Abstract
Exosomes are extracellular vesicles which are released from healthy and tumor cells into blood circulation. Unique biomolecular cargos such as RNA and protein are loaded in these vesicles. These molecules may have biological functions such as signaling, cell communications and have the potential to be analyzed as biomarkers. In this initial study, we describe the analysis of exosomes in the serum of healthy subjects, intraductal papillary mucosal neoplasms and pancreatic ductal adenocarcinoma including the characterization of their RNA cargos by next generation sequencing (EXO-NGS). Results indicate the presence of a wide variety of RNAs including mRNA, miRNA, lincRNA, tRNA and piRNA in these vesicles. Based on the differential mRNA expression observed upon EXO-NGS analysis, we independently evaluated two protein coding genes, matrix metalloproteinase-8 (MMP-8) and transcription factor T-Box 3 (TBX3) by qRT-PCR for selective expression in the serum samples. Results indicate a variable expression pattern of these genes across serum samples between different study groups. Further, qRT-PCR analysis with the same serum exosomes processed for EXO-NGS, we observed two long non-coding RNAs, malat-1 and CRNDE to be variably expressed. Overall, our observations emphasize the potential value of different exosome components in distinguishing between healthy, premalignant and malignant conditions related to the pancreas.
Subject terms: Gastrointestinal diseases, Oncology
Introduction
Currently, only a small number of useful biomarkers for the clinical diagnosis of different types of cancer. These have been used for several years without any considerable changes. This results in patients undergoing unnecessary medical procedures. This is further compounded by the fact that a single cancer type may be heterogeneous, presumably due to disparate genetic defects in their tumors1. Recently, studies have been directed towards identification of biomarkers for cancer and other diseases, through non-invasive means utilizing components in blood such as circulating tumor cells (CTCs), cell free DNA (cfDNA) and very recently extracellular vesicles (EVs) which includes microvesicles and exosomes2–4. These liquid biopsy (noninvasive) based analysis present an alternate to conventional tumor biopsies (invasive) and may facilitate biomarker identification to detect early stages of a disease.
EVs such as exosomes are released into blood circulation and may have important functions in physiological and pathological conditions. These vesicles have recently invoked interest due to their potential in disease diagnosis and treatment3,4. They originate from a multivesicular body and upon fusing with the plasma membrane of the cell, they are released into the extracellular environment5. Exosomes range between 30–150 nm in size, are secreted by different cell types including tumor cells6,7 and are heterogeneous which could reflect the phenotypic state of the cell that generates them8. The presence of exosomes has been reported in different biological fluids such as saliva, semen, urine, cerebrospinal fluid, breast milk and blood9–12. Due to the presence of exosomes in variety of pathological conditions, it has been investigated as a source of novel biomarkers based on analysis of their protein and RNA content. Nucleic acids such as mRNA, miRNA, transfer RNAs and long non-coding RNAs (lncRNAs) have been detected in exosomes13,14. Further, nucleic acid inside exosomes could be more resistant to RNA degrading enzymes such as RNases compared to those present in free circulation, which allows for a higher detection sensitivity and specificity12. The utility of exosomes and their cargos, especially the protein content has been previously reported for early detection of pancreatic cancer and predicting organotropic metastasis15,16. However, the RNA elements in exosomes and their probable utility as biomarkers are less explored in this disease.
PDAC is projected to become the second leading cause of cancer-related mortality in the United States by 203017. Due to the lack of screening imaging modalities and specific biomarkers, early diagnosis of this disease is difficult and about 50% of all patients are diagnosed at advanced stages18 and have limited therapeutic options. Unique challenges associated with PDAC include aggressive etiology and deep anatomic location. Currently, serum C19-9 is used as a circulating biomarker for PDAC, although its sensitivity and specificity are less than enough for its use as a diagnostic or early stage biomarker3. Recently, exosome associated protein glypican has been reported as an early detection marker for PDAC15. However, this was disputed by different studies which indicated that glypican may not be an ideal marker to detect early PDAC19,20 because of its presence in non-malignant conditions.
Profiling of RNAs in circulation, specifically miRNAs, has been reported to identify biomarkers for pathologies including cancer21–23. A majority of these RNAs appears to be associated with exosomes. In this initial study, our underlying objective was the isolation and characterization of exosomes in blood serum along with the analysis of RNA cargos present in these vesicles by next generation sequencing (EXO-NGS). Further, we wanted to evaluate any variation in gene expression identified by EXO-NGS across healthy subjects, premalignant and malignant conditions associated with the pancreas. Such comparisons would offer a minimally invasive means of screening exosome RNA biomarkers that lay the groundwork in identifying potential biomarkers for early disease detection.
Results and Discussion
Isolation and characterization of serum exosomes
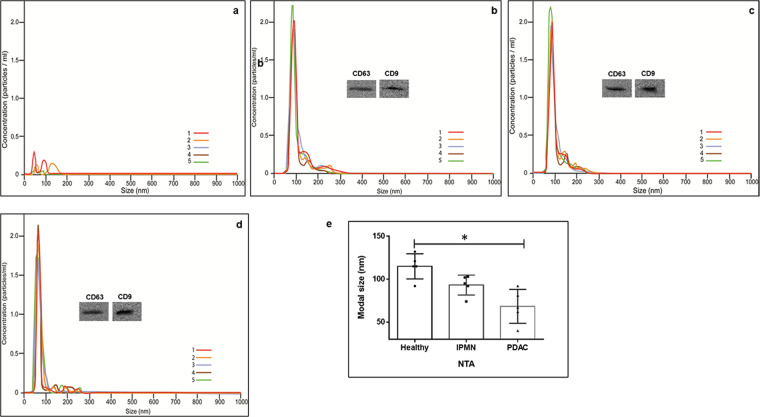
Several commercial kits are available to isolate exosomes from biological fluids which typically consists of polymeric additives to induce precipitation of exosomes24. Use of these kits along with a standard centrifugation process (10,000 × g) allows for a faster separation of exosomes compared to the gold-standard technique of ultracentrifugation which is both laborious and time-consuming. Moreover, studies have reported that the yields and purity using these kits were higher compared to ultracentrifugation25,26. While several options are available to isolate exosomes, depending on the source of exosomes and the purpose of the study, the protocol needs to be optimized to obtain a reasonably high yield and purity of these vesicles for downstream applications. We obtained exosomes (Fig. 1) from different serum samples within the size range (30–150 nm) which is in keeping with the previous studies27.
Figure 1.
Isolation and characterization of exosomes from healthy, IPMN and PC serum. Exosomes from serum were separated using ExoquickTC and purified further using gel exclusion chromatography as mentioned in methods. Exosome size was measured using Nanosight NS300 (Malvern Panalytical, Malvern, UK). (a) Buffer only; (b) Healthy serum; (c) IPMN serum; (d) PDAC serum; Five replicates were performed and modal size distribution was analyzed to assess significance in size differences. (e) Figure depicting the modal size of exosomes in the serum. Size differences between healthy and PDAC serum exosomes was significant (*p < 0.05). Immunoblot analysis were done to identify protein markers such as CD63 and CD9 with respective antibodies. Immunoblots are cropped images from different gels. Uncropped images are shown in Supplementary Fig. 1.
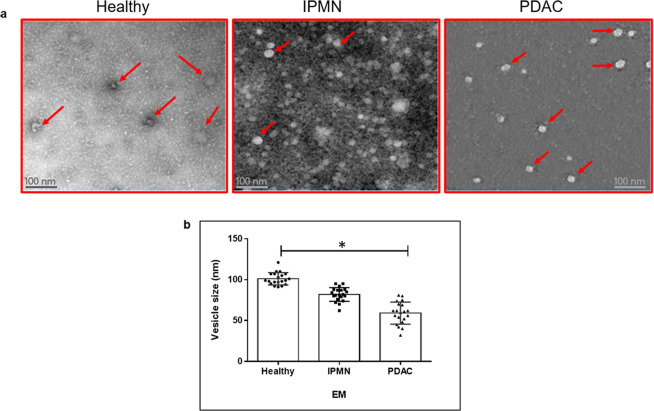
In our study, exosomes isolated from the serum of healthy subjects and different pancreatic conditions were analyzed for their size distribution and quantity using Nanosight NS300 nanoparticle tracking analysis (NTA). Instruments such as dynamic light scattering and NTA have been used previously to measure the size of these vesicles28. NTA instrument in addition to size distribution also measures the nanoparticle concentrations29. The size distribution of exosomes analyzed by NTA between serum samples were different. The normal serum exosomes were predominantly 115 nm ± 20 nm in size while IPMN and PDAC serum exosomes exhibited sizes in the range of 94 nm ± 10 nm and 67 nm ± 25 nm, respectively (Fig. 1). While NTA was useful in analyzing exosome size and concentration the technique may have drawbacks. For instance, the size distributions obtained with this method may depend on set-up-parameters, and the concentration measurements based on refractive index of the material under examination. Further, underestimation of small size particles may be observed due to material-dependent lower detection limit. Hence more than one method may be necessary to analyze exosome morphology and size distribution. Further examination of purified vesicles by transmission electron microscopy (TEM) revealed that these vesicles possess varying size (35–100 nm) and morphology (Fig. 2a) resembling that of exosomes. The average size of serum exosomes as observed by EM was found to be 100 nm (range 90–120 nm) in healthy, 85 nm (range 62–95 nm) in IPMN and 59 nm (range 32–75 nm) in PDAC (Fig. 2b). There appears to be no consensus with regards to the minimal or maximal size range of exosomes in the literature. The lower size range reported varied between 30–50 nm and the upper size range varied between 100–150 nm24,30. This discrepancy could be due to the various exosome isolation methods employed during the studies as well as their source of origin. Our own studies indicated that exosomes isolated from cell culture supernatants tend to be of relatively larger size (~125 nm) compared to liquid biopsy samples such as serum but well within the upper range of 150 nm (data not shown). Collectively, in our current study we observed that serum exosomes appear to be in the size range of 32–130 nm.
Figure 2.
Ultrastructure analysis of exosomes in healthy, IPMN and PDAC serum by TEM. Exosomes were separated from serum samples using ExoquickTC, and subsequently purified by gel exclusion chromatography mentioned in methods. The samples were prepared for TEM and negative staining (n = 3 each). Images were acquired using JEOL JEM 1400 at 120 kV on a Gatan Ultrascan 1000 CCD. Red arrows depict exosomes. (a) Exosome vesicles in serum samples (n = 3 each) from healthy, IPMN and PDAC were measured (twenty vesicles/sample) randomly for their size distribution, and the median values are depicted. Vesicle size were lower in PDAC compared to healthy serum samples (*p = 0.032) (b).
The concentration of exosomes was found to be different in the serum of healthy subjects (7 × 108 particles/ml), patients with IPMN (7.06 × 1010 particles/ml) and patients diagnosed with PDAC (4 × 1011 particles/ml). Exosome concentrations in the serum from PDAC patients tend to be higher than healthy or IPMN samples. Previous reports indicate that exosome concentrations can vary between 1–3 × 1012 particles per ml serum27,31. To confirm the vesicles were indeed exosomes, protein markers for exosomes such as CD63 and CD9 were analyzed using immunoblot studies. Serum exosomes from different conditions exhibit both the presence of CD63 and CD9 (Fig. 1 inset), widely reported to function as exosome markers32,33. Uncropped gel images are shown in supplementary (Fig. S1).
RNA recovery from exosomes
Total RNA was isolated from different serum exosomes and the RNAs were purified using a phenol-free lysis buffer and rapid spin columns. RNA from serum exosomes isolated from each sample category was verified for integrity using a Bioanalyzer. A representative analysis of the small RNAs in serum samples are shown in supplementary (Fig. S2). Exosomes derived from serum typically yielded total RNA in the range of 2.4–12 ng/ml. RNA concentrations up to 10 ng were reported earlier in exosomes27. However, depending on the source of the starting material and the concentration of exosomes, the RNA concentrations could vary. Moreover, exosomes are a heterogeneous population of vesicles with widely varying cargo, in that some vesicles may have many RNAs while some carry only the protein cargo and no RNA.
Analysis of RNA in exosomes by sequencing
Two donors were used for each sample type. The library construction was made by ligating the adaptor sequences to the RNA in the samples. Subsequently, PCR amplification was performed34. Once the library construction was complete, the samples were subjected to sequencing using the NextSeq. 500/550 instrument in high output configuration. The adaptor sequences were trimmed after sequencing exosome libraries and single-end RNA reads were generated. An example of the number of reads pre and post quality filtering is depicted in supplementary (Fig. S3) for different samples. The total number of reads in millions (M) retained after quality assessment were approximately 10 M for healthy serum, 25 M for IPMN and 19 M for PDAC. Further, the reads retained or discarded after quality assessment is depicted in supplementary (Fig. S4). About 75–80% of the reads in different serum samples were of good quality reads. Our analysis also included spike-in-DNA controls35 to avoid any misinterpretations during data normalization, based on the presumption that different serum samples will not yield identical amounts of RNA for analysis.
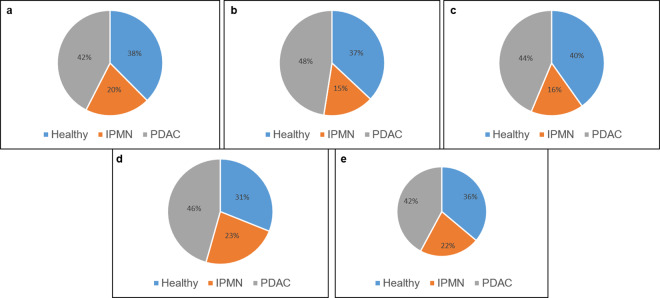
Based on the mapped sequence read counts, the RNA content of different serum exosomes was quantified. The whole summary of reads mapping to gene annotation types are depicted in supplementary (Fig. S5). As illustrated in the pie chart graphs, serum exosomes contained very diverse RNA ‘cargo’. The following total gene types with varying abundance were present in exosomes from individual serum types:(mRNA (20,596), (lincRNA, 8660), ribosomal RNAs (rRNAs,3275), piwi interacting RNAs (piRNA, 2298), microRNAs (miRNA,1521) and transfer RNAs (tRNA/tRNA-like, 391). Also, other contents such as antisense elements to exons, introns, and small lncRNA, small Cajal body-specific RNAs (scaRNAs) including unannotated genes were also present in exosomes. Pie charts depicting the percentage of various RNAs between different serum exosomes are shown in Fig. 3. Overall, the abundance of these RNAs in exosomes are relatively high in PDAC serum compared to healthy and IPMN conditions. We will further focus only on three types of RNAs in this study (mRNA, piRNA and tRNA). Theoretically, it has been indicated that each exosome can accommodate 70–25,000 small RNA or protein molecules27. Given the size and high concentration of exosomes present in blood (1010–1012 particles per ml of blood)31,36 it is not surprising that exosomes could hold a variety of biological molecules within its boundaries and be capable of evoking considerable biological effects in vivo.
Figure 3.
Pie charts depicting different RNAs in exosomes. The pie chart represents the mean percentage of each RNA biotype relative to mapped reads within exosomes obtained from healthy, IPMN and PDAC serum. mRNA (a), piRNA (b), tRNA (c), miRNA (d) and lincRNA (e).
Differentially expressed genes in serum exosomes
Total differentially expressed genes in exosomes between the serum types are shown as volcano plots in supplementary (Fig. S6). Overall, several genes were significantly up or down regulated in IPMN and PDAC compared to healthy serum exosomes. However, such differential expression of genes between two different healthy samples was very minimal. Often exosome studies involve reporting miRNA as biomarker candidates for cancer detection including that of the pancreas37. Other non-coding RNA types such as piRNA and tRNA are underrepresented and should be considered as an important source for differentiating the disease conditions independently or in combination with other RNA types. The ten differentially expressed mRNAs, piRNAs, and five tRNAs in different serum samples are listed in Tables 1–3. Heatmaps indicating the differences in various RNA expression are depicted in supplementary (Fig. S7). RNA-Seq raw data pertained to the depicted genes are shown in supplementary (File S1).
Table 1.
RNA representation in serum derived exosomes. Transcripts organized by RNA types, mRNA, (1) piRNA (2) and tRNA (3). For each differentially expressed RNAs (FDR < 0.05), gene fold changes between groups are depicted. Raw data pertained to these different RNA types are shown in the supplementary excel file.
| mRNA –Gene ID | IPMN/Healthy | PDAC/Healthy | PDAC/IPMN |
|---|---|---|---|
| MMP8 | 3.752300693 | 11.22053302 | 2.990307531 |
| TBX3 | 2.495344919 | 6.110038834 | 2.448574859 |
| PDX1 | 1.862313018 | 4.650163632 | 2.496982831 |
| CTSL | 0.940116753 | 9.621165889 | 10.23401175 |
| SIGLEC15 | 1.139847435 | 3.261413452 | 2.861271914 |
| IL32 | 1.126054868 | 0.538110379 | 0.477872255 |
| SIGLEC11 | 2.197075703 | 0.387841497 | 0.176526233 |
| DCN | 1.039535435 | 0.587693174 | 0.565342127 |
| HOXA5 | 0.531177671 | 0.066935876 | 0.126014101 |
| KLRB1 | 0.82795046 | 0.308809226 | 0.372980318 |
Table 3.
RNA representation in serum derived exosomes.
| tRNA–Gene ID | IPMN/Healthy | PDAC/Healthy | PDAC/IPMN |
|---|---|---|---|
| tRNA125-Thr CGT | 17.23077001 | 24.37797911 | 1.414793366 |
| tRNA21- Ser TGA | 2.930639525 | 19.74772768 | 6.738368027 |
| tRNA15-Cys GCA | 1.657407286 | 2.041150703 | 1.231532357 |
| tRNA55-Ile-TAT | 0.56605197 | 0.149507902 | 0.264123985 |
| tRNA5-ILE TAT | 0.260427893 | 0.331645749 | 1.273464778 |
Among the different mRNA transcripts, MMP8 was high in PDAC exosomes. MMP8, a member of the matrix metalloproteinase family, has been implicated in several cancer types and reported to have conflicting roles in cancer as a promoter and suppressor of metastasis38. However, the role of MMP8 in pancreatic disease is less known. While MMP 8 has been implicated in acute pancreatitis39 its function in PDAC is unclear. A different study suggested that MMP8 could also function as a predictive biomarker in serum for colorectal cancer40. Another coding transcript, TBX3 is also highly represented in PDAC exosome compared to healthy and IPMN serum. TBX3 protein product suppress E-cadherin and enhances melanoma invasiveness41 and is also correlated with advanced stages of gastric cancer42. While these genes were studied previously in direct plasma or tissues, their presence in exosomes have not been reported. Interestingly, we also observed increased representation of the pancreatic and duodenal homeobox-1 (PDX1) transcript in IPMN and tumor exosomes. The protein product from this gene was reported to be involved in the transcriptional activation of insulin and has been implicated in the pathological and clinical features of invasive PDAC43. Conversely, we also observed that some of the coding transcripts such as SIGLEC1144 which is implicated in anti-inflammatory function and DCN which functions as a tumor suppressor and anti-metastatic protein45 were low in PDAC and IPMN exosomes. Also, KLRB1 (CD161) transcript appears to be low in IPMN and PDAC serum. KLRB1 transcript has been reported to be suppressed in lung tumors and esophageal squamous cell carcinoma46 although the exact biological function of this protein is unclear.
Like the protein coding transcripts, the presence of other small ncRNAs such as piRNA and tRNA transcripts were also observed in the exosomes. Previously, these ncRNAs were studied in either pancreatic tissues47 or cells but not in exosomes. For instance, tRNAs have been shown to interact with MEK2 in pancreatic carcinoma cells and alter cell behavior48. Likewise, piRNAs may have tumorigenic or suppressive roles in cancer and are likely involved in regulation of DNA methylation49. While piRNAs have been reported in variety of cancers49, reports are sparsely available for pancreatic cancer. One study, however, indicated that piR-017061 was downregulated in pancreatic cancer tissues47. We observed several piRNA transcripts increased or decreased in PDAC exosomes relative to healthy or IPMN conditions (Table 2). It is unclear regarding the significance of above mentioned ncRNAs in exosomes. However, their expression seems to vary in pathological conditions. We speculate that these RNAs could be exchanged between the exosome target cells and may have functional significance.
Table 2.
RNA representation in serum derived exosomes.
| piRNA–Gene ID | IPMN/Healthy | PDAC/Healthy | PDAC/IPMN |
|---|---|---|---|
| hsa-piR-52959 | 0.737581056 | 48.80631338 | 66.17077944 |
| hsa-piR-53108 | 0.737581056 | 48.70232593 | 66.029795 |
| hsa-piR-30690 | 0.910145955 | 9.543237165 | 10.48539206 |
| hsa-piR-54479 | 3.434405108 | 7.023957071 | 2.045174302 |
| hsa-piR-56621 | 0.23389614 | 12.03718339 | 51.4637967 |
| hsa-piR-54888 | 0.01989337 | 0.021437229 | 1.077606739 |
| hsa-piR-42185 | 0.039457419 | 0.042519581 | 1.077606739 |
| hsa-piR-46410 | 0.031030153 | 0.033438302 | 1.077606739 |
| hsa-piR-58897 | 0.047468386 | 0.051152253 | 1.077606739 |
| hsa-piR-43043 | 0.90149582 | 0.027407666 | 0.030402433 |
Validation of differentially expressed exosome mRNAs by qRT-PCR analysis
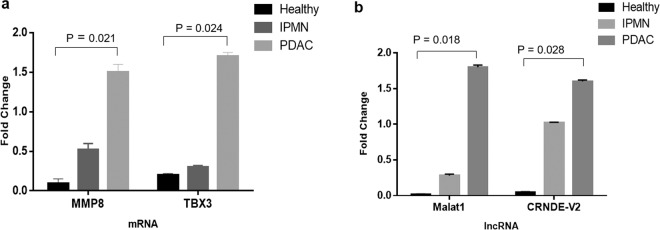
To validate the EXO-NGS sequencing results, the expression of candidate genes MMP8 and TBX3 were analyzed in the serum exosomes under study. Consistent with NGS-EXO observations, both MMP8 and TBX3 were higher in tumor exosomes compared to healthy or IPMN samples with Ct values ranging from 27.6–30.8 with a median Ct of 29.2 (MMP8), and Ct values (28.0–31.7) with a median Ct of 29.8 (TBX3) (Fig. 4a).
Figure 4.
qRT-PCR analysis for RNA expression. Gene expression for MMP8 and TBX3 (a) including malat-1 and CRNDE lncRNA (b) were analyzed by qRT-PCR in the same serum exosomes used in EXO-NGS studies. Two biological replicates were used for each normal, IPMN and PDAC samples and the experiments were performed in duplicates. The dissociation curves for the qRT-PCR are shown in supplementary (Fig. 8).
Both lncRNAs malat1 and CRNDE have been reported to be increased in pancreatic cancer cells or tissues50,51. However, their presence in exosomes are less known. Due to limitation in analysis of gene size during EXO-NGS, lncRNA analysis in exosomes were conducted by qPCR analysis in exosomes isolated directly using the serum samples under investigation. The differences in the expression of lncRNAs malat1 and CRNDE in serum exosomes are depicted in (Fig. 4b). Both lncRNAs were expressed higher in PDAC or IPMN vs healthy samples. The Ct values ranges were between 27 and 31 with a median Ct of 28.7 for malat1 and Ct values between 29–32 with a median Ct of 30.5 for CRNDE. The dissociation curves corresponding to each gene is depicted in supplementary (Fig. S8).
While our studies indicate differences in various RNAs between serum types, the presence of exosome RNAs could be differentially regulated in pancreatic tumor subtypes. For instance, previous RNA sequence analysis in tissues from different pancreatic tumor subtypes52 which varied in their neoplastic cellularity53, indicated that an individual mRNA could be differentially regulated (up or down) within these subtypes. Therefore, it is reasonable to anticipate that exosomes and their components representing different cellular origins could likely mimic these changes. For instance, as observed in this study the exosome mRNA transcripts PDX1 and CTSL (CTSL1) both were found to be high in PDAC. Previous studies52 indicated the presence of these genes in pancreatic tumor tissues but high in progenitor and immunogenic subtypes compared to the basal type. Alternately, the DCN transcript which we observed to be low in PDAC serum exosomes was also absent or decreased in most tumor subtypes except immunogenic subtype. The mRNA distribution of these genes in PDAC serum exosomes from the present study, compared to tumor tissues of different subtypes reported earlier52 are depicted in Table 4.
Table 4.
Gene expression comparison between exosomes and tumor subtypes. The transcripts expressed in exosomes of PDAC serum in the current study was compared with previously reported results obtained in tissues from different PDAC tumor subtypes. The data for PDAC tumor subtypes* are from ref. 52. The transcripts in exosomes appear to indicate the most likely PDAC tumor subtypes a particular gene may be up or down regulated.
| mRNA –Gene ID | PDAC serum exosomes | PDAC Tumor subtypes* | |||
|---|---|---|---|---|---|
| Basal | Classical/Progenitor | ADEX | Immunogenic | ||
| PDX1 | Up | Down | Up | Up | Down |
| CTSL | Up | None | Down | None | Up |
| DCN | Down | None | Down | None | Up |
In the current study, we were able to investigate the RNAs in the patient serum exosomes, however, a limitation in this study is the exosome isolation technique. While various methods have been reported in the literature regarding exosome isolation54, each has its own advantages and disadvantages. Thus far, the established methods appear to lack robustness in obtaining pure exosome population. However, a previous report indicate that both ultracentrifugation and commercially available total exosome isolation reagents have been shown to recover exosomes in a comparable manner55. Another study, indicated that techniques such as precipitation (comparable to the method used in our studies) and membrane affinity are suitable for miRNA discovery in exosomes4. Overall, challenges in isolating exosomes include reproducibility and consistency between techniques. Both technical (e.g. sample collection) and biological (e.g. ideal matched control samples) challenges need to be considered to develop a reliable method for exosome analysis. Going forward, a standard and reproducible exosome enrichment method is necessary for its use in downstream clinical applications.
We identified several RNA types in serum exosomes, which may have the potential to be biomarkers and translatable for clinical applications to detect pancreatic disease early and differentiate among subtypes. Exosome RNA transcripts identified in this study such as PDX1, CTSL and DCN are reported to be variably expressed in different pancreatic tumor subtypes52 and could form the basis for investigating further in precursor lesions such as IPMN. Further validation of these biomolecules needs screening for more specific disease category and larger sample cohorts. Investigations of these biomolecules as markers should also consider biological diversity between patients. Also, any specific and abundantly expressed RNA transcripts in exosomes should exist within detectable levels for further validation as biomarkers in larger cohorts using routinely used technology platforms (e.g. qRT-PCR).
Conclusion
Our studies indicate the presence of exosome vesicles of varying size in different serum samples. While NTA method may be used in analyzing exosomes, additional methods such as EM will be necessary to analyze exosomes. Unlike NTA, EM not only allows to visualize exosome morphology but enables to measure even small size vesicles below 40 nm. By performing EXO-NGS analysis we identified several RNA transcripts which have not been previously reported in exosomes associated with different pancreatic conditions. While most previous RNA studies have been done with pancreatic tumor cells or tissues, liquid biopsy sources such as exosomes would facilitate the noninvasive discovery of useful biomarkers for early detection. The presence of RNAs such a piRNA and tRNA, hitherto sparsely reported in pancreatic exosomes, suggested the need for further studies to gain insights into their biological effects on target cells. While our studies involve limited sample analysis, validation in large prospective cohorts which include more cases of premalignant lesions will be extremely useful in detecting the early disease. Our future studies are directed toward such validation efforts in patients to identify prognostic or predictive biomarkers using RNA signatures.
Methods
Collection of serum samples
After approval of the Institutional Review Board (IRB), Office of Research, University of Missouri, and informed consent from the human subjects for study participation, blood samples without anticoagulants from healthy individuals, intraductal papillary mucinous neoplasms (IPMN) and PDAC (stage III-IV) were collected. An hour after the blood collection, serum was separated by centrifugation (2300 rpm, 15 min) and used immediately for isolation of exosomes or stored at −80 °C for long term. If stored, 500 μl of serum was centrifuged at 1500 × g for 5 min to remove residual cells and debris. The supernatant was transferred to a new 1.5 ml Eppendorf tube before exosome isolation. All methods were performed in accordance with relevant guidelines and regulations.
Exosome isolation from serum
ExoQuick Ultra (System Biosciences, Palo Alto, CA) was added to the supernatant from the above step at a 1:4 ratio, mixed gently, and incubated for 30 min at 4 °C. After the incubation, the admixture was centrifuged at 1500 × g for 30 min to recover exosomes for total RNA isolation. For gel purification, serum was diluted with PBS (two-fold) and processed using qEV size exclusion columns (IZON, Medford, MA) according to manufacturer’s instructions.
Exosome particle size analysis
Exosomes obtained from serum were diluted in PBS (dilution depending on the concentration of exosomes) and subsequently analyzed using a NanoSight NS300. The instrument is equipped with a 405 nm laser (NanoSight, Amesbury, UK), and an electron multiplying charge‐coupled device (EMCCD) was used to determine the histogram of particle size by tracking the Brownian motion of single particles at 25 °C for size and concentration determination. Videos were recorded for 60 seconds during which the movement of the nanoparticles were tracked by NTA software (version 2.3, NanoSight) with low refractive index corresponding to serum derived vesicles. The Stokes-Einstein equation was employed to determine the size distribution and number of particles within the sample.
Transmission electron microscopy
Exosomes isolated from the serum were processed for transmission electron microscopy (TEM) and negative staining. All specimen preparation was performed at the Electron Microscopy Core Facility, University of Missouri. Exosomes were suspended in PBS (pH 7.2), and placed on a negatively charged carbon coated copper grid and allowed to settle at room temperature for 20 minutes. Excess sample was removed by wicking with filter paper, then fixed for two minutes in 2% paraformaldehyde and, 2% glutaraldehyde in 0.05 M phosphate buffer (pH 7.0) solution. Grids were rinsed with three consecutive washes of distilled water and negative staining was performed by placing 5 μL of Nano Tungsten (Nanoprobes, Inc.) on each grid for two minutes prior to wicking to dryness with filter paper and allowed to dry at room temperature. Images were acquired with a JEOL JEM 1400 transmission electron microscope (JEOL, Peabody, MA) at 120 kV on a Gatan Ultrascan 1000 CCD (Gatan, Inc, Pleasanton, CA).
Total RNA isolation from exosomes
Total RNA from exosomes was isolated using the SeraMir Exosome RNA Purification kit (System Biosciences) according to the manufacturer’s instructions. Subsequently, to measure RNA concentration and integrity of each sample, 1 μl of the final RNA eluate was used for measurement of small RNA concentration by Agilent Bioanalyzer Small RNA Assay using Bioanalyzer 2100 Expert instrument (Agilent Technologies, Santa Clara, CA).
Immunoblotting
Exosomes isolated from serum were suspended in 50 μl M-PER reagent (Thermo Scientific) with HALT protease inhibitor cocktail (Thermo Scientific). After determining the protein concentration using Bradford analysis, the samples were fractionated on a 4–12% gradient SDS-PAGE gels (Invitrogen, Carlsbad, CA). Subsequently, the gels were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). The membrane was incubated with anti-CD63 (H5C6) (Novus Biologicals, Centennial, CO) or anti-CD9 (D3H4P) antibodies (Cell Signaling Technology, Danvers, MA) overnight at 4 °C, followed by incubation with peroxidase conjugated anti-mouse/-rabbit antibody. The proteins were visualized using an Immobilon Forte Western HRP blotting substrate (Millipore, Burlington, MA).
NGS Library generation and sequencing
Small RNA libraries were constructed with the Clean Tag Small RNA Library Preparation Kit (TriLink, Cat# L-3206) according to the manufacturer’s protocol. The final purified library was quantified with High Sensitivity DNA Reagents (Agilent Technologies, PO# G2933-85004) and High Sensitivity DNA Chips (Agilent Technologies, PO# 5067–4626). The libraries were pooled, and the 140 bp to 300 bp region was size selected on an 8% TBE gel (Invitrogen, Life Technologies). The size selected library was quantified with High Sensitivity DNA 1000 Screen Tape (Agilent Technologies, PO # 5067–5584), High Sensitivity D1000 reagents (Agilent Technologies, PO# 5067–5585), and the TailorMix HT1 qPCR assay (SeqMatic, Cat# TM-505), followed by a NextSeq High Output single-end sequencing run at SR75 using NextSeq. 500/550 High Output v2 kit (Cat #FC-404-2005, Illumina, San Diego, CA) according to the manufacturer’s instructions.
Bioinformatic analysis
Raw reads filtering
The Exosome RNA-seq analysis initiates with a data quality check of the input sequence using FastQC, an open-source quality control (QC) tool for high-throughput sequence data56. FastQC runs analyses of the uploaded raw sequence reads that reveals the quality of the data and inform the subsequent preprocessing steps in the analysis. Following the initial assessment, Bowtie257 was used to map the spike-in DNA before the trimming and filtering steps where RNA-seq reads are preprocessed to improve the quality of data input for read mapping. The open-source tools used for trimming of adapters are FastqMcf56 and Cutadapt58, while PRINSEQ59 was used in the quality filtering step.
Reads mapping
Sequencing adapters were trimmed from reads, and filtered for quality and length in the preprocessing steps. FastQC is then re-run to analyze the trimmed reads, allowing for a before and after comparison. The processed set of input data was used in the subsequent read mapping step. The improved set of sequence reads are merged, if needed, using SeqPrep60 then mapped to the reference genome assembly (GRCh37-hg19) using Bowtie257. Using the open-source software BEDTools61 and SAMtools62, read alignment and read coverage tracks are generated and deployed to the genome browser.
RNA abundance analysis
Abundance determination and differential expression analysis was performed by DEseq63. Abundance levels for ncRNAs (miRNAs, tRNAs, rRNAs, lincRNAs, piRNAs, snoRNAs), antisense transcripts, coding genes and repeat elements (LTR, LINE, SINE, and tandem repeats) were determined. A summary of reads overlapping each of these annotations in the reference genome is created using SAM tools.
cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Exosome RNA was converted to cDNA using a High Capacity Reverse Transcription kit (Applied Biosystems, Foster City, CA) with RNAse inhibitor treatment according to manufacturer’s protocol. qRT-PCR was performed using SYBR Green assay for exosome mRNA expression (n = 3 each sample) of matrix metalloproteinase-8 (MMP-8), TBX3 and long non-coding RNAs, malat-1 and CRNDE. β-actin was used as internal control. Relative quantification (fold change) was determined using 2−ΔΔCt method. Following primers were used in this study: MMP8, F-CCTGGTGCCTTGATGTA, R- GGTCCAGTAGGTTGGATAG; TBX3, F- GTGACTGCATACCAGAAT, R-TTCTCTTCGGCCATTTC; Malat1, F-GTTCTGATCCCGCTGCTATT, R-TCCTCAACACTCAGCCTTTATC; CRNDE, F- GTTGTCACGCAGAAGAAG, R-TCCTATACCTTGGCTAAACA. Two replicate serum samples each from healthy, IPMN and PDAC were used. Analysis was done in duplicates.
Statistical analyses
Differences in gene expression levels were analyzed using either a paired t-test or nonparametric two-tailed Mann Whitney –U test. P value < 0.05 was considered significant.
Supplementary information
Acknowledgements
A core faculty grant from College of Veterinary Medicine to S.R.K. is kindly acknowledged. The authors are grateful for the help of Maren Fleer and Sydney Hackwell for their assistance in this study. S.R.K. receives support from a Grant #5UL1TR002345.
Author contributions
S.R.K. conceived the study and wrote the manuscript. E.T.K. coordinated patients for this study. S.G. performed the experiments along with S.R.K. Y.M. along with E.T.K. had direct contact with the patients to collect blood samples and subsequent processing for downstream studies. S.R.K. and A.J.S. performed the bioinformatic analysis. J.T.K. as the responsible IRB primary investigator coordinated patient recruitment and subjects’ consenting, blood sample acquisition and processing/analysis in the laboratory. All authors reviewed the manuscript.
Data availability
The data pertained to this manuscript are available to readers upon publication. Certain materials as a part of this data but not associated with this manuscript may not be shared due to either ongoing analysis or publication restrictions.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/16/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
is available for this paper at 10.1038/s41598-020-59523-0.
References
- 1.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 2.Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J. Mol. Diagn. 2015;17:209–224. doi: 10.1016/j.jmoldx.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Buschmann D, et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles. 2018;7:1481321. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braccioli L, van Velthoven C, Heijnen CJ. Exosomes: a new weapon to treat the central nervous system. Mol. Neurobiol. 2014;49:113–119. doi: 10.1007/s12035-013-8504-9. [DOI] [PubMed] [Google Scholar]
- 6.Caivano A, et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumour Biol. 2015;36:9739–9752. doi: 10.1007/s13277-015-3741-3. [DOI] [PubMed] [Google Scholar]
- 7.Moldovan L, Batte K, Wang Y, Wisler J, Piper M. Analyzing the circulating microRNAs in exosomes/extracellular vesicles from serum or plasma by qRT-PCR. Methods Mol. Biol. 2013;1024:129–145. doi: 10.1007/978-1-62703-453-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi DS et al., Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles. 1 (2012). [DOI] [PMC free article] [PubMed]
- 9.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Toro J, Herschlik L, Waldner C, Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front. Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lässer C. Identification and analysis of circulating exosomal microRNA in human body fluids. Methods Mol. Biol. 2013;1024:109–128. doi: 10.1007/978-1-62703-453-1_9. [DOI] [PubMed] [Google Scholar]
- 13.Gusachenko ON, Zenkova MA, Vlassov VV. Nucleic acids in exosomes: disease markers and intercellular communication molecules. Biochem. 2013;78:1–7. doi: 10.1134/S000629791301001X. [DOI] [PubMed] [Google Scholar]
- 14.Valadi H, et al. Exosome mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 15.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa-Silva B. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 18.Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in Pancreatic cancer and their application to treatment. Nat. Rev. Gastroenterol. Hepatol. 2012;9:435–444. doi: 10.1038/nrgastro.2012.119. [DOI] [PubMed] [Google Scholar]
- 19.Frampton AE, et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget. 2018;9:19006–19013. doi: 10.18632/oncotarget.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucien F, et al. Glypican-1 and glycoprotein 2 bearing extracellular vesicles do not discern pancreatic cancer from benign pancreatic diseases. Oncotarget. 2019;10:1045–1055. doi: 10.18632/oncotarget.26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 22.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2017;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer Detection. Proc. Natl Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab. Chip. 2017;17:3558–3577. doi: 10.1039/c7lc00592j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane RE, Korbie D, Anderson W, Vaidyanathan R, Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci. Rep. 2015;5:7639. doi: 10.1038/srep07639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobb RJ, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, et al. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:1652. doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranyai T, et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One. 2015;10:e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Pol E, Coumans F, Varga Z, Krumrey M, Nieuwland R. Innovation in detection of microparticles and exosomes. J. Thromb. Haemost. 2013;1:36–45. doi: 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- 30.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caradec J, et al. Reproducibility and efficiency of serum-derived exosome extraction Methods. Clin. Biochem. 2014;47:1286–1292. doi: 10.1016/j.clinbiochem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 34.Zeringer E, et al. Methods for the extraction and RNA profiling of exosomes. World J. Methodol. 2013;3:11–18. doi: 10.5662/wjm.v3.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen K, et al. The Overlooked Fact: Fundamental Need for Spike-In Control for Virtually All Genome-Wide Analyses. Mol. Cell Biol. 2015;36:662–667. doi: 10.1128/MCB.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melo SA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chronopoulos, Lieberthal, T. J. & del Río Hernández, A. E. Exosomes as a platform for ‘liquid biopsy’ in pancreatic cancer. Converg. Sci. Phys. Oncol. 3:013005.
- 38.Thirkettle S, et al. Matrix metalloproteinase 8 (collagenase 2) induces the expression of interleukins 6 and 8 in breast cancer cells. J. Biol. Chem. 2013;288:16282–16294. doi: 10.1074/jbc.M113.464230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nukarinen E, et al. Association of Matrix Metalloproteinases -7, -8 and -9 and TIMP -1 with Disease Severity in Acute Pancreatitis. A Cohort Study. PLoS One. 2016;11:e0161480. doi: 10.1371/journal.pone.0161480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Böckelman C, et al. Serum MMP-8 and TIMP-1 predict prognosis in colorectal Cancer. BMC Cancer. 2018;18:679. doi: 10.1186/s12885-018-4589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008;68:7872–7881. doi: 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- 42.Miao ZF, et al. Tbx3 overexpression in human gastric cancer is correlated with advanced tumor stage and nodal status and promotes cancer cell growth and invasion. Virchows Arch. 2016;469:505–513. doi: 10.1007/s00428-016-2007-9. [DOI] [PubMed] [Google Scholar]
- 43.Quint K, et al. The expression pattern of PDX-1, SHH, Patched and Gli-1 is associated with pathological and clinical features in human pancreatic cancer. Pancreatology. 2009;9:116–126. doi: 10.1159/000178882. [DOI] [PubMed] [Google Scholar]
- 44.Angata T, et al. Cloning and characterization of human Siglec-11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J. Biol. Chem. 2002;277:24466–24474. doi: 10.1074/jbc.M202833200. [DOI] [PubMed] [Google Scholar]
- 45.Sofeu Feugaing DD, Götte M, Viola M. More than matrix: the multifaceted role of decorin in cancer. Eur. J. Cell Biol. 2013;92:1–11. doi: 10.1016/j.ejcb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Pleshkan VV, Zinov’eva MV, Vinogradova TV, Sverdlov ED. Transcription of the KLRB1 gene is suppressed in human cancer tissues. Mol. Gen. Mikrobiol. Virusol. 2007;4:3–7. [PubMed] [Google Scholar]
- 47.Müller S, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol. Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, et al. Interaction of tRNA with MEK2 in pancreatic cancer cells. Sci. Rep. 2016;6:28260. doi: 10.1038/srep28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim. Biophys. Acta Rev. Cancer. 2019;1871:160–169. doi: 10.1016/j.bbcan.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Tang X, Shi M, Wen C, Shen B. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2017;483:816–822. doi: 10.1016/j.bbrc.2016.12.167. [DOI] [PubMed] [Google Scholar]
- 51.Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017;50:e12389. doi: 10.1111/cpr.12389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Raphael BJ, et al. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood LD, Hruban RH. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012;18:492–501. doi: 10.1097/PPO.0b013e31827459b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control. Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schageman J, et al. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed. Res. Int. 2013;2013:253957. doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrews, S. FastQC: A quality control tool for high throughput sequence data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2005).
- 57.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 59.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.John, J. S. SeqPrep, https://github.com/jstjohn/SeqPrep (2011).
- 61.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data pertained to this manuscript are available to readers upon publication. Certain materials as a part of this data but not associated with this manuscript may not be shared due to either ongoing analysis or publication restrictions.