Abstract
Colorectal cancer (CRC), the second leading cause of cancer mortality, constitutes a significant global health burden. An accurate, noninvasive detection method for CRC as complement to colonoscopy could improve the effectiveness of treatment. In the present study, SureSelectXT Methyl-Seq was performed on cancerous and normal colon tissues and CLDN1, INHBA and SLC30A10 were found as candidate methylated genes. MethyLight assay was run on formalin-fixed paraffin-embedded (FFPE) and fresh case and control tissues to validate the methylation of the selected gene. The methylation was significantly different (p-values < 2.2e-16) with a sensitivity of 87.17%; at a specificity cut-off of 100% in FFPE tissues. Methylation studies on fresh tissues, indicated a sensitivity of 82.14% and a specificity cut-off of 92% (p-values = 1.163e-07). The biomarker performance was robust since, normal tissues indicated a significant 22.1-fold over-expression of the selected gene as compared to the corresponding CRC tissues (p-value < 2.2e-16) in the FFPE expression assay. In our plasma pilot study, evaluation of the tissue methylation marker in the circulating cell-free DNA, demonstrated that 9 out of 22 CRC samples and 20 out of 20 normal samples were identified correctly. In summary, there is a clinical feasibility that the offered methylated gene could serve as a candidate biomarker for CRC diagnostic purpose, although further exploration of our candidate gene is warranted.
Subject terms: Cancer epigenetics, Cancer epigenetics
Introduction
Colorectal cancer (CRC) constitutes a significant global health burden, leading to over 862,000 deaths globally in 2018. It is the second main cause of cancer mortality in the world and currently stands as the third most common cancer, with a yearly incidence of over one million and eight thousand cases worldwide1. Its leading cause of death is due to liver metastasis with a median survival rate of approximately 30 months. Generally, half of the patients with CRC develop tumor recurrences2. For early-diagnosed CRC patients, the 5-year survival rates are approximately 90% but this lowers to less than 10% in patients with extensive metastases. Thus, the most effective approach to reduce CRC incidence and its mortality is early detection of colonic lesions3. Fortunately, because of implementation and growth of wide spread cancer screening assays, such as colonoscopy as well as increasingly effective therapies, the mortality rate of CRC is lowering in many countries2.
Flexible colonoscopy is considered as the golden screening option for this type of cancer. This procedure is an invasive method and has the highest rate of complications among other screening methods. Besides, it has about 5% error probability, and is relatively expensive. A large number of people are reluctant to undergo colonoscopy because of the fear of pain, and discomfort or even embarrassment4. Altogether, colonoscopy includes patient non-compliance, invasiveness, and uncertain cost-effectiveness, which poses challenges in population-based screening5. All other available CRC screening methods seem to be used less frequently for various reasons. An ideal screening tool should be inexpensive, noninvasive, easy to perform, and accurate with high sensitivity for advanced adenomas or early cancer. It should be widely available and specific enough to avoid unnecessary second level tests. Finally, it should reduce mortality rate significantly above the figure of approximately 30–40%. The fecal occult blood test (FOBT), used in population-based screening, was the first and cheapest noninvasive option, which is widely available now. However, this method requires several attempts and has a low sensitivity (59.7%). In recent years, more specific and sensitive methods such as the fecal immunochemical test (FIT) have replaced FOBT by the guidelines for CRC screening panel4.
In 2017, Wu et al. proposed a miRNA-based approach to quantify fecal blood levels over a broad, clinically relevant range as a new marker class for FOBT. Candidate miRNA markers (hsa-miR-144-3 p, 144-5 p, 451 a, 486-5 p, 363-3 p, 20b-5 p) were identified by small RNA sequencing of human whole blood compared with colorectal epithelia6. In addition, gut microbiota changes in the intestinal microbiota composition in CRC patients have also been reported as CRC biomarkers in several studies7. In the last two decades, many research concerning CRC potential biomarkers have focused on analyzing the fecal DNA8. Stool DNA test was approved by US Food and Drug Administration (FDA) in August 2014 and was available under the commercial name of Cologuard® (Exact Sciences, Madison, WI, USA). It is recommended for the average risk screening in asymptomatic 50–85 years old9. Later, in April 2016 Epi proColon was introduced as the first FDA-approved blood-based CRC screening test. Epi proColon 2.0 CE is based on methylated septin 9 (SEPT9) gene from the circulating cell-free DNA (cfDNA) in the blood. It is now accessible in Europe and different nations such as China10,11. This test offers improved sensitivity and specificity over the first generation Epi proColon test11 although it has not been recommended by the US Preventive Services Task Force in their most recent guidelines due to its very low sensitivity for cancer12. Based on a meta-analysis conducted by Song et al., the performance of the blood methylated SEPT9 assay is superior to the serum protein biomarkers and FIT in symptomatic patients, while it seems to be less potent than FIT in asymptomatic patients13. In a recent meta-analysis comparing CRC patients with healthy subjects, the pooled sensitivity and specificity of SEPT9 methylation were 0.74 (95% CI: 0.61–0.84) and 0.96 (95% CI: 0.95–0.97), respectively14.
During recent years, the non-protein biomarkers so-called “liquid biopsy” of circulating tumor cells and circulating DNA or exosome products has drawn great attention4,15. Developing new biomarkers is relied on understanding the mechanisms exploited by cancer. A variety of biomarkers such as diagnostic, preventive and prognostic has been offered for CRC based the cellular and molecular tumorigenesis.
CRC represents a group of molecularly heterogeneous diseases characterized by genetic and epigenetic alterations such as genetic mutations and DNA methylation, along with a tumorigenesis sequence2. DNA methylation alteration is a hallmark of not only CRC, but also virtually all tumor types. It represents a sophisticated molecular mechanism for annotating genetic information16, and in most cases, modulates the genetic expression level. Although the mechanism and the role of DNA methylation is not completely understood, it is assumed that DNA methylation could affect the binding of the transcription factors to their DNA target sites and subsequently, alter the expression of downstream genes17. In tumor cells, abnormal DNA methylation could be commonly classified into two groups: (1) site specific CpG island promoter hypermethylation, (2) global DNA hypomethylation. Several studies in tumor cells have elucidated site specific CpG island promoter hypermethylation in tumor suppressor genes and global DNA hypomethylation in repetitive sequences2. Besides, the gene body DNA hypermethylation in oncogenes is concerned with gene overexpression, suggesting that the genes regulated by DNA methylation are driving elements in tumorigenesis18. CpG islands (CGIs), shores and shelves that are the 2 kb and 4 kb regions immediately upstream and downstream of the CGI boundaries respectively, are also subjected to methylation. A recent study has shown that cancer specific differentially methylation occurs more often within CGI shores with low CpG densities, rather than CGIs with high CpG densities, indicating a distinct methylation profile in cancer19.
Today, DNA methylation could be mapped by whole-genome bisulfite sequencing (WGBS), which is currently the state-of-the-art technology to achieve a comprehensive, nucleotide-resolution view of the entire epigenome. The methylated differences among samples are generally determined by differentially methylated regions (DMRs). The detection of methylation alterations between cancer patients and normal individuals requires to take the variation of relative methylation within each group into consideration. Such variations could be associated with several technical and biological issues including unequal cytosine conversion rates, different library preparation protocols, different technical methylation assays and the existence of the natural epigenetic variations among individuals17.
In this study, we sought to identify candidate genes for the accurate detection of CRC as novel biomarkers based on the methylation profile and gene expression. There is a clinical feasibility that the methylation biomarkers could be used for detection of CRC. An accurate, noninvasive method based on the methylation biomarkers for early detection and screening of CRC as a complement to colonoscopy has the potential to improve patient satisfaction and the overall effectiveness.
Results
Discovery
Methylation sequencing data processing and quality control
The platform of this study is illustrated in Fig. 1. To perform a genome-wide analysis of DNA methylation in CRC, we applied SureSelectXT Human Methyl-Seq approach with 101 read length that generates 57–76 million Illumina sequencing reads for each sample. Of these, 88.5% to 89.8% were successfully mapped to either strand of the human genome (GRCh37/19). The average number of times that each CpG has been sequenced per sample was between 19.26X and 24.43X (Supplementary Table S1).
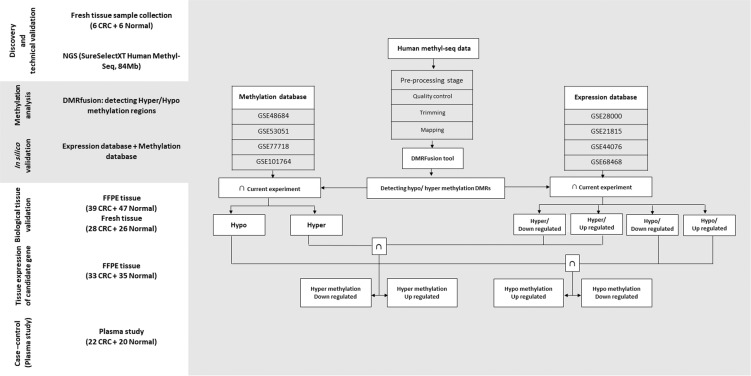
Figure 1.
Project workflow. Discovery and technical validation set: CRC and normal DNA samples collected for the study of NGS. Bioinformatic analysis set: Bioinformatic analysis among Methylation and Expression database and current expriment for detecting robust hypo- hyper methylation genes. Biological validation and tissue expression set: CRC and normal FFPE samples used for Biological validation and candidate gene expression by real time PCR. Plasma study sets: CRC and normal plasma samples used for methylation study of candidate gene.
DMR detection
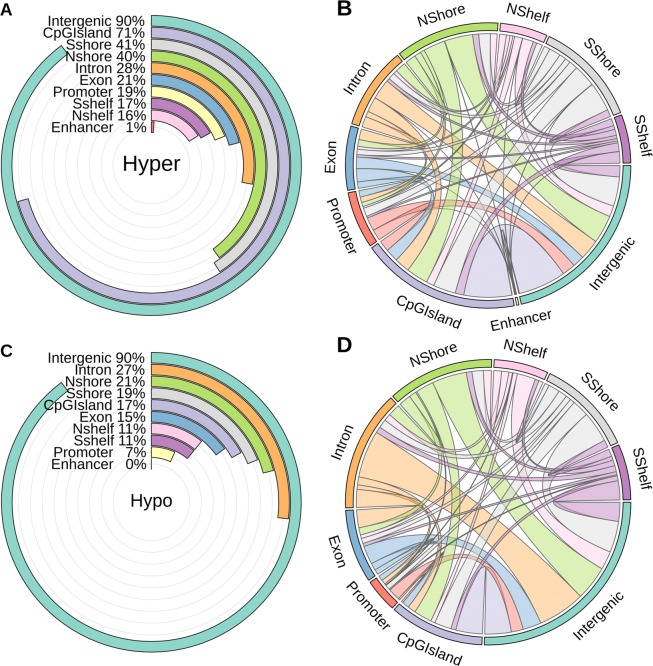
Several thousand hyper and hypo methylation DMRs were detected in multi-samples by comparing CRC and normal groups. In total, we identified 5780 hyper and 8909 hypo DMRs with the length of more than 200 bp, with the highest fold difference score between these two groups, and also with the p-value and false discovery rate (FDR) less than 0.05. Figure 2A,C indicate that the majority of detected DMRs in both hyper and hypo categories were located in the intergenic regions (90%). Seventy one percent of the hyper DMRs were annotated in CpG islands, in comparison with only 17% of the hypo DMRs allocated in these regions. It is worth noting that, the most percentage of the hyper DMRs regions were located in CGI shores, exons, promoters and CGI shelves.
Figure 2.
Statistic information of Hyper-Hypo Methylation DMRs annotation.
As the detected DMRs were larger than 200 bp, some of them were expanded in several regions of the genome, possessing more than one annotation feature. Figure 2B,D illustrate the expansion of the detected hyper and hypo DMRs’ annotation, respectively. To clarify, the majority of the DMRs located in the intergenic regions were expanded to the intronic regions both in hyper and hypo categories (Fig. 2).
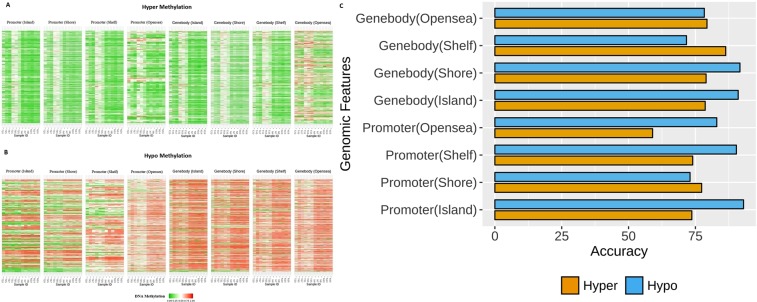
We assessed the distribution of DNA methylation changes across the genomic features based on fold difference score in hyper- and hypo-methylation (Supplementary Fig. S1). The distribution of DNA methylation changes in promoter (opensea) and gene body (opensea) were lower than other features in hyper-methylation, while promoter (island, shore, shelf) and gene body (island, shore, shelf) had similar variation. The distribution of DNA methylation changes in promoter (opensea) and gene body (island, shore, shelf and opensea) were lower than promoter (island, shore, shelf) in the hypo-methylation state. The variation of fold difference score in hyper- and hypo-methylation ranged from 0.2 to 6.6 and 0.3 to 6.3, respectively.
In the present study, we evaluated the overall DNA methylation patterns across different genomic features in hyper- and hypo-methylation states (Fig. 3A,B). We assessed the prediction performance with support vector machine classification method (SVM)20 towards the discrimination between CRC and normal groups on DNA methylation patterns from different genomic features. The most accuracy predictors were the gene body (shelf) 86.33% followed by the gene body (opensea) 79.33%, whereas the promoter (opensea) was 54% that is the least accuracy in hyper-methylation patterns. In hypo-methylation patterns, promoter (island) and gene body (shore) had the most accuracy predictors in the values of 93% and 91.66%, respectively. However, the gene body (shelf) was 71.66%, which is the least accuracy in hypo-methylation patterns. The accuracy of SVM20 classification on DNA methylation patterns across different genomic features were shown in (Fig. 3C).
Figure 3.
Heatmap representation of Human Methyl-seq data between CRC and normal samples in (A) hyper- methylation (B) hypo- methylation DMRs on different genomic features [Promoter (Island), Promoter (Shore), Promoter (Shelf), Promoter (Opensea), Genebody (Island), Genebody (Shore), Genebody (Shelf),Genebody (Opensea)], (C) Accuracy of SVM classification model on different subsets of genes.
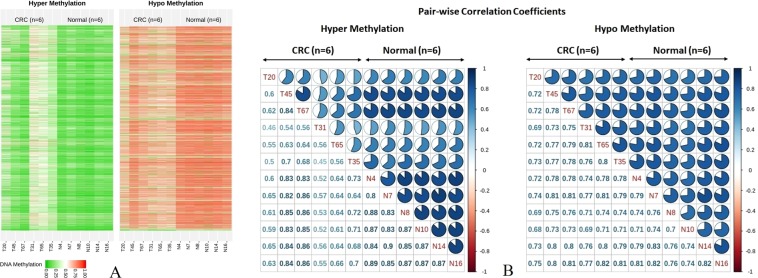
We performed comprehensive DNA methylation profiling based on the detected hyper and hypo DMRs between our CRC and normal groups, illustrated in Fig. 4A. We also assessed the mutation status of KRAS and BRAF in all samples. Three out of 6 CRC samples (T31, T35 and T65) were detected as positive- KRAS somatic mutation but no mutation was detected in normal samples. BRAF somatic mutation was detected in only T20. A hierarchical model (hCluster) clustering approach was used on the 315606 variable CpGs. We identified 3 distinct CRC subgroups, described as cluster 1 (N = 1, T20), cluster 2 (N = 2, T45 & T67) and cluster 3 (N = 3, T31, T65 & T35), having mutation status of positive BRAF and negative KRAS in cluster 1, negative BRAF and negative KRAS in cluster 2 and negative BRAF and positive KRAS in cluster 3. Cluster 1 and cluster 3 were represented as intermediate- and high-relative methylation among CRC samples, while cluster 2 with low-relative methylation, was similar to the normal relative methylation pattern. DNA methylation changes’ pattern among CRC samples in hyper- and hypo-methylation were higher and lower than normal samples, respectively. Figure 4B shows a pair-wise correlation coefficients of DNA methylation changes’ pattern in hyper- and hypo-methylation between CRC and normal samples. The overall CpG methylation profiles between cluster 2 (T45 & T65) and normal samples were highly correlated as pairwise correlation coefficients ranged from 0.80 to 0.89 in hyper-methylation, while the correlation between cluster 1 and cluster 3 were low, ranged between 0.4 and 0.5 in hyper-methylation. Furthermore, the correlation between CRC and normal samples were ranged from 0.6 to 0.8 in hypo-methylation.
Figure 4.
(A) Heatmap representation of DNA Human Methyl-seq data between CRC and normal samples on whole genes. Each column represents one sample and each row represents CpGs methylation status in hyper- hypo Methylation DMRs identified by DMRFusion. (B) Pair-wise correlation coefficients matrix comparing DNA methylation between CRC and normal groups in hyper- hypo methylation.
Robust genes in CRC and gene ontology
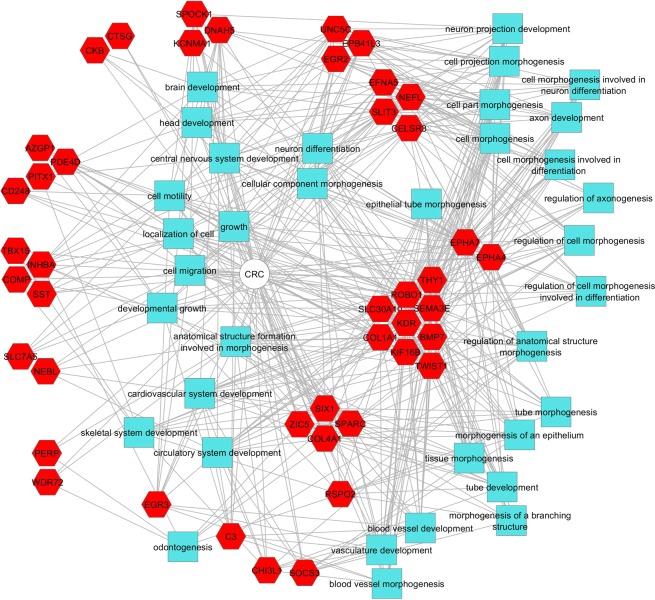
Online analysis was performed by GEO2R software to identify DEGs (differentially expressed genes) or DMGs (differentially methylated genes). By comparing DEGs from the expression datasets with the current experiments, we detected 14 common DEGs including 4 hyper-methylated/down-regulated, 2 hyper-methylated/up-regulated, 4 hypo- methylated/down-regulated and 4 hypo-methylated/up-regulated. The detailed information regarding expression levels were shown in Supplementary Table S2. Common DMGs between the current experiments and the methylation datasets were 827 genes in 2 categories including 449 hyper-methylated genes and 378 hypo-methylated genes. According to the flowchart that was presented in Fig. 1, by comparing these differentially methylated genes with gene expression datasets from microarray datasets, 78 common genes were recognized as functionally methylated genes. Gene ontology of these genes demonstrated that the majority of terms were related to cell differentiation, tissue development and embryonic organ morphogenesis (Fig. 5 and Supplementary Fig. S2).
Figure 5.
Functional enrichment analysis of regulatory network of hyper/hypo methylated genes with significant changes in transcription level.
Besides, comparing these 78 genes with methylation arrays resulted in only 3 common genes SLC30A10 (hyper- methylation/down-regulated), INHBA (hypo-methylation/up-regulated) and CLDN1 (hypo-methylation/up-regulated) that were robustly appeared among previous studies and our current experiment. Statistics information from expression and methylation values for candidate genes were shown in Table 1.
Table 1.
Statistical information of expression and methylation values in candidate genes (SLC30A10, CLDN1 and INHBA).
| Group | Gene Abbreviation | Gene Full Name | Current experiment (FDS) | Expression array (logFC) | |||
|---|---|---|---|---|---|---|---|
| GSE28000 | GSE21815 | GSE44076 | GSE68468 | ||||
| Hyper Methylation Down regulated | SLC30A10 | Solute Carrier Family 30 Member 10 | 0.37 | −2.04 | −2.87 | −2.16 | −2.38 |
| Methylation array (logFC) | |||||||
| GSE48684 | GSE53051 | GSE77718 | GSE101764 | ||||
| 0.28 | 0.44 | 0.25 | 0.19 | ||||
| Hypo Methylation Up regulated | CLDN1 | Claudin 1 | 2.29 | Expression array (logFC) | |||
| GSE28000 | GSE21815 | GSE44076 | GSE68468 | ||||
| 2.29 | 3.14 | 5.09 | 4.98 | ||||
| Methylation array (logFC) | |||||||
| GSE48684 | GSE53051 | GSE77718 | GSE101764 | ||||
| −0.17 | −0.38 | −0.22 | −0.31 | ||||
| Hypo Methylation Up regulated | INHBA | Inhibin beta A subunit | 1.54 | Expression array (logFC) | |||
| GSE28000 | GSE21815 | GSE44076 | GSE68468 | ||||
| 2.88 | 4.64 | 3.51 | 5.12 | ||||
| Methylation array (logFC) | |||||||
| GSE48684 | GSE53051 | GSE77718 | GSE101764 | ||||
| −0.168 | −0.24 | −0.21 | −0.23 | ||||
Technical and biological validations
The technical validation was performed by methylation sensitive high-resolution melting (MS-HRM) on the discovery sample set in CRC and normal control groups. They were accordingly discriminated based on methylation sequencing results.
Methylated DNA markers (MDMs) were identified for our candidate genes and primers and probes were designed and synthesized for the top MDM considered as a diagnostic biomarker.
The candidate MDMs, hyper- methylated/down-regulated, were tested by MethyLight assay on DNA extracted from 39 and 47 formalin-fixed paraffin-embedded (FFPE) case and control tissues, respectively. We also performed the experiment on 28 case and 26 control fresh tissues. The results showed that the methylation at MDM was significantly different (t-test, p-values < 2.2e-16; mean of Cq case 29.58 and mean of Cq control 36.27 for FFPE tissues and t-test, p-values = 1.163e-07; mean of Cq case 29.69 and mean of Cq control 34.96 for fresh tissues) with the sensitivity of 87.17% and 82.14% at a specificity cut-off of 100% and 92% for FFPE and fresh tissues, respectively (Table 2).
Table 2.
Patients and the tumor characteristics of CRC adenocarcinoma in FFPE, fresh tissue and plasma studies. *N/A: Not applicable.
| Type | Biological FFPE tissue validation | Biological fresh tissue validation | Tissue expression | Plasma study | ||||
|---|---|---|---|---|---|---|---|---|
| Cases | Control | Cases | Control | Cases | Control | Cases | Control | |
| Number | (N = 39) | (N = 47) | (N = 28) | (N = 26) | (N = 33) | (N = 35) | (N = 22) | (N = 20) |
| Gender | ||||||||
| Male | 23 (59%) | 27 (57%) | 19 (68%) | 18 (69) | 19 (57%) | 17 (48%) | 13 (59%) | 11(55%) |
| Female | 16 (41%) | 20 (43%) | 9 (32%) | 8 (31) | 14 (43) | 18 (52%) | 9 (41%) | 9 (45%) |
| Age at Diagnosis (Year) | ||||||||
| Median | 58 | 61 | 67 | 59 | 59 | 56 | 67 | 56 |
| Range | [24–85] | [24–85] | [44–82] | [45–84] | [24–85] | [35–77] | [44–82] | [45–84] |
| Tumor site | ||||||||
| Anal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — |
| Rectum | 3 (8%) | 4 (8%) | 10 (36%) | 9 (35%) | 2 (6%) | 0 | 8 (36%) | — |
| Sigmoid | 20(51%) | 24 (51%) | 6 (21%) | 12 (46%) | 18 (54%) | 21 (60%) | 4 (18%) | — |
| Transverse colon | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — |
| Descending colon | 4 (10%) | 5 (11%) | 1 (3%) | 0 | 3 (9%) | 7 (20%) | 1 (5%) | — |
| Ascending colon | 7 (18%) | 7 (15%) | 3 (11%) | 1 (4%) | 5 (15%) | 0 | 3 (14%) | — |
| Cecum | 2 (5%) | 2 (4%) | 8 (29%) | 4 (15%) | 2 (6%) | 0 | 6 (27%) | — |
| Entire colon | 3 (8%) | 5 (11%) | 0 | 0 | 3 (9%) | 7 (20%) | 0 | — |
| TNM stage | ||||||||
| I | 7 (18%) | — | N/A | — | 6 (18%) | — | N/A | — |
| IIA | 8 (21%) | — | N/A | — | 6 (18%) | — | N/A | — |
| IIB | 2 (5%) | — | N/A | — | 2 (6%) | — | N/A | — |
| IIC | 1 (2%) | — | N/A | — | 1 (3%) | — | N/A | — |
| IIIA | 2 (5%) | — | N/A | — | 2 (6%) | — | N/A | — |
| IIIB | 10 (26%) | — | N/A | — | 7 (21%) | — | N/A | — |
| IIIC | 9 (23%) | — | N/A | — | 9 (27%) | — | N/A | — |
| IVA | 0 | — | N/A | — | 0 | — | N/A | — |
| IVB | 0 | — | N/A | — | 0 | — | N/A | — |
| Tumor size | ||||||||
| ≥4 | 15 (39%) | — | N/A | — | 10 (30%) | — | N/A | — |
| 4.1–7.9 | 20 (51%) | — | N/A | — | 20 (61%) | — | N/A | — |
| 8–11.9 | 2 (5%) | — | N/A | — | 1 (3%) | — | N/A | — |
| ≤12 cm | 2 (5%) | — | N/A | — | 2 (6%) | — | N/A | — |
| Mean of Cq | 29.7 | 35.5 | 29.7 | 35 | 32.5 | 27.8 | 36.3 | 0 |
| KRAS mutation | 19 (49%) | — | 9 (32%) | — | 12 (36%) | — | 6 (27%) | — |
| BRAF mutation | 9 (23%) | — | 3 (11%) | — | 5 (15%) | — | 2 (9%) | — |
| Diagnostic | ||||||||
| Sensitivity | 87% | 82% | — | 41% | ||||
| Specificity | 100% | 92% | — | 100% | ||||
To evaluate the ability of the entire biomarker to correctly classify between the two possible conditions (cancerous vs. control), we built a support vector machine learning model (SVM)20, using the GSE42752 dataset as a training set and the GSE52270 as a test set with 10-fold cross validation. The sensitivity and specificity results of classification methods were 91.93% and 82.85%, respectively. In total, 329 DMRs were recognized with an impact on transcription.
Tissue expression of candidate genes
We performed RT-qPCR to compare the expression profile of candidate gene (SLC30A10) between case and control FFPE tissues. The reference gene of GAPDH was used as a reference control in the experiment. The expression pattern revealed that the candidate gene was expressed in both case and control FFPE tissues with a significant difference. As shown in Supplementary Fig. S3, normal tissue mRNA elucidated a significant 22.1-fold upregulation as compared to the CRC tissues (p-value < 2.2e-16 mean of Cq case 32.51 and mean of Cq control 27.84) (Table 2). Hence, the expression level was significantly lower in cancerous tissues.
Tissue-specific methylation study
In order to study the specificity of the candidate MDM (hyper- methylated/down-regulated), it was also tested by MethyLight assay on gastric, liver and esophagus cancer tissues. The results were statistically not significant (p value > 0/05) indicating that the levels of methylation in these cancers were similar to the normal colon tissue.
Plasma pilot study
Methylation of the top MDM was assessed in circulating DNA of CRC patients and normal individuals. Nine out of 22 CRC samples and 20 out of 20 normal samples were identified correctly (Table 2). Besides, the methylation pattern of the top MDM was studied in BLUEPRINT Epigenome database. Based on the BLUEPRINT Epigenome results, the methylation percentage of blood-borne cells for the MDM was lower than 0.10%, indicating a poorly methylation level of hematopoietic cells. It is worth mentioning that the methylation profile of normal blood was similar to the normal colon (data not shown).
Discussion
DNA CpG methylation is usually associated with a closed state of chromatin and has been widely accepted as an important mechanism to maintain gene repression21. In addition to control gene expression, it is known to be a cancer driver mechanism. DNA methylation is negatively correlated with gene expression but so far this association can only be detected for hundreds of genes, and the correlation direction is both positive and negative22.
In the current study, we investigated the interplay between CpG methylation and gene expression. Common DMGs between the current experiments and the methylation datasets were 827 genes in 2 categories including 449 hyper-methylated genes and 378 hypo-methylated genes. Gene ontology analysis for methylated genes showed that the majority of pathways were involved in cell differentiation including smooth muscle cell differentiation (GO: 0051145), positive regulation of animal organ morphogenesis (GO: 0110110), embryonic hind limb morphogenesis (GO: 0035116), and positive regulation of cell junction assembly (GO:1901890). Any aberration in these pathways could result in abnormal cell division or differentiation23. Three genes: SLC30A10, CLDN1 and INHBA were eventually selected. SLC30A10 was hyper-methylated and down-regulated, CLDN1 and INHBA were hypo-methylated and up-regulated. Based on the annotation, the DMR were located in CpG island/shore, shore and shore of SLC30A10, CLDN1 and INHBA, respectively. These results are in agreement with other reports showing that in cancerous tissue distinct methylation profiles are exhibited more frequently within CpG island and shores19.
From the candidate genes, MDMs were tested by MethyLight assay on the DNA extracted from 39 and 47 FFPE case and control tissues, respectively. The results showed that the methylation at MDM was significantly different (p-values < 2.2e-16) with a sensitivity of 87.17% at a specificity cut-off of 100%. Methylation studies on fresh tissues (28 CRC vs 26 control samples), indicated very similar results (p-values = 1.163e-07) with a sensitivity of 82.14% at a specificity cut-off of 92%.
In the current study, significant differences in RNA levels were seen in our candidate gene when comparing cases with controls. We reported a negative correlation in methylation and gene expression in SLC30A10 gene, which was hyper-methylated and down-regulated in the same tissue samples.
SLCs are the second largest family of membrane proteins in the human genome, which transport a broad spectrum of substrates such as nutrients and drugs. The SLC proteins control key physiological functions, including nutrient uptake, ion transport, and could also function as tumor suppressors24–26. The tumor suppressive function of SLCs relates to the inhibition of histone deacetylases (HDAC) and intracellular pH regulation27. This superfamily are located in all cellular and intracellular organelle membranes, except the nuclear membrane28.
In recent years, increasing attention has been given to the role of plasma membrane transporters such as SLCs in cancer. Some SLCs are upregulated in tumor cells due to higher energy and nutritional requirements29.
The SLC30 family is comprised of 10 members. They are involved in transport of zinc (Zn) and manganese (Mn), which are so important in resisting programmed cell death, since Zn has been demonstrated to play a role in a number of cancers30 such as prostate cancer27. Besides, Mn is an essential metal that is required for various cellular enzymatic activities31,32. It may inhibit the acetylation of histone H3 and H4 by increasing and decreasing the activity of HDAC and histone acetyltransferase (HAT), respectively, which eventually causes cell damage and apoptosis33. Shangkuan et al. identified SLC30A10 as a candidate gene among the ten recommended candidate genes correlated with CRC using bioinformatics analysis of microarray data from the GEO database34. In a recent study from Yagi et al. they reported SLC30A10 as a methylation marker in the CRC. Furthermore, they suggested that methylation epigenotype significantly correlated with KRAS and BRAF mutation35.
The claudin (CLDN) family, consists of at least 24 members, and their expression depends on the cell type. Recent reports have shown that the expression of CLDN genes is often altered in cancers. CLDN1 is a capital component of tight junctions (Tjs), which plays an essential role in tumorigenesis36. The role of CLDN in cancer has not been clearly identified, but a recent study suggested that the CLDN1-dependent pathway might be involved in the suppression of CRC expression and correlated to tumor invasiveness and prognostic factors37.
INHBA (Inhibin β A) is a member of the transforming growth factor β (TGF-β) superfamily. It is able to grossly induce embryonic stem cell differentiation. Notwithstanding, the INHBA’s role in cancer has not been fully explained. Recent studies have shown that INHBA gene is overexpressed in cancer38 and correlates with cell proliferation and outcomes in several tumors such as lung39, gastric40, esophageal41, and colorectal38 cancers. Wildi et al. found that activing A, a homodimer of INHBA, is also up-regulated in human CRC42. However, only a few studies have investigated the epigenetically regulation of this gene in cancer. Seder et al. investigated the role of epigenetic regulation of INHBA gene expression in esophageal cancer cells. They suggested that overexpression of INHBA may promote cell proliferation and may be affected by promoter methylation and histone acetylation41. The MDM identified in this study, presenting a specific methylated region in the SLC30A10 gene, has not previously been reported.
To suggest the candidate MDM as a blood biomarker, we assessed the epigenetic signatures of white blood cells, which compose the largest portion of circulating DNA in plasma43. The methylation pattern of the top MDM was studied in BLUEPRINT Epigenome and based on this database, the methylation percentage of blood-borne cells for the MDM was very low (<0.10%), suggesting our candidate MDM as a serum diagnostic biomarker. In addition, in the plasma pilot study we were able to display the methylation pattern of our MDM in plasma cfDNA in CRC patients although the sensitivity was not relatively high. It remains to be determined if technical refinements can improve this issue. Reassessment of the pre-analytical procedures including collecting larger blood volumes, given that the Epi proColon kit requires at least 3.5 ml of plasma for the evaluation of gene methylation11 and utilization of more specific collection tubes designed for ctDNA recovery and standardization of blood processing could ameliorate the plasma results. In addition, advanced technology assay implementation including BEAMing (beads emulsion amplification magnetics), digital droplet PCR, and Target Enrichment Long-probe Quantitative Amplified Signal (TELQAS) could progressively enhance the ability to detect aberrant methylated ctDNA among the total cfDNA in a clinical sample43. The performance outcome observed in the pilot plasma testing would need to be corroborated in further plasma studies in order to compare its results with available blood tests such as septin 9 blood test. There is only one FDA-approved plasma-based DNA methylation biomarker test for CRC in the market44. These days, finding novel biomarkers that are simple, cost-effective, highly specific and sensitive has led to a large clinical interest.
Despite these encouraging early results, there are several potential limitations to the present study. First, patients were enrolled from a limited referral centers. More centers should be involved. Sample sizes especially in plasma phase were not sufficiently large to study the actual performance of the MDM in plasma. Since the main purpose of this study was to find new biomarkers for CRC detection, a plasma pilot study was performed only to demonstrate the eligibility of the candidate gene as a biomarker in plasma. This plasma study was performed in order to evaluate the feasibility, and improve on the study design prior to conducting the entire research project (an ongoing project by the authors’ group). To establish a sensitive approach for solving the technical problems of cfDNA methylation detection in liquid biopsy, we developed and optimized a single tube methylation detection method by selective capture and bisulfite conversion of cfDNA on magnetic beads (unpublished data). This method could significantly improve pre-analytical phase of the diagnostic test leading to a higher sensitivity and specificity for the candidate biomarker in plasma. Thus, further plasma phase studies are needed. The overall sensitivity will be improved with optimized methods as described earlier. Last but not least, further studies are needed to evaluate the organ site specificity of the MDM candidate. In this initial study, we explored a novel MDM for the detection of CRC. There is a clinical feasibility that the candidate MDM could be used for early detection of CRC. Besides, our candidate MDM could be also studied in precancerous lesions, along with the oncogenic cascade from metaplasia through adenoma to CRC.
Materials and Methods
Study design
This study had multiple components illustrated in Fig. 1. Following sample collection, it began with a discovery step based on unbiased methylome sequencing using SureSelectXT Methyl-Seq. Regions demonstrating significant differential methylation were identified and technically validated as candidate DMRs. Candidate methylated genes were chosen based on the shared genes with several other methylation (GSE48684, GSE53051, GSE77718, GSE101764) and expression (GSE28000, GSE21815, GSE44076, GSE68468) datasets. MDMs were chosen based on several criteria including methylation fold change, absolute methylation difference, area under the receiver-operator curve and percentage of control sample methylation.
Biospecimen sources
The current study was approved by Mashhad University of Medical Sciences (MUMS) ethics committee (approval number: 975011) and all methods were performed in accordance with the relevant guidelines and regulations of MUMS. Informed written consent had been obtained from all participants in this study. Patient samples with adenocarcinoma of CRC (N = 6) had a tentative diagnosis determined by colonoscopy. The lesions were removed during endoscopy and confirmed by pathology evaluation at Reza Radiotherapy and Oncology Center (RROC) and Mashhad Pathobiology laboratory by two gastroenterology expert pathologists. Paraffin embedded tissue samples were obtained from Razavi Hospital, Mashhad, Iran. CRC patients with stages I, II & III diseases were included. Excluding criteria of the study were patients with previous CRC, other cancers, positive familial history of adenoma polyposis, inflammatory bowel disease, hereditary CRC and patients with incomplete colonoscopy and documentations. Colorectal tissue controls (N = 6) were taken from individuals who underwent CRC screening by colonoscopy who were negative for adenomatous polyps and CRC through the entire colon and rectum. Demographic characteristics, colonoscopy reports, history of drug, and smoking as well as medical history were collected. The location of lesion was defined as anal, rectum, sigmoid, transverse colon, descending colon, ascending colon, and cecum. The data is presented in Supplementary Table S3.
Discovery
SureSelectXT Methyl-Seq
First, the purity and quantity of DNA tissue samples were evaluated. For passing sample quality control, we included the minimum condition [concentration ≥ 50 ng/μ, purity (A260/A280) ≥ 1.7, volume ≥ 20 ng/μ, total amount ≥ 3.0μg]. The global methylation profiles of cancer and normal colon tissues were analyzed using SureSelectXT Human Methyl-Seq. This platform assesses 84 mega bases (MB) of genome, 3.7 million CpGs, 19.6 Mb CpG islands, 9.8 Mb cancer- and tissue- specific DMRs, 37 MB GENCODE promoters, 48 MB enhancers, CpG island shores/shelves ± 4 Kb and DNase I hypersensitive sites45. Sequencing was performed on the Illumina HiSeq. 4000 (Macrogen Co., South Korea).
Identification of DMRs
To detect DMRs, we used the DMRFusion tool which could assess the optimal and significant hyper- and hypo-methylation DMRs in the genome with minimum redundancy and maximum relevance between cancer and control groups. DMRFusion describes the annotation of DMRs such as the nearest transcription start site (TSS), CGIs/shores/shelves, and regions within genome and visualizes DMRs with high fold difference score (p-value and FDR < 0.05 and type I error < 0.01) as described earlier17. The Human Methyl-Seq analysis was composed of three steps in the pre-processing stage before detecting DMRs. Firstly, the total reads were assessed by Quality Control (QC) tool46 in order to provide informative global and graphical representations of methylation sequencing read quality, prior and after alignment. In this study, our data had high quality in raw sequencing reads among all samples. Secondly, the raw sequencing reads were cleaned by Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to clip sequencing adapters (Illumina universal adapter) and low quality bases (Q < 67 in Illumina) in the 3′ and ambiguous bases in both reads. Thirdly, these raw bisulfite sequencing data were converted into a number of methylated reads and covered reads of cytosines (including unmethylated/methylated reads) by aligning them to the human reference genome (GRCh37/19) using the Bismark tool47.
Identification of shared differentially regions
In the present study, gene expression profiling datasets (GSE28000, GSE21815, GSE44076, GSE68468) and gene methylation profiling datasets (GSE48684, GSE53051, GSE77718, GSE101764) were obtained from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), of the National Center for Biotechnology Information (NCBI). Gene expression included data from 509 CRC and 154 normal mucosa tissue samples. Hundred and ninety three CRC and 161 normal mucosa specimens were enrolled in gene methylation profiling datasets. Data from each microarray and methylation were separately analyzed by online software GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), in order to analyze the DMGs or DEGs by comparing two groups of samples (CRC and normal mucosa tissue) across setup conditions in a GEO series. These DEGs and DMGs were compared with our current experiment results (hypo- or hyper-methylation DMRs) in order to detect robust hypo- or hyper- methylation genes existing in different populations.
In this study, we used p-value and adjusted p-value < 0.05 as the cut-off standard to define DEGs with absolute |fold change| > 2 and DMGs with absolute |fold change| > 0.1. Common hypo-, or hyper- methylation genes were suggested by comparing gene expression and methylation datasets with the result of our current experiment. Thus, the methylation status of the candidate genes were in accordance with gene regulation through gene-specific mechanisms. The overlapping down-regulated and hyper-methylation genes were identified as hyper-methylated, and poorly expressed. Similarly, overlapping upregulated and hypo-methylation genes were considered hypo-methylated, and highly expressed genes. The flowchart illustrating our bioinformatics analysis was presented in Fig. 1.
According to the following criteria: (1) [case/control] methylation fold change (FC) > 20, (2) [case – control] absolute methylation difference (AMD) > 0.10, (3) area under the receiver-operator curve (AUC) > 0.80, p-value < 0.01, (4) control sample methylation < 1.0%, the candidate MDM was identified among the candidate genes derived from Fig. 1.
Technical validation
Methyl-Seq data reliability was validated by MS-HRM analysis. The MS-HRM technical validation was performed on the discovery sample set. Briefly, primers specific for bisulfite converted sequences were designed (MethPrime 2.0 software package) and synthesized (Metabion, Germany). Prior to use, MS-HRM assays were evaluated on methylated and un-methylated bisulfite converted control DNA. Optimal annealing temperatures were determined empirically. Genomic DNA isolated from fresh cancerous and normal colon tissue samples were quantitated by Epoch Microplate Spectrophotometer (Winooski, Vermont, USA). Subsequently, DNA was bisulfite-converted using EpiTect Fast Bisulfite Conversion Kit (Qiagen, Germany) according to the manufacturer’s instructions and amplified using the LightCycler 96 (Roche, Mannheim, Germany).
Biological validation
Tissue validation
Specific probe and primers for MethyLight assay were designed (MethPrime 2.0 software package) and synthesized (Bioneer Corporation, South Korea). The MethyLight assay was run on DNA extracted from 39 and 47 independent FFPE and 28 and 26 fresh case and control tissues, respectively. The DNA was extracted using the QiaAmp FFPE Tissue Mini kit (Qiagen, Germany) and QIAamp® Fast DNA Tissue Kit (Qiagen, Germany) for FFPE (5 sections, 5–10 µm thick) and fresh tissues (5–25 mg, each specimen), respectively. They were bisulfite treated with the EpiTect Fast Bisulfite Conversion Kit (Qiagen, Germany) as described above. For MethyLight assay, the multiplex PCR reactions were performed on bisulfite converted DNA using QuantiTect Multiplex PCR (Qiagen, Germany). All amplifications were assayed on the LightCycler 96 (Roche, Mannheim, Germany), and the results were normalized to the β-actin from the same sample.
KRAS and BRAF mutation detection
All samples were tested for the most common and known KRAS and BRAF mutations for exon 12 and 15, respectively based on a protocol previously published48,49. The details are described in Supplementary Fig. S4 and Table 2.
Tissue expression of candidate gene
Total RNA from 33 and 35 FFPE case and control tissues were isolated using RNeasy FFPE kit (Qiagen, Germany), according to the manufacturer’s protocol. The RNA was quantified by measuring absorbance at 280, 260 and 230 nm in Epoch Microplate Spectrophotometer (Winooski, Vermont, USA). cDNA was synthesized using the RocketScript RT premix (Bioneer, Korea). The gene specific primer targeting the candidate, SLC30A10, and GAPDH genes were designed (by primer premier 6 software) and synthesized (Eurofins, Germany). Quantitative real-time RT-PCR reaction was carried out using HOT FIREPol qPCR mix with EvaGreen (Solis BioDyne- Estonia) on the LightCycler 96 (Roche, Mannheim, Germany) and the experiment was conducted in duplicate for each sample. Five point-standard curve was tested for each primer using the pooled cDNA from all samples. The pooled cDNA, serially diluted in nuclease free water by 2 fold, was used as template for real time RT PCR. After the PCR, mRNA levels were normalized to GAPDH and the relative expression was determined using the 2-ΔΔCT method. All experiments were carried out according to the digital MIQE guidelines50.
Tissue-specific methylation study
We also tested the MDM identified in this study (hyper-methylated/down-regulated) in FFPE tissues of other cancers, including gastric (N = 12), liver (N = 7) and esophagus (N = 8) cancers. The DNAs were extracted using the QiaAmp FFPE Tissue Mini kit (Qiagen, Germany) and bisulfite treated with the EpiTect Fast Bisulfite Conversion Kit (Qiagen, Germany). For MethyLight assay, the multiplex PCR reactions were performed as described above.
Plasma pilot study
Prior to plasma pilot study, the methylation behavior of the top MDM in blood was explored using online BLUEPRINT tool (Software release: 1.0.5, EnsEMBL version: 79, http://www.blueprint-epigenome.eu/). In the plasma study, the top MDM was assessed with MethyLight assay on cfDNA obtained from CRC patients and normal individuals. The cfDNAs were extracted from 1–3 mL plasma of 22 cases and 20 controls using QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany) according to the manufacturer’s protocol and bisulfite-converted (as above). A multiplex PCR reaction was performed on bisulfite converted DNA for the candidate MDM and β-actin gene. The MethyLight assay was normalized to the products of β-actin gene. The PCR run was performed in triplicates and similar to the conditions of tissue validation.
Gene ontology analysis
Hyper- and hypo-methylation DMRs with a significant impact on transcription were selected to draw a network of GO terms and pathways by ToppCluster51 and ClueGo (2.5.3), a Cytoscape application52. Both networks were visualized by Cytoscape (version 3.7.1). Molecular Function, Cellular Components and Biological Process terms were selected as standard GO terms in both applications.
Supplementary information
Acknowledgements
Our sincere thanks go to Mr. Ebrahim Pouladin and Mrs. Nafiseh Shalchi for their close support in colorectal cancer research programs. We would give special thanks to Reza Radiotherapy and Oncology Center staffs and also doctors including Aliakbar Masoomian, Mahdi Zardadi, Mahdi Afzal Aghaei, Elham Mokhtari, Homira Ommati, Ali Reza Radfar, and Hamed Golmohammadzadeh, Fatemeh Varshovee Tabrizi and Mahdieh Dayyani for their cooperation in this study. This study was supported by Reza Radiotherapy and Oncology Center, Mashhad University of Medical Sciences (Proposal No. 975011) and Iran National Science Foundation (INSF; Proposal No. 93048371).
Author contributions
M.A.K. and A.J. conceived and designed the experiments. M.A., A.M.S., F.K. performed the experiments. M.Y., M.A. and E.S.D. analyzed the data. A.T. and K.G. provided the pathology reports. G.S. and A.H. contributed in sampling. M.A.K., A.J., M.Y. and M.A. participated in preparation of the drafted manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59690-0.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kerachian, M. A. & Kerachian, M. Long interspersed nucleotide element-1 (LINE-1) methylation in colorectal cancer. Clinica Chimica Acta. 209–214 (2019). [DOI] [PubMed]
- 3.Bagerabad MB, Tavakolian S, Abbaszadegan MR, Kerachian MA. Promoter hypermethylation of the eyes absent 4 gene is a tumor-specific epigenetic biomarker in Iranian colorectal cancer patients. Acta Medica Iranica. 2018;56(1):21–7. [PubMed] [Google Scholar]
- 4.Kormi SMA, Ardehkhani S, Kerachian MA. New insights into colorectal cancer screening and early detection tests. Colorectal Cancer. 2017;6(2):63–8. doi: 10.2217/crc-2017-0007. [DOI] [Google Scholar]
- 5.Kisiel JB, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology. 2019;69(3):1180–92. doi: 10.1002/hep.30244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu CW, et al. Novel approach to fecal occult blood testing by assay of erythrocyte-specific MicroRNA markers. Digestive diseases and sciences. 2017;62(8):1985–94. doi: 10.1007/s10620-017-4627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Q, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clinical Cancer Research. 2017;23(8):2061–70. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 8.Prince M, Lester L, Chiniwala R, Berger B. Multitarget stool DNA tests increases colorectal cancer screening among previously noncompliant Medicare patients. World journal of gastroenterology. 2017;23(3):464. doi: 10.3748/wjg.v23.i3.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang TJ, Lehmann LS, Kesselheim AS. Precision medicine and the FDA’s draft guidance on laboratory-developed tests. Nature biotechnology. 2015;33(5):449. doi: 10.1038/nbt.3221. [DOI] [PubMed] [Google Scholar]
- 10.Jin P, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. Journal of gastroenterology and hepatology. 2015;30(5):830–3. doi: 10.1111/jgh.12855. [DOI] [PubMed] [Google Scholar]
- 11.Lamb YN, Dhillon S. Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Molecular diagnosis & therapy. 2017;21(2):225–32. doi: 10.1007/s40291-017-0259-y. [DOI] [PubMed] [Google Scholar]
- 12.Rex DK, et al. Colorectal cancer screening: recommendations for physicians and patients from the US Multi-Society Task Force on Colorectal Cancer. The American journal of gastroenterology. 2017;112(7):1016. doi: 10.1038/ajg.2017.174. [DOI] [PubMed] [Google Scholar]
- 13.Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Scientific reports. 2017;7(1):3032. doi: 10.1038/s41598-017-03321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun, G., Meng, J., Duan, H., Zhang, D. & Tang, Y. Diagnostic Assessment of septin9 DNA Methylation for Colorectal Cancer Using Blood Detection: A Meta-Analysis. Pathology & Oncology Research. 1–10 (2018). [DOI] [PubMed]
- 15.Rokni P, Shariatpanahi AM, Sakhinia E, Kerachian MA. BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Genes & genomics. 2018;40(4):423–8. doi: 10.1007/s13258-017-0644-2. [DOI] [PubMed] [Google Scholar]
- 16.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. The Lancet. 2018;392(10149):777–86. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 17.Yassi M, et al. DMRFusion: A differentially methylated region detection tool based on the ranked fusion method. Genomics. 2018;110(6):366–74. doi: 10.1016/j.ygeno.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Weisenberger D, Liang G, Lenz H. DNA methylation aberrancies delineate clinically distinct subsets of colorectal cancer and provide novel targets for epigenetic therapies. Oncogene. 2018;37(5):566. doi: 10.1038/onc.2017.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong M, Goodell MA. New answers to old questions from genome-wide maps of DNA methylation in hematopoietic cells. Experimental hematology. 2014;42(8):609–17. doi: 10.1016/j.exphem.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–97.. [Google Scholar]
- 21.Lande-Diner L, Cedar H. Silence of the genes—mechanisms of long-term repression. Nature Reviews Genetics. 2005;6(8):648. doi: 10.1038/nrg1639. [DOI] [PubMed] [Google Scholar]
- 22.Kim M-C, Kim N-Y, Seo Y-R, Kim Y. An integrated analysis of the genome-wide profiles of dna methylation and mRNA expression defining the side population of a human malignant mesothelioma cell line. Journal of Cancer. 2016;7(12):1668. doi: 10.7150/jca.15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonda DJ, et al. Review: cell cycle aberrations and neurodegeneration. Neuropathol Appl Neurobiol. 2010;36(2):157–63. doi: 10.1111/j.1365-2990.2010.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhutia YD, et al. SLC transporters as a novel class of tumour suppressors: identity, function and molecular mechanisms. Biochemical Journal. 2016;473(9):1113–24. doi: 10.1042/BJ20150751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.César-Razquin A, et al. A call for systematic research on solute carriers. Cell. 2015;162(3):478–87. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Höglund PJ, Nordström KJ, Schiöth HB, Fredriksson R. The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Molecular biology and evolution. 2010;28(4):1531–41. doi: 10.1093/molbev/msq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng P, Li TL, Guan ZX, Franklin RB, Costello LC. Direct effect of zinc on mitochondrial apoptogenesis in prostate cells. The Prostate. 2002;52(4):311–8. doi: 10.1002/pros.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Vasiliou K, Nebert DW. Analysis and update of the human solute carrier (SLC) gene superfamily. Human genomics. 2009;3(2):195. doi: 10.1186/1479-7364-3-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Gebali S, Bentz S, Hediger MA, Anderle P. Solute carriers (SLCs) in cancer. Molecular aspects of medicine. 2013;34(2–3):719–34. doi: 10.1016/j.mam.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Franklin RB, Costello LC. The important role of the apoptotic effects of zinc in the development of cancers. Journal of cellular biochemistry. 2009;106(5):750–7. doi: 10.1002/jcb.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, P., Bowman, A. B., Mukhopadhyay, S. & Aschner, M. editors. SLC30A10: A novel manganese transporter. Worm,Taylor & Francis.2015; p. e1042648. [DOI] [PMC free article] [PubMed]
- 32.Huang L, Tepaamorndech S. The SLC30 family of zinc transporters–a review of current understanding of their biological and pathophysiological roles. Molecular aspects of medicine. 2013;34(2–3):548–60. doi: 10.1016/j.mam.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Guo Z, et al. Manganese chloride induces histone acetylation changes in neuronal cells: Its role in manganese-induced damage. Neurotoxicology. 2018;65:255–63. doi: 10.1016/j.neuro.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Shangkuan WC, et al. Risk analysis of colorectal cancer incidence by gene expression analysis. PeerJ. 2017;5:e3003. doi: 10.7717/peerj.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagi K, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clinical Cancer Research. 2010;16(1):21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 36.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer research. 2005;65(21):9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa S, et al. Expression of CLDN1 in colorectal cancer: a novel marker for prognosis. International journal of oncology. 2011;39(4):791–6. doi: 10.3892/ijo.2011.1102. [DOI] [PubMed] [Google Scholar]
- 38.Okano M, et al. Significance of INHBA expression in human colorectal cancer. Oncology reports. 2013;30(6):2903–8. doi: 10.3892/or.2013.2761. [DOI] [PubMed] [Google Scholar]
- 39.Seder CW, et al. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia (New York, NY). 2009;11(4):388–96. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katayama Y, et al. Clinical Significance of INHBA Gene Expression in Patients with Gastric Cancer who Receive Curative Resection Followed by Adjuvant S-1 Chemotherapy. In vivo (Athens, Greece). 2017;31(4):565–71. doi: 10.21873/invivo.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seder CW, et al. INHBA overexpression promotes cell proliferation and may be epigenetically regulated in esophageal adenocarcinoma. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2009;4(4):455–62. doi: 10.1097/JTO.0b013e31819c791a. [DOI] [PubMed] [Google Scholar]
- 42.Wildi S, et al. Overexpression of activin A in stage IV colorectal cancer. Gut. 2001;49(3):409–17. doi: 10.1136/gut.49.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun K, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proceedings of the National Academy of Sciences. 2015;112(40):E5503–E12. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song L, et al. The quantitative profiling of blood mSEPT9 determines the detection performance on colorectal tumors. Epigenomics. 2018;10(12):1569–83. doi: 10.2217/epi-2017-0154. [DOI] [PubMed] [Google Scholar]
- 45.Soto J, Rodriguez-Antolin C, Vallespin E, De Castro Carpeno J, De Caceres II. The impact of next-generation sequencing on the DNA methylation–based translational cancer research. Translational Research. 2016;169:1–18. e1. doi: 10.1016/j.trsl.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Wingett, S. W. & Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. 7, F1000Research (2018). [DOI] [PMC free article] [PubMed]
- 47.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. bioinformatics. 2011;27(11):1571–2. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamzehzadeh L, Khadangi F, Karimiani EG, Pasdar A, Kerachian MA. Common KRAS and NRAS gene mutations in sporadic colorectal cancer in Northeastern Iranian patients. Current Problems in Cancer. 2018;42(6):572–81. doi: 10.1016/j.currproblcancer.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Thierry AR, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nature medicine. 2014;20(4):430. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 50.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55(4):611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 51.Kaimal, V., Bardes, E. E., Tabar, S. C., Jegga, A. G. & Aronow, B. J. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic acids research. 38(Web Server issue), W96–102 (2010). [DOI] [PMC free article] [PubMed]
- 52.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.