Abstract
Brahma-related gene 1 (BRG1), an ATPase subunit of the SWItch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex controls multipotent neural crest formation by regulating epithelial-mesenchymal transition (EMT)-related genes with adenosine triphosphate-dependent chromodomain-helicase DNA-binding protein 7 (CHD7). The expression of BRG1 engages in pre-mRNA splicing through interacting RNPs in cancers; however, the detailed molecular pathology of how BRG1and CHD7 relate to cancer development remains largely unveiled. This study demonstrated novel post-transcriptional regulation of BRG1 in EMT and relationship with FIRΔexon2, which is a splicing variant of the far-upstream element-binding protein (FUBP) 1-interacting repressor (FIR) lacking exon 2, which fails to repress c-myc transcription in cancers. Previously, we have reported that FIR complete knockout mice (FIR−/−) was embryonic lethal before E9.5, suggesting FIR is crucial for development. FIRΔexon2 acetylated H3K27 on promoter of BRG1 by CHIP-sequence and suppressed BRG1 expression post-transcriptionally; herein BRG1 suppressed Snai1 that is a transcriptional suppressor of E-cadherin that prevents cancer invasion and metastasis. Ribosomal proteins, hnRNPs, splicing-related factors, poly (A) binding proteins, mRNA-binding proteins, tRNA, DEAD box, and WD-repeat proteins were identified as co-immunoprecipitated proteins with FIR and FIRΔexon2 by redoing exhaustive mass spectrometry analysis. Furthermore, the effect of FIRΔexon2 on FGF8 mRNA splicing was examined as an indicator of neural development due to impaired CHD7 revealed in CHARGE syndrome. Expectedly, siRNA of FIRΔexon2 altered FGF8 pre-mRNA splicing, indicated close molecular interaction among FIRΔexon2, BRG1 and CHD7. FIRΔexon2 mRNA was elevated in human gastric cancers but not in non-invasive gastric tumors in FIR+/ mice (K19-Wnt1/C2mE x FIR+/−). The levels of FIR family (FIR, FIRΔexon2 and PUF60), BRG1, Snai1, FBW7, E-cadherin, c-Myc, cyclin-E, and SAP155 increased in the gastric tumors in FIR+/− mice compared to those expressed in wild-type mice. FIR family, Snai1, cyclin-E, BRG1, and c-Myc showed trends toward higher expression in larger tumors than in smaller tumors in Gan-mice (K19-Wnt1/C2mE). The expressions of BRG1 and Snai1 were positively correlated in the gastric tumors of the Gan-mice. Finally, BRG1 is a candidate substrate of F-box and WD-repeat domain-containing 7 (FBW7) revealed by three-dimensional crystal structure analysis that the U2AF-homology motif (UHM) of FIRΔexon2 interacted with tryptophan-425 and asparate-399 (WD)-like motif in the degron pocket of FBW7 as a UHM-ligand motif. Together, FIRΔexon2 engages in multi-step post-transcriptional regulation of BRG1, affecting EMT through the BRG1/Snai1/E-cadherin pathway and promoting tumor proliferation and invasion of gastric cancers.
Subject terms: High-throughput screening, Cancer genomics, Focal adhesion, Cell signalling, High-throughput screening
Introduction
The brahma-related gene 1 (BRG1), an ATPase subunit of the SWItch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex, controls multipotent neural crest formation by regulating epithelial-mesenchymal transition (EMT)-related genes with adenosine triphosphate-dependent chromodomain-helicase DNA-binding protein 7 (CHD7)1,2. Integrative analysis identifies co-dependent gene expression regulation of BRG1 and CHD7 at distal regulatory sites in embryonic stem cells2. The expression of BRG1 engages in pre-mRNA splicing through interacting RNPs in cancers;3–5 however, the detailed molecular pathology of how BRG1and CHD7 relate to cancer development remains largely unknown.
The far-upstream element (FUSE) is a sequence required for the proper expression of the human c-myc gene6. Yeast two-hybrid analysis revealed that FUBP1 binds to a protein that has transcriptional inhibitory activity, termed FUBP1-interacting repressor (FIR)7. FIR is a splicing variant lacking exon 5 of poly (U)-binding-splicing factor (PUF60)8. FIRΔexon2, a splicing variant of FIR that lacks exon 2, failed to repress c-myc as a dominant-negative form of FIR in cancers7. This study examined a novel mechanism how FIRΔexon2 regulates BRG1 through the epigenome, transcription and alternative splicing in cancer development. To explore this purpose, following functional analyses of FIR and FIRΔexon2 were performed with several cancer cell lines and animal models having non-invasive gastric tumor. First, FIR and FIRΔexon2-binding proteins were revaluated among the data previously identified by exhaustive mass spectrometry analysis9. In fact, an autoantibody against FIRΔexon2 was detected in the sera of various cancer patients10,11, showing that FIRΔexon2 protein authentically expresses in cancers. Given that c-Myc activates ribosome protein synthesis, FIRΔexon2 is crucial for carcinogenesis in terms of ribosome protein synthesis in cancers. Second, the effect of FIRΔexon2 on FGF8 mRNA splicing was examined as an indicator of neural development in CHARGE syndrome which is an autosomal-dominant, multiple congenital anomaly condition that is characterized by vision and hearing loss, congenital heart disease, and malformations of craniofacial and other structures. The PUF60 gene, as well as CHD7, is considered responsible for CHARGE syndrome, which cooperatively translocates nucleosomes to permit transcription of FGF8 by RNA pol II in neural development12. Further, the U2AF-homology motif (UHM) of PUF60 has been reported to interact with the WD-repeat of SAP155 (SF3B1)13. Additionally, expression of E-cadherin (encoded by the CDH1/Cadherin 1 gene) prevents cancer invasion and metastasis14,15. E-cadherin suppresses initiation and epithelial-mesenchymal transition (EMT) in early-stage gastric carcinogenesis16,17. Previous studies indicated that loss of F-box and WD-repeat domain-containing 7 (FBW7) induced EMT in cancers18,19. The Snai1 transcriptionally represses E-cadherin and promotes tumor proliferation and invasion20,21. The regulation of BRG1 by FBW7 has been studied that BRG1 is a substrate of FBW7 and was found to suppress E-cadherin through the FBW7/BRG1/Snai1 axis in gastric cancer4. Since FIRΔexon2 was co-immunoprecipitated with WD-repeat proteins9, a potential interaction was assessed between FIRΔexon2 and the WD-like motif in the degron pocket of FBW7 by three-dimension crystal analysis22,23. Finally, low molecular weight chemicals that bind to FIRΔexon2 were identified from natural chemical libraries (RIKEN, Wako, Saitama, Japan) and we investigated their effect on BRG1/Snai1 pathway for clinical applications. Collectively, this study proposed that FIRΔexon2 engages in multi-step post-transcriptional regulation of BRG1, affecting EMT through the BRG1/Snai1/E-cadherin pathway and promoting tumor proliferation and invasion of gastric cancers.
Results
Ribosomal proteins and splicing factors were co-immunoprecipitated with FIR and FIRΔexon2
The FIR/PUF60 gene is located at 8q24.3 and contains 12 exons. Genomic structure of PUF60, FIR, and FIRΔexon2 are indicated (Fig. 1a). This study revaluated the co-immunoprecipitated proteins with FIR or FIRΔexon2 previously identified by a direct nanoflow liquid chromatography-tandem mass spectrometry analysis of the 293 T cells (Fig. 1b, top) or by GeLC-MS of Flag-conjugated bead pull down with LC-MS in HeLa cells (Fig. 1b, botom) according to the previously established cell culture systems9. Particularly, ribosomal proteins, hnRNPs, splicing-related factors, poly(A) binding proteins, mRNA-binding proteins, tRNA, WD-repeat proteins or DEAD box proteins were commonly co-immunoprecipitated with FIR or FIRΔexon2 (Table 1 and Supplementary Table S1), indicating that both FIR and FIRΔexon2 participate in post-transcriptional or translational processes. Consequently, FIR families, PUF60, FIR, and FIRΔexon2, potentially link among epigenetic modification, transcription, post-transcription, and alternative splicing in gene regulation. Recently, the ribosomal RPL10 R98S mutation was associated with T-cell type acute lymphocytic leukemia (T-ALL) pathogenesis24–26. Notably, FIR+/−TP53−/− generated T-ALL27, indicating that FIRΔexon2 modifies ribosomal protein synthesis in cancers. Moreover, transformation/transcription domain-associated protein (TRRAP) that is pivotal for rDNA transcription28 was co-immunoprecipitated with FIRΔexon2, but not with FIR9. Recently, the BRG1/Snai1 pathway has been reported to regulate E-cadherin in gastric cancers4, the effect of FIRΔexon2 on the BRG1/Snai1 pathway was investigated in this study. Comprehensive RNA-sequencing and ChIP-sequencing analysis with FIR and FIRΔexon2 overexpressed in HeLa cells were examined to reveal the mechanism. As result, FIRΔexon2, but not FIR, reduced the level of H3K27ac at the BRG1 promotor (Fig. 1c). Notably, the overexpression of FIRΔexon2 reduced the level of H3K27ac by 37% compared to that in the untreated sample in the BRG1 genome region, but its mRNA level remained unchanged (Fig. 1c). The protein expression of BRG1 was decreased by FIRΔexon2 overexpression (Fig. 1d), suggesting that FIRΔexon2 partly affects nucleosome remodeling. Possibly, the histone modification of translational processes in BRG1 expression were interrupted by FIRΔexon2 and its complex.
Fig. 1. FIRΔexon2, but not FIR, reduced the level of H3K27ac of BRG1.
a Genome structure of PUF60, FIR, and FIRΔexon2 are indicated. The FIR/PUF60 gene is located at 8q24.3 and contains 12 exons. Primers and probes used for RT-PCR are indicated. PUF60 consists of 559 amino acids (a.a.) and FIR, lacking exon5, is 542 a.a. FIRΔexon2 lacking exon2 of transcriptional repression domain, is 513 a.a. RNA recognition motif and U2AF-homology motif (UHM) are indicated. The 6B4 (Supplementary Table S5) monoclonal antibody recognizes the amino-terminus of FIR family (arrow). FIR family, consisting of FIR, PUF60 and FIRΔexon2, could not be identified separately by the monoclonal antibody (6B4) used in this study. The list of co-immunoprecipitated proteins with FIR and FIRΔexon2 detected by a direct nanoflow liquid chromatography-tandem mass spectrometry analysis in 293 T cells and GeLC-MS of Flag-conjugated bead pull down with LC-MS in HeLa cells (b), c The overexpression of FIRΔexon2, dominant-negative of FIR, reduced the level of H3K27ac by 37% as compared to that in untreated sample in the BRG1 genome region, but its mRNA level remained unchanged. One possibility is that translational processes were interfered by FIRΔexon2 and its counterparts. d The protein expression of BRG1 was decreased by FIRΔexon2 overexpression.
Table 1.
List of FIR or FIRΔexon2 binding proteins detected by GeLC-MS of Flag-conjugated beads pull down with LC-MS in HeLa cells.
| IPI | kDa | Unique peptide | Unique peptide | |
|---|---|---|---|---|
| Identified protein name | Accession number | Molecular weight (kDa) | FIR_FlagIP | FIRΔexon2_FlagIP |
| Isoform 1 of Poly(U)-binding-splicing factor PUF60 | IPI00069750 | 60 | 5 | 10 |
| Actin, alpha cardiac muscle 1 | IPI00023006 | 42 | 5 | 7 |
| LIM domain and actin-binding 1 isoform a | IPI00883896 | 85 | 13 | 6 |
| 60 kDa heat shock protein, mitochondrial | IPI00784154 | 61 | 4 | 5 |
| Isoform 1 of U5 small nuclear ribonucleoprotein 200 kDa helicase | IPI00420014 | 245 | 1 | 5 |
| Actin-related protein 3 | IPI00028091 | 47 | 4 | 4 |
| Isoform 2 of Suppressor of SWI4 1 homolog | IPI00219793 | 52 | 3 | 4 |
| Isoform 1 of Drebrin | IPI00003406 | 71 | 7 | 3 |
| 60 S ribosomal protein L18 | IPI00215719 | 22 | 5 | 3 |
| ATP-dependent RNA helicase DDX50 | IPI00031554 | 83 | 4 | 3 |
| WD-repeat-containing protein 3 | IPI00009471 | 106 | 3 | 3 |
| Isoform 1 of RNA-binding protein 39 | IPI00163505 | 59 | 3 | 3 |
| RNA-binding protein 28 | IPI00304187 | 86 | 2 | 3 |
| WD-repeat-containing protein 36 | IPI00169325 | 105 | 1 | 3 |
| WD-repeat-containing protein 75 | IPI00217240 | 95 | 1 | 3 |
| cDNA FLJ56443, highly similar to Putative ATP-dependent RNA helicase DHX33 | IPI00302860 | 85 | 1 | 3 |
| Glioma tumor suppressor candidate region gene 2 protein | IPI00024567 | 54 | 1 | 3 |
| CCAAT/enhancer-binding protein zeta | IPI00306723 | 121 | 1 | 3 |
| Protein MAK16 homolog | IPI00332428 | 35 | 1 | 3 |
| Actin-related protein 2/3 complex subunit 2 | IPI00005161 | 34 | 6 | 2 |
| Isoform 1 of Guanine nucleotide-binding protein G(i), alpha-2 subunit | IPI00748145 | 40 | 3 | 2 |
| F-actin-capping protein subunit alpha-2 | IPI00026182 | 33 | 3 | 2 |
| Poly [ADP-ribose] polymerase 1 | IPI00449049 | 113 | 2 | 2 |
| ATP-dependent RNA helicase DDX24 | IPI00006987 | 96 | 2 | 2 |
| Isoform 1 of Transformer-2 protein homolog beta | IPI00301503 | 34 | 2 | 2 |
| RNA-binding protein, autoantigenic (HnRNP-associated with lethal yellow homolog (Mouse)), isoform CRA_a (Fragment) | IPI00011268 | 33 | 2 | 2 |
| Apolipoprotein A-I | IPI00021841 | 31 | 2 | 2 |
| 40 S ribosomal protein S2 | IPI00013485 | 31 | 2 | 2 |
| 60 S ribosomal protein L10a | IPI00412579 | 25 | 2 | 2 |
| 60 S ribosomal protein L15 | IPI00470528 | 24 | 2 | 2 |
| 40 S ribosomal protein S15 | IPI00479058 | 17 | 2 | 2 |
| WD-repeat-containing protein 43 | IPI00055954 | 79 | 1 | 2 |
| Isoform 1 of Fragile X mental retardation syndrome-related protein 1 | IPI00016249 | 70 | 1 | 2 |
| Isoform 8 of Fragile X mental retardation 1 protein | IPI00645666 | 66 | 1 | 2 |
| 60 S ribosomal protein L21 | IPI00247583 | 19 | 1 | 2 |
| Thyroid hormone receptor-associated protein 3 | IPI00104050 | 109 | 1 | 2 |
| Fragile X mental retardation syndrome-related protein 2 | IPI00016250 | 77 | 1 | 2 |
| TDP43 | IPI00025815 | 45 | 1 | 2 |
| Forty-two-three domain-containing protein 1 | IPI00289907 | 36 | 1 | 2 |
| Isoform 1 of Uncharacterized protein C1orf77 | IPI00300990 | 26 | 1 | 2 |
| 40 S ribosomal protein S14 | IPI00026271 | 16 | 1 | 2 |
| Heat shock protein HSP 90-beta | IPI00414676 | 83 | 8 | 1 |
| Actin-related protein 2 | IPI00005159 | 45 | 7 | 1 |
| Uncharacterized protein C19orf21 | IPI00217121 | 75 | 6 | 1 |
| Isoform 4 of Myosin-XIX | IPI00062809 | 87 | 4 | 1 |
| myosin VA isoform 2 | IPI00873959 | 212 | 4 | 1 |
| Splicing factor 3B subunit 1(SAP155) | IPI00026089 | 146 | 4 | 1 |
| Ribosome biogenesis protein BOP1 | IPI00028955 | 84 | 3 | 1 |
| 60 S ribosomal protein L34 | IPI00219160 | 13 | 3 | 1 |
| Actin-related protein 2/3 complex subunit 1B | IPI00005160 | 41 | 3 | 1 |
| protein phosphatase 1, catalytic subunit, alpha isoform 3 | IPI00027423 | 39 | 3 | 1 |
| 40 S ribosomal protein S16 | IPI00221092 | 16 | 3 | 1 |
| Actin-related protein 2/3 complex subunit 1 A | IPI00333068 | 42 | 2 | 1 |
| Isoform 1 of 60 S ribosomal protein L11 | IPI00376798 | 20 | 2 | 1 |
| Histone H2A.V | IPI00018278 | 14 | 2 | 1 |
| Enhancer of rudimentary homolog | IPI00029631 | 12 | 2 | 1 |
| Isoform 1 of DNA-dependent protein kinase catalytic subunit | IPI00296337 | 469 | 2 | 1 |
| protein phosphatase 1, regulatory subunit 9B | IPI00045550 | 89 | 2 | 1 |
| cDNA FLJ37875 fis, clone BRSSN2018771, highly similar to Poly(A)-binding protein 1 | IPI00796945 | 71 | 2 | 1 |
| Isoform A of Phosphate carrier protein, mitochondrial | IPI00022202 | 40 | 2 | 1 |
| Isoform Long of Transformer-2 protein homolog alpha | IPI00013891 | 33 | 2 | 1 |
| similar to beta-actin | IPI00739464 | 17 | 2 | 1 |
| Isoform 1 of Synaptic glycoprotein SC2 | IPI00100656 | 36 | 2 | 1 |
| ADP/ATP translocase 3 | IPI00291467 | 33 | 2 | 1 |
| 60 S ribosomal protein L19 | IPI00025329 | 23 | 2 | 1 |
| Cystatin-A | IPI00032325 | 11 | 2 | 1 |
| Isoform 1 of Spectrin beta chain, brain 2 | IPI00012645 | 271 | 16 | 0 |
| epiplakin 1 | IPI00010951 | 556 | 12 | 0 |
| Src substrate cortactin | IPI00029601 | 62 | 11 | 0 |
| Isoform 1 of Myosin-XVIIIa | IPI00760846 | 233 | 8 | 0 |
| Isoform 2 of Myosin-VI | IPI00008455 | 146 | 6 | 0 |
| Pre-mRNA-processing-splicing factor 8 | IPI00007928 | 274 | 5 | 0 |
| Isoform 2 of Supervillin | IPI00018370 | 201 | 5 | 0 |
| Isoform 1 of Actin-binding LIM protein 1 | IPI00329495 | 88 | 5 | 0 |
| Tropomodulin-1 | IPI00002375 | 41 | 5 | 0 |
| Isoform 1 of Probable ATP-dependent RNA helicase DDX31 | IPI00043990 | 94 | 4 | 0 |
| 40 S ribosomal protein S3 | IPI00011253 | 27 | 4 | 0 |
| Myosin-Ie | IPI00329672 | 127 | 3 | 0 |
| Isoform 1 of Nexilin | IPI00180404 | 81 | 3 | 0 |
| D-3-phosphoglycerate dehydrogenase | IPI00011200 | 57 | 3 | 0 |
| EF-hand domain-containing protein D1 | IPI00031091 | 27 | 3 | 0 |
| Cofilin-1 | IPI00012011 | 19 | 3 | 0 |
| Filamin A | IPI00909642 | 246 | 2 | 0 |
| Isoform 2 of Nuclear mitotic apparatus protein 1 | IPI00006196 | 237 | 2 | 0 |
| Isoform 1 of Chromodomain-helicase-DNA-binding protein 4 | IPI00000846 | 218 | 2 | 0 |
| Importin subunit beta-1 | IPI00001639 | 97 | 2 | 0 |
| Nuclear cap-binding protein subunit 1 | IPI00019380 | 92 | 2 | 0 |
| ATP-dependent RNA helicase DDX51 | IPI00217541 | 72 | 2 | 0 |
| Plastin-3 | IPI00216694 | 71 | 2 | 0 |
| Isoform 1 of Heterogeneous nuclear ribonucleoprotein Q | IPI00018140 | 70 | 2 | 0 |
| cDNA FLJ41552 fis, clone COLON2004478, highly similar to Protein Tro alpha1 H,myeloma | IPI00647704 | 53 | 2 | 0 |
| Pyruvate kinase | IPI00847989 | 50 | 2 | 0 |
| Isoform 1 of Sequestosome-1 | IPI00179473 | 48 | 2 | 0 |
| Tricarboxylate transport protein, mitochondrial | IPI00294159 | 34 | 2 | 0 |
| 60 S ribosomal protein L6 | IPI00329389 | 33 | 2 | 0 |
| Isoform 2 of Voltage-dependent anion-selective channel protein 2 | IPI00024145 | 30 | 2 | 0 |
| Metaxin-2 | IPI00025717 | 30 | 2 | 0 |
| 60 S ribosomal protein L7-like 1 | IPI00456940 | 29 | 2 | 0 |
| 14-3-3 protein epsilon | IPI00000816 | 29 | 2 | 0 |
| Isoform 1 of Splicing factor, arginine/serine-rich 7 | IPI00003377 | 27 | 2 | 0 |
| Chloride intracellular channel protein 1 | IPI00010896 | 27 | 2 | 0 |
| Isoform 1 of 60 S ribosome subunit biogenesis protein NIP7 homolog | IPI00007175 | 20 | 2 | 0 |
| Profilin-1 | IPI00216691 | 15 | 2 | 0 |
| Ribosomal protein 26 (RPS26) pseudogene | IPI00401819 | 13 | 2 | 0 |
| Periodic tryptophan protein 2 homolog | IPI00300078 | 102 | 0 | 3 |
| Isoform 1 of Transformation/transcription domain-associated protein | IPI00069084 | 438 | 0 | 2 |
| hypothetical LOC731605 | IPI00886987 | 100 | 0 | 2 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 isoform 1 | IPI00152510 | 99 | 0 | 2 |
| Isoform Long of Splicing factor, proline- and glutamine-rich | IPI00010740 | 76 | 0 | 2 |
| Ribosome biogenesis protein WDR12 | IPI00304232 | 48 | 0 | 2 |
FGF8 splicing variant analysis by up- or downregulated FIR or FIRΔexon2 in cancer cells
FIR complete knockout C57BL/6 mice (FIR−/−) was embryonic lethal before E9.5; strongly suggests that FIR is crucial for development27. Recently, the germ line mutations or deletions in the RNA recognition motifs (RRMs) of the PUF60 gene were reported in Verheij or CHARGE syndrome12,29–31. In CHARGE syndrome patients, protein interactions are spoiled in alternative splicing of FGF8 mRNA pre-mRNA processing in neural development due to dysfunction among BRG1, SWI/SNF and CHD7. Furthermore, CHD7 binds to unmethylated active rDNA and promotes rRNA transcription32. Five gastric cancer cell lines, MNK7, MNK45, MNK74, NUGC3, and NIGC4, were examined FGF8 expression at protein and mRNA levels (Supplementary Figs. S1a, S1b). NUGC4 showed highest expression of FGF8 expression among them, however, the transfection efficiency of FIR-FLAG and FIRΔexon2-FLAG plasmids were low in NUGC4, adherent round cells (Supplementary Fig. S1c), or Jurkat cells; therefore, MCF7 were used in this study33. Expectedly, FIR or FIRΔexon2 affected to the alternative splicing of FGF8 mRNA expression revealed by PCR analysis in cancer cell lines, MCF-7, HeLa, and Jurkat cells (Lanes 1 and 7; 100 bp marker, lane 2: HeLa cells, lane 3; MCF-7 cells, lane 4; Jurkat cells, lane 5; negative control, lane 6; FGF8 positive control; Fig. 2a). The internal control was β-actin (Fig. 2b). Western blotting analysis of knockdown of FIR or FIRΔexon2 in MCF-7 cells and normalized intensity of bands were indicated (Fig. 2c). qRT-PCR of FIR and FIRΔexon2 mRNAs are shown in Fig. 2d. Overexpression or knockdown of FIR or FIRΔexon2 changed the alternative splicing of FGF8 pre-mRNA in MCF-7 cells (Fig. 2e). These results indicated that FIR or FIRΔexon2 surely engages in the alternative splicing process of FGF8 pre-mRNA at least in cancer cells, in vitro.
Fig. 2. FIR and FIRΔexon2 affected alternative splicing of FGF8 mRNA.
a Expression of FGF8 analysis was confirmed by PCR using cancer cell lines, HeLa, MCF-7 and Jurkat cells. Lane 1 and 7: 100 bp marker, lane 2: HeLa cells, lane 3: MCF-7 cells, lane 4: Jurkat cells, lane 5: negative control, lane 6: FGF8 positive control. b MCF-7 cells were transfected with FIR-FLAG, FIRΔexon2-FLAG (left). Bar graph was showed the relative intensity of bands (right). The internal control is β-actin. c Western blotting analysis of knockdown of FIR or FIRΔexon2 in MCF-7 cells (left). Bar graph is normalized intensity of bands (right). Internal control protein is β-actin. siRNA concentration of FIR-1, FIR-2 and FIRΔexon2 is 25 and 50 pmol respectively. d qRT-PCR of FIR, FIRΔexon2 mRNAs. siGL2 is control siRNA. Internal control gene is HPRT. e Splicing variants analysis of FGF8 using bioanalyzer electrophoresis in MCF-7 cells. siRNA concentration of FIR-1, FIR-2 and FIRΔexon2 was 50 pmol.
Elevated expression of BRG1 and FIR, but not FIRΔexon2, in Gan-mice as a non-invasive early gastric cancer model
To study the effect of FIRΔexon2 on BRG1 expression in tumor development, Gan-mice (K19-Wnt1/C2mE) that develop non-invasive gastric tumors at a frequency of 100% were examined34. They are considered as a model of non-invasive gastric cancer (Fig. 3a, top panels). Gan x FIR+/− mice (K19-Wnt1/C2mE x FIR+/− mice) were prepared to employ an invasive gastric tumor model; however, the gastric tumors remained non-invasive (Fig. 3a, middle panels). No wild-type mouse developed a gastric tumor during the period of observation (Fig. 3a, bottom panels, Supplementary Table S2). Additionally, there was no apparent difference in tumor growth speed and histological type between the Gan and FIR+/− mice (data not shown). Expression profiles of the FIR or FIRΔexon2-related proteins9 were examined in gastric tumors of Gan and FIR+/− mice. The levels of FIR family, BRG1, Snai1, FBW7, E-cadherin, c-Myc, cyclin-E, and SAP155 increased in the gastric tumors in FIR+/− mice compared to those expressed in wild-type mice. Interestingly, FIR family, Snai1, cyclin-E, BRG1, and c-Myc showed trends toward higher expression in larger tumors (Fig. 3b, lanes 7–9) than in smaller tumors (Fig. 3b, lanes 4–6) in Gan-mice. An alternative splicing variant forms of FIR, FIRΔexon2, were indicated by qRT-PCR (Fig. 3b, bottom). In contrast, the expressions of cyclin-E, Snai1, and BRG1 were decreased, whereas FBW7 and E-cadherin expressions were sustained in FIR+/− mice tumors. Although the TP53 gene mutation affects the expression of various cellular proteins, the TP53 gene mutation status did not significantly affect the expression levels of FIR, FBW7, and SAP155 in esophageal cancer cells23. The expression levels of proteins were quantified by densitometry (Fig. 3c), and the statistical significance of these protein expressions were confirmed (Fig. 3c and Supplementary Fig. S2a). In mice fibroblast NIH3T3 cells, FIR and PUF60 mRNAs were detected, but FIRΔexon2 mRNA was not detected by RT-PCR (Fig. 3d). The expressions of BRG1 and Snai1 were positively correlated in the gastric tumors of the Gan-mice (Supplementary Fig. S2b). The epitope of the anti-FIR monoclonal antibody, 6B4, is located in the UHM at the carboxyl terminus (a thick arrow, Fig. 1a) in FIR, PUF60, and FIRΔexon235.
Fig. 3. Tumorigenesis and histological type of Gan-mouse and FIR+/− mouse.
a, top panels Picture of Gan-mouse gastric tumor and FIR+/− mouse gastric tumor. a, middle panels hematoxylin-eosin staining photo (40 × ) of Gan-mouse and FIR+/− mouse gastric tumor. Note, lamina muscularis mucosae was intact (a, top and middle panels). a, bottom panels Wild-type mice showed no gastric tumors during the period of this experiment. Normal gastric mucosa was used as a negative control. b Expression of FIR and related proteins was examined by western blotting in wild-type mice tissues, as well as in Gan-mouse and FIR+/− mice gastric tumor tissues. The expressions of FIR family, Snai1, BRG1, E-cadherin, FBW7, c-Myc, cyclin-E, SAP155, and hnRNPA1 of wild-type, Gan-mice, and FIR+/− mice were examined. Frozen tissues samples of mice gastric tumors were obtained and proteins expression profiles examined by western blotting in three different genotypes: wild-type mouse (lanes 1–3), Gan-mice (gastric tumor wet weight < 0.5 g) (lanes 4–6), Gan-mice (gastric tumor wet weight > 2.0 g) (lanes 7–9) and FIR+/− mice (lanes 10–12). c The extent of the signals detected by western blotting revealed in Fig. 1 was quantified by densitometry. d Alternative splicing forms of PUF60 and FIR mRNAs were detected in NIH3T3 cells by reverse-transcription-polymerase chain reaction (RT-PCR).
Expression of BRG1, FIR and FIRΔexon2 in human invasive gastric cancers
Next, the effect of FIRΔexon2 on BRG1 expression in human invasive gastric cancers was examined by western blot analysis for various molecules expressed in the gastric tissues of 14 representative patients (cases no 1–5 in Fig. 4a and cases no 6–14 in Supplementary Fig. S3). Unlike Gan-mice (Fig. S2b), the expressions of BRG1 and Snai1 were not significantly correlated (Supplementary Fig. S4a, b). Snai1 expression was increased but BRG1 was decreased in (T) than those in (N) in human gastric cancer tissues (Supplementary Fig. S4c). Given the FBW7/BRG1 signaling axis governs E-cadherin expression by modulates Snai1 in gastric cancer metastasis4, FIRΔexon2 is a candidate that affects FBW7/BRG1 signaling axis. Invasive gastric cancer showed that the expressions of E-cadherin and FBW7 were decreased in human gastric tumor tissues relative to those of their non-tumorous counterparts (Fig. 4a). FIRΔexon2 mRNA was significantly highly expressed in cancers compared to their non-cancerous counterpart tissues (Fig. 4a, bottom panel)27,35. The ratio of mRNA expressions of FIRΔexon2/FIR was significantly higher in (T) than in (N) (Fig. 4b). Furthermore, stably transfected pcDNA-3.1/FIRΔexon2 clones of NIH3T3 cells showed colony formation in a soft-agar assay (Fig. 4c). The expressions of FIR family and Snai1 were significantly higher in tumor tissues (T) than in their non-cancerous counterparts (N) (Fig. 4d). Protein expression profiles in Gan-mice and human gastric tumor tissue were indicated (Supplementary Table S3). These results indicated that FIRΔexon2 was expressed in human gastric cancers but not in FIR+/− mice, and was engaged in tumor invasive behavior to a certain extent.
Fig. 4. Protein expression profiles of human gastric cancer tissue samples.
a Expressions of FIR family and related proteins were examined by western blotting in five paired-tumor (T) and adjacent non-tumor (N) tissue samples from human gastric cancer tissues. a, bottom Four splicing variants of the FIR/PUF60 gene PUF60, FIR, PUF60Δexon2, and FIRΔexon2 were detected in gastric cancer tissues in five paired tumors (T) and adjacent non-tumors (N). tub1: well-differentiated tubular adenocarcinoma, tub2: moderately differentiated tubular adenocarcinoma; por1: poorly differentiated adenocarcinoma, solid type; muc: mucinous adenocarcinoma. b The ratio of mRNA expression of FIRΔexon2/FIR was significantly higher in gastric cancer tissues (T) that in corresponding non-cancer tissues (N). N = 14, P = 0.007 by Student’s t-test. c Clones of stably transfected pcDNA3.1-FIRΔexon2 plasmids (clones 1,2,3,7,9,10,12,14, and 15 among 30 clones). At least 30 clones were screened by immunoblotting and immunostaining with anti-FLAG and anti-FIR antibodies (6B4) to find FIR-FLAG-expressing clones for FIR-FLAG stably expressing cells, or with anti-c-Myc antibody to examine c-Myc expression for FIRΔexon2 stably expressing cells. Soft-agar colony formation assay of clone12. The cells of clone12 (2 × 103) were inoculated in 0.3% low-melting-temperature agarose (FMC Bio Products, Rockland, ME, USA) in DMEM supplemented with 10% FCS, and colonies were scored after incubating for 2 weeks. 1 A cells was a positive control cells provided by Dr Ariga27. d Comparison of protein expressions from five paired tumors determined by statistical analysis in gastric cancer tissues in 14 paired tumors (T) and adjacent non-tumors (N). N = 5, P < 0.01, R < 1.0 was obtained by Student’s t-test.
Potential interaction between a novel WD-like motif (W425 and D399) of FBW7 and UHM of FIR protein: a three-dimensional crystal structure analysis
Finally, possibility of the inhibition of FIRΔexon2 in the BRG1 degradation by FBW7 was investigated4. FBW7 is a member of the Skp1-Cull-F-box (SCF) type ubiquitin ligase complex and participates in proteasomal degradation of various tumor-promoting molecules and is considered to be a bona fide tumor suppressor36. WD-repeat proteins were co-immunoprecipitated with FIR and FIRΔexon2 (Table 1). If FIR and FIRΔexon2 interact with FBW7, FBW7 need to contain the WD-like domain. In this study, the interaction between UHM of FIR or PUF60 and the WD-like motif (W425 and D399) of FBW7 was found with three-dimensional crystal structure analysis23. The FIR family contains the UHM at the carboxyl terminus (Fig. 1a, arrow) that potentially interacts with WD-like motifs, such as the UHM-ligand motif, in the degron pocket of FBW7 (Fig. 5a), and thus might interfere with ubiquitination of its substrate proteins22,23,37,38. To research this possibility, the interaction between UHM of FIR or PUF60 and the WD-like motif (W425 and D399) of FBW7 was examined by three-dimensional crystal structure analysis22. The binding structure between SF3B1 and one of the splicing factors containing UHM and SPF45 (human splicing factor 45) has previously been clarified by X-ray crystal analysis (PDB code: #2PEH)13 (Fig. 5a). In the 2PEH structure, the crystal unit cell contains two SPF45 recombinant proteins (amino acids: 301–401) and two SF3B1 partial peptides (aa: 333–342). The amino acid sequence LNGRYFGGRVVKA in SPF45 (aa: 372–384) is similar to sequences at the C-terminal domains of FIR-wild (aa: 505–517) and U2AF65 (aa: 449–510; Fig. 5b). A comparison of the two crystal structures indicates that the positions and configurations of W425 and D399 in FBW7 are considerably similar to those in SAP155 (Fig. 5c). Therefore, FBW7 possibly binds to FIR in a similar manner as the way SAP155 binds to SPF45. The extra pair of W and D at the WD-repeated domain (molecules indicated in yellow, Fig. 5c -right) cannot be involved in the ligand recognition of the phosphorylated peptides, but the WD pair can interact with the peptide with the above-mentioned conserved sequence based on their structures (molecules indicated in magenta, Fig. 5c -right). This result is compatible with the importance of Asp of FBW7 in the molecular binding because Asp usually contributes to hydrophilic interactions, such as hydrogen bond formation between molecules (Fig. 5d). Therefore, FIRΔexon2 interferes with the WD-like motif of FBW7 and potentially inhibits its function. Thereafter, small molecular weight chemicals were screened that inhibit the FIRΔexon2-FWB7 interaction23.
Fig. 5. Crystal structure analysis of the interaction between FBW7 and FIR/PUF60-UHM.
a The amino sequence LNGRYFGGRVVKA in SPF45 is similarto sequences in the C-terminal domains of FIR and U2AF65. A comparison of two crystal structures indicates that the positions and configurations of W425 and D399 in FBW7 are considerably similar to those of SAP155. FBW7 has many WD-motifs, and most of the motifs are involved in the conformational stabilization of the WD-repeated domain (molecules indicated in yellow). The binding structure between SF3B1 and one of the splicing factors containing UHM, SPF45 (human splicing factor 45), has previously been clarified by X-ray crystal analysis (PDB code: #2PEH)29. In the 2PEH structure, the crystal unit cell contains two SPF45 recombinant proteins (amino acids: 301–401) and two SF3B1 partialpe ptides (aa: 333–342). b Amino acids sequence of U2AF65 (475aa). Similarity among SPF45, FIR, and U2AF65. c There is an extra pair of W425 and D399 at the center of the WD-repeated domain (molecules indicated in magenta). Since the binding pocket of FBW7 contains the WD-motif that is expected to interact with FIRΔexon2, the chemical skeleton of the two synthesized compounds is regarded as a WD-mimicking form. A low molecular weight artificial chemical, BK697, that inhibits FIRΔexon2, as shown by in silico analysis, was synthesized. d The isothermal titration calorimetry (ITC) measurement of FBW7 with FIRΔexon2 suggested the molecular interaction between two proteins. The exothermic peaks were observed in the initial 17 injections.The peak level was decreased in the later injections. Due to the quick upward change in the ITC thermogram, the association constant is >108 M−1. Since the binding reaction is exothermic, the binding of FBW7 and FIRΔexon2 is enthalpically driven.This result is compatible with the importance of Asp of FBW7 in the molecular binding because Asp usually contributes to hydrophilic interactions, such as hydrogen bond formation, between protein molecules.
Decreased E-cadherin expression promoted migration of gastric cancer cells
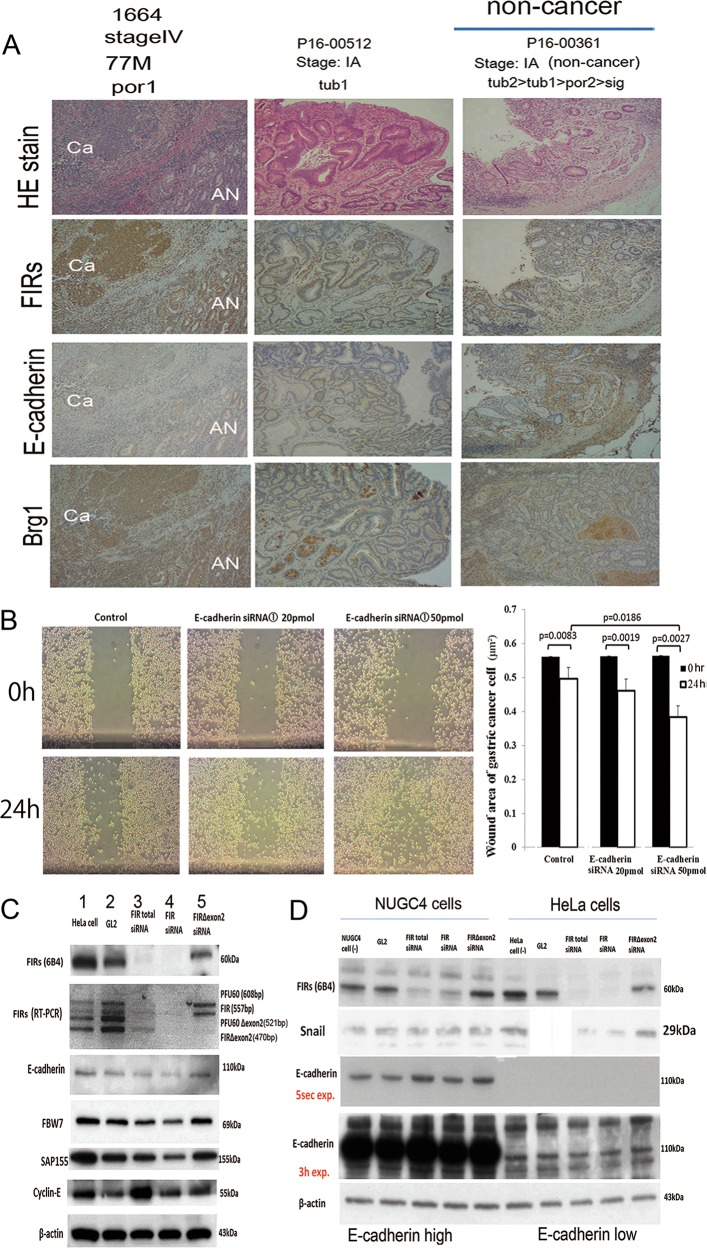
Immunohistochemical staining showed that FIR and FIRΔexon2 (FIR family), Snai1, and BRG1 expressions were increased but FBW7 and E-cadherin expressions were decreased in human gastric cancers (Fig. 6a, Supplementary Figs. S3 and S4a-c); however, the expression of BRG1 was reduced in some cases (cases 1 and 4 in Fig. 2a; cases 6,8,10,12, and 14 in Supplementary Fig. S3). Given that E-cadherin is involved in cell–cell adhesion and the invasion of cancers39, knockdown of E-cadherin by siRNA may affect the migration of cancer cells according to an MTS assay in NUGC cells (cell proliferation assay; Fig. 6b). Firefly luciferase gene (GL2) siRNA was transfected as a negative control. As expected, a wound-healing assay showed that the migration of NUGC cells was enhanced by transfection of E-cadherin-specific siRNA (Fig. 6b). SiRNA of FIRΔexon2 was slightly increased but siRNA of FIR decreased E-cadherin expression (Fig. 6c), indicating FIRΔexon2 was decreases E-cadherin expression in HeLa cells. The level of E-cadherin expression was much higher in NUGC4 than that of HeLa cells (Fig. 6d).
Fig. 6. E-cadherin siRNA promotedg astric cancer cells migration in the wound-healing assay.
a E-cadherin expression decreased, whereas FIR family and BRG1 expressions increased in cancer tissues (Ca) relative to those in adjacent normal tissues revealed by immunohistochemical staining. Case numbers 2832, 1621, and 1664 are listed in Suppl. Table S4. In the non-invasive early stage (IA) of differentiated cancers, the expressions of E-cadherin and FBW7 were decreased. Case number P16-00361 are listed in Suppl. Table S8. b Migration of E-cadherin siRNAs transfected into NUGC4 cells and corresponding control cells was measured by wound-healing assay. P < 0.01 was obtained by Student’s t-test. c FIR family and related protein expressions were examined aftertreatment of HeLa cells withthe FIRsiRNAs. GL2 siRNA was transfected as the negativecontrol. After 48 h of transfection, whole-cell extracts were analyzed by western blotting. Three types of FIR siRNAs were transfected into HeLa cells. Lane 2 is GL2 siRNA control transfection, lane 3 is 20 pmol of total FIR siRNA transfection, lane 4 is 20 pmol of FIRsiRNA transfection, and lane 5 is 20 pmolof FIRΔexon2 siRNA transfection. d E-cadherin expression in HeLa cells. The level of E-cadherin expression was much higher in NUGC4 than that of HeLa cells. Three types of FIR siRNAs were transfected into NUGC4 and HeLa cells. GL2 siRNA is internal control. 20 pmol of total FIR siRNA, 20 pmol of FIR siRNA, and 20 pmol of FIRΔexon2 siRNA transfection were performed. After 48 h of transfection, whole-cell extracts were analyzed by western blotting.
A low molecular weight chemical, BK697, identified as a FIRΔexon2 inhibitor, suppressed tumor growth by decreasing FIR family and E-cadherin and increasing Snai1 expression
Expectedly, small molecular weight chemicals that interacted with FIRΔexon2 have a WD-like motif and were identified with NPDepo screening23,40. Computer screening using the Namiki database (Namiki Shoji Co., Ltd., Tokyo, Japan) was used to search for synthesized chemicals that mimic the structure of the identified compound that consisted of commercially available chemicals. Actually, the chemical skeletons of the two synthesized compounds were regarded as WD-mimicking forms (Fig. 7a and Supplementary Fig. S6)23. On the basis of the tests with the synthesized compounds, we modified the chemical structure and finally identified BK697 (Fig. 7a and Supplementary Fig. S6). BK697 suppressed the growth of HeLa cells (Fig. 7b) and affected FIR family, E-cadherin, Snai1, and BRG1 expression (Fig. 7c, d). Notably, the dose of BK697 necessary to increase E-cadherin and SAP155 mRNAs was smaller (50 μM) than that required to increase FIR and FIRΔexon2 mRNAs (100 μM; Fig. 7e). Given the U2AF-homology motif (UHM) of PUF60 directly interacting with the WD-repeat of SAP155 (SF3B1)13, FIRΔexon2 is expressed in cancers when FIR pre-mRNA is affected by an autocatalytic mechanism through inhibiting the FIR-SAP155 splicing complex9. Overexpression of FIRΔexon2 decreased the level of histone acetylation in the BRG1 genome region but not its mRNAs (Fig. 1c); however, FIRΔexon2 can suppress BRG1 (Fig. 1c, d). Together, FIRΔexon2 suppressed BRG1 post-transcriptional processes (Fig. 8).
Fig. 7. Chemical structure of BK697 and its effect to HeLa and NUGC4 cells.
a The conformations of the two inhibitory compounds were found to resemble the WD-motif. b Among the chemical compounds, BK697 showed significant inhibition of cell growth by MTT assay. Chemicals were diluted in DMSO at a concentration of 10 mM, and we used 1 and 2 μL/well/100 μL medium (final concentrations in the medium were 0.1 mM and 0.2 mM, respectively). The same volume of DMSO was used as a negative control. The same volume of 3% H2O2 was used as a positive control. Untreated cells were used as the blank. c, d BK697 suppressed FIR family as well as SAP155 and hnRNPA1 expression in HeLa cells in a dose-dependent manner. e FIR and FIRΔexon2 mRNAs were examined by quantitative reverse-transcription-polymerase chain reaction after 24 h treatment of BK697. HPRT mRNA was used as an internal control.
Fig. 8. Alternative splicing product of FIR/PUF60 gene, FIRΔexon2, contributes to E-cadherin suppression through post-transcriptional regulation of BRG1.

FIRΔexon2 is an alternative splicing variant form of FIR. FIRΔexon2 is expressed in cancers when FIR pre-mRNA is disturbed by an autocatalytic mechanism through inhibiting the FIR-SAP155 splicing complex9. Ribosomal proteins, hnRNPs, splicing-related factors, poly(A) binding proteins, mRNA-binding proteins, tRNA, or DEAD box proteins (DDX) were commonly co-immunoprecipitated with FIR or FIRΔexon2, indicating that both FIR and FIRΔexon2 participate in post-transcriptional or translational processes. FIRΔexon2, but not FIR, reduced the level of H3K27ac at the BRG1 promotor. The protein expression of BRG1 was decreased by FIRΔexon2 overexpression, suggesting that FIRΔexon2 partly affects nucleosome remodeling. FIRΔexon2 potentially inhibits the accession of substrate proteins of FBW7 to its degron pocket (c.f. Fig. 6). BK697 is a small molecular weight chemical containing a WD-like domain that inhibits FIR/FIRΔexon2. Snai1 is a transcriptional repressor of E-cadherin. Together, the suppression of E-cadherin by BK697 was at least partly post-transcriptional, including inhibiting FBW7 function. A novel chemical inhibitor of FIR/PUF60 and its splicing variants were revealed to target EMT through FBW7 and E-cadherin in this study. Three-dimensional crystal structure analysis revealed that the U2AF-homology motif (UHM) of FIR or FIRΔexon2 interacted with the WD-like motif in the degron pocket of FBW7. Therefore, the interaction between FIRΔexon2 and FBW7 inhibits FBW7-mediated proteasomal degradation of BRG1 and Snai1. BK697 is a novel, low molecular weight compound containing a WD-like domain targeting FIRΔexon2. Overall, our study suggests that FIRΔexon2-mediated suppression of E-cadherin via the FBW7/BRG1/Snai1 axis prevents EMT and invasion in gastric cancer.
Discussion
Revaluation of co-immunoprecipitated proteins with FIRΔexon2 revealed that FIRΔexon2 engages in post-transcriptional processes with ribosomal proteins, splicing-related factors, mRNA-binding proteins, poly (A) binding proteins, hnRNPs, tRNA, DEAD box and WD-repeat proteins. The transformation/transcription domain-associated protein (TRRAP) that acetylates histones in rDNA transcription was also co-immunoprecipitated with FIRΔexon2 (Fig. 1b)9. H3K27ac modification is induced by histone acetyl transferase (HAT), such as GCN5. GCN5 interacts with TRRAP and forms a complex25. FIRΔexon2 increases c-Myc expression that activates rDNA transcription7 then potentially promotes ribosome synthesis41. As expected, FIRΔexon2, but not FIR, acetylated H3K27 on the BRG1 promoter revealed by ChIP-seq and suppressed BRG1 expression. Further, FIRΔexon2 suppressed BRG1 protein expression, but not BRG1 mRNA expression, indicating that FIRΔexon2 post-transcriptionally regulates BRG1 (Fig. 1c). BRG1 controls multipotent neural crest formation by regulating EMT-related genes with CHD71,2. BRG1 also engages in pre-mRNA splicing by interacting with RNPs in cancers3–5. In addition to cancers, PUF60 and CHD7 are responsible for the CHARGE syndrome that shows developmental disorders12; herein, cooperatively translocating nucleosomes to permit transcription of FGF8 by RNA pol II in neural development. Since siRNA of FIR or FIRΔexon2 surely changed alternative splicing of FGF8 pre-mRNA (Fig. 2e), FIRΔexon2 possibly cooperates with BRG1 in neural development through alternative splicing of FGF8 pre-mRNA processing12. FGF8 has been reported as a prognostic factor in adenocarcinoma of esophago-gasric junction42 and contributes to radiation resistance in rectal cancer43. Furthermore, FIRΔexon2 was detected in human gastric cancers but not in mice tumor cells (Fig. 3). The simultaneous increases in BRG1, Snai1, and E-cadherin did not induce EMT in Gan-mice (Fig. 3b, c); however, the expressions of FBW7, BRG1, and E-cadherin decreased in the human gastric cancer tissues (Fig. 4a, b). The expressional change of FIR and FIRΔexon2 increases c-Myc expression that activates rDNA transcription7 and then potentially promotes ribosome synthesis41. FIRΔexon2 altered ribosomal protein expression by RNA-seq analysis and H3K27ac increased specifically in FIR/PUF60 genome regions (data not shown) because FIRΔexon2 may directly or indirectly affect the TRRAP complex9. These results indicated that FIRΔexon2 possibly relates to rDNA transcription in the nucleolus and to FIR/PUF60 gene regulation in the nucleoplasm as well. Therefore, autocatalytic production of FIRΔexon2 is crucial for carcinogenesis in terms of ribosome protein synthesis. Together, E-cadherin suppression by FIRΔexon2 at least partly via the FBW7/BRG1/Snai1 axis was shown to promote invasion of gastric cancer cells (Fig. 8).
Given the U2AF-homology motif (UHM) of PUF60 directly interacting with the WD-repeat of SAP155 (SF3B1)13, FIRΔexon2 expressed in cancer cells by alternative splicing of FIR pre-mRNA, which is dysregulated by the FIR-SAP155 interaction9. FIRΔexon2 transgenic mice were prepared with C57BL/6 mice (UNITECH Co., Ltd., Kashiwa, Chiba, Japan). Body weight curve of FIRΔexon2 transgenic mice was significantly lower than that of wild mice (Supplementary Fig. S7). There was no apparent tumor formation in organs of FIRΔexon2 transgenic mice (data not shown). These results indicated that certain level of sustained FIRΔexon2 expression is required for carcinogenesis. One possible mechanism of persistent FIRΔexon2 activation is the disturbance of FIR pre-mRNA by an autocatalytic mechanism through inhibiting the FIR-SAP155 splicing complex. To test the potential causal relationship between FIRΔexon2 and BRG1 and invasive gastric cancer, an introduction of FIRΔexon2 by CRISPR editing in FIR+/− mouse or PDX (Patient Derived Xenograft) could be expected to induce an invasive tumor in future experiment.
FIR family (FIR, FIRΔexon2, and PUF60), Snai1, and BRG1 increased, but FBW7 and E-cadherin decreased in human gastric cancers, as shown by immunohistochemical staining (Fig. 5a, Supplementary Figs. S4 and S5); however, the expression of BRG1 was reduced in some cases (Fig. 4a and Supplementary Fig. S4). The expression profiles of other proteins were indicated in a mouse-model and in human gastric cancers (Supplementary Figs S2, S4a, b). A wound-healing assay by transfection of E-cadherin-specific siRNA showed enhanced migration of NUGC cells (Fig. 5b). The interaction between UHM of FIR or PUF60 and the WD-like motif (W425 and D399) of FBW7 was examined by three-dimensional crystal structure analysis17. The titration curve of FBW7 with FIR∆exon2 suggested a molecular interaction between these two proteins (Fig. 6d). Therefore, the interaction of UHM of FIRΔexon2 could interfere with the WD-like motif of FBW7 and potentially inhibit its function. For clinical applications, BK697 (Fig. S6), a chemical inhibitor of FIRΔexon2, suppressed FIR family expression and tumor cell growth with SAP155 and E-cadherin suppression (Fig. 7c) and increased Snai1 expression (Fig. 7d). All compounds that chemically interact with FIRΔexon2, including BK697, have an aromatic ring connected to a carboxyl group by a short linker. Hence, FIRΔexon2 potentially binds to analogs of the WD-motif of FBW7. Possible interactions between FIRΔexon2 and FBW7 caused the FBW7’s disability to further destabilize Snai1 proteins and resulted in decreased E-cadherin levels that promoted gastric cancers through EMT (Figs. 6 and 7). Therefore, FIRΔexon2 appears to suppress E-cadherin expression at least in part through the FBW7/BRG1/Snai1 axis for promoting EMT and invasion in gastric cancer. The expression of BRG1 was not necessarily increased nor positively correlated with Snai1 in some human cancers (Fig. 4 and Supplementary Fig. S4), which indicated that the FBW7/BRG1/Snai1 axis is perturbed in gastric cancers. Actually, the level of H3K27 histone acetylation in the BRG1 genome region was found to be decreased by FIRΔexon2 overexpression in vitro (Fig. 1c). Since the expression of cyclin-E is a substrate of FBW723 and E-cadherin is regulated by the FBW7/BRG1/Snai1 pathway3, altered or disturbed expression of FIR family possibly affects FBW7 function in gastric cancers. Further study is required to explore alternative pathways in which FIRΔexon2 directly or epigenetically affects BRG1 gene regulation.
Additionally, FIR deficiency promoted alternative splicing to increase pyruvate kinase M2 that engages in the glucose metabolism of cancers in mice thymic lymphoma tissues44. Moreover, the long non-coding RNAs (lncRNAs) are a novel class of regulatory genes that have critical roles in cancer progression, and translational regulatory lncRNA (treRNA) downregulates the expression of the epithelial marker E-cadherin by suppressing the translation of its mRNA45. A novel mechanism of treRNA is dependent on the 3′UTR of the E-cadherin mRNA, and a novel ribonucleoprotein (RNP) complex, including FIR or PUF60, is required for this treRNA function45. The altered FIRΔexon2 expression changes the specific ribonucleoprotein (RNP) complex revealed by RNA-sequencing analysis (data not shown) and, accordingly, FIRΔexon2 potentially generates lncRNAs in cancers.
In conclusion, autocatalytic regulation in alternative splicing of the FIR/PUF60 gene by the SAP155-FIRΔexon2 complex9 simultaneously downregulated FBW7 and E-cadherin via Snai1. This pathway is potentially pivotal for the invasion or metastasis of gastric cancer through EMT. Clinically, BK697 and its derivatives are potential FIRΔexon2 inhibitors and candidate drugs for the treatment of gastric cancer.
Materials and methods
Human samples and cell lines
Human gastric cancer tissues were obtained from 14 patients who underwent gastrectomy in the Department of General Surgery, Chiba University Hospital, Chiba, Japan (Supplementary Table S4). Written informed consent was obtained from each patient before surgery after gaining approval by the ethics committee of the Graduate School of Medicine, Chiba University. Fresh tissue samples were frozen immediately in liquid nitrogen and stored at −80 °C until analysis. The definition of clinical stages of gastric cancer patients was determined according to the Japanese Classification of Gastric Carcinoma46. Mouse fibroblast NIH3T3 cells, human gastric cancer cell lines MNK7, MNK45, MNK74, NUGC3, and NUGC4 and HeLa (human cervical squamous cell carcinoma cells) cells (purchased from ATCC) were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) and supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. Cells were grown at 37 °C in a 5% CO2 incubator.
Animal experiments
The fertilized eggs of a Gan-mouse (K19-Wnt1/C2 mE) were obtained from Kumamoto University and matured in Chiba University. We crossbred a male Gan-mouse and female FIR+/− mouse female. The bred FIR+/− mice were maintained in the animal research facility of the Graduate School of Medicine, Chiba University in accordance with institutional guidelines. FIRΔexon2 transgenic mice were prepared with C57BL/6 mice by UNITECH Co., Ltd. (Kashiwa, Chiba, Japan).
Protein extraction and western blotting
Human frozen tissues (25 mg each) of paired tumor and corresponding adjacent mucosa from the same patients with gastric cancer were pulverized for extraction of proteins. The tissue in the extraction buffer (2 M Thiourea,7 M Urea, 30 mM Tris CL2%, CHAPSProtease inhibitor complete 1 tablet/50 μL, 1% DTT, and 1% Pharmalyte TM 3-1 in dH2O) was homogenized three times with homogenizer (Polytron, Tokyo, Japan) for 30 sec to 1 min per homogenization. The homogenate was then centrifuged using an ultra-high speed centrifuge for 1 h at 50 K. The supernatant was collected and stored at −80 °C following measurement of the protein concentration with a Bio-Rad protein assay (Bio-Rad, CA, USA). Western blotting was performed as previously described9. The other antibodies used in this study are listed in (Supplementary Table S5).
Quantitative reverse-transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cancer cells using a QIAgene Miniprep Kit (Qiagen, Tokyo, Japan). cDNA was synthesized from total RNA using a First Strand cDNA Synthesis Kit for RT-PCR (Roche, Mannheim, Germany). PUF60, FIR and FIRΔexon2 [FIR splicing variants] cDNA were amplified from the cDNA with the respective sets of forward and reverse primer pairs10,27 (Fig. 1d, Supplementary Table S6) by using the qRT-PCR reagents (Supplementary Table S7)9.
Chromatin immunoprecipitation (ChIP) and ChIP-qPCR
ChIP assays were performed on approximately 107 cells as previously described47. Briefly, the cells were crosslinked with 1% formaldehyde at room temperature for 10 min and formaldehyde was quenched by the addition of 2.5 M glycine to a final concentration of 0.125 M. Crosslinked chromatin was sonicated to a size of 0.2–1 kb using an ultrasonic disruptor [BRANSON Digital Sonifier (BRANSON)]. A total of 1 μg of H3K27ac antibodies and 20 μL of Dynabeads Protein G (Thermo Fisher Scientific Diagnostics) were mixed in IP dilution buffer and incubated at 4 °C overnight. After washing with IP dilution buffer, antibody-binding beads were added to the sonicated-chromatin sample and incubated at 4 °C overnight. The beads were washed and the chromatin was eluted, followed by reversal of crosslinking and DNA purification. Chromatin-immunoprecipitated DNA was dissolved in the elution buffer. Enrichment of ChIP samples were further confirmed with ChIP-qPCR using specifically targeted positive and negative region primer sets (Supplementary Table S6).
ChIP-seq and data analysis
Libraries were constructed with a KAPA Hyper Prep Kit (Kapa Biosystems) according to the manufacturer’s instructions. ChIP-seq libraries were quantified using a Bioanalyzer (Agilent, USA) and sequenced at a concentration of 6.5 pM on a Hiseq1500 (Illumina, USA) platform or at a concentration of 1.5 pM on a NextSeq500 (Illumina, USA) platform. Sequenced reads in the ChIP-seq experiment were mapped to the UCSC human genome (hg19) using bowtie software. Duplicated reads were removed with Picard tools. Peak calling and motif analysis were performed with HOMER software (http://homer.salk.edu/homer/index.html). HOMER was also used to obtain differential peaks. The H3K27ac peaks in FIR-FLAG or FIRΔexon2-FLAG were compared with untreated HeLa cells.
Small interfering (si) RNA transfection
E-cadherin siRNA duplexes were purchased from Sigma–Aldrich. Transient transfection of siRNAs was performed using Lipofectamine 2000 (Invitrogen, Japan) according to the manufacturer’s instructions. The transfected cells were cultured at 37 °C for 48 h in a CO2 incubator. The target sequences for the siRNAs are listed in (Supplementary Table S7).
Immunohistochemical staining
Immunohistochemical staining was performed as previously described5,9. Human sample list are indicated (Supplementary Tables S4 and S8).
Wound-healing assay
A wound-healing assay was performed as previously described9.
Stable transfection of pcDNA3.1-FIRΔexon2 plasmids
Stable transfection of pcDNA3.1-FIRΔexon2 plasmids was performed described previously9.
Soft-agar colony formation assay
Soft-agar colony formation assay was performed as previously described48. The positive control cells (1 A cells) were kindly provided by Dr. Ariga H49.
Display of the three-dimensional structure of FBW7
The crystal structures, Protein Data Bank (PDB) code: 2QVR50 and 2PEH37, were visualized with PyMOL (DeLano, W. L.; The PyMOL Molecular Graphics System, Schrödinger, LLC).
Expression and purification of FIR∆exon2 and FBW7
The FIR∆exon2 gene was inserted into a pET-50b (+) DNA plasmid vector. An Escherichia coli strain, Rosetta (DE3) pLysS (competent cells), that was transformed with the pET-50b-FIR∆exon2 vector, was cultured in 1 L Luria-Bertani (LB) medium at 30°C until the O.D. 600 value reached 0.6. This was followed by 12 h of incubation after an addition of 0.2 mM isopropyl β-D-1 thiogalactopyranoside (IPTG). A cell pellet was obtained by centrifugation of the cultured medium. The pellet was resuspended in a buffer of 50 mM Tris-HCl at pH8.0 and 500 mM NaCl containing 10 mM imidazole and 1 mM phenylmethylsulfonyl fluoride (PMSF). After disrupting the bacterial cell membrane by sonication, the protein was purified with a co-affinity column with a gradient rise of the imidazole concentration. The eluted fraction was dialyzed overnight against the buffer without imidazole. The Nus-tag was cleaved by HRV-3C protease and the protein was purified with a Ni-affinity column to remove the cleaved Nus-tag and HRV-3C protease. The protein was finally purified by gel filtration with a running buffer of 10 mM Tris-HCl at pH 8.0 and 300 mM NaCl. FBW7 was expressed as a complex with Skp-1 using the pCDF-2 plasmid vector. A competent cell, Rosetta (DE3) pLysS, was transformed with the pCDF-2 Nus-tag-fused FBW7-Skp1 vector that was cultured in 1 L of LB medium. After an addition of 0.2 mM IPTG, the competent cells were incubated for 12 h. After resuspension of the cell pellet, the bacterial membrane was disrupted with a French press. Protein purification was performed with a co-affinity column, followed by Nus-tag cleavage with HRV-3C, a Ni-affinity column, and gel filtration.
Isothermal titration calorimetry (ITC) measurement
The binding affinity between FIR∆exon2 and FBW7 was measured with an isothermal titration calorimetry (ITC) technique using the MicroCal VP-ITC system (Malvern Panalytical, UK). The sample cells were filled with 1400 μL of 50 mM phosphate buffer, pH 7.4, containing 15 μM of the purified Skp1-FWB7 complex. The measurement of binding affinity was performed at 30 °C. The solution of 50 mM phosphate buffer, pH 7.4, containing 300 μM FIR∆exon2 was injected into the sample cells from the syringe for titration. The injection volumes were 10 μL each, the injection time was 20 s, and the injection was 150 s. The titration was repeated 25 times.
Binding affinity between FIR∆exon2 and FBW7
The titration curve of FBW7 with FIR∆exon2 suggested a molecular interaction between two proteins. The exothermic peaks were observed with the earlier injections. The exothermicity of the injection was decreased in the later injections. The dissociation constant was calculated to be 1.3 × 106 M−1 from the ITC thermogram. The binding of FBW7 and FIR∆exon2 was enthalpically favorable because the ITC measurement showed the interaction was exothermic.
In silico screening for inhibitory compounds against FIRΔexon2 protein function
Small molecular chemical compounds against His-tagged FIR (His-FIR) and His-tagged FIRΔexon2 were screened among 23,275 natural chemicals of NPDepo (RIKEN Natural Products Depository) at RIKEN institutes (Wako, Saitama, Japan) as previously described51. In silico computer screening was performed to search fo synthesized chemicals that mimicked the structure of the identified compound using the Namiki database that was composed of commercially available chemicals23. Based on the computer screening, 16 compounds were selected and purchased from chemical suppliers (Namiki Shoji Co., Ltd., Tokyo, Japan). Since the WD-motif of SAP155 (SF3B1) directly binds to PUF6013, chemical skeletons of the two synthesized compounds contained a WD-like motif (Fig. 7a). Several compounds were selected from the synthesized chemicals40. Consequently, we identified a highly active compound, BK697, competing with FIRΔexon2 functions, which suppressed the proliferation of cancer cells22.
Biological effect of BK697, a candidate FIRΔexon2 inhibitor
HeLa and NUGC4 cells were treated with different concentrations of BK697 at different time intervals22. Briefly, on day one, NUGC4 or HeLa cells were prepared in IMDM supplemented with 10% FBS. On day two, candidate chemicals that inhibited FIRΔexon2 were diluted in DMSO at a concentration of 10 mM and added as 10 or 20 μL/well/2 mL in the medium (the final concentration in the medium was 50 and 100 μM, respectively) or added as 20 or 60 μL/well/2 mL medium (the final concentration in the medium was 100 and 300 μM, respectively). A total of 100 or 300 μM of BK697 was exposed to NUGC4 cells for 24 h, and 50 or 100 μM of BK697 was exposed to NUGC4 cells or HeLa cells for 6 h, 24 h and 48 h at 37 °C in a CO2 incubator.
Statistical analysis
Statistical significance of the differences in numerical data was assessed with a Student’s t-test and the Wilcoxon test. All tests were two-tailed and a P-value below 0.05 was considered significant.
Supplementary information
Acknowledgements
The authors appreciate Dr. Hiroyuki Osada and his coworkers (RIKEN, Wako, Saitama, Japan) for screening compounds bound to FIR or FIRΔexon2 from a chemical library, which was a collection of the isolates from natural products and Dr. Tohru Natsume (Biomedicinal Information Research Center, National Institute of Advanced Industrial Science and Technology, Tokyo, Japan) for detecting FIR or FIRΔexon2-binding proteins by a direct nanoflow liquid chromatography-tandem mass spectrometry analysis in 293 cells. We thank Dr. Masanobu Oshima for giving us the chance to use Gan-mouse, and Drs. Masahiko Hatano and Takashi Miki for preparing some transgenic mice in this experiment. This study was supported in part by Grant-in-Aid 26460667 for priority areas in cancer research from “the Ministry of Education, Science, Sports and Culture of Japan” (KAKENHI), AMED (Japan Agency for Medical Research and Development), “Chiba Foundation for Health Promotion & Disease Prevention” and was partly supported by Extramural Collaborative Research Grant of Cancer Research Institute, Kanazawa University to K.M.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41389-020-0205-4).
References
- 1.Bajpai R, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang P, et al. Integrative analysis identifies co-dependent gene expression regulation of BRG1 and CHD7 at distal regulatory sites in embryonic stem cells. Bioinformatics. 2017;33:1916–1920. doi: 10.1093/bioinformatics/btx092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyagi A, et al. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5:e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang LY, et al. SCFFBW7-mediated degradation of BRG1 suppresses gastric cancer metastasis. Nat. Commun. 2018;9:3569–3577. doi: 10.1038/s41467-018-06038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyo JS, Son BK, Oh D, Kim EK. BRG1 is correlated with poor prognosis in colorectal cancer. Hum. Pathol. 2018;73:66–73. doi: 10.1016/j.humpath.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/S0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita K, et al. An essential role of alternative splicing of c-myc suppressor fuse-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006;66:1409–1417. doi: 10.1158/0008-5472.CAN-04-4459. [DOI] [PubMed] [Google Scholar]
- 8.Page-McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA. 1999;5:1548–1560. doi: 10.1017/S1355838299991938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsushita K, et al. SAP155-mediated splicing of FUSE-binding protein-interacting repressor (FIR) serves as a molecular switch for c-myc gene expression. Mol. Cancer Res. 2012;10:787–799. doi: 10.1158/1541-7786.MCR-11-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, et al. Anti-FIR (PUF60) auto-antibodies are detected in the sera of early-stage colon cancer patients. Oncotarget. 2016;7:82493–82503. doi: 10.18632/oncotarget.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi S, et al. Anti-FIRΔexon2, a splicing variant form of PUF60, autoantibody is detected in the sera of esophageal squamous cell carcinoma. Cancer Sci. 2019;110:2004–2013. doi: 10.1111/cas.14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moccia A, et al. Genetic analysis of CHARGE syndrome identifies overlapping molecular biology. Genet. Med. 2018;20:1022–1029. doi: 10.1038/gim.2017.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corsini L, et al. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J. Biol. Chem. 2009;284:30–39. doi: 10.1074/jbc.M805395200. [DOI] [PubMed] [Google Scholar]
- 14.Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J. Gastroenterol. 2014;20:5403–5410. doi: 10.3748/wjg.v20.i18.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao, Y. L., Shepard, C. R., Wells, A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol. Cancer9, 179–196 (2010). [DOI] [PMC free article] [PubMed]
- 16.Zhou F, et al. Expression profile of E-cadherin, estrogen receptors, and P53 in early-onset gastric cancers. Cancer Med. 2016;5:3403–3411. doi: 10.1002/cam4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano-Gomez, S. J., Maziveyi, M., Alahari, S. K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer15, 18–31 (2016). [DOI] [PMC free article] [PubMed]
- 18.Wang Y, et al. Rapamycin inhibits FBXW7 loss-induced epithelial–mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem. Biophys. Res. Commun. 2013;434:352–356. doi: 10.1016/j.bbrc.2013.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. FBW7 loss promotes epithelial-to-mesenchymal transition in non-small cell lung cancer through the stabilization of Snai1 protein. Cancer Lett. 2018;419:275–283. doi: 10.1016/j.canlet.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 20.Olmeda D, Jordá M, Peinado H, Fabra A, Cano A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene. 2007;26:1862–1874. doi: 10.1038/sj.onc.1209997. [DOI] [PubMed] [Google Scholar]
- 21.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushita K, Hoshino T. Novel diagnosis and therapy for hepatoma targeting HBV-related carcinogenesis through alternative splicing of FIR (PUF60)/FIRΔexon2. Hepatoma Res. 2018;4:2394–5079. doi: 10.20517/2394-5079.2018.81. [DOI] [Google Scholar]
- 23.Ogura Y, et al. Disturbed alternative splicing of FIR (PUF60) directed cyclin E overexpression in esophageal cancers. Oncotarget. 2018;9:22929–22944. doi: 10.18632/oncotarget.25149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kampen, K. R. et al. Translatome analysis reveals altered serine and glycine metabolism in T-cell acute lymphoblastic leukemia cells. Nat. Commun. 10, 2542–2557 (2019). [DOI] [PMC free article] [PubMed]
- 25.Sulima SO, et al. Ribosomal lesions promote oncogenic mutagenesis. Cancer Res. 2019;79:320–327. doi: 10.1158/0008-5472.CAN-18-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girardi T, et al. The T-cell leukemia-associated ribosomal RPL10 R98S mutation enhances JAK-STAT signaling. Leukemia. 2018;32:809–819. doi: 10.1038/leu.2017.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushita K, et al. Haploinsufficiency of the c-myc transcriptional repressor FIR, as a dominant negative-alternative splicing model, promoted p53-dependent T-cell acute lymphoblastic leukemia progression by activating Notch1. Oncotarget. 2015;6:5102–5117. doi: 10.18632/oncotarget.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu-Shui D, Xiao-Xin S, Hua L. Ribosomal Protein L11 Associates with c-Myc at 5 S rRNA and tRNA Genes and Regulates Their Expression. J. Biol. Chem. 2010;285:12587–12594. doi: 10.1074/jbc.M109.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verheij JB, et al. An 8.35 Mb overlapping interstitial deletion of 8q24 in two patients with coloboma, congenital heart defect, limb abnormalities, psychomotor retardation and convulsions. Eur. J. Med. Genet. 2009;52:353–357. doi: 10.1016/j.ejmg.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Dauber A, et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 2013;93:798–811. doi: 10.1016/j.ajhg.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low KJ, et al. PUF60 variants cause a syndrome of ID, short stature, microcephaly, coloboma, craniofacial, cardiac, renal and spinal features. Eur. J. Hum. Genet. 2017;25:552–559. doi: 10.1038/ejhg.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zentner GE, et al. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum. Mol. Genet. 2010;19:3491–3501. doi: 10.1093/hmg/ddq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsh SK, et al. Increased expression of fibroblast growth factor 8 in human breast cancer. Oncogene. 1999;18:1053–1060. doi: 10.1038/sj.onc.1202392. [DOI] [PubMed] [Google Scholar]
- 34.Oshima H, et al. Prostaglandin E2 signaling and bacterial infection recruit tumor-promoting macrophages to mouse gastric tumors. Gastroenterology. 2011;140:596–607.e7. doi: 10.1053/j.gastro.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Kajiwara T, et al. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. 2013;104:149–156. doi: 10.1111/cas.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corsini L, et al. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 38.Stepanyuk GA, et al. UHM-ULM interactions in the RBM39-U2AF65 splicing-factor complex. Acta Crystallogr. D. Struct. Biol. 2016;72:497–511. doi: 10.1107/S2059798316001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison OJ, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanoh N, et al. Immobilization of natural products on glass slides by using a photoaffinity reaction and the detection of protein-small-molecule interactions. Angew. Chem. Int. Ed. Engl. 2003;42:5584–5587. doi: 10.1002/anie.200352164. [DOI] [PubMed] [Google Scholar]
- 41.Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 42.Jomrich G, et al. Expression of FGF8, FGF18, and FGFR4 in Gastroesophageal Adenocarcinomas. Cells. 2019;8:piiE1092. doi: 10.3390/cells8091092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harpain F, et al. FGF8 induces therapy resistance in neoadjuvantly radiated rectal cancer. J. Cancer Res Clin. Oncol. 2019;145:77–86. doi: 10.1007/s00432-018-2757-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura A, et al. FIR haplodeficiency promotes splicing to pyruvate kinase M2 in mice thymic lymphoma tissues revealed by six-plex tandem mass tag quantitative proteomic analysis. Oncotarget. 2017;8:67955–67965. doi: 10.18632/oncotarget.19061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gumireddy K, et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma. Gastric Cancer. 2011;1:101–112. doi: 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- 47.Kaneda A, et al. Activation of Bmp2-Smad1 signal and its regulation by coordinated alteration of H3K27 trimethylation in Ras-induced senescence. PLoS Genet. 2011;7:e1002359. doi: 10.1371/journal.pgen.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita K, et al. Interactions between SAP155 and FUSE-binding protein-interacting repressor bridges c-myc and P27Kip1 expression. Mol. Cancer Res. 2013;11:689–698. doi: 10.1158/1541-7786.MCR-12-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagakubo D, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 50.Hines, J. K., Chen, X., Nix, J. C., Fromm, H. J. & Honzatko, R. B. Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition. J. Biol. Chem.282, 36121–36131 (2007). [DOI] [PubMed]
- 51.Yanagita H, et al. Structural modulation study of inhibitory compounds for RNase H activity of HIV-1 reverse transcriptase. Chem. Pharm. Bull. (Tokyo) 2012;60:764–771. doi: 10.1248/cpb.60.764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.