Fig 3). The facial nucleus was not seen, but its fascicles (67) were identified in the midpons within the respective genu and colliculus bordering the abducens nucleus (17). The superior olivary complex (31) was identified at the lower pons level, posterolateral to the medial lemniscus (9) (Fig 2). Within the medulla, the inferior olivary nucleus (35) was clearly seen, but the dorsal and medial accessory olivary nuclei and ambiguous nucleus were not. Cochlear (19) and vestibular (36) nuclear group positions were seen along the lateral and dorsal medullary surface, but their subnuclei could not be discerned. The dorsal motor nucleus of the vagus (57) and the hypoglossal nucleus (40) could be identified on axial images and further directly distinguished on a parasagittal image at the level of the medial longitudinal fasciculus (Fig 5). The cunate (41) and gracile (42) nuclei in the caudal dorsal medulla were identified giving rise to the internal arcuate fibers (37).
Discussion
This modified TSE sequence provided detailed images of brain stem anatomy using whole postmortem brains and a widely available clinical 3T MR imaging system. Previous studies have used ultra-high-field MR imaging, dissected and isolated brain stem samples, specialized radiofrequency coils, and/or relatively long acquisition times.34–36 Anatomic image contrast was generated directly from the MR imaging sequence without mathematically complex, off-line, model-based reconstructions as required for relaxation-mapping9,37 or advanced diffusion-based contrasts.15,38 Such techniques have been difficult to validate.18 This postmortem MR imaging protocol directly visualizes many small brain stem structures such as the MLF (<1 mm in transverse dimension) that are beyond the spatial resolution or detection limits of current state-of-the-art diffusion-weighted imaging techniques.9,15 Furthermore, direction-encoded color images of diffusion anisotropy cannot discriminate adjacent structures with parallel craniocaudal orientations (eg, vertical columns within the midbrain; Fig 5B). Conversely, T2-weighted contrast reported here cannot discriminate all brain stem structures identified with histology such as distinguishing the dentatorubrothalamic projections from the red nucleus they envelope (On-line Fig 3C). T2-weighted contrast detects but cannot resolve the individual crossing or interdigitating fiber bundles of the sensory, motor, or superior cerebellar peduncle decussations (Figs 2F, 3D, and On-line Fig 3B, respectively). Future work will evaluate potential synergies for brain stem structure resolution when this TSE contrast is combined with diffusion, susceptibility, and other MR imaging contrasts at 3T (or ultra-high-field MR imaging). This optimized TSE sequence also produces exquisite contrast resolution of subthalamic, thalamic, and basal ganglia structures that will be described in a separate companion report.
For clinicians, it is challenging to learn and retain brain stem anatomy because internal structures are only discriminated on stained histology slides, unlike imaging performed in clinical practice. We must mentally juxtapose structures discriminated by specific histology stains onto MR images on the basis of mostly the craniocaudal position and brain stem surface features. Here, knowledge and mental maps of brain stem neuroanatomy may be facilitated because this postmortem protocol provides anatomic discrimination of brain stem structures comparable with histology atlases,28–31 yet it is derived from a commonly used clinical MR imaging sequence and contrast mechanism (albeit with higher spatial resolution). MR imaging data facilitate the creation of user-controllable videos to evaluate the orientation and evolution of specific pathways throughout the brain stem (On-line Videos 1–3). Furthermore, multiplanar images or series can illustrate specific brain stem tracts or key anatomic relationships in novel ways—that is, the oblique coronal plane perpendicular to the long axis of the hippocampus, just deep to the rhomboid fossa, illustrates functional cell columns of cranial nuclei V, VI, VII, VIII, X, and XII (On-line Fig 4). It would be technically challenging and resource- and time-intensive to obtain such images from histologic sections of individual human brain stem samples; hence, previous histologic or MR imaging–based brain stem images emphasized idealized axial views.19,20,33 The postmortem MR imaging protocol can be applied quickly and inexpensively across many samples without tissue consumption. This feature should enhance the experiential component of learning by exposing trainees to more individual variations in brain stem anatomy (On-line Fig 1).
The ability to directly visualize specific brain stem structures in multiple individual brains also facilitates creation of normative coordinates for structures in specific fiducial planes and surfaces that can be used in clinical studies. For example, our data from SUDC brains predict that a lesion in the midbrain tegmentum, 0.1, 0.3, or 0.8 cm lateral to the midsagittal plane, would involve the MLF, CTT, or ML, respectively. We estimated the size and cross-sectional areas of several major brain stem tracts (Table). We observed a trend (P < .10) toward ∼20% larger cross-sectional areas for the left corticospinal tract in the pons and medulla (Table). While handedness is less established in young children,39 functional asymmetries in brain stem structures may alter the numbers of axons, degree of myelination, and/or myelin compaction that could affect TSE MR imaging contrast. These asymmetries may change during childhood. The potential corticospinal tract asymmetries and brain stem pathway coordinates and sizes will require further future investigation in adult brains without neurologic disease and documented handedness. Future work could also produce a group-based brain atlas and/or a normative data base of brain stem structures across different ages and sex. These data could assess changes to brain stem structure with aging or subcortical dementias40–43 or could be used as a structural template for extracting other forms of quantitative MR imaging data in postmortem investigations.
The use of pediatric brains from an SUDC study is a limitation for the measurements reported in this study. Deformity or relaxation of the posterior fossa structures from procurement, agonal hydration status, or brain changes associated with formaldehyde fixation also may affect the external validity of these results. While repeatability measures of the cross-sectional area in this preliminary study were lower than the differences observed among tracts or between the right and left CST, manual measurements are prone to error from image noise, slice orientation, and rater biases. Measurements in the sagittal plane may also be confounded by variable posterior angulation of the lower brain stem created during specimen procurement. Assignments of brain stem structures were made by consensus between 2 board-certified neuroradiologists using standard reference texts based on histologic staining28–31; inter- or intraobserver variability for structure identification was not assessed. TSE signal intensity correlated inversely with myelin staining in the histology of different brain samples; however, the biophysical basis for gradations of T2-weighted signal variation in the brain stem will require further investigation. Histology sampling and specific stains were restricted to the SUDC forensic investigation. MR imaging relaxation parameters of these ex vivo brains differ from those in vivo due to the postmortem interval,22 formaldehyde fixation and tissue penetration,23–25 incomplete myelination,44,45 or subtle unrecognized SUDC pathology.46 Preliminary experiments suggested that true 3D T2-weighted MR imaging acquisitions47 did not produce such exquisite contrast resolution of the brain stem, but this will be the subject of future investigation.
Conclusions
An optimized TSE T2 sequence applied to washed postmortem brain samples revealed exquisite and reproducible brain stem anatomic MR imaging contrast comparable with histologic atlases. The current results suggest that intrinsic nervous tissue T2 differences could potentially generate sufficient contrast to also identify brain stem structures in vivo. It will be challenging to feasibly adapt this MR imaging protocol to living subjects, yet this would greatly enhance its applicability to neuroanatomy training, clinical practice, and future research.
Acknowledgments
The authors thank the medical examiners, coroners, and the SUDC families for their support of this research.
ABBREVIATIONS:
- ACPC
anterior/posterior commissure
- CST
corticospinal tract
- CTT
central tegmental tract
- ML
medial lemniscus
- MLF
medial longitudinal fasciculus
- SUDC
sudden unexplained death of childhood
Footnotes
Disclosures: Laura Crandall—RELATED: Grant: SUDC Foundation, Comments: The SUDC Foundation of which I am President and the volunteer Executive Director provided a grant to New York University School of Medicine to perform this study. I have an agreed management plan with New York University whereby I am not involved with any grant negotiations between the Foundation and New York University. No grant funds were allocated to my work on the study*; UNRELATED: Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: SUDC Foundation, Comments: The SUDC Foundation supports my travel to medical-related meetings and awareness events not related to this study, but to SUDC in general.* Thomas Wisniewski—RELATED: Grant: National Institutes of Health, Comments: funding from National Institutes of Health National Institute on Aging grant AG008051.* Orrin Devinsky—RELATED: Grant: SUDC Foundation*; UNRELATED: Employment: New York University School of Medicine; Other: National Institutes of Health Center for Sudden Unexpected Death in Epilepsy Research on separate projects.* Timothy M. Shepherd—RELATED: Grant: National Institutes of Health National Institute on Aging, K23 AG048622*; UNRELATED: Expert Testimony: medicolegal expert testimony; Grants/Grants Pending: Brainlab, Comments: Principal Investigator, multiparametric MRI study of metastases treated with gamma knife irradiation*; OTHER RELATIONSHIPS: scientific advisor for Velona Technologies (devices for CT-guided image interventions) and MICroStruture Imaging (postprocessing tools for advanced MRI acquisitions). No payments were involved. *Money paid to the institution.
This study was funded by the SUDC Foundation and the Finding a Cure for Epilepsy and Seizures fund. T.M. Shepherd received research support from the National Institute of Aging (grant AG048622). T. Wisniewski and A. Faustin received research support from the National Institute of Aging (grant AG008051). This work was supported, in part, by the Center for Advanced Imaging Innovation and Research, a National Institutes of Health National Institute of Biomedical Imaging and Bioengineering Biomedical Technology Resource Center (grant P41EB017183).
References
- 1. Carpenter MB, Strong OS, Truex RC. Human Neuroanatomy: (Formerly Strong and Elwyn's Human Neuroanatomy). 7th ed Baltimore: Lippincott Williams & Wilkins; 1976 [Google Scholar]
- 2. Tintore M, Rovira A, Arrambide G, et al. . Brainstem lesions in clinically isolated syndromes. Neurology 2010;75:1933–38 10.1212/WNL.0b013e3181feb26f [DOI] [PubMed] [Google Scholar]
- 3. Donaldson SS, Laningham F, Fisher PG. Advances toward an understanding of brainstem gliomas. J Clin Oncol 2006;24:1266–72 10.1200/JCO.2005.04.6599 [DOI] [PubMed] [Google Scholar]
- 4. Tan IL, Mowry EM, Steele SU, et al. . Brainstem encephalitis: etiologies, treatment, and predictors of outcome. J Neurol 2013;260:2312–19 10.1007/s00415-013-6986-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grinberg LT, Rueb U, Heinsen H. Brainstem: neglected locus in neurodegenerative diseases. Front Neurol 2011;2:42 10.3389/fneur.2011.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mazzone P, Vilela Filho O, Viselli F, et al. . Our first decade of experience in deep brain stimulation of the brainstem: elucidating the mechanism of action of stimulation of the ventrolateral pontine tegmentum. J Neural Transm (Vienna) 2016;123:751–67 10.1007/s00702-016-1518-5 [DOI] [PubMed] [Google Scholar]
- 7. Golfinos JG, Roland JT Jr, Rodgers SD. Auditory brainstem implants. J Neurosurg 2014;120:543–44 10.3171/2013.5.JNS13430 [DOI] [PubMed] [Google Scholar]
- 8. Manova ES, Habib CA, Boikov AS, et al. . Characterizing the mesencephalon using susceptibility-weighted imaging. AJNR Am J Neuroradiol 2009;30:569–74 10.3174/ajnr.A1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deistung A, Schäfer A, Schweser F, et al. . High resolution MR imaging of the human brainstem in vivo at 7 Tesla. Front Hum Neurosci 2013;7:710 10.3389/fnhum.2013.00710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gizewski ER, Maderwald S, Linn J, et al. . High-resolution anatomy of the human brain stem using 7-T MRI: improved detection of inner structures and nerves? Neuroradiology 2014;56:177–86 10.1007/s00234-013-1312-0 [DOI] [PubMed] [Google Scholar]
- 11. Cho ZH, Calamante F, Chi JG, eds. 7.0 Tesla MRI Brain White Matter Atlas. 2nd ed Heidelberg: Springer-Verlag; 2015 [Google Scholar]
- 12. Eapen M, Zald DH, Gatenby JC, et al. . Using high-resolution MR imaging at 7T to evaluate the anatomy of the midbrain dopaminergic system. AJNR Am J Neuroradiol 2011;32:688–94 10.3174/ajnr.A2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calamante F, Tourneir JD, Jackson GD, et al. . Track density imaging (TDI): super-resolution white matter imaging using whole-brain track density mapping. Neuroimage 2010;53:1233–43 10.1016/j.neuroimage.2010.07.024 [DOI] [PubMed] [Google Scholar]
- 14. Naganawa S, Yamazaki M, Kawai H, et al. . Anatomical details of the brainstem and cranial nerves visualized by high resolution read- out-segmented multi-shot echo-planar diffusion-weighted images using unidirectional MPG at 3 T. Magnetic Resonance in Medical Sciences 2011;10:269–75 10.2463/mrms.10.269 [DOI] [PubMed] [Google Scholar]
- 15. Hoch MJ, Chung S, Ben-Eliezer N, et al. . New clinically feasible 3T MRI protocol to discriminate internal brain stem anatomy. AJNR Am J Neuroradiol 2016;37:1058–65 10.3174/ajnr.A4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI. Neuroimage 2013;73:239–54 10.1016/j.neuroimage.2012.06.081 [DOI] [PubMed] [Google Scholar]
- 17. Jones DK, Cercignani M. Twenty-five pitfalls in the analysis of diffusion MRI data. NMR Biomed 2010;23:803–20 10.1002/nbm.1543 [DOI] [PubMed] [Google Scholar]
- 18. Maier-Hein KH, Neher PF, Houde J, et al. . The challenge of mapping the human connectome based on diffusion tractography. Nature Comm 20177;8:1349 10.1038/s41467-016-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soria G, De Notaris M, Tudela R, et al. . Improved assessment of ex vivo brainstem neuroanatomy with high-resolution MRI and DTI at 7 Tesla. Anat Rec (Hoboken) 2011;294:1035–44 10.1002/ar.21383 [DOI] [PubMed] [Google Scholar]
- 20. Naidich TP, Duvernoy HM, Delman BN, et al. . Duvernoy's Atlas of the Human Brain Stem and Cerebellum. New York: Springer-Verlag/Wien; 2009 [Google Scholar]
- 21. de Graaf RA, Brown PB, McIntyre S, et al. . High magnetic field water and metabolite proton T1 and T2 relaxation in rat brain in vivo. Magn Reson Med 2006;56:386–94 10.1002/mrm.20946 [DOI] [PubMed] [Google Scholar]
- 22. Shepherd TM, Flint JJ, Thelwall PE, et al. . Postmortem interval alters the water relaxation and diffusion properties of rat nervous tissue: implications for MRI studies of human autopsy samples. Neuroimage 2009;44:820–26 10.1016/j.neuroimage.2008.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shepherd TM, Thelwall PE, Stanisz GJ, et al. . Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn Reson Med 2009;62:26–34 10.1002/mrm.21977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dawe RJ, Bennett DA, Schneider JA, et al. . Postmortem MRI of human brain hemispheres: T2 relaxation times during formaldehyde fixation. Magn Reson Med 2009;61:810–18 10.1002/mrm.21909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yong-Hing CJ, Obenaus A, Stryker R, et al. . Magnetic resonance imaging and mathematical modeling of progressive formalin fixation of the human brain. Magn Reson Med 2005;54:324–32 10.1002/mrm.20578 [DOI] [PubMed] [Google Scholar]
- 26. Miller S, Goldberg J, Bruno M, et al. . Intrinsic T2-weighted MRI contrast of the subcortical human brain. In: Proceedings of the Scientific Assembly and National Meeting of the Radiological Society of North America, Chicago, Illinois November 26 to December 1, 2017; Abstract ID: 27997 [Google Scholar]
- 27. Hesdorffer DC, Crandall LA, Friedman D, et al. . Sudden unexplained death in childhood: a comparison of cases with and without a febrile seizure history. Epilepsia 2015;56:1294–300 10.1111/epi.13066 [DOI] [PubMed] [Google Scholar]
- 28. Haines DE. Neuroanatomy: An Atlas of Structures, Sections and Systems. 6th ed Philadelphia: Lippincott Williams & Wilkins; 2004 [Google Scholar]
- 29. Warner JJ. Atlas of Neuroanatomy: With Systems Organization and Case Correlations. Boston: Butterworth-Heinemann; 2001 [Google Scholar]
- 30. DeArmond SJ, Fusco MM, Dewey MM. Structure of the Human Brain: A Photographic Atlas. 3rd ed New York: Oxford University Press; 1989 [Google Scholar]
- 31. Olszewski J, Baxter D. Cytoarchitecture of the Human Brain Stem. 2nd ed Basel: Karger; 1982 [Google Scholar]
- 32. Crandall LA, Devinsky O. Sudden unexplained death in children. Lancet Child Adolesc Health 2017;1:8–9 10.1016/S2352-4642(17)30003-2 [DOI] [PubMed] [Google Scholar]
- 33. Hirsch WL, Kemp SS, Martinez AJ, et al. . Anatomy of the brainstem: correlation of in vitro MR images with histologic sections. AJNR Am J Neuroradiol 1989;10:923–28 [PMC free article] [PubMed] [Google Scholar]
- 34. Miller KL, Stagg CJ, Douaud G, et al. . Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. Neuroimage 2011;57:167–81 10.1016/j.neuroimage.2011.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallay MN, Jeanmonod D, Liu J, et al. . Human pallidothalamic and cerebellothalamic tracts: anatomical basis for functional stereotactic neurosurgery. Brain Struct Funct 2008;212:443–63 10.1007/s00429-007-0170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morel A, Magnin M, Jeanmonod D. Multiarchitectonic and stereotactic atlas of the human thalamus. J Comp Neurol 1997;387:588–630 [DOI] [PubMed] [Google Scholar]
- 37. Lambert C, Chowdhury R, Fitzgerald TH, et al. . Characterizing aging in the human brainstem using quantitative multimodal MRI analysis. Front Hum Neurosci 2013;7:462 10.3389/fnhum.2013.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang Y, Sun W, Toga AW, et al. . A probabilistic atlas of human brainstem pathways based on connectome imaging data. Neuroimage 2018;169:227–39 10.1016/j.neuroimage.2017.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scharoun SM, Bryden PJ. Hand preference, performance abilities, and hand selection in children. Front Psychol 2014;5:82 10.3389/fpsyg.2014.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Urban PP, Caplan LR, eds. Brainstem Disorders. Berlin: Springer-Verlag; 2011 [Google Scholar]
- 41. Janzen J, van 't Ent D, Lemstra AW. The pedunculopontine nucleus is related to visual hallucinations in Parkinson's disease: preliminary results of a voxel-based morphometry study. J Neurol 2012;259:147–54 10.1007/s00415-011-6149-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rolland Y, Vérin M, Payan CA, et al. ; NNIPPS Study Group. A new MRI rating scale for progressive supranuclear palsy and multiple system atrophy: validity and reliability. J Neurol Neurosurg Psychiatry 2011;82:1025–32 10.1136/jnnp.2010.214890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Makino T, Ito S, Kuwabara S. Involvement of pontine transverse and longitudinal fibers in multiple system atrophy: a tractography-based study. J Neurol Sci 2011;303:61–66 10.1016/j.jns.2011.01.014 [DOI] [PubMed] [Google Scholar]
- 44. Gay CT, Hardies LJ, Rauch RA, et al. . Magnetic resonance imaging demonstrates incomplete myelination in 18q- syndrome: evidence for myelin basic protein haploinsufficiency. Am J Med Genet 1997;74:422–31 [DOI] [PubMed] [Google Scholar]
- 45. Baierl P, Förster Ch, Fendel H, et al. . Magnetic resonance imaging of normal and pathological white matter maturation. Pediatr Radiol 1988;18:183–89 10.1007/BF02390391 [DOI] [PubMed] [Google Scholar]
- 46. Krous HF, Chadwick AE, Crandall LA, et al. . Sudden unexpected death in childhood: a report of 50 cases. Pediatr Dev Pathol 2005;8:307–19 10.1007/s10024-005-1155-8 [DOI] [PubMed] [Google Scholar]
- 47. Mugler JP., 3rd Optimized three-dimensional fast-spin-echo MRI. J Magn Reson Imaging 2014;39:745–67 10.1002/jmri.24542 [DOI] [PubMed] [Google Scholar]