Abstract
In stress conditions, as neoplastic transformation, amino acids serve not only as nutrients to maintain the cell survival but also as mediators of several regulatory pathways which are involved in apoptosis and autophagy. Especially, under glucose deprivation, in order to maintain the cell survival, proline and glutamine together with other glutamine-derived products such as glutamate, alpha-ketoglutarate, and ornithine serve as alternative sources of energy. They are substrates for production of pyrroline-5-carboxylate which is the product of conversion of proline by proline dehydrogenase/ proline oxidase (PRODH/POX) to produce ATP for protective autophagy or reactive oxygen species for apoptosis. Interconversion of proline, ornithine, and glutamate may therefore regulate PRODH/POX-dependent apoptosis/autophagy. The key amino acid is proline, circulating between mitochondria and cytoplasm in the proline cycle. This shuttle is known as proline cycle. It is coupled to pentose phosphate pathway producing nucleotides for DNA biosynthesis. PRODH/POX is also linked to p53 and AMP-activated protein kinase (AMPK)-dependent pathways. Proline availability for PRODH/POX-dependent apoptosis/autophagy is regulated at the level of collagen biosynthesis (proline utilizing process) and prolidase activity (proline supporting process). In this review, we suggest that amino acid metabolism linking TCA and Urea cycles affect PRODH/POX-dependent apoptosis/autophagy and the knowledge might be useful to targeted cancer therapy.
Keywords: Apoptosis, Autophagy, Proline dehydrogenase/proline oxidase, Proline, Glutamine
Introduction
In stress conditions, cellular homeostasis is maintained by alteration of anabolic and catabolic processes. Anabolic processes are regulated by several factors affecting biosynthesis of cellular components. Major catabolic processes are mediated by the ubiquitin–proteasome system and autophagy [1]. In some cases, autophagy and apoptosis simultaneously occur in the same cell or autophagy precedes apoptosis via p53-dependent pathways or AMP-activated protein kinase (AMPK) [1]. Alternatively, autophagy can directly activate cell death pathway [1, 2]. Both p53 and AMPK are potent stimulators of proline dehydrogenase/proline oxidase (PRODH/POX) that has been implicated in the induction of autophagy and apoptosis [3–10]. Since PRODH/POX is linked to conversion of proline to pyrroline-5-carboxylate (P5C) [11], the availability of proline to this process is of critical importance. Proline and P5C are intermediates of interconversion of glutamine, glutamate, ornithine, and α-ketoglutarate suggesting the key role of these amino acids in the regulation of PRODH/POX-dependent apoptosis/autophagy. Therefore, this review aims to discuss the contribution of proline, glutamine, and its metabolites in regulation of PRODH/POX-dependent apoptosis/autophagy.
Regulatory mechanism of autophagy and apoptosis
Autophagy
Autophagy is a homeostatic, intracellular degradation process in which dispensable, long-lived, or aberrant proteins and damaged organelles are digested in lysosomes. The digestion products are recycled in cellular metabolism. It usually happens under stress conditions such as amino acid starvation [12–14]. Besides the removal of useless components retained in the cell, the other function of autophagy is to generate energy for synthesis of new building blocks in the process of homeostasis and cellular renovation [12, 13]. It suggests that autophagy has a profound impact on cancer cell survival [15]. Autophagy may also contribute to the suppression of cancer cell growth. The activation of autophagy explains a resistance mechanism in the course of cancer therapy. Therefore, the inhibition of autophagy was suggested as a potential pharmacotherapeutic approach for tumor growth suppression [13, 16].
A variety of proteins have been considered as autophagy markers for the assessment of presence or absence of autophagy in the cell. The first autophagy markers were found in yeast and identified more than 30 autophagy-related (ATG) genes, many of which have known orthologs in higher eukaryotes [17, 18]. Atg proteins have been classified into different groups based on their function in autophagy: (1) the Atg1/ULK complex (Atg1, Atg11, Atg13, Atg17, Atg29, and Atg31) regulates the induction of autophagosome formation; (2) the Atg9 complex (Atg2, Atg9, and Atg18), involved in membrane delivery to the expanding phagophore; (3) the PtdIns 3-kinase (PtdIns3K) complex (Vps34, Vps15, Vps30/Atg6, and Atg14) functions to recruit PtdIns3P-binding proteins; (4) two ubiquitin-like (Ubl) conjugation systems including the Atg12 complex (Atg5, Atg7, Atg10, Atg12, and Atg16) and a Atg8 complex (Atg3, Atg4, Atg7, and Atg8) that plays crucial role in vesicle expansion [19, 20] (Table 1). The mammalian ULK1/2 complex comprises ULK1/2 (mammalian homologs of Atg1), ATG13 (a homolog of yeast Atg13), RB1CC1/FIP200 (a putative Atg17 homolog), and C12orf44/ATG101 [21, 22]. The other study provided evidence that ULK1 kinase can be activated by AMP-activated protein kinase (AMPK) under glucose or amino acid starvation [23]. The ULK1/2 complex is inhibited by the phosphorylation of mTORC1 preventing interaction between ULK1 and AMPK. However, during induction of autophagy, the suppression of mTOR occurs and the protein complex of ULK1/2, ATG13, and RB1CC1 is formed to initiate the autophagy. Moreover, the autophagy process is mediated by Beclin-1 (autophagy-related gene, Atg 6) which codes for another autophagy protein [24, 25]. Some of these markers were linked to the PRODH/POX-dependent apoptosis/autophagy [3–10, 26, 27]. Since it has been proved that there is a cross-talk between autophagy and apoptosis [28], it cannot be excluded that the mechanism of this process may involve PRODH/POX.
Table 1.
Classification of biomarkers of autophagy
| ATG complex | Yeast | Mammals | Functions | References |
|---|---|---|---|---|
|
Atg/ULK complex (regulates the class III phosphatidylinositol (PtdIns) 3-kinase complex) |
Atg1 | ULK1/2 | Ser/Thr protein kinase; phosphorylated by M/TORC1; recruitment of Atg proteins to the PAS | [22] |
| Atg13 | ATG13 | Regulatory subunit through phosphorylation by M/TORC1 and/or PKA, linker between Atg1 and Atg17 | ||
| Atg17 | RB1CC1/FIP200 (functional homolog) | Scaffold protein, ternary complex with Atg29 and Atg31. Phosphorylation by ULK1; scaffold for ULK1/2 and ATG13 | ||
| C12orf44/Atg101 | Component of the complex with ATG13 and RB1CC1 | |||
| Atg2-Atg18/Atg9 complex (maintenance of mitochondrial integrity) | Atg2 | ATG2 | Regulates Atg9 recycling from phagophore assembly site | [79] |
| Atg18 | WIPI1/2 | |||
| Atg9 | ATG9A/B | Required for autophagosome formation; Required for the efficient recruitment of Atg8 and Atg18 | ||
| Atg23 |
Interaction with Atg9 Required for the biosynthetic cytoplasm to vacuole targeting (Cvt) pathway and efficient autophagy |
[80] | ||
|
PtdIns3K complex (Beclin1-Atg14-Ambra1- Vps15- Vps34) |
Vps34 | PIK3C3/VPS34 | PtdIns 3-kinase | [18] |
| Vps15 | PIK3R4/VPS15 | Ser/Thr protein kinase | ||
| Vps30/Atg6 | BECN 1/Beclin 1 |
Component of PtdIns3K complex I and II Forms a complex with ER-associated Bcl-2 under nutrient-rich conditions and is released upon phosphorylation of Bcl-2 by JNK1 |
||
| AMBRA1 | Interacts with Beclin 1 | |||
| Atg14 | ATG14 | Component of PtdIns3K complex I | ||
|
Atg8 complex (Ubiquitin-like conjugation system) |
Atg8 | LC3A/B/C, GABARAP, GABARAPL1/2 | A unique ubiquitin-like conjugation to phosphatidylethanolamine on the autophagic membrane | [18, 81] |
| Atg7 | ATG7 | E1-like enzyme | ||
| Atg3 | ATG3 | E2-like enzyme | ||
| Atg4 | ATG4A-D |
Cysteine proteinase LC3/Atg8 C-terminal hydrolase; deconjugating enzyme |
||
|
Atg12-Atg5-Atg16 Complex (Ubiquitin-like conjugation system) |
Atg12 | ATG12 | Ubiquitin-like | [18] |
| Atg7 | ATG7 | E1-like enzyme | ||
| Atg10 | ATG10 | E2-like enzyme | ||
| Atg16 | ATG16L1 | Activate Atg5; Interacts Atg12 | ||
| Atg5 | ATG5 |
Conjugated by Atg12 Directly binds membranes |
||
Apoptosis
A concept of apoptosis was initially reported by Karl Vogt in 1872 then described by Walther Flemming who was the first to explain the mechanism of programmed cell death in 1885. Several studies suggested this mechanism as a program of cellular suicide where the cell destroys itself to maintain tissue homeostasis [29]. The machinery of apoptosis is mediated by a family of proteases, namely caspases which contain a cysteine at their active site and cleave the target proteins at a residue of aspartic acids [30]. Their precursors are called procaspases which are expressed as inactive forms in normal condition. These proteins, however, are cleaved to become active caspases triggering the apoptosis via energy-dependent cascade pathways [30]. The apoptosis is recruited through 3 different pathways: the extrinsic pathway, the intrinsic pathway, and Granzyme B-dependent pathway [31]. Among these pathways, the intrinsic and extrinsic pathways are the major mechanisms of apoptosis.
The intrinsic apoptosis pathway is activated by damages taking place within the cell. This mechanism involves the presence of pro-apoptotic proteins, BAX, and BID in the outer membrane of the mitochondria. They interact with the other protein, BAK to activate cytochrome c that binds to apoptotic protease activating factor-1 (Apaf-1) [32]. This binding activates active caspase 9 that triggers cascade downstream of effector caspases (such as caspase 3, caspase 7, and caspase 6), finally resulting in cell death [33]. The p53 protein is a key factor to activate the intrinsic pathway due to its contribution to activate BAX protein [34].
In contrast, the extrinsic pathway is initiated from extracellular events, triggered by ligand binding to plasma membrane death receptors, leading to activation of initiator caspase 8 [31]. Death receptors such as Fas/CD95 and tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptors DR-4 and DR-5 are transmembrane proteins that function to detect specific extracellular death signals [35, 36]. For instance, Adapter molecules like Fas Associated via Death Domain (FADD) contain death domain (DD) and a death effector domain (DED) which activate an active caspase-8 via a sequential action of a homotypic DED–DED interaction. Active caspase-8 generates a downstream of effector caspases contributing to cell death. However, they have the same execution pathway which is initiated by the activation of caspase-3 [31]. Typical biomarkers of apoptosis are listed in Table 2. Most of them were linked to PRODH/POX-dependent apoptosis [3–10].
Table 2.
Typical biomarkers of apoptosis
| Biomarker | Testing sample | Function | Method of detection | References |
|---|---|---|---|---|
| Activated caspase 2, 3, 7, 8 and 9 | Tissue | Primary modulators of apoptosis | IHC, ELISA, flow cytometry, cytometric bead arrays | [82] |
| Caspase-3 | Myocardial injury and cardiovascular disease | Responsible for chromatin condensation and DNA fragmentation | IHC, ELISA, flow cytometry, cytometric bead arrays | [82, 83] |
| Caspase 3/7 | Hypothalamic cell model | Primary modulators of apoptosis | Multiplexing fluorescent and luminescent assays | [84] |
| Caspase 6 | Neurodegenerative disorders (Alzheimer’s and Huntington disease) | Primary modulators of apoptosis | Electrochemiluminescence-based ELISA assay | [85] |
| Cytochrome C |
Tissue, serum HL-60 cells and thymocytes |
Transfer electrons from the cytochrome bc1 complex to cytochrome oxidase membrane | ELISA, flow cytometry | [82, 86] |
| CK18 | Hepatocellular Carcinoma Treated with Sorafenib | M30- and M65-based sandwich ELISAs | [87] | |
| Cytokeratins | Tissue, serum plasma | IHC, ELISA, flow cytometry, | [82] | |
| Nucleosomal DNA | Tissue, serum | ELISA, DNA array, PCR | [82] | |
|
Apo-I/Fas, Fas ligand (sFAsL) Expressed on B and T cells as well as in normal and tumor tissue |
Granulomatous disease | Increase the antigen-specific CD8( +) T-cell responses during viral infection | IHC, ELISA, flow cytometry | [82, 88, 89] |
| Bcl-2/Bcl-xl/Mcl-I | Cells, tissue | IHC, ELISA, flow cytometry | [82] | |
| TRAIL | Inducing the autoimmune inflammation | Induces apoptosis through an extrinsic pathway, | [90] | |
|
Tumor protein p53 (TP53) |
Colorectal cancer and other cancers | TP53 activation is capable of inducing apoptosis by intrinsic pathway | IHC, ELISA, flow cytometry | [82, 91] |
ELISA enzyme-linked immunosorbent assay; IHC immunohistochemistry; PCR polymerase chain reaction
PRODH/POX-dependent pathways relevant to apoptosis and autophagy
A variety of approaches to the inhibition of autophagy or activation of apoptosis have recently focused on proline dehydrogenase (PRODH), known also as proline oxidase (POX). PRODH/POX, a mitochondrial enzyme, converts proline to pyrroline-5-carboxylate (P5C) with the concomitant transfer of electrons to cytochrome c producing ATP or directly on oxygen generating reactive oxygen species (ROS) [5]. There are two human genes annotated as PRODH: PRODH1 (chromosome 22q11.21; NCBI Accession NM_016335) and PRODH2 (chromosome 19q13.12; NCBI Accession NM_021232). It has been suggested that the function of the enzyme may depend on substrate availability, proline. The main source of this amino acid is collagen which comprises 25% of total protein mass in animals [10, 30].
Briefly, these proteins are classified into major types which are type I in the skin, tendon, and bone, type II in cartilage, and type IV in basal laminae. Up to date, 28 types of collagen with 46 distinct polypeptide chains were found in vertebrates, as well as many other proteins containing collagenous domains [37, 38]. The predominant amino acids in collagen are proline and glycine, which enable triple-helical collagen structure. Extracellular degradation of collagens by tissue collagenases and further intracellular degradation of collagen degradation products in lysosomes release imidopeptides that are cleaved by cytoplasmic prolidase releasing a large amount of proline, the substrate for PRODH/POX.
After the conversion of proline to P5C, further proline metabolism is catalyzed by pyrroline-5-carboxylate dehydrogenase (P5CDH), transforming P5C into glutamate which is a precursor of α-ketoglutarate (α-KG) involved in the tricarboxylic acid (TCA) cycle. When the TCA cycle is overloaded by metabolites, the reversible reaction of conversion of P5C into proline by pyrroline-5-carboxylate reductase (P5CR) may occur, using NADPH or NADH as a cofactor. This interconversion of P5C-proline called proline cycle was first introduced in 1986 [39]. It has been demonstrated that the cellular proline, glutamine, and glutamate are linked to the proline pathway [40] regulating apoptosis/autophagy. The cycle is coupled to pentose phosphate shunt through NADPH from pentose pathway and NADP + from the proline cycle [4, 41]. Base on this mechanism, the role of PRODH/POX in the regulation of cellular metabolism has recently studied as an approach to cancer treatment. This cycle is responsible for the regulation of gene expression, purine biosynthesis, cellular redox state, apoptosis, and cell proliferation [3]. Moreover, PRODH/POX has a variety of regulatory functions, such as osmotic adjustment, protection against metabolic stress, and signaling in bacteria, plants, and mammals [10]. However, the most important function of PRODH/POX is donating electrons through flavin adenine dinucleotide (FAD) into the electron transport chain to generate ROS or ATP depending on environmental conditions [10].
PRODH/POX-induced apoptosis
Both intrinsic and extrinsic pathways of apoptosis may be induced by PRODH/POX [42]. Especially, in the extrinsic pathway (death receptor), PRODH/POX stimulates the expression of tumor necrosis factor-related apoptosis-activated ligand (TRAIL), DR5, and cleavage of caspase-8 [42, 43], and also activates caspase-9 and caspase-3 [44, 45]. In cancer cells, PRODH/POX is upregulated by a variety of factors, for example tumor suppressor p53 and inflammatory factor peroxisome proliferator-activated receptor gamma (PPARγ) [7, 10]. However, its level in cancer tissue is much lower than that in normal tissues from the patients [46, 47]. Regarding the overexpression of POX, the ROS generation is integrated with the p53-dependent mechanisms [5, 48], switching the apoptotic cell death in a variety of cancer cell types [5, 48–51]. The supporting evidence showed that the PRODH/POX coding gene induced the expression of p53 [52]. On the other hand, inactivation of proline oxidase reduced p53-induced upregulation of proline oxidase, a release of cytochrome c from mitochondria, and apoptosis in cancer cells [42, 49]. PRODH/POX acting as a driver of apoptosis was clearly evaluated in a model of PRODH/POX knockdown cancer cells [53].
PRODH/POX-induced autophagy
The recent study of Zareba et al., (2018) showed that in knocked down PRODH/POX MCF-7 breast cancer cells, cytoplasmic proline accumulation induced autophagy. However it was established that environmental conditions such as hypoxia or glucose deficiency may affect PRODH/POX-dependent autophagy/apoptosis [9]. It seems that proline availability may determine PRODH/POX-dependent apoptosis/autophagy. Although the mechanism of this process is not known, it has been suggested that hypoxia-inducible factor-1 alpha (HIF-1α) plays an important role in cancer cell metabolism. The availability of proline in the cell facilitates generation of α-KG that inhibits the transcriptional activity of HIF-1α. An increase in αKG concentration leads to an increase in the activity of a prolyl hydroxylase domain (PHD) of HIF-1α inducing proteasomal degradation of HIF-1α [43, 45, 54]. In contrast, proline through the same mechanism inhibits the activity of PHD, contributing to a decrease in HIF-1α proteasomal degradation and increase in its transcriptional activity.
It is well established that glutamine and proline metabolism, as well as other non-essential amino acids, are involved in oncometabolism of cells [9]. This process is called as “parametabolic pathway”. Particularly, the proline biosynthetic pathway was linked to glucose metabolism and POX-dependent apoptosis that is under the regulation of oncogene MYC.
Depending on the metabolic situation, proline can either be used for protein synthesis or oxidized in the mitochondria for energy production. Under nutrient deficiency and hypoxia, cancer cells may adopt to switch a survival mechanism which is the degradation of proline to produce the energy [26]. Therefore, hypoxia, glucose depletion, or treatment with rapamycin stimulated degradation of proline and POX-dependent autophagy.
The impact of amino acids on cell re-programming
Several amino acids have been linked to activation or inhibition of apoptosis/autophagy [55]. It is well recognized that they participate in the mTORC1 and GCN2/eIF2 pathways which function to regulate protein translation and control the cellular demand for amino acids by concomitantly regulating autophagy-dependent catabolism [56–58]. For instance, non-essential amino acids (NEA) as proline in condition of glucose deprivation activate anti-apoptotic pathways in cancer cells by inducing the expression of anti-apoptotic members of the Bcl-2 gene family and preventing the expression of pro-apoptotic proteins [59]. The study suggested that although under low glucose condition apoptosis could be induced in cancer cells, the non-essential amino acids may counteract the process. It was supported by the upregulation of amino acid transporter gene LAT1 in the membranes of cancer cells [27, 60, 61] under glucose stress [59].
Glutamine was proved to be a sustainable source of energy. Early findings indicated that tumor formation is significantly due to the mitochondrial vulnerability through the alteration of glycolysis [62]. The proliferation of cancer cells is mostly maintained by energy products derived from the TCA cycle [63, 64]. A larger majority of tumor suppressors and oncogenes have been linked to metabolic pathways [64–67]. Glutamine is an integral metabolite in the proliferation of mammalian cells. The consumption rate of glutamine in cancer cells is compared to that of other amino acids. However, the demand for glutamine was observed to be tenfold higher than that for other amino acids [68]. Glutamine has profound impact on the functional activity of mammalian target of rapamycin (mTOR) kinase, mitochondrial membrane potential, and NADPH production [69]. Glutamine is a nitrogen source both for purine and pyrimidine synthesis [70, 71]. In the non-essential amino acid synthetic pathways, glutamine-derived glutamic acid continues donating its amine group to accelerate the tricarboxylic acid (TCA) cycle metabolites for the production of α-ketoglutarate, serine, alanine, aspartate, and ornithine. Glutamine acts as a source of carbon and nitrogen for the synthesis of proline, ornithine, and arginine as well as a donor for the synthesis of asparagine from aspartic acid [69]. Lack of exogenous glutamine is one of the major causes for the death of cancer cells [72]. Several tumor cell lines, generated from pancreatic cancer, glioblastoma multiforme, acute myelogenous leukemia, and small cell lung cancer, are substantially vulnerable due to glutamine starvation [73]. The study suggested that derivatives of glutamine like glutamate, α-ketoglutarate, and glutathione are involved in the apoptotic pathway [74]. Similarly, proline interconvertibility with glutamate and arginine [3, 75] may play an important role in cell programming. However, recent data linked glutamine metabolism and apoptosis/autophagy through P5C to urea cycle.
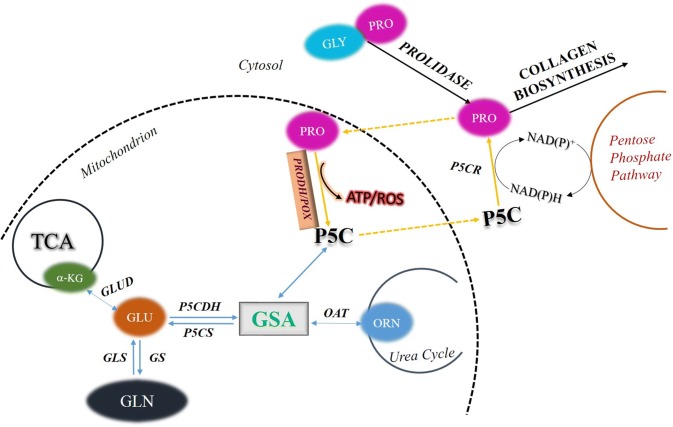
Ornithine and glutamate are important sources of P5C. Ornithine is converted into P5C in a reaction catalyzed by mitochondrial vitamin B6-dependent ornithine-δ-aminotransferase (OAT), while glutamate through a reduction reaction catalyzed by mitochondrial ATP- and NAD(P)H-dependent P5C synthase (P5CS) [76, 77]. This reaction can be reversed by mitochondrial P5C dehydrogenase (P5CDH) [76]. The role of this metabolic pathway in apoptosis/ autophagy was supported by data showing that degradation of ornithine by ornithine decarboxylase (ODC) play an important role in cell proliferation, differentiation, and cell death. It has been demonstrated that decreasing the activity of ODC by difluoromethylornithine (DFMO) causes accumulation of intracellular reactive oxygen species (ROS) and cell arrest, thus inducing cell death. These findings indicate that urea cycle contributes to the regulation of apoptosis and autophagy [78]. Since ornithine is easily convertible into P5C (products of catalytic activity of PRODH/POX), it may affect PRODH/POX-dependent apoptosis/autophagy. The results of these studies allow us to present a hypothesis on the regulation of PRODH/POX-dependent apoptosis/autophagy by key amino acids (Fig. 1). During conversion of PRO into P5C by PRODH/POX, ATP or ROS is generated inducing autophagy or apoptosis. PRO availability for this process is critical requirement for PRODH/POX-dependent function. PRO comes from collagen degradation products (last step of the degradation is catalyzed by prolidase) or proline convertible amino acids, mainly GLU and ORN. Conversion of PRO into P5C takes place in mitochondria, while P5C into PRO mainly in cytoplasm. This process is known as a “proline cycle” and is coupled to pentose phosphate pathway generating nucleotides for DNA biosynthesis. Interconversion of PRO, GLU, and ORN through intermediate GSA to P5C may represent an interface regulating PRODH/POX-dependent P5C generation and ATP/ROS for autophagy/apoptosis. The process links TCA and Urea cycles to proline cycle providing complex regulatory mechanism of PRODH/POX-dependent functions. Understanding the interplay between key amino acids and TCA/Urea metabolites and their role in the regulation of PRODH/POX-dependent apoptosis/autophagy might be a promising approach to targeted cancer therapy.
Fig. 1.
Regulation of PRODH/POX-dependent apoptosis/autophagy by key amino acids. PRO proline; GLU glutamate; ORN ornithine; GLN glutamine; GLYPRO glycyl-proline; PRODH/POX proline dehydrogenase (PRODH)/proline oxidase (POX); ROS reactive oxygen species; P5C pyrroline-5-carboxylate; P5CR pyrroline-5-carboxylate reductase; P5CDH pyrroline-5-carboxylate dehydrogenase; P5CS pyrroline-5-carboxylate synthase; OAT ornithine aminotransferase; GSA glutamic gamma-semialdehyde; αKG α-ketoglutarate; TCA tricarboxylic acid cycle; GS glutamine synthase; GLS glutaminase; GLUD glutamate dehydrogenase
Conclusion
Studies of last decade provided several lines of evidence for the regulatory role of proline availability in PRODH/POX-dependent apoptosis/autophagy in cancer cells. The enzyme expression is often downregulated in various tumors, limiting mitochondrial proline degradation and PRODH/POX-dependent apoptosis. Critical factor for the process is proline availability that depends on the activity of prolidase (enzyme supporting cytoplasmic proline level) and the rate of proline utilization in the process of collagen biosynthesis. However, proline also represents an energy-sensing molecule that reprograms cellular metabolism. Interconversion of proline, glutamate, and ornithine links TCA cycle, urea cycle, and amino acid metabolism to PRODH/POX-dependent apoptosis/autophagy. Deregulation of energetic metabolism in cancer cells due to Warburg’s effect facilitates protein degradation as an alternative source of energy. Therefore, when glucose supply is limited, cancer cells may select proline as an alternative energy source. Therefore, amino acid metabolism in specific environmental cellular conditions may represent interface of PRODH/POX-dependent apoptosis and autophagy. The hypothesis is outlined in Fig. 1.
Funding
This work was supported by the National Science Center [grant number 2017/25/B/NZ7/02183]; the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie [grant agreement number 754432].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen S, Kepp O, Kroemer G. The end of autophagic cell death? Autophagy. 2012;8:1–3. doi: 10.4161/auto.8.1.16618. [DOI] [PubMed] [Google Scholar]
- 3.Phang JM. The regulatory functions of proline and pyrroline-5-carboxylic acid. Curr Top Cell Regul. 1985;25:91–132. doi: 10.1016/B978-0-12-152825-6.50008-4. [DOI] [PubMed] [Google Scholar]
- 4.Pandhare J, Donald SP, Cooper SK, Phang JM. Regulation and function of proline oxidase under nutrient stress. J Cell Biochem. 2009;107:759–768. doi: 10.1002/jcb.22174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810–1815. [PubMed] [Google Scholar]
- 6.Liu Y, Borchert GL, Surazynski A, Phang JM. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene. 2008;27:6729–6737. doi: 10.1038/onc.2008.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phang JM, Liu W, Zabirnyk O. Proline metabolism and microenvironmental stress. Annu Rev Nutr. 2010;30:441–463. doi: 10.1146/annurev.nutr.012809.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phang JM, Liu W. Proline metabolism and cancer. Front Biosci (Landmark Ed) 2012;17:1835–1845. doi: 10.2741/4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Phang JM. Proline dehydrogenase (oxidase), a mitochondrial tumor suppressor, and autophagy under the hypoxia microenvironment. Autophagy. 2012;8:1407–1409. doi: 10.4161/auto.21152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Phang JM. Proline dehydrogenase (oxidase) in cancer. BioFactors. 2012;38:398–406. doi: 10.1002/biof.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicencio JM, Galluzzi L, Tajeddine N, Ortiz C, Criollo A, Tasdemir E, Morselli E, Ben Younes A, Maiuri MC, Lavandero S, Kroemer G. Senescence, apoptosis or autophagy? When a damaged cell must decide its path–a mini-review. Gerontology. 2008;54:92–99. doi: 10.1159/000129697. [DOI] [PubMed] [Google Scholar]
- 15.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chude CI, Amaravadi RK. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int J Mol Sci. 2017;18:1279. doi: 10.3390/ijms18061279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 21.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.e08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Česen MH, Pegan K, Spes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Lee JS, Kim YJ, Kim CL, Lee GM. Differential induction of autophagy in caspase-3/7 down-regulating and Bcl-2 overexpressing recombinant CHO cells subjected to sodium butyrate treatment. J Biotechnol. 2012;161:34–41. doi: 10.1016/j.jbiotec.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Phang JM, Pandhare J, Liu Y. The metabolism of proline as microenvironmental stress substrate. J Nutr. 2008;138:2008S–2015S. doi: 10.1093/jn/138.10.2008S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichinoe M, Mikami T, Yoshida T, Igawa I, Tsuruta T, Nakada N, Anzai N, Suzuki Y, Endou H, Okayasu I. High expression of L-type amino-acid transporter 1 (LAT1) in gastric carcinomas: comparison with non-cancerous lesions. Pathol Int. 2011;61:281–289. doi: 10.1111/j.1440-1827.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 28.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 29.Goodman SR. Medical cell biology. Amsterdam: Elsevier/Academic Press; 2007. [Google Scholar]
- 30.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- 31.Logue SE, Martin SJ. Caspase activation cascades in apoptosis. Biochem Soc Trans. 2008;36:1–9. doi: 10.1042/BST0360001. [DOI] [PubMed] [Google Scholar]
- 32.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/S0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 33.Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, Green DR, Martin SJ. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maximov GK, Maximov KG. The role of p53 tumor-supressor protein in apoptosis and cancerogenesis. Biotechnol Biotechnol Equipment. 2008;22:664–668. doi: 10.1080/13102818.2008.10817532. [DOI] [Google Scholar]
- 35.Fossati S, Ghiso J, Rostagno A. TRAIL death receptors DR4 and DR5 mediate cerebral microvascular endothelial cell apoptosis induced by oligomeric Alzheimer's Aβ. Cell Death Dis. 2012;3:e321. doi: 10.1038/cddis.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 37.Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R. Collagen XXVIII, a novel von Willebrand factor A domain-containing protein with many imperfections in the collagenous domain. J Biol Chem. 2006;281:3494–3504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- 38.Brinckmann J. Collagens at a Glance. Berlin, Heidelberg: Springer; 2005. [Google Scholar]
- 39.Hagedorn CH, Phang JM. Catalytic transfer of hydride ions from NADPH to oxygen by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch Biochem Biophys. 1986;248:166–174. doi: 10.1016/0003-9861(86)90413-3. [DOI] [PubMed] [Google Scholar]
- 40.Cappelletti P, Tallarita E, Rabattoni V, Campomenosi P, Sacchi S, Pollegioni L. Proline oxidase controls proline, glutamate, and glutamine cellular concentrations in a U87 glioblastoma cell line. PLoS ONE. 2018;13:e0196283. doi: 10.1371/journal.pone.0196283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25:5640–5647. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- 43.Kononczuk J, Czyzewska U, Moczydlowska J, Surażyński A, Palka J, Miltyk W. Proline oxidase (POX) as a target for cancer therapy. Curr Drug Targets. 2015;16:1464–1469. doi: 10.2174/138945011613151031150637. [DOI] [PubMed] [Google Scholar]
- 44.Cooper SK, Pandhare J, Donald SP, Phang JM. A novel function for hydroxyproline oxidase in apoptosis through generation of reactive oxygen species. J Biol Chem. 2008;283:10485–10492. doi: 10.1074/jbc.M702181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zareba I, Celinska-Janowicz K, Surazynski A, Miltyk W, Palka J. Proline oxidase silencing induces proline-dependent pro-survival pathways in MCF-7 cells. Oncotarget. 2018;9:13748–13757. doi: 10.18632/oncotarget.24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Borchert GL, Donald SP, Diwan BA, Anver M, Phang JM. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res. 2009;69:6414–6422. doi: 10.1158/0008-5472.CAN-09-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W, Zabirnyk O, Wang H, Shiao YH, Nickerson ML, Khalil S, Anderson LM, Perantoni AO, Phang JM. miR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene. 2010;29:4914–4924. doi: 10.1038/onc.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Borchert GL, Donald SP, Surazynski A, Hu CA, Weydert CJ, Oberley LW, Phang JM. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis. 2005;26:1335–1342. doi: 10.1093/carcin/bgi083. [DOI] [PubMed] [Google Scholar]
- 49.Maxwell SA, Rivera A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. J Biol Chem. 2003;278:9784–9789. doi: 10.1074/jbc.M210012200. [DOI] [PubMed] [Google Scholar]
- 50.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 51.Hu CA, Donald SP, Yu J, Lin WW, Liu Z, Steel G, Obie C, Valle D, Phang JM. Overexpression of proline oxidase induces proline-dependent and mitochondria-mediated apoptosis. Mol Cell Biochem. 2007;295:85–92. doi: 10.1007/s11010-006-9276-6. [DOI] [PubMed] [Google Scholar]
- 52.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 53.Zareba I, Surazynski A, Chrusciel M, Miltyk W, Doroszko M, Rahman N, Palka J. Functional Consequences of Intracellular Proline Levels Manipulation Affecting PRODH/POX-Dependent Pro-Apoptotic Pathways in a Novel in Vitro Cell Culture Model. Cell Physiol Biochem. 2017;43:670–684. doi: 10.1159/000480653. [DOI] [PubMed] [Google Scholar]
- 54.Myllyharju J. Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol (Oxf) 2013;208:148–165. doi: 10.1111/apha.12096. [DOI] [PubMed] [Google Scholar]
- 55.Seglen PO, Gordon PB. Amino acid control of autophagic sequestration and protein degradation in isolated rat hepatocytes. J Cell Biol. 1984;99:435–444. doi: 10.1083/jcb.99.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meijer AJ, Dubbelhuis PF. Amino acid signalling and the integration of metabolism. Biochem Biophys Res Commun. 2004;313:397–403. doi: 10.1016/j.bbrc.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 59.Wang G, Dai L, Luo L, Xu W, Zhang C, Zhu Y, Chen Z, Hu W, Xu X, Pan W. Non-essential amino acids attenuate apoptosis of gastric cancer cells induced by glucose starvation. Oncol Rep. 2014;32:332–340. doi: 10.3892/or.2014.3205. [DOI] [PubMed] [Google Scholar]
- 60.Fukumoto S, Hanazono K, Komatsu T, Ueno H, Kadosawa T, Iwano H, Uchide T. L-type amino acid transporter 1 (LAT1): a new therapeutic target for canine mammary gland tumour. Vet J. 2013;198:164–169. doi: 10.1016/j.tvjl.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 61.Kaira K, Oriuchi N, Takahashi T, Nakagawa K, Ohde Y, Okumura T, Murakami H, Shukuya T, Kenmotsu H, Naito T, Kanai Y, Endo M, Kondo H, Nakajima T, Yamamoto N. L-type amino acid transporter 1 (LAT1) expression in malignant pleural mesothelioma. Anticancer Res. 2011;31:4075–4082. [PubMed] [Google Scholar]
- 62.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 63.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 66.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 67.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eagle H, Oyama VI, Levy M, Horton CL, Fleischman R. The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J Biol Chem. 1956;218:607–616. [PubMed] [Google Scholar]
- 69.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young VR, Ajami AM. Glutamine: the emperor or his clothes? J Nutr. 2001;131:2449S–S2459. doi: 10.1093/jn/131.9.2449S. [DOI] [PubMed] [Google Scholar]
- 71.Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Metabolism and action of amino acid analog anti-cancer agents. Pharmacol Ther. 1990;46:243–271. doi: 10.1016/0163-7258(90)90094-I. [DOI] [PubMed] [Google Scholar]
- 72.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 73.Wu MC, Arimura GK, Yunis AA. Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase. Int J Cancer. 1978;22:728–733. doi: 10.1002/ijc.2910220615. [DOI] [PubMed] [Google Scholar]
- 74.Matés JM, Segura JA, Alonso FJ, Márquez J. Natural antioxidants: therapeutic prospects for cancer and neurological diseases. Mini Rev Med Chem. 2009;9:1202–1214. doi: 10.2174/138955709789055180. [DOI] [PubMed] [Google Scholar]
- 75.Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- 76.Hu CA, Lin WW, Valle D. Cloning, characterization, and expression of cDNAs encoding human delta 1-pyrroline-5-carboxylate dehydrogenase. J Biol Chem. 1996;271:9795–9800. doi: 10.1074/jbc.271.16.9795. [DOI] [PubMed] [Google Scholar]
- 77.Hu CA, Lin WW, Obie C, Valle D. Molecular enzymology of mammalian Delta1-pyrroline-5-carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J Biol Chem. 1999;274:6754–6762. doi: 10.1074/jbc.274.10.6754. [DOI] [PubMed] [Google Scholar]
- 78.Liu GY, Hung YC, Hsu PC, Liao YF, Chang WH, Tsay GJ, Hung HC. Ornithine decarboxylase prevents tumor necrosis factor alpha-induced apoptosis by decreasing intracellular reactive oxygen species. Apoptosis. 2005;10:569–581. doi: 10.1007/s10495-005-1891-2. [DOI] [PubMed] [Google Scholar]
- 79.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, Lee SS, Brezovich A, Lou JH, Turk BE, Aebersold R, Ammerer G, Peter M, Kraft C. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tucker KA, Reggiori F, Dunn WA, Klionsky DJ. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J Biol Chem. 2003;278:48445–48452. doi: 10.1074/jbc.M309238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ward TH, Cummings J, Dean E, Greystoke A, Hou JM, Backen A, Ranson M, Dive C. Biomarkers of apoptosis. Br J Cancer. 2008;99:841–846. doi: 10.1038/sj.bjc.6604519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 84.Butterick TA, Duffy CM, Lee RE, Billington CJ, Kotz CM, Nixon JP. Use of a caspase multiplexing assay to determine apoptosis in a hypothalamic cell model. J Vis Exp. 2014 doi: 10.3791/51305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ehrnhoefer DE, Skotte NH, Savill J, Nguyen YT, Ladha S, Cao LP, Dullaghan E, Hayden MR. A quantitative method for the specific assessment of caspase-6 activity in cell culture. PLoS ONE. 2011;6:e27680. doi: 10.1371/journal.pone.0027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campos CB, Paim BA, Cosso RG, Castilho RF, Rottenberg H, Vercesi AE. Method for monitoring of mitochondrial cytochrome c release during cell death: Immunodetection of cytochrome c by flow cytometry after selective permeabilization of the plasma membrane. Cytometry A. 2006;69:515–523. doi: 10.1002/cyto.a.20273. [DOI] [PubMed] [Google Scholar]
- 87.Godin C, Louandre C, Bodeau S, Diouf M, Saidak Z, Conte MA, Chauffert B, Barbare JC, Barget N, Trinchet JC, Ganne N, Galmiche A. Biomarkers of apoptosis and necrosis in patients with hepatocellular carcinoma treated with sorafenib. Anticancer Res. 2015;35:1803–1808. [PubMed] [Google Scholar]
- 88.Weant AE, Michalek RD, Khan IU, Holbrook BC, Willingham MC, Grayson JM. Apoptosis regulators Bim and Fas function concurrently to control autoimmunity and CD8+ T cell contraction. Immunity. 2008;28:218–230. doi: 10.1016/j.immuni.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 89.Montes-Berrueta D, Ramírez L, Salmen S, Berrueta L. Fas and FasL expression in leukocytes from chronic granulomatous disease patients. Invest Clin. 2012;53:157–167. [PubMed] [Google Scholar]
- 90.El-Karaksy SM, Kholoussi NM, Shahin RM, El-Ghar MM, Rl-S G. TRAIL mRNA expression in peripheral blood mononuclear cells of Egyptian SLE patients. Gene. 2013;527:211–214. doi: 10.1016/j.gene.2013.05.084. [DOI] [PubMed] [Google Scholar]
- 91.Zeestraten EC, Benard A, Reimers MS, Schouten PC, Liefers GJ, van de Velde CJ, Kuppen PJ. The prognostic value of the apoptosis pathway in colorectal cancer: a review of the literature on biomarkers identified by immunohistochemistry. Biomark Cancer. 2013;5:13–29. doi: 10.4137/BIC.S11475. [DOI] [PMC free article] [PubMed] [Google Scholar]