The hyperdiverse arthropods often harbor maternally transmitted bacteria that protect against natural enemies. In many species, low-diversity communities of heritable symbionts are common, providing opportunities for cooperation and conflict among symbionts, which can impact the defensive services rendered. Using the pea aphid, a model for defensive symbiosis, we show that coinfections with two common defensive symbionts, the antipathogen Regiella and the antiparasite Hamiltonella, produce outcomes that are highly variable compared to single infections, which consistently protect against designated enemies. Compared to single infections, coinfections often reduced defensive services during enemy challenge yet improved aphid fitness in the absence of enemies. Thus, infection with multiple symbionts does not necessarily create generalist aphids with “Swiss army knife” defenses against numerous enemies. Instead, particular combinations of symbionts may be favored for a variety of reasons, including their abilities to lessen the costs of other defensive symbionts when enemies are not present.
KEYWORDS: insect, symbiosis, protective mutualism, parasitoid, pathogen, evolution, coinfection, defense, endosymbionts, symbiont
ABSTRACT
Animal-associated microbes are highly variable, contributing to a diverse set of symbiont-mediated phenotypes. Given that host and symbiont genotypes, and their interactions, can impact symbiont-based phenotypes across environments, there is potential for extensive variation in fitness outcomes. Pea aphids, Acyrthosiphon pisum, host a diverse assemblage of heritable facultative symbionts (HFS) with characterized roles in host defense. Protective phenotypes have been largely studied as single infections, but pea aphids often carry multiple HFS species, and particular combinations may be enriched or depleted compared to expectations based on chance. Here, we examined the consequences of single infection versus coinfection with two common HFS exhibiting variable enrichment, the antiparasitoid Hamiltonella defensa and the antipathogen Regiella insecticola, across three host genotypes and environments. As expected, single infections with either H. defensa or R. insecticola raised defenses against their respective targets. Single infections with protective H. defensa lowered aphid fitness in the absence of enemy challenge, while R. insecticola was comparatively benign. However, as a coinfection, R. insecticola ameliorated H. defensa infection costs. Coinfected aphids continued to receive antiparasitoid protection from H. defensa, but protection was weakened by R. insecticola in two clones. Notably, H. defensa eliminated survival benefits conferred after pathogen exposure by coinfecting R. insecticola. Since pathogen sporulation was suppressed by R. insecticola in coinfected aphids, the poor performance likely stemmed from H. defensa-imposed costs rather than weakened defenses. Our results reveal a complex set of coinfection outcomes which may partially explain natural infection patterns and suggest that symbiont-based phenotypes may not be easily predicted based solely on infection status.
IMPORTANCE The hyperdiverse arthropods often harbor maternally transmitted bacteria that protect against natural enemies. In many species, low-diversity communities of heritable symbionts are common, providing opportunities for cooperation and conflict among symbionts, which can impact the defensive services rendered. Using the pea aphid, a model for defensive symbiosis, we show that coinfections with two common defensive symbionts, the antipathogen Regiella and the antiparasite Hamiltonella, produce outcomes that are highly variable compared to single infections, which consistently protect against designated enemies. Compared to single infections, coinfections often reduced defensive services during enemy challenge yet improved aphid fitness in the absence of enemies. Thus, infection with multiple symbionts does not necessarily create generalist aphids with “Swiss army knife” defenses against numerous enemies. Instead, particular combinations of symbionts may be favored for a variety of reasons, including their abilities to lessen the costs of other defensive symbionts when enemies are not present.
INTRODUCTION
Long-term associations with specific microbial communities are common in animals, with the importance of the resident microbiota in the evolution and ecology of hosts increasingly recognized (1, 2). Infections with maternally transmitted bacterial endosymbionts, especially heritable facultative symbionts (HFS), are widespread across terrestrial arthropods (3, 4). HFS infections are major sources of phenotypic diversity, providing their animal hosts with a range of nutritional and defensive services (5–8). While most studies examining HFS roles are based on assays isolating the effects of infection by a single symbiont species (9), in many insect groups, inherited symbionts naturally occur in low-diversity multispecies communities (10–14). The factors underlying the maintenance of multiple HFS, including consequences of coinfection for infected hosts, have received considerably less attention. However, prior work suggests that infection by multiple symbionts or symbiont strains may affect the within-host abundance of symbionts (15, 16), the fidelity of vertical transmission (12, 17), and the conferred phenotypes (18, 19) while impacting the costs of symbiont infection experienced by the animal host (19, 20). The relative simplicity of heritable microbial communities in insects, combined with the ability to engineer specific microbial compositions, provides a tractable model for investigating host–heritable-microbiome interactions.
Aphids represent a well-developed model for studying the phenotypic effects of infection with HFS (21, 22). As a group, aphids are infected with at least nine HFS species, and their distributions vary both within and among aphid species (22–25). Most HFS have been studied in isolation and are known to confer conditional benefits on their aphid hosts, including heat tolerance, protection against natural enemies, and modifications in dietary breadth (26–33). For protective symbionts, phenotypes can vary depending on the symbiont strain (34–39) and temperature (40–42), and particular strains may target specific natural enemies (43–46). Infection costs, although not typically large, are often observed in the absence of enemy challenge and can also vary with the symbiont strain or host genotype (20, 30, 36, 47–49).
The phenotypic effects of individual HFS species have been best explored in the pea aphid, Acyrthosiphon pisum, in which seven commonly occurring heritable symbionts have been implicated in mediating ecological interactions (8). However, population level surveys of European and North American pea aphids indicate that multispecies infections are common, if not the norm, in some populations (11, 13). For example, a recent paper reported that most HFS-infected pea aphids from New York carried two or more HFS, and when viewed across six locales in the United States, some HFS combinations were enriched, while others were depleted, relative to expectations based on chance (12). At particular times, coinfections can be very common and average as high as 3.73 HFS per aphid (50). Many selective and nonselective factors potentially contribute to the maintenance of multiple infections in nature (9, 51). While stable combinations of HFS coinfections can be maintained in laboratory-held lines (20, 52, 53), early studies observed lower fidelity in the vertical transmission of double infections (17). More recent field efforts have confirmed this pattern for some HFS communities while also showing that common HFS partners may improve each others’ transmission (12). Multiple infections may or may not modify conferred phenotypes. For instance, some groupings enhance protection relative to single infection (19), while others exhibit neutral or detrimental effects on protective phenotypes (52, 54). Multispecies HFS communities may also produce generalist aphids capable of responding to multiple threats (20). In the absence of such threats, the benefits of these pairings appear variable, with costs increasing for some pairings and declining for others (19, 20, 54).
Two of the best-characterized pea aphid defensive symbionts are Hamiltonella defensa and the closely related Regiella insecticola (both in the Yersiniaceae [Enterobacteriales]) (55–59). In North America, these two HFS occur at intermediate infection frequencies within most populations and are geographically widespread (12, 13, 50). Infection by H. defensa protects the pea aphid from parasitism by disrupting development of the braconid wasp Aphidius ervi (37), the most common parasitoid of the aphid in North America (60). However, H. defensa cannot provide this defensive effect on its own; associated temperate bacteriophages, called APSEs (A. pisum secondary endosymbionts), are required for protection (61–64). R. insecticola, on the other hand, is protective against the specialized entomopathogenic fungus Pandora neoaphidis (Entomophthorales [Entomophthoraceae]), which is an important natural enemy of aphids. Symbiont infection increases aphid survival and reproduction while decreasing fungal spore production from deceased aphids (27, 31, 65). R. insecticola has also been associated with changes in the propensity for wing development, possibly with host plant utilization, and with the timing of sexual reproduction in pea aphids (66–69). Currently, there are no reports that it confers protection against parasitoids in pea aphids (29, 35, 47). However, one R. insecticola strain from the green peach aphid, Myzus persicae, confers protection against parasitoids when transferred to the black bean aphid, Aphis fabae (33). This raises the possibility that some as yet unassayed strains from pea aphids may also provide this benefit.
Coinfections between H. defensa and R. insecticola provide a compelling system for investigating the potential costs and benefits of coinfection. One field-based study found that, while each symbiont protected against its targeted enemy, benefits were largely eliminated due to infection costs and mortality from the enemy not targeted by a single-symbiont infection (70). This suggests that aphids may benefit from coinfections with particular combinations, including H. defensa and R. insecticola, that create generalist aphids capable of withstanding attack from both parasitoids and fungal pathogens. A recent laboratory study supported this idea (20), demonstrating that aphids coinfected with H. defensa and R. insecticola received the multiple defensive benefits conferred by these symbionts under single infection. In the same study, the strengths of symbiont-conferred protection against the parasitoid A. ervi and, separately, the fungal pathogen Pandora were similar in coinfected versus singly infected lines harboring these symbionts, while fitness costs were not altered by single infection versus coinfection. In the present study, our goal was to expand the study of infection with multiple HFS using the same general interaction: pea aphids infected with H. defensa and/or R. insecticola presented with challenge by the same enemies, Pandora and A. ervi. Our study used different aphid genotypes and symbiont strains, including strains of H. defensa that are protective against parasitoids and those that are not, each with and without coinfection with R. insecticola. We also examined these HFS combinations across multiple aphid genotypes varying in their endogenous resistance to parasitoids (71, 72), as significant host-genotype interactions have been found (36, 73).
RESULTS
Natural-enemy challenge. (i) The parasitoid wasp A. ervi.
Overall, we found significant variation in successful parasitism (Fig. 1) (generalized linear model [GLzM]; χ2 = 815.2; df = 10; P ≤ 0.0001) with only protective H. defensa strains (χ2 = 298.0; P ≤ 0.0001), aphid genotype (χ2 = 58.5; P ≤ 0.0001), and their interaction (i.e., H. defensa effects were reduced in endogenously resistant genotypes relative to the susceptible one; χ2 = 16.1; P = 0.0003) having significant effects. Since time blocks did not contribute to variation in outcomes of aphid-parasitoid interactions, subsequent within-genotype comparisons lumped data from separate time blocks into single analyses.
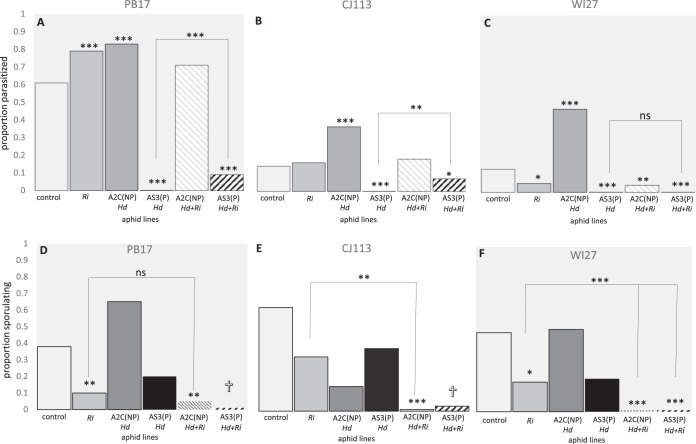
FIG 1.
Proportion of aphids parasitized (A to C) or sporulating with the pathogen Pandora (D to F). Assays compared singly infected or coinfected lines across three aphid genotypes: PB17, CJ113, and WI27. The asterisks indicate significance levels (***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; ns, not significant) relative to controls (uninfected with symbionts) of the same genotype, except those above brackets, which contrast aphids infected with H. defensa AS3 with those coinfected with R. insecticola. Hd, H. defensa; Ri, R. insecticola; Hd+Ri, coinfection; NP, nonprotective H. defensa strain; P, protective H. defensa strain. The crosses indicate high mortality, contributing to lack of significance in these lines.
Consistent with a priori predictions (Table 1), the uninfected line PB17 was susceptible to attack (i.e., more likely to be killed) by the wasp A. ervi, while lines CJ113 and WI27 were similarly and significantly more resistant (i.e., had a higher likelihood of survival) than PB17 (Fig. 1 and Table 2, group A). In the susceptible line (PB17), H. defensa strain AS3 conferred high levels of protection compared to uninfected controls. We also found that infection with the AS3 strain significantly increased resistance to parasitism in both endogenously resistant aphid genotypes CJ113 and WI27 (Table 2, groups B to D), completely eliminating successful parasitism. Surprisingly the phage-free H. defensa strain A2C resulted in significantly less protection than uninfected controls in both susceptible and resistant lines (Table 2, groups B to D). The 5AU strain of R. insecticola exhibited the full range of effects on parasitism, depending on the aphid genotype, significantly increasing successful parasitism in PB17, having no significant effect in CJ113, and reducing successful parasitism in WI27. In all three genotypes, when R. insecticola infected aphids along with the protective AS3 strain of H. defensa, the aphids continued to receive significant protection compared to uninfected controls (Table 2, groups B to D). However, in two of three lines, coinfection with R. insecticola slightly reduced the strength of protection relative to infection with only the AS3 strain of H. defensa (Fig. 1, bracketed comparisons; Table 2, groups B to D, coinfection effects), with successful parasitism rates increasing from 0% to 7% and 9% in genotypes PB17 and CJ113, respectively.
TABLE 1.
Host genotypes and symbiont strains used in this studya
| Aphid or symbiont | Collection state, yr | Level of endogenous resistance to A. ervi or symbiont-based resistance |
|---|---|---|
| Aphids | ||
| WI27 | WI, 2011 | High |
| PB17 | PA, 2011 | Low |
| CJ113 | UT, 2011 | High |
| Symbionts | ||
| H. defensa strain AS3 with phage APSE3 | UT, 2007 | Wasp, high; fungus, unknown |
| H. defensa phage-free strain A2C | UT, 2003 | Wasp, none; fungus, unknown |
| R. insecticola strain 5AU | NY, 2000 | Wasp, none; fungus, high |
Each aphid clone was used to create six subclones: (i) one that was left uninfected, (ii) one infected with AS3, (iii) one infected with A2C, (iv) one infected with 5AU, (v) one infected with AS3 and 5AU, and (vi) one infected with A2C and 5AU.
TABLE 2.
Logistic regression analyses for parasitism and fungal pathogen challenge assays
| Comparison group | Infection | Line | Variable | Contrast | Likelihood ratio test,a χ2; P value | Logistic regression equationb |
|---|---|---|---|---|---|---|
| A | None | Uninfected lines | Mummification | PB17 | ↑ FCJ113 = 59.6, P < 0.0001 | YM = −0.43 + 2.21CJ113 + 2.31WI27 |
| ↑ FWI27 = 59.6, P < 0.0001 | ||||||
| Dual mortality | CJ113 | ↔ FWI27 = 1.46, P = 0.22 | YDM = 0.36 − 0.24WI27 + 0.30PB17 | |||
| ↔ FPB17 = 2.09, P = 0.15 | ||||||
| B | Single and double infections | Susceptible line PB17 | Mummification | PB17 | ↓ F5AU = 11.4, P = 0.0007 | YM = −0.43 − 0.915AU − 1.15A2C + 9.7AS3 + 2.7AS3-5AU − 0.49A2C-5AU |
| ↓ FA2C = 13.1, P = 0.0003 | ||||||
| ↑ FAS3 = 133.1, P < 0.0001 | ||||||
| ↑ FAS3-5AU = 83.2, P < 0.0001 | ||||||
| ↔ FA2C-5AU = 3.2, P < 0.06 | ||||||
| Coinfection effects | AS3-PB17 | ↓ FAS3-5AU = 15.4, P < 0.0001 | ||||
| Dual mortality | PB17 | ↓ FA2C = 66.4, P < 0.0001 | YDM = 0.66 + 0.185AU − 1.54A2C − 0.38AS3 + 0.02AS3-5AU + 0.03A2C-5AU | |||
| ↔ all other lines | ||||||
| C | Single and double infections | Resistant line CJ113 | Mummification | CJ113 | ↔ F5AU = 0.07, P = 0.79 | YM = −1.78 + 0.105AU − 1.20A2C + 7.5AS3 + 0.88AS3-5AU − 0.23A2C-5AU |
| ↓ FA2C = 14.0, P = 0.0002 | ||||||
| ↑ FAS3 = 24.6, P < 0.0001 | ||||||
| ↑ FAS3-5AU = 4.1, P = 0.04 | ||||||
| ↔ FA2C-5AU = 0.5, P = 0.43 | ||||||
| Coinfection effects | AS3-CJ113 | ↓ FAS3-5AU = 10.8, P < 0.001 | ||||
| Dual mortality | ↔ All lines | YDM = 0.36 + 0.215AU − 0.30A2C − 0.08AS3 + 0.10AS3-5AU + 0.14A2C-5AU | ||||
| D | Single and double infections | Resistant line WI27 | Mummification | WI27 | ↑ F5AU = 4.0, P = 0.05 | YM = 1.88 + 1.095AU − 1.73A2C + 7.6AS3 + 7.4AS3-5AU + 1.25A2C-5AU |
| ↓ FA2C = 31.1, P < 0.0001 | ||||||
| ↑ FAS3 = 22.9, P < 0.0001 | ||||||
| ↑ FAS3-5AU = 20.9, P < 0.0001 | ||||||
| ↑ FA2C-5AU = 6.1, P = 0.01 | ||||||
| Coinfection effects | AS3-WI27 | ↔ FAS3-5AU = 0.0, P = 0.99 | ||||
| Dual mortality | WI27 | ↓ F5AU = 5.8, P = 0.02 | YDM = 0.12 − 0.485AU − 0.02A2C + 0.63AS3 + 0.10AS3-5AU + 0.36A2C-5AU | |||
| ↑ FAS3 = 9.5, P = 0.002 | ||||||
| ↔ all other lines | ||||||
| E | None | Uninfected lines | Fungal sporulation | CJ113 | ↔ FWI27 = 0.06, P = 0.79 | YS = −4.9 + 2.5PB17 − 0.14WI27 |
| ↔ FPB17 = 1.74, P = 0.19 | ||||||
| F | Single and double infections | Susceptible line PB17 | Fungal sporulation | PB17 | ↑ F5AU = 7.6, P = 0.005 | Ys = 3.83 − 3.625AU + 0.27A2C − 0.24AS3 − 3.76AS3-5AU − 3.97A2C-5AU |
| ↔ FA2C = 0.0, P = 0.97 | ||||||
| ↔ FAS3 = 0.0, P = 0.98 | ||||||
| ↔ FAS3-5AU = 0.1, P = 0.76 | ||||||
| ↑ FA2C-5AU = 10.2, P = 0.002 | ||||||
| Coinfection effects | 5AU-PB17 | ↔ FA2C-5AU = 0.3, P = 0.56 | ||||
| ↔ FAS3-5AU = 0.0, P = 0.99 | ||||||
| G | Single and double infections | Susceptible line CJ113 | Fungal sporulation | CJ113 | ↔ F5AU = 3.4, P = 0.07 | Ys = −3.05 − 0.785AU + 2.02A2C + 2.57AS3 + 1.15AS3-5AU − 4.44A2C-5AU |
| ↔ FA2C = 0.6, P = 0.42 | ||||||
| ↔ FAS3 = 1.8, P = 0.19 | ||||||
| ↔ FAS3-5AU = 0.1, P = 0.75 | ||||||
| ↑ FA2C-5AU = 16.1, P < 0.0001 | ||||||
| Coinfection effects | 5AU-CJ113 | ↑ FA2C-5AU = 8.4, P = 0.004 | ||||
| ↔ FAS3-5AU = 0.6, P = 0.45 | ||||||
| H | Single and double infections | Susceptible line WI27 | Fungal sporulation | WI27 | ↑ F5AU = 4.6, P = 0.03 | Ys = 4.94 − 0.965AU + 0.37A2C + 2.32AS3 − 4.43AS3-5AU − 0.37A2C-5AU |
| ↔ FA2C = 0.4, P = 0.55 | ||||||
| ↔ FAS3 = 1.3, P = 0.26 | ||||||
| ↑ FAS3-5AU = 19.5, P < 0.0001 | ||||||
| ↑ FA2C-5AU = 19.5, P < 0.0001 | ||||||
| Coinfection effects | 5AU-WI27 | ↑ FA2C-5AU = 7.7, P = 0.006 | ||||
| ↑ FAS3-5AU = 7.7, P = 0.006 | ||||||
↑, increased aphid survival relative to control; ↓, decreased aphid survival relative to control; ↔, not significantly different from control.
Logistic regression equation, Y = β0 + β1X1 + β2X2 (providing the strength and direction of the superscripted clonal lines β1X1 [e.g., + 2.21CJ113] compared to the contrast β0).
(ii) The entomopathogenic fungus P. neoaphidis.
We also found significant variation in fungal sporulation (Fig. 1) (GLzM; χ2 = 93.0; df = 7; P ≤ 0.0001), with R. insecticola infection contributing the largest effect (χ2 = 15.7; P ≤ 0.0001), followed by coinfection status (χ2 = 8.7; P ≤ 0.0001). In this assay, neither time blocks, H. defensa infection, nor R. insecticola-genotype interactions influenced sporulation rates, and subsequent within-genotype analyses lumped time blocks. We found no significant variation in Pandora sporulation among aphid genotypes (Table 2, group E). Aphids singly infected with R. insecticola exhibited a decrease in total sporulation relative to uninfected genotypes (although not significantly for line CJ113 [P = 0.07]) (Fig. 1). Most surprisingly, coinfection by H. defensa with R. insecticola often enhanced protection against Pandora as measured by reductions in sporulation. For instance, both H. defensa strains (A2C and AS3) significantly reduced fungal sporulation in coinfections relative to R. insecticola-only infections in aphid line WI27 (Table 2, group H), whereas only H. defensa A2C did so in aphid line CJ113 (Table 2, group G). In aphid line PB17, H. defensa strain A2C with R. insecticola also reduced sporulation relative to the uninfected control, although not significantly more than R. insecticola alone (Table 2, group F). High mortality in coinfections by H. defensa AS3 with R. insecticola contributed to lack of significance despite low sporulation rates. Single infections with either strain of H. defensa alone did not impact sporulation relative to symbiont-free controls (Table 2, groups F to H).
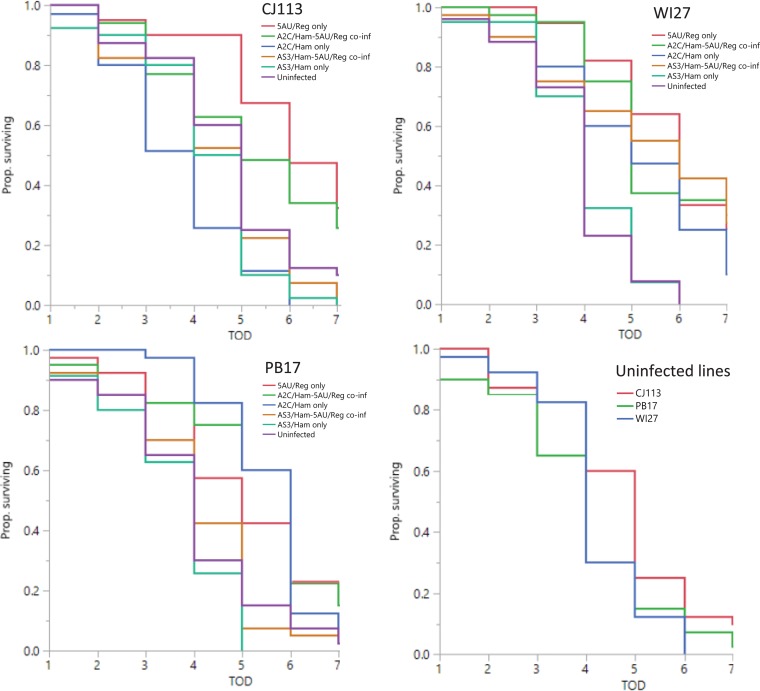
Aphid lines also varied in percent survival to day 7 following enemy challenge (Fig. 2) (Wilcoxon test; df = 17; χ2 = 154.4; P < 0.0001), with R. insecticola-infected aphids showing significantly higher survival probabilities (ca. 27%) than aphids lacking the symbiont (6%) (Wilcoxon test; df = 1; χ2 = 49.0; P < 0.0001). Lines carrying no HFS also varied in probability of survival to day 7, although this was only marginally significant (Wilcoxon test; df = 2; χ2 = 7.2; P = 0.03); uninfected line CJ113 performed best (16% survival), while only 4% of aphids from line PB17 and less than 1% of WI27 aphids survived without R. insecticola. We also found significant variation within each aphid genotype, with consistent patterns throughout. For example, in all three genotypes, infection with only R. insecticola significantly increased the probability of survival to day 7 compared to uninfected control lines sharing the same genotype (Table 3, group A). We also found that coinfection with H. defensa strain A2C did not significantly affect survival to day 7 following enemy challenge with Pandora in any of the three aphid genotypes (Table 3, group B), but coinfection with strain AS3 significantly reduced survival across all the aphid genotypes (Table 3, group C). Interestingly, single infection with H. defensa strain A2C (nonprotective against parasitoids) significantly increased survival in two aphid genotypes (PB17 and WI17) while reducing it in a third (CJ113), while H. defensa strain AS3 did not affect survival relative to uninfected controls in any line (Table 3, group D). For logistical reasons, we did not measure aphid survival in the absence of fungal challenge, which also exposes aphids to modest increases in humidity (85 to 100% versus 100%) over a 24-h period, so it is possible that some symbiont effects on survival are related to these differences in humidity rather than the fungal challenge.
FIG 2.
The top panels and the bottom left panel show aphid survival to day 7 following exposure to the fungal pathogen Pandora for each of the three aphid genotypes (CJ113, WI27, and PB17) infected with zero, one, or two facultative symbionts. The bottom right panel shows aphid survival after Pandora challenge for uninfected aphid lines. Prop., proportion; TOD, time of death.
TABLE 3.
Survival to day 7 following Pandora challenge
| Comparison group | Line | Comparison | Probability (%) of survival to day 7a | Wilcoxon χ2, P value |
|---|---|---|---|---|
| A | CJ113 | R. insecticola | 45 ↑ | 15, 0.0001 |
| Uninfected control | 16 | |||
| PB17 | R. insecticola | 22 ↑ | 7.9, 0.005 | |
| Uninfected control | 4 | |||
| WI27 | R. insecticola | 39 ↑ | 25.0, <0.0001 | |
| Uninfected control | 0 | |||
| B | CJ113 | R. insecticola | 45 ↔ | 3.1, 0.08 |
| A2C-R. insecticola | 30 | |||
| PB17 | R. insecticola | 22 ↔ | 0.9, 0.35 | |
| Coinfection with A2C | 26 | |||
| WI27 | R. insecticola | 39 ↔ | 0.9, 0.34 | |
| A2C-R. insecticola | 35 | |||
| C | CJ113 | R. insecticola | 45 | 22.2, <0.0001 |
| AS3-R. insecticola | 5 ↓ | |||
| PB17 | R. insecticola | 22 | 7.9, 0.005 | |
| Coinfection with AS3 | 3 ↓ | |||
| WI27 | R. insecticola | 39 | 14.6, 0.0001 | |
| AS3-R. insecticola | 5 ↓ | |||
| D | CJ113 | A2C | 0 ↓ | 12.2, 0.002 |
| AS3 | 5 ↔ | |||
| Uninfected control | 16 | |||
| PB117 | H. defensa A2C | 11 ↑ | 37.0, <0.0001 | |
| H. defensa AS3 | 0 ↔ | |||
| Uninfected control | 4 | |||
| WI27 | A2C | 19 ↑ | 10.5, 0.005 | |
| AS3 | 0 ↔ |
↑, increased aphid survival relative to control; ↓, decreased aphid survival relative to control; ↔, not significantly different from control.
Aphid fitness in the absence of natural enemies.
In all three aphid genotypes, single infection with H. defensa strain AS3, which is highly protective against the wasp A. ervi, resulted in large and significant reductions in lifetime reproductive output relative to uninfected controls, indicating clear infection costs in the absence of enemies (Table 4). Most interestingly, coinfection with R. insecticola ameliorated fecundity costs associated with H. defensa strain AS3 to varying degrees in all three aphid genotypes. In lines PB17 and WI27, coinfected cumulative fecundity did not differ significantly from that of uninfected controls, while in CJ113, coinfection resulted in only partial recovery (Table 4). We used Dunnett’s multiple-comparison test to compare cumulative fecundities across all lines within each genotype, finding that only in line PB17 was the coinfected line marginally significantly different ( = 160 versus 100) from the single H. defensa AS3 infection (P = 0.055), although a post hoc t test was significant (F1,19 = 14.0; P = 0.002). Mean fecundity increased in coinfection relative to single AS3 infection for the other two lines (Table 4), but not significantly.
TABLE 4.
Component fitness assays in the absence of enemy challenge
| Aphid line | Symbiont infection status | No. of offspring by day 26 ± SE | Day of 50% survival (95% CI)a |
|---|---|---|---|
| CJ113 | Uninfected | 194 ± 15.5 | 23.2 (21.5–25.3) |
| 5AU-CJ113 | R. insecticola only | 187.8 ± 16.9 (P = 0.997) | 23.6 (21.3–25.1) (P = 0.200) |
| AS3-CJ113 | Protective H. defensa only | 61.1 ± 14.1 (P < 0.0001) | 15.1 (13.6–16.7) (P < 0.0001) |
| A2C-CJ113 | Nonprotective H. defensa only | 168.6 ± 8.7 (P = 0.540) | 21.8 (20.0–23.7) (P = 0.870) |
| AS3-5AU-CJ113 | Protective H. defensa + R. insecticola | 91.8 ± 11.5 (P < 0.0001) | 18.0 (16.2–20.1) (P = 0.030) |
| A2C-5AU-CJ113 | Nonprotective H. defensa + R. insecticola | 137.7 ± 13.6 (P = 0.021) | 22.2 (20.5–24.1) (P = 0.690) |
| PB17 (control) | Uninfected | 214.4 ± 28.1 | 23.6 (21.7–25.6) |
| 5AU-PB17 | R. insecticola only | 183.1 ± 17.6 (P = 0.596) | 21.6 (19.5–3.9) (P = 0.71) |
| AS3-PB17 | Protective H. defensa only | 100.0 ± 10.5 (P = 0.0001) | 18.8 (17.2–20.6) (P = 0.02) |
| A2C-PB17 | Nonprotective H. defensa only | 194.6 ± 16.0 (P = 0.892) | 22.3 (20.4–24.4) (P = 0.500) |
| AS3-5AU-PB17 | Protective H. defensa + R. insecticola | 163.6 ± 13.3 (P = 0.170) | 22.0 (20.3–23.9) (P = 0.867) |
| A2C-5AU-PB17 | Nonprotective H. defensa + R. insecticola | 160.4 ± 14.9 (P = 0.130) | 23.0 (21.4–24.8) (P = 0.482) |
| WI27 (control) | Uninfected | 109.0 ± 10.3 | 21.4 (19.5–23.7) |
| 5AU-WI27 | R. insecticola only | 175.6 ± 15.9 (P = 0.0001) | 24.9 (22.9–26.9) (P = 0.053) |
| AS3-WI27 | Protective H. defensa only | 53.3 ± 9.6 (P = 0.008) | 17.3 (16.2–18.5) (P = 0.002) |
| A2C-WI27 | Nonprotective H. defensa only | 112.4 ± 9.8 (P = 0.999) | 21.4 (19.5–23.4) (P = 0.894) |
| AS3-5AU-WI27 | Protective H. defensa + R. insecticola | 81.7 ± 14.1 (P = 0.368) | 18.6 (16.6–20.9) (P = 0.136) |
| A2C-5AU-WI27 | Nonprotective H. defensa + R. insecticola | 97.0 ± 11.2 (P = 0.930) | 19.7 (17.9–21.6) (P = 0.291) |
CI, confidence interval. Boldface indicates statistical significance.
The A2C strain of H. defensa, which does not protect against wasps, did not significantly reduce cumulative fecundity in any line, and coinfection of A2C with R. insecticola did not significantly affect cumulative fecundity. In slight contrast, the 5AU line of R. insecticola had no effect on cumulative fecundity in genotype PB17 or CJ113, but surprisingly, it increased fecundity in line WI27 (Table 4).
Mortality (measured as the date of 50% survival) in the absence of enemy challenge also varied among lines (Cox proportional hazards; df = 5; χ2 = 30.3; P < 0.0001). Across all lines, genotype, infection with R. insecticola, and coinfection status did not contribute significantly to mortality; only infection with H. defensa exhibited significant effects (effects Wald test; χ2 = 14.2; P < 0.0002). Most notably, infection with the antiparasitoid AS3 strain (and not the nonprotective A2C strain) contributed to significantly increased mortality in all three genotypes. As seen with other assays, coinfection with R. insecticola reduced the costs of infection with H. defensa AS3 in all the genotypes. In genotypes PB17 and WI27, coinfection reduced mortality so that it was not significantly different from that of the control. In line CJ113, fitness was only partially recovered and was still reduced significantly relative to uninfected controls. Pairwise comparisons indicated the coinfected lines (AS3-5AU-CJ113) suffered significantly less mortality than the H. defensa AS3-only line for two lines (effects Wald test; CJ113, χ2 = 15.4, P < 0.0005; PB17, χ2 = 4.7, P = 0.03), but not a third (WI27, χ2 = 1.2, P < 0.27).
DISCUSSION
Experimentation on single-symbiont infections and comparisons to uninfected controls of the same aphid clone have provided a powerful tool for characterizing symbiont-mediated phenotypes in aphids and other insects. However, this approach obscures a more complex reality in which HFS often occur in structured, multispecies communities (12). In this study, we investigated associations between H. defensa and R. insecticola, given their varying tendencies to coinfect in natural pea aphid populations and their well-characterized roles involving distinct defensive benefits under single infection (9, 12, 13, 50). While HFS produce considerable variation in aphid phenotypes, we were nonetheless surprised at the amount of variation observed among a limited set of aphid genotypes and symbiont strains. Across all three aphid genotypes, protection against parasitoids was maintained in H. defensa AS3-plus-R. insecticola coinfections compared to uninfected controls, but in two of three aphid clones, protection levels decreased relative to H. defensa-only infections (Fig. 1 and 2; Tables 2 and 3). Compared to single infections with R. insecticola, protection against the fungal pathogen Pandora was generally maintained when aphids were coinfected with the phage-free, nondefensive strain of H. defensa (A2C), yet coinfection with the antiparasitoid APSE3 H. defensa (line AS3) sharply reduced aphid survival in all aphid genotypes (Table 3). Thus, no strain combinations resulted in “generalist” aphids capable of responding to multiple common threats, as hypothesized. For example, while the 5AU-plus-AS3 combination maintained antiparasitoid defenses, antifungal protection decreased. And while the 5AU-plus-A2C combination maintained protection against Pandora, there was no antiparasitoid function, owing to the lack of APSE in this H. defensa strain. The aphid genotype also impacted the protective phenotypes of coinfections. For example, aphids of clone WI27 received stronger antifungal protection from coinfection than the other two clones (Table 2). Coinfection also affected aphid fitness in the absence of natural enemies. Rather than resulting in additional infection costs in the absence of parasitism relative to single infections, as predicted, coinfection with R. insecticola partially reduced fecundity and longevity costs induced by a protective strain of H. defensa (Table 4).
With respect to symbiont-based resistance against the parasitic wasp A. ervi, we found that the AS3 strain of H. defensa conferred significant protection in both susceptible (PB17) and endogenously resistant (CJ113 and WI27) aphid genotypes. While dual (endogenous plus symbiont) defenses may generate more robust protection (74), the high costs of infection with the AS3 strain (see below) likely outweigh the modest increases in protection. Hence, such dual defenses may not be maintained in natural populations (36). Coinfections by H. defensa with R. insecticola maintained significant antiparasitoid protection relative to uninfected controls in all three aphid genotypes (Fig. 1). In one genotype (WI27) levels of resistance of singly infected and coinfected aphids were identical (0% successful parasitism), but protection levels were significantly reduced (while still protective relative to uninfected controls) in the other two genotypes (Table 2).
Prior studies showed that harboring APSE-free H. defensa strains resulted in the complete loss of antiparasitoid protection, so that aphids became mummified at levels equal to those of uninfected controls sharing the same aphid genotype (62, 63). In the present study, the phage-free strains not only eliminated protection, but unexpectedly rendered each aphid genotype more susceptible to parasitism than uninfected controls (Fig. 1), arguing further as to why we rarely see phage-free Hamiltonella in the field (13). While unexpected in this system, there are reports of other symbiont strains, including Wolbachia, enhancing rather than preventing parasite and pathogen infections (75). The above-mentioned reports examining this phage-free strain used a highly susceptible aphid genotype (ca. 80% successful parasitism), and this may have masked further increases in parasitism success due to limited phenotypic space. Interestingly, coinfection by R. insecticola ameliorated increases in wasp susceptibility associated with infection with the A2C strain in all three genotypes. While phage-free H. defensa should be rapidly removed from aphid populations (76), coinfections with R. insecticola potentially lengthen persistence times and thus the window to acquire new APSEs (62).
Interestingly, increases in successful parasitism associated with the A2C strain of H. defensa were eliminated in two genotypes (PB17 and CJ113) and completely reversed in line WI27 via coinfection with R. insecticola. The latter may be explained by the unexpected result where single infection with R. insecticola conferred antiparasitoid protection in this aphid genotype. While one strain of R. insecticola from M. persicae (green peach aphid) was shown to protect against parasitoids when transferred to pea aphids and black bean aphids (33, 35), antiparasitoid defenses by native Regiella strains from pea aphids have not been reported. If protective effects of particular HFS strains are manifested only in occasional aphid genotypes, this may represent additional cryptic phenotypic diversity in the system, but further assays are needed to confirm there are direct benefits to infection (i.e., increased fecundity after parasitism) with particular strain-genotype combinations in the presence of parasitism.
In terms of symbiont-mediated resistance to the fungal pathogen P. neoaphidis, we found that single infection with R. insecticola resulted in consistent reductions in sporulation and increases in aphid survival across all three genotypes, as expected (Fig. 1 and 2; Tables 2 and 3). In contrast, H. defensa produced variable outcomes. Unexpectedly, single infection with H. defensa strain A2C (but not AS3) also improved post-Pandora challenge survival (but did not affect sporulation) in two of three aphid genotypes, but further work is needed to confirm that some H. defensa strains are truly antifungal protectors. Then, as a coinfection with R. insecticola, neither strain (A2C or AS3) improved aphid survival after Pandora challenge (Table 3), while the effects on sporulation were variable (Table 2).
We also conducted component fitness assays in the absence of natural-enemy challenge to gauge the constitutive costs of single infections versus coinfections relative to uninfected controls. Our results showed that infection with the antiparasitoid H. defensa strain AS3 resulted in substantial costs to survival and offspring production in all three aphid genotypes (Table 4). Coinfection with R. insecticola ameliorated H. defensa (AS3)-associated infection costs to varying degrees, depending on the aphid genotype. The consistently negative effects of the protective H. defensa AS3 in all three are consistent with a prior study that also found the strain to be costly in its native aphid genotype (36). The severe costs associated with the strain may at least partially explain the reduced aphid survival in coinfection (AS3 plus 5AU) compared to R. insecticola-only infection when challenged with Pandora. In other words, infection costs with H. defensa AS3 may override increases in survival owing to R. insecticola infection. In aphid genotype WI27, which reproduced at far lower rates than the other two genotypes used in the experiment, R. insecticola drastically improved host reproduction in the absence of enemy challenge as a single infection, but not when it shared a host with either strain of H. defensa. The overall lack of deleterious effects on component fitness measures resulting from infection with the bacteriophage-free, nonprotective H. defensa strain (A2C) contrasts with previous findings that bacteriophage loss has costs for aphid fitness (76). In two of the three aphid genotypes (CJ113 and PB17), we saw trends toward lower fecundity and higher mortality relative to uninfected controls, but they were not significant. Whole-genome sequencing indicated that the presence/absence of the APSE3 phage is one of the very few differences between H. defensa strains A2C and AS3 (55). One potential explanation is that A2C has been maintained phage free in the laboratory since 2003 (i.e., hundreds of aphid generations), and virulence has attenuated over time. We also did not measure the fecundity of parasitized aphids, which may vary with the coinfection context and hence may be an important determinant of the costs and benefits of coinfections. While parasitized, symbiont-protected aphids generally produce significantly more offspring than parasitized, uninfected controls, this can vary substantially with the aphid genotype and symbiont strain (19, 36).
Aphid HFS, including R. insecticola and H. defensa, are primarily vertically transmitted, but occasional horizontal transmission through food plants or parasitoid ovipositors potentially creates novel coinfections, with rates of symbiont establishment that are likely influenced by aphid and symbiont genotypes (22, 48, 49, 77–79). Once established, our results revealed a complex suite of coinfection outcomes that vary across symbiont strains and aphid genotypes, impacting protective phenotypes and infection costs, and hence likely impact the maintenance of these symbionts in natural populations. Importantly, by pairing protective symbionts, we did not create generalist aphids capable of responding to multiple threats. These findings contrast with those of a previous study (20) using the same two HFS, which found that coinfections resulted in protection against fungal pathogens and parasitoids at levels similar to those of single infections. The symbiont strains and aphid genotypes used varied between the studies, which likely accounts for some of the variability. This variability is consistent with findings that this particular HFS pairing is variably enriched across space and time in field populations (12, 13, 50). One possibility is that particular R. insecticola and H. defensa strains may successfully pair while others are selected against, depending on the aphid genotype and environmental exposure. In addition to spatially varying selective pressures from enemies, geographically divergent strain differences, rather than HFS species identity, may drive coinfection patterns. Hence, studies examining single populations or comparing species level coinfection patterns across populations may miss critical variation (11, 13, 50). More generally, this study contributes to the emerging picture in which particular combinations of HFS may be favored for a variety of reasons, including protection levels, infection costs, and transmission dynamics, that vary between single-infection and coinfection contexts. For example, enhanced protection in the presence of enemies may be overridden by severe costs in their absence (19), or the benefits of two protective symbionts may accrue primarily in the absence of a particular enemy (this study) or during transmission (12).
Our results also question whether defensive phenotypes of aphids can be easily predicted by combinations of HFS infecting single aphid hosts. These findings contribute to a small but emerging body of work on coinfection showing highly variable phenotypes (i.e., coinfections may enhance or reduce defensive services and/or infection costs or leave them unaffected) depending on the specific interacting participants. Furthermore, host level selection may act in concert with nonselective factors, including variation in maternal-transmission rates, that favor particular HFS combinations (12). The presence of enriched or depleted combinations of particular heritable symbionts in natural populations provides ample opportunities for parsing the effects of multiple infection at among-host and within-host levels of selection and for studying the impacts of these complex interactions at the broader community level (80, 81).
MATERIALS AND METHODS
Aphid subclone collection and maintenance.
A. pisum is a cyclical parthenogen across most of its temperate range, and clonally reproducing aphids can be maintained indefinitely in the laboratory using summer-like lighting conditions (21). All the aphids in this study were maintained at 20°C on a 16-h light/8-h dark light cycle at a relative humidity of ca. 70% on fava bean (Vicia faba) plants unless otherwise specified. The three aphid genotypes used here were collected from alfalfa (Medicago sativa) and naturally uninfected by any HFS. Their uninfected status was confirmed using universal primers with denaturing gradient gel electrophoresis (DGGE), as well as diagnostic PCR with specific primers for the known aphid HFS (the DGGE protocol, PCR primers, and reaction conditions are described in reference 13). In addition, microsatellite analyses confirmed they each represented a distinct clonal line (the methods are described in reference 37). Together, the three chosen lines represented a range of endogenous immunity to the parasitoid A. ervi (Table 1): lines CJ113 and WI27 are resistant, while line PB17 is susceptible (71). Endogenous resistance to fungal pathogens has also been reported (72), but host-encoded antifungal phenotypes had not been studied for these aphid lines. We used hemolymph-to-hemolymph transinfection via glass needles to produce experimental lines for use in bioassays (82) (raw data from the bioassays has been deposited in figshare [https://doi.org/10.6084/m9.figshare.11503296]). We also used two H. defensa strains that vary in protective effects: the first, strain AS3, is infected by bacteriophage variant APSE3 and provides nearly total protection against the parasitoid A. ervi, while the second, strain A2C, lacks APSE and confers no protection but is otherwise nearly identical to strain AS3 based on whole-genome sequencing (55, 63). R. insecticola strain 5AU was expected to protect against Pandora, but not A. ervi (29). In all of the coinfected experimental lines, the R. insecticola strain 5AU infection was established first, and then one of the two H. defensa strains was introduced into the 5AU-infected subclone. All experiments occurred a minimum of 10 generations after the establishment of the coinfection. The experimental lines were rescreened using diagnostic PCR (as described above) to confirm the expected infection status prior to the bioassays.
Natural-enemy challenge. (i) The parasitoid wasp A. ervi.
Aphidius ervi (Hymenoptera: Braconidae) is a solitary endoparasitic wasp. The wasps used in this study were derived from a mixed colony of commercially produced (Syngenta Bioline Ltd.) and field-collected A. ervi wasps reared on susceptible aphid lines. Mated female wasps were allowed to make a single oviposition into 2nd- or 3rd-instar aphids, and then the parasitized aphids were placed onto fresh fava bean plants in cohorts of 20 and reared under standard conditions (see above). Nine days after parasitism, all the aphids were scored as living, mummified, or exhibiting dual mortality (i.e., both aphid and wasp died) (63). Ten replicates were conducted for each of the 18 subclones, with the exception of A2C-5AU-PB17, for which there were 11. For logistical reasons, these replicates were conducted over three time blocks. Across-line comparisons were conducted with GzLM models with a binomial distribution and canonical logit link function; the model factors included H. defensa infection, R. insecticola infection, genotype, time block, and coinfection status. Within-genotype comparisons were conducted using logistic regression (proportion mummified/mummified plus surviving). All the statistical tests described here and below were carried out using JMP 14.1.0 (SAS Institute, Inc.).
(ii) The entomopathogenic fungus P. neoaphidis.
We used P. neoaphidis genotype ARSEF 2588 from the U.S. Department of Agriculture Agricultural Research Service (USDA ARS) Collection of Entomopathogenic Fungal Cultures (obtained from N. Gerardo, Emory University) to measure symbiont, coinfection, and host genotype impacts on antipathogen defense. The aphid exposure methods were adopted from prior studies (72–74, 83) and modified as described below. Spore-containing aphid corpses were removed from 4°C storage and placed on fresh 1.5% agar plates and then sealed with parafilm and held in the dark at 20°C for 14 h to initiate sporulation. Each plate with large visible spore showers was then inverted above a 35-mm petri dish containing 20 apterous 10- ± 1-day-old adult aphids from each of the 18 experimental lines. After 15 min of exposure, the fungal plates were rotated within each aphid genotype (WI27, PB17, or CJ113) to randomize the number of spores across within-aphid-genotype subclones: this was repeated until the total exposure time reached 90 minutes per treatment (i.e., every subclone of the same aphid genotype in a single time block was exposed to the same set of sporulating corpses for the same length of time). The exposed aphids were then placed in groups of five on fresh fava bean plants at 100% humidity (accomplished via an unvented cup lid) under otherwise-standard rearing conditions (as described above) for 24 h. The unvented cup was then replaced with a standard vented cup (the humidity ranged from 85 to 100%), and the aphids were checked every 24 hours for 10 days after fungal exposure to assess survival, mortality, and sporulation. Corpses were left in place unless/until they sporulated, at which point they were removed to prevent secondary fungal infections. Aphids not found on daily checks were marked as alive or assigned an age at death on the basis of presence/absence on subsequent checks. Offspring were removed at every checkpoint to minimize crowding, and plants were changed on an as-needed basis. The assay was conducted over two time blocks, producing 8 replicates of each treatment, with the exceptions of AS3-PB17, A2C-PB17, and A2C-5AU-PB17, for which only 7 replicates were produced due to a shortage of apterous adults in the source cultures. Total sporulation proportions over the course of the 10-day treatment were then compared within aphid genotypes using logistic regression. Survival data were analyzed within and among aphid genotypes using Kaplan-Meier plots, with a probability of survival (calculated after Weilbull fit) to 7 days after fungal exposure (α = 0.05). This point was chosen because aphids reproduce at this age, which potentially leads to direct benefits of symbiont infection. A GzLM model was performed as described for wasp parasitism above for the proportion of aphids sporulating by day 10; the factors included were H. defensa infection, R. insecticola infection, genotype, time block, and coinfection status.
Aphid fitness in the absence of natural enemies.
To assess the constitutive costs of infection in single infection versus coinfection versus uninfected controls, we estimated cumulative fecundity and mortality in the absence of natural enemies with protocols adapted from reference 19. To do this, 7-day- ± 12-h-old aphids were placed in cohorts of three on single fava bean plants. Starting at a maternal age of 11 days, all offspring were removed and counted; counts then proceeded every 3 days until day 26, enabling estimates of cumulative fecundity. Mortality among the reproductive adults was also recorded on the basis of the first noted absence. Goodness-of-fit (Shapiro-Wilk) tests were performed to ensure that the reproductive output satisfied assumptions of normality. Given that the uninfected control lines varied substantially in cumulative fecundity (analysis of variance [ANOVA]; F2,29 = 8.3; P = 0.002), we focused on within-genotype analyses, which had the most power to directly address our focal hypotheses. To do this, we performed ANOVA with Dunnett’s tests for the three aphid genotypes, with the uninfected subclone of each genotype acting as the control. Survival in the absence of enemy challenge was analyzed with the Cox semiparametric regression model to fit proportional hazards.
ACKNOWLEDGMENTS
We thank Adam J. Martinez, Clesson Higashi, Laura Kraft, Pooja Patel, Kyungsun Kim, Nicole Lynn-Bell, Alex Hedaya, Matthew Doremus, Khin Khine, and Nhu-y Tan Phan for technical assistance with setting up and carrying out assays.
Funding support was provided by USDA-NIFA award 2015-67011-22789 to S.R.W. and NSF award 1754302 to K.M.O., and J.A.R.
S.R.W., J.A.R., and K.M.O. conceived the project and wrote the manuscript. S.R.W. carried out the experiments.
REFERENCES
- 1.Douglas AE. 2015. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol 60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 4.Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. 2015. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc Biol Sci 282:20150249. doi: 10.1098/rspb.2015.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 6.Duron O, Hurst GD. 2013. Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol 11:45. doi: 10.1186/1741-7007-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haine ER. 2008. Symbiont-mediated protection. Proc Biol Sci 275:353–361. doi: 10.1098/rspb.2007.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver KM, Martinez AJ. 2014. How resident microbes modulate ecologically-important traits of insects. Curr Opin Insect Sci 4:1–7. doi: 10.1016/j.cois.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Oliver KM, Smith AH, Russell JA. 2014. Defensive symbiosis in the real world—advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol 28:341–355. doi: 10.1111/1365-2435.12133. [DOI] [Google Scholar]
- 10.Chiel E, Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Katzir N, Inbar M, Ghanim M. 2007. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull Entomol Res 97:407–413. doi: 10.1017/S0007485307005159. [DOI] [PubMed] [Google Scholar]
- 11.Ferrari J, West JA, Via S, Godfray H. 2012. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66:375–390. doi: 10.1111/j.1558-5646.2011.01436.x. [DOI] [PubMed] [Google Scholar]
- 12.Rock DI, Smith AH, Joffe J, Albertus A, Wong N, O'Connor M, Oliver KM, Russell JA. 2018. Context-dependent vertical transmission shapes strong endosymbiont community structure in the pea aphid, Acyrthosiphon pisum. Mol Ecol 27:2039–2056. doi: 10.1111/mec.14449. [DOI] [PubMed] [Google Scholar]
- 13.Russell JA, Weldon S, Smith AH, Kim KL, Hu Y, Łukasik P, Doll S, Anastopoulos I, Novin M, Oliver KM. 2013. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol Ecol 22:2045–2059. doi: 10.1111/mec.12211. [DOI] [PubMed] [Google Scholar]
- 14.Zchori-Fein E, Lahav T, Freilich S. 2014. Variations in the identity and complexity of endosymbiont combinations in whitefly hosts. Front Microbiol 5:310. doi: 10.3389/fmicb.2014.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto S, Anbutsu H, Fukatsu T. 2006. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl Environ Microbiol 72:4805–4810. doi: 10.1128/AEM.00416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo N, Shimada M, Fukatsu T. 2005. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol Lett 1:488–491. doi: 10.1098/rsbl.2005.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandstrom JP, Russell JA, White JP, Moran NA. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228. doi: 10.1046/j.1365-294x.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 18.Narita S, Nomura M, Kageyama D. 2007. Naturally occurring single and double infection with Wolbachia strains in the butterfly Eurema hecabe: transmission efficiencies and population density dynamics of each Wolbachia strain. FEMS Microbiol Ecol 61:235–245. doi: 10.1111/j.1574-6941.2007.00333.x. [DOI] [PubMed] [Google Scholar]
- 19.Oliver KM, Moran NA, Hunter MS. 2006. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc Biol Sci 273:1273–1280. doi: 10.1098/rspb.2005.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLean AHC, Parker BJ, Hrcek J, Kavanagh JC, Wellham PAD, Godfray H. 2018. Consequences of symbiont co-infections for insect host phenotypes. J Anim Ecol 87:478–488. doi: 10.1111/1365-2656.12705. [DOI] [PubMed] [Google Scholar]
- 21.Brisson JA, Stern DL. 2006. The pea aphid, Acyrthosiphon pisum: an emerging genomic model system for ecological, developmental and evolutionary studies. Bioessays 28:747–755. doi: 10.1002/bies.20436. [DOI] [PubMed] [Google Scholar]
- 22.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 23.Guo JQ, Hatt S, He KL, Chen JL, Francis F, Wang ZY. 2017. Nine facultative endosymbionts in aphids. A review. J Asia-Pacific Entomol 20:794–801. doi: 10.1016/j.aspen.2017.03.025. [DOI] [Google Scholar]
- 24.Henry LM, Maiden MCJ, Ferrari J, Godfray H. 2015. Insect life history and the evolution of bacterial mutualism. Ecol Lett 18:516–525. doi: 10.1111/ele.12425. [DOI] [PubMed] [Google Scholar]
- 25.Zytynska SE, Weisser WW. 2016. The natural occurrence of secondary bacterial symbionts in aphids. Ecol Entomol 41:13–26. doi: 10.1111/een.12281. [DOI] [Google Scholar]
- 26.Leybourne DJ, Bos JIB, Valentine TA, Karley AJ. 2020. The price of protection: a defensive endosymbiont impairs nymph growth in the bird cherry-oat aphid, Rhopalosiphum padi. Insect Sci 27:69–85. doi: 10.1111/1744-7917.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Łukasik P, van Asch M, Guo HF, Ferrari J, Godfray H. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett 16:214–218. doi: 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 28.Montllor CB, Maxmen A, Purcell AH. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 29.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell JA, Moran NA. 2006. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc Biol Sci 273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scarborough CL, Ferrari J, Godfray H. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 32.Schmid M, Sieber R, Zimmermann YS, Vorburger C. 2012. Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct Ecol 26:207–215. doi: 10.1111/j.1365-2435.2011.01904.x. [DOI] [Google Scholar]
- 33.Vorburger C, Gehrer L, Rodriguez P. 2010. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol Lett 6:109–111. doi: 10.1098/rsbl.2009.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis AB, Patel V, Oliver KM, Vorburger C. 2017. Parasitoid gene expression changes after adaptation to symbiont-protected hosts. Evolution 71:2599–2617. doi: 10.1111/evo.13333. [DOI] [PubMed] [Google Scholar]
- 35.Hansen AK, Vorburger C, Moran NA. 2012. Genomic basis of endosymbiont-conferred protection against an insect parasitoid. Genome Res 22:106–114. doi: 10.1101/gr.125351.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez AJ, Doremus MR, Kraft LJ, Kim KL, Oliver KM. 2018. Multi-modal defences in aphids offer redundant protection and increased costs likely impeding a protective mutualism. J Anim Ecol 87:464–477. doi: 10.1111/1365-2656.12675. [DOI] [PubMed] [Google Scholar]
- 37.Martinez AJ, Weldon SR, Oliver KM. 2014. Effects of parasitism on aphid nutritional and protective symbioses. Mol Ecol 23:1594–1607. doi: 10.1111/mec.12550. [DOI] [PubMed] [Google Scholar]
- 38.Oliver KM, Moran NA, Hunter MS. 2005. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci U S A 102:12795–12800. doi: 10.1073/pnas.0506131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandrock C, Gouskov A, Vorburger C. 2010. Ample genetic variation but no evidence for genotype specificity in an all-parthenogenetic host-parasitoid interaction. J Evol Biol 23:578–585. doi: 10.1111/j.1420-9101.2009.01925.x. [DOI] [PubMed] [Google Scholar]
- 40.Doremus MR, Smith AH, Kim KL, Holder AJ, Russell JA, Oliver KM. 2018. Breakdown of a defensive symbiosis, but not endogenous defences, at elevated temperatures. Mol Ecol 27:2138–2151. doi: 10.1111/mec.14399. [DOI] [PubMed] [Google Scholar]
- 41.Guay JF, Boudreault S, Michaud D, Cloutier C. 2009. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J Insect Physiol 55:919–926. doi: 10.1016/j.jinsphys.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Heyworth ER, Ferrari J. 2016. Heat stress affects facultative symbiont-mediated protection from a parasitoid wasp. PlosOne 11:e0167180. doi: 10.1371/journal.pone.0167180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asplen MK, Bano N, Brady CM, Desneux N, Hopper KR, Malouines C, Oliver KM, White JA, Heimpel GE. 2014. Specialisation of bacterial endosymbionts that protect aphids from parasitoids. Ecol Entomol 39:736–739. doi: 10.1111/een.12153. [DOI] [Google Scholar]
- 44.Cayetano L, Vorburger C. 2015. Symbiont-conferred protection against Hymenopteran parasitoids in aphids: how general is it? Ecol Entomol 40:85–93. doi: 10.1111/een.12161. [DOI] [Google Scholar]
- 45.Martinez AJ, Kim KL, Harmon JP, Oliver KM. 2016. Specificity of multi-modal aphid defenses against two rival parasitoids. PlosOne 11:e0154670. doi: 10.1371/journal.pone.0154670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLean AHC, Godfray HCJ. 2015. Evidence for specificity in symbiont-conferred protection against parasitoids. Proc Biol Sci 282. doi: 10.1098/rspb.2015.0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jamin AR, Vorburger C. 2019. Estimating costs of aphid resistance to parasitoids conferred by a protective strain of the bacterial endosymbiont Regiella insecticola. Entomol Exp Appl 167:252–260. doi: 10.1111/eea.12749. [DOI] [Google Scholar]
- 48.Niepoth N, Ellers J, Henry LM. 2018. Symbiont interactions with non-native hosts limit the formation of new symbioses. BMC Evol Biol 18:27. doi: 10.1186/s12862-018-1143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker BJ, McLean AHC, Hrček J, Gerardo NM, Godfray HCJ, Parker BJ, McLean AHC. 2017. Establishment and maintenance of aphid endosymbionts after horizontal transfer is dependent on host genotype. Biol Lett 13:20170016. doi: 10.1098/rsbl.2017.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith AH, Łukasik P, O'Connor MP, Lee A, Mayo G, Drott MT, Doll S, Tuttle R, Disciullo RA, Messina A, Oliver KM, Russell JA. 2015. Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol 24:1135–1149. doi: 10.1111/mec.13095. [DOI] [PubMed] [Google Scholar]
- 51.Jaenike J. 2012. Population genetics of beneficial heritable symbionts. Trends Ecol Evol 27:226–232. doi: 10.1016/j.tree.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Doremus MR, Oliver KM. 2017. Aphid heritable symbiont exploits defensive mutualism. Appl Environ Microbiol 83:e03276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heyworth ER, Ferrari J. 2015. A facultative endosymbiont in aphids can provide diverse ecological benefits. J Evol Biol 28:1753–1760. doi: 10.1111/jeb.12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leclair M, Polin S, Jousseaume T, Simon JC, Sugio A, Morliere S, Fukatsu T, Tsuchida T, Outreman Y. 2017. Consequences of coinfection with protective symbionts on the host phenotype and symbiont titres in the pea aphid system. Insect Sci 24:798–808. doi: 10.1111/1744-7917.12380. [DOI] [PubMed] [Google Scholar]
- 55.Chevignon G, Boyd BM, Brandt JW, Oliver KM, Strand MR. 2018. Culture-facilitated comparative genomics of the facultative symbiont Hamiltonella defensa. Genome Biol Evol 10:786–802. doi: 10.1093/gbe/evy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Degnan PH, Leonardo TE, Cass BN, Hurwitz B, Stern D, Gibbs RA, Richards S, Moran NA. 2010. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol 12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A 106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moran NA, Russell JA, Koga R, Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol 71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliver KM, Higashi C. 2019. Variations on a protective theme: Hamiltonella defensa infections in aphids variably impact parasitoid success. Curr Opin Insect Sci 32:1–7. doi: 10.1016/j.cois.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Angalet GW, Fuester R. 1977. The Aphidius parasites of the pea aphid Acyrthosiphon pisum in the eastern half of the United States. Ann Entomol Soc Am 70:87–96. doi: 10.1093/aesa/70.1.87. [DOI] [Google Scholar]
- 61.Brandt JW, Chevignon G, Oliver KM, Strand MR. 2017. Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proc Biol Sci 284:20171925. doi: 10.1098/rspb.2017.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynn-Bell NL, Strand MR, Oliver KM. 2019. Bacteriophage acquisition restores protective mutualism. Microbiology 65:985–989. doi: 10.1099/mic.0.000816. [DOI] [PubMed] [Google Scholar]
- 63.Oliver KM, Degnan PH, Hunter MS, Moran NA. 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weldon SR, Oliver KM. 2016. Diverse bacteriophage roles in an aphid-bacterial defensive mutualism, p 173–206. In Hurst CJ. (ed), Mechanistic benefits of microbial symbionts. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 65.Parker BJ, Spragg CJ, Altincicek B, Gerardo NM. 2013. Symbiont-mediated protection against fungal pathogens in pea aphids: a role for pathogen specificity? Appl Environ Microbiol 79:2455–2458. doi: 10.1128/AEM.03193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leonardo TE, Mondor EB. 2006. Symbiont modifies host life-history traits that affect gene flow. Proc Biol Sci 273:1079–1084. doi: 10.1098/rspb.2005.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McLean AHC, van Asch M, Ferrari J, Godfray H. 2011. Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc Biol Sci 278:760–766. doi: 10.1098/rspb.2010.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reyes ML, Laughton AM, Parker BJ, Wichmann H, Fan M, Sok D, Hrcek J, Acevedo T, Gerardo NM. 2019. The influence of symbiotic bacteria on reproductive strategies and wing polyphenism in pea aphids responding to stress. J Anim Ecol 88:601–611. doi: 10.1111/1365-2656.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuchida T, Koga R, Fukatsu T. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989–1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- 70.Hrcek J, McLean AHC, Godfray H. 2016. Symbionts modify interactions between insects and natural enemies in the field. J Anim Ecol 85:1605–1612. doi: 10.1111/1365-2656.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez AJ, Ritter SG, Doremus MR, Russell JA, Oliver KM. 2014. Aphid-encoded variability in susceptibility to a parasitoid. BMC Evol Biol 14:127. doi: 10.1186/1471-2148-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parker BJ, Garcia JR, Gerardo NM. 2014. Genetic variation in resistance and fecundity tolerance in a natural host-pathogen interaction. Evolution 68:2421–2429. doi: 10.1111/evo.12418. [DOI] [PubMed] [Google Scholar]
- 73.Łukasik P, Guo H, Asch M, Ferrari J, Godfray H. 2013. Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co‐infection with another symbiont. J Evol Biol 26:2654–2661. doi: 10.1111/jeb.12260. [DOI] [PubMed] [Google Scholar]
- 74.Hrček J, Parker BJ, McLean AHC, Simon J-C, Mann CM, Godfray H. 2018. Hosts do not simply outsource pathogen resistance to protective symbionts. Evolution 72:1488–1499. doi: 10.1111/evo.13512. [DOI] [PubMed] [Google Scholar]
- 75.Graham RI, Grzywacz D, Mushobozi WL, Wilson K. 2012. Wolbachia in a major African crop pest increases susceptibility to viral disease rather than protects. Ecol Lett 15:993–1000. doi: 10.1111/j.1461-0248.2012.01820.x. [DOI] [PubMed] [Google Scholar]
- 76.Weldon SR, Strand MR, Oliver KM. 2013. Phage loss and the breakdown of a defensive symbiosis in aphids. Proc Biol Sci 280:20122103. doi: 10.1098/rspb.2012.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gehrer L, Vorburger C. 2012. Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol Lett 8:613–615. doi: 10.1098/rsbl.2012.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henry LM, Peccoud J, Simon JC, Hadfield JD, Maiden MJC, Ferrari J, Godfray H. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol 23:1713–1717. doi: 10.1016/j.cub.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oliver KM, Campos J, Moran NA, Hunter MS. 2008. Population dynamics of defensive symbionts in aphids. Proc Biol Sci 275:293–299. doi: 10.1098/rspb.2007.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Christian N, Whitaker BK, Clay K. 2015. Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front Microbiol 6:869. doi: 10.3389/fmicb.2015.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McLean A. 2019. Cascading effects of defensive endosymbionts. Curr Opin Insect Sci 32:42–46. doi: 10.1016/j.cois.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Chen DQ, Purcell AH. 1997. Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol 34:220–225. doi: 10.1007/s002849900172. [DOI] [PubMed] [Google Scholar]
- 83.Ferrari J, Muller CB, Kraaijeveld AR, Godfray H. 2001. Clonal variation and covariation in aphid resistance to parasitoids and a pathogen. Evolution 55:1805–1814. doi: 10.1111/j.0014-3820.2001.tb00829.x. [DOI] [PubMed] [Google Scholar]