In recent years, nonthermal plasma (NTP) technology has received a lot of attention as a promising alternative to thermal pasteurization in the food industry. However, little is known about the microbial stress response toward NTP, which could be a potential risk to food safety and impede the development of NTP. A viable but nonculturable (VBNC) state is one of the most common survival strategies employed by microorganisms against external stress. This study investigated the mechanisms of the formation of VBNC Staphylococcus aureus by NTP in a more comprehensive and systematic aspect than had been done before. Our work confirmed that the NTP-generated oxidative stress induced changes in energy allocation as a driving force for the formation of VBNC S. aureus. This study could provide better knowledge for controlling the occurrence of VBNC S. aureus induced by NTP, which could lead to more rational design and ensure the development of safe foods.
KEYWORDS: nonthermal plasma, Staphylococcus aureus, viable but nonculturable, molecular mechanisms, energy allocation
ABSTRACT
As a novel nonthermal technology, nonthermal plasma (NTP) has attracted a lot of attention. However, it could induce microorganisms into a viable but nonculturable (VBNC) state, posing a potential risk to food safety and public health. In this study, the molecular mechanisms of VBNC Staphylococcus aureus induced by NTP were investigated. With the use of a propidium monoazide quantitative PCR (PMA-qPCR) technique combined with a plate count method, we confirmed that 8.1 to 24.3 kJ NTP induced S. aureus into a VBNC state at a level of 7.4 to 7.6 log10 CFU/ml. The transcriptomic analysis was conducted and revealed that most energy-dependent physiological activities (e.g., metabolism) were arrested in VBNC S. aureus, while the oxidative stress response-related genes (katA, dps, msrB, msrA, and trxA) were significantly upregulated. In addition, this study showed that the ATP depletion by carbonyl cyanide m-chlorophenyl hydrazone (CCCP) pretreatment could accelerate the formation of VBNC S. aureus. The NTP-generated oxidative stress triggers the staphylococcal oxidative stress response, which consumes part of cellular energy (e.g., ATP). The energy allocation is therefore changed, and the energy assigned for other energy-dependent physiological activities (cell growth and division, etc.) is reduced, subsequently forcing S. aureus into a VBNC state. Therefore, the alterations of energy allocation should be some of the major contributors to the induction of VBNC S. aureus with NTP exposure. This study provides valuable knowledge for controlling the formation of VBNC S. aureus during NTP treatment.
IMPORTANCE In recent years, nonthermal plasma (NTP) technology has received a lot of attention as a promising alternative to thermal pasteurization in the food industry. However, little is known about the microbial stress response toward NTP, which could be a potential risk to food safety and impede the development of NTP. A viable but nonculturable (VBNC) state is one of the most common survival strategies employed by microorganisms against external stress. This study investigated the mechanisms of the formation of VBNC Staphylococcus aureus by NTP in a more comprehensive and systematic aspect than had been done before. Our work confirmed that the NTP-generated oxidative stress induced changes in energy allocation as a driving force for the formation of VBNC S. aureus. This study could provide better knowledge for controlling the occurrence of VBNC S. aureus induced by NTP, which could lead to more rational design and ensure the development of safe foods.
INTRODUCTION
Consumers’ increasing demand for freshness and nutrition places increasing demand on industry to achieve the minimum processing of food (1). The shorter shelf life of fresh produce has associated costs related to food waste and environmental loads. Consequently, a significant objective for the global food industry involves innovative intervention technologies to ensure the safety and sustainability of food processes and production. Nonthermal plasma (NTP), an emerging decontamination technology, has gained increasing attention for its efficient antimicrobial activities (2–4). NTP treatment has been employed to inactivate foodborne pathogens and extend the shelf life of various foods, such as milk, nuts, vegetables, meat, poultry, and so on (5–8). Plasma, considered the fourth state of matter, is produced from the ionization of gases into various species, including molecules, atoms, ions, electrons, UV photons, and free radicals (e.g., reactive oxygen species and reactive nitrogen species [RONS]) (9, 10). In NTP, ions, electrons, and uncharged particles exist in a thermodynamic nonequilibrium state, which means that the cooling of the ions and uncharged molecules is more effective than that achieved by electrons. Therefore, NTP can work at a relatively low temperature (<60°C). The abundant reactive species in NTP are the major contributors to its efficient inactivation activity on various microorganisms, including bacteria, fungi, spores, and biofilms (11). Many researchers have applied NTP for food decontamination due to its advantages of high microbicidal efficiency, low working temperature, negligible effect on the food matrix, freedom from water, solvent, and residues, and suitability for both solid and liquid foods (12).
A better understanding of the microbial stress response toward a novel nonthermal technology can help optimize the design and improve the application of these emerging technologies (13). It is well known that microorganisms employ survival strategies in response to external stressors (14). A viable but nonculturable (VBNC) state is one of the most common microbial stress responses against external stimuli, including oxidative stress, temperature shifts, nutrient starvation, and high osmolarity (15). A VBNC state describes a special physiological state when the microorganisms fail to grow on the medium while maintaining part of their metabolic activities (16). The greatest challenge created by VBNC microorganisms is that these VBNC cells cannot be detected with the conventional plate count method, but still they could retain their virulence and might be resistant to various stresses. Therefore, the presence of VBNC microorganisms during food processing could result in a big threat to food safety and public health. In past decades, some nonthermal technologies have been found to produce VBNC pathogens during treatments. Zhao et al. (16) reported that approximately 6 log VBNC Escherichia coli O157:H7 was induced by high-pressure CO2. Said et al. (17) found that the pulsed UV light treatment could force Salmonella enterica serotype Typhimurium into a VBNC state. Kramer and Muranyi (18) observed a large proportion of VBNC Listeria innocua and E. coli cells after the pulsed-light treatment. Liao et al. (19) confirmed that ultrasound exposure combined with mild heat (53°C) caused a VBNC state subpopulation of S. Typhimurium.
The available literature has mentioned that NTP can induce microorganisms into a VBNC state (20–22). Cooper et al. (21) observed that Bacillus stratosphericus had increased respiratory activity by 8-fold but was nonculturable by colony assays within 24 h post-plasma treatment. The authors attributed the formation of VBNC states to the effect of reactive oxygen species (ROS) produced by plasma. Ziuzina et al. (22) found that 60 s of plasma exposure could result in an undetectable level of E. coli, while a 300-s treatment was required for the completed loss of metabolic activity from the 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt (XTT) assay. The authors explained that the oxidative stress induced by reactive species from NTP might induce VBNC E. coli. All these studies have shown the induction of VBNC microorganisms by NTP. However, no further investigation of the underlying molecular mechanisms of NTP-induced VBNC cells was conducted in any of the aforementioned works. A clear description of the formation of VBNC microorganisms during the NTP process is essential for the development of safe NTP-treated food.
In this study, we employed the conventional cultured-based method and quantitative PCR (qPCR) combined with a viability dye-propidium iodide monoazide (PMA) for the quantification of the VBNC Staphylococcus aureus organisms induced by NTP treatment. Furthermore, the molecular mechanisms underlying NTP-induced VBNC S. aureus were explored through the RNA-sequencing (RNA-seq) transcriptomics and molecular biological methods.
RESULTS
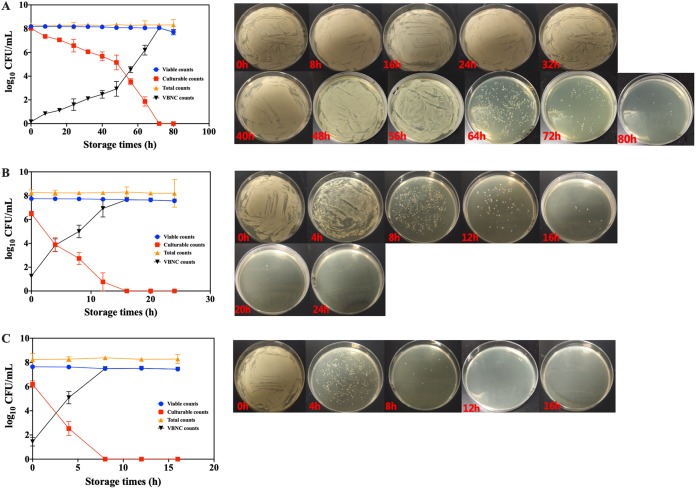
The induction of VBNC S. aureus by NTP with various applied energies.
After 8.1 kJ NTP treatment, the culturable counts of S. aureus decreased as the storage time increased (Fig. 1A). Based on direct observation of the colonies on the medium, S. aureus became smaller and finally invisible at the end of the storage period. When the storage time extended to 80 h, the culturable counts of S. aureus were undetectable on the medium, while the viable counts of S. aureus remained high (7.56 log10 CFU/ml) as determined by the PMA-qPCR method. The results confirmed that NTP potentially induced the formation of VBNC S. aureus. After treatment with NTP with the applied energies of 16.2 and 24.3 kJ, the culturable and viable counts of S. aureus exhibited similar changes during storage (Fig. 1B and C). The times required for S. aureus to completely enter into VBNC states are 24 and 16 h after 16.2 and 24.3 kJ NTP treatment, respectively. The higher the plasma energy applied, the faster the S. aureus population entered into a VBNC state.
FIG 1.
The viable counts, culturable counts, total counts, and VBNC counts (left) and the medium photographs (right) of S. aureus cells under various NTP energies of (A) 8.1 kJ, (B) 16.2 kJ, and (C) 24.3 kJ.
RNA-seq transcriptomics reveals the gene expression in NTP-induced VBNC S. aureus.
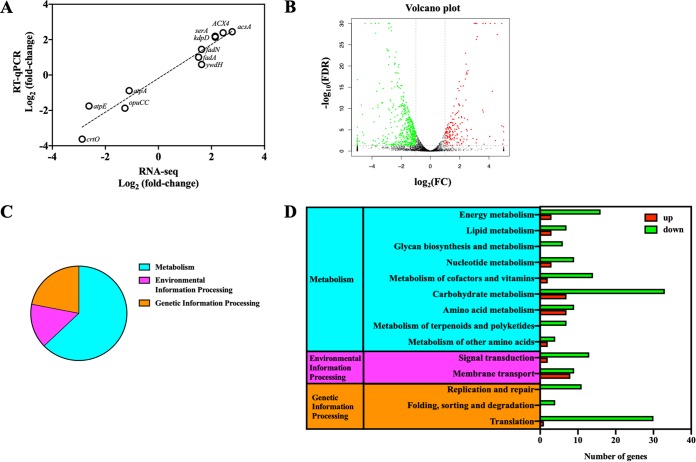
RNA-seq transcriptomics was employed to investigate the physiological changes of NTP-induced VBNC S. aureus. VBNC S. aureus induced by 8.1 kJ NTP treatment with 40-h storage (4°C) was used for the analysis of RNA-seq transcriptomics. The exponential-phase S. aureus without NTP treatment was stored at 4°C for 40 h as the control group (CK). To verify the performance of RNA-seq analysis, a quantitative reverse transcription-PCR (RT-qPCR) assay of 11 randomly selected genes was employed. As Fig. 2A shows, RNA-seq results were consistent with RT-qPCR data with a Pearson correlation coefficient value of 0.969 (P < 0.05). With RNA-seq analysis validated, 616 differential expression genes (DEGs) were found between VBNC and CK S. aureus, among which were 159 upregulated DEGs and 457 downregulated DEGs (Fig. 2B). We further conducted the enriched analysis of these DEGs based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fig. 2C and D) and obtained the following results.
FIG 2.
(A) Validation of RNA sequencing (RNA-seq) data by quantitative reverse transcription-PCR (RT-qPCR). Eleven genes were randomly selected for the validation. (B) The volcano plot showing the upregulated (red points) and downregulated (green points) differential expression genes (DEGs) between CK and NTP-induced VBNC Staphylococcus aureus groups. FDR, false-discovery rate; FC, fold change. (C) The percentages for the DEGs in metabolism, environmental information processing and genetic information processing KEGG pathways of NTP-induced VBNC S. aureus. (D) The number of the upregulated (red bar) and downregulated (green bar) DEGs in metabolism, environmental information processing, and genetic information processing KEGG pathways of NTP-induced VBNC S. aureus.
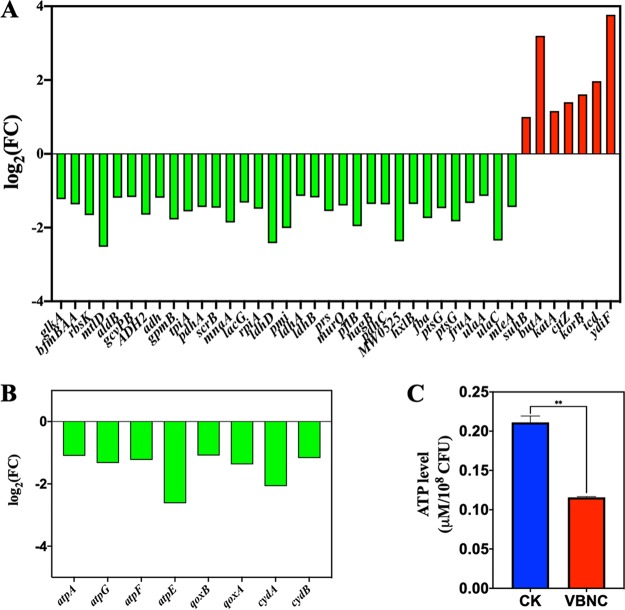
(i) Low metabolic activity. According to the enriched KEGG pathways, the largest proportion of DEGs between VBNC and CK S. aureus was associated with metabolism (Fig. 2C). The downregulated DEGs (79.55%) were dominant over upregulated DEGs (20.45%) in the metabolism pathways, indicating the downregulation in metabolism of NTP-induced VBNC S. aureus. Carbohydrate and energy metabolism associated with energy production was downregulated in VBNC S. aureus (Fig. 2D and 3A and B). Consistently, the ATP level in VBNC S. aureus was decreased approximately 2-fold compared with that in CK cells (Fig. 3C). For lipid metabolism, genes related to fatty acid biosynthesis (accC1, accB, fabG, and fabZ) were significantly (P < 0.05) downregulated in VBNC S. aureus, while fatty acid degradation-β-oxidation genes (fadA, fadN, and ACX4) were upregulated (Fig. 4A). As shown in Fig. 4B, all the DEGs (murA1, ddl, ponA, femX, murA2, and pbpH) related to glycan biosynthesis and metabolism were also downregulated in VBNC S. aureus.
FIG 3.
The expression level for differential expression genes (DEGs) related to (A) carbohydrate metabolism and (B) energy metabolism in NTP-induced VBNC Staphylococcus aureus compared with the control group (CK). Downregulation, green bar; upregulation, red bar. (C) The ATP level in VBNC and CK S. aureus cells. **, P < 0.01.
FIG 4.
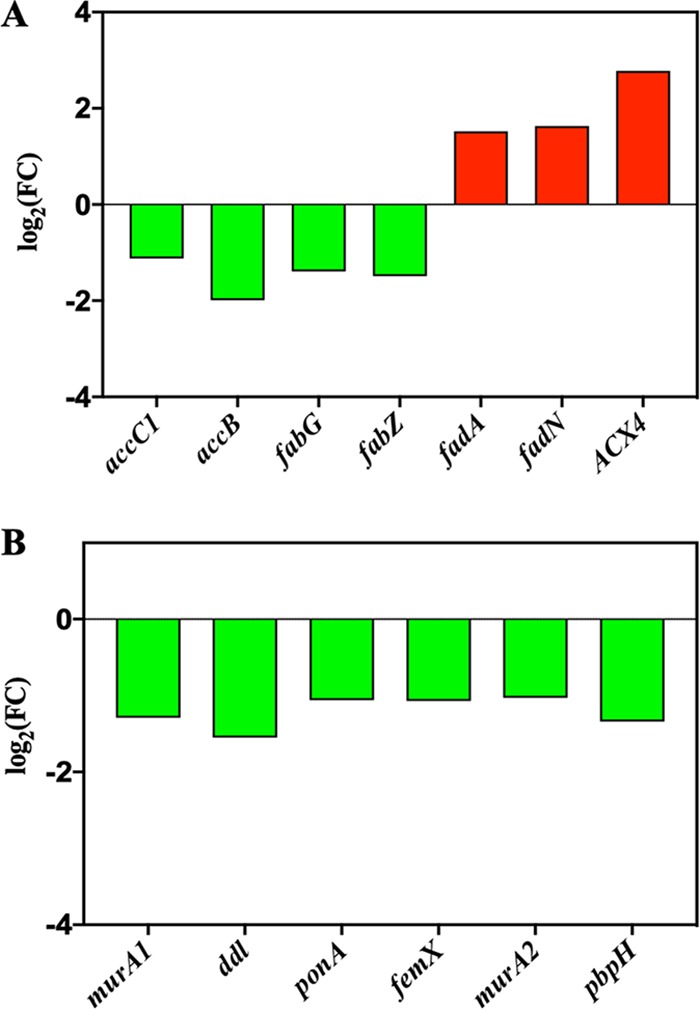
The expression level for differential expression genes (DEGs) related to (A) lipid metabolism and (B) glycan biosynthesis and metabolism in NTP-induced VBNC S. aureus compared with the CK group. Downregulation, green bar; upregulation, red bar.
(ii) Genetic information processing. Several genetic information processing pathways in VBNC S. aureus were downregulated (P < 0.05). Most of the DEGs (e.g., rpsF, rplY, and rplK) associated with ribosomal proteins were repressed (Fig. 5A), which was consistent with the transmission electron microscopy (TEM) observations of an obvious decrease in the electron density of the cytoplasm in S. aureus organisms with intact cell membranes (Fig. 5B). The electron-dense granules are mainly composed of intracellular ribosomes (23). The genes related to the folding, the sorting and degradation, and the replication and repair pathways were also downregulated in VBNC S. aureus (Fig. 2D).
FIG 5.
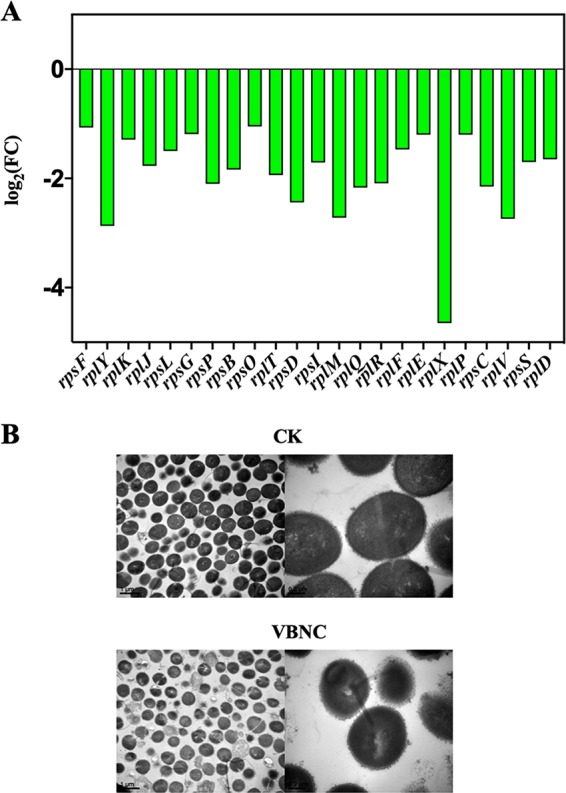
(A) The expression level for differential expression genes (DEGs) related to ribosomal proteins in genetic information processing pathways in NTP-induced VBNC Staphylococcus aureus compared with the CK group. Downregulation, green bar. (B) The transmission electron microscope (TEM) photographs of VBNC and CK S. aureus cells.
(iii) Environmental information processing. This category includes signal transduction and membrane transport (Fig. 2D). As Fig. 6A shows, most of the downregulated genes related to membrane transport are the ATP-dependent transporters, such as sapF for the peptide/nickel transport system ATP-binding protein, yfmC for heme ABC type transporter HtsABC, and so on. In addition, nearly all the genes related to signal transduction were downregulated in VBNC S. aureus, except for kdpD and kdpA (Fig. 6B). The proteins encoded by kdpA and kdpD are members of the KdpD/E two-component system, which has been reported to increase microbial resistance to the oxidative stressors (24, 25).
FIG 6.
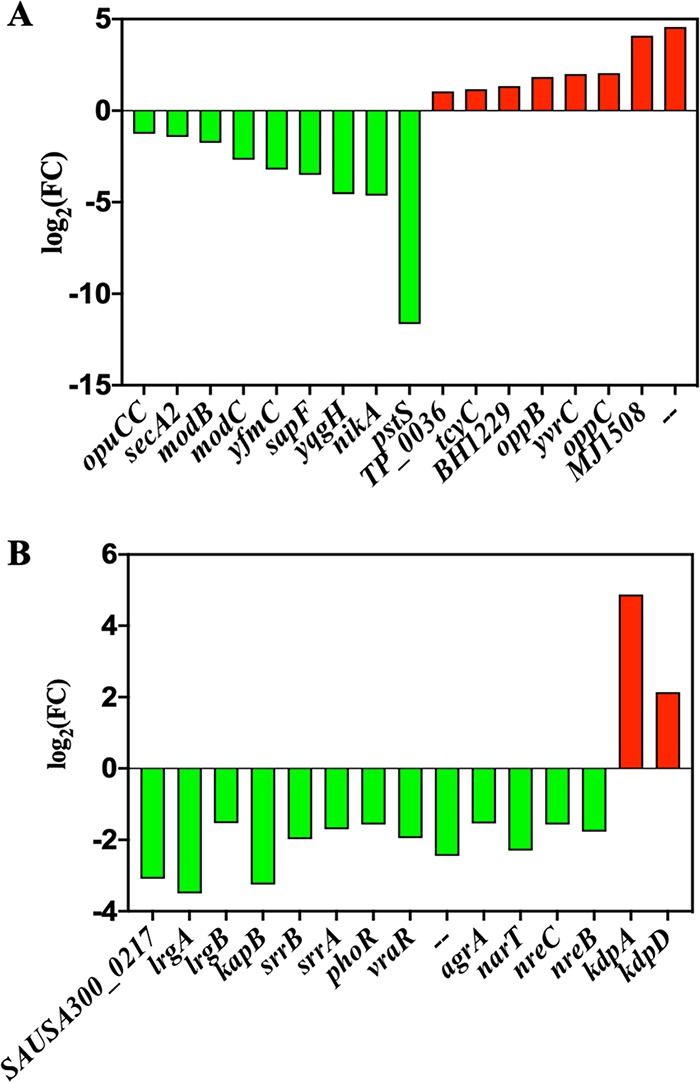
The expression level for differential expression genes (DEGs) related to (A) membrane transport and (B) signal transduction pathways in NTP-induced VBNC S. aureus compared with the CK group. Downregulation, green bar; upregulation, red bar.
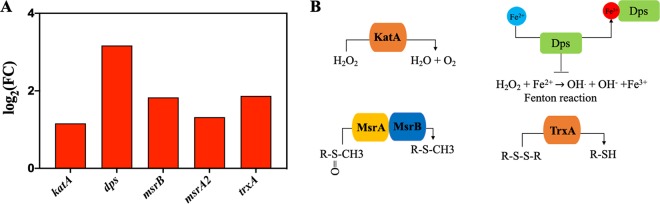
(iv) Oxidative stress response induced in VBNC S. aureus. From the KEGG analysis of RNA-seq, most of the energy-dependent physiological activities were found to be downregulated in VBNC S. aureus cells. However, the links between NTP and the changes in the energy-dependent physiological activities of VBNC S. aureus were still unclear. Further analysis of RNA-seq data revealed that the oxidative stress response cells were induced in VBNC S. aureus. As shown in Fig. 7A, some antioxidant-related genes (katA, dps, msrB, msrA2, and trxA) were significantly upregulated in VBNC cells.
FIG 7.
(A) The expression level for differential expression genes (DEGs) related to oxidative stress response in NTP-induced VBNC Staphylococcus aureus compared with the CK group. Upregulation, red bar. (B) The roles of the related proteins encoded by the DEGs shown in panel A for oxidative stress response.
Higher oxidative stress of NTP is associated with lower ATP levels in S. aureus cells.
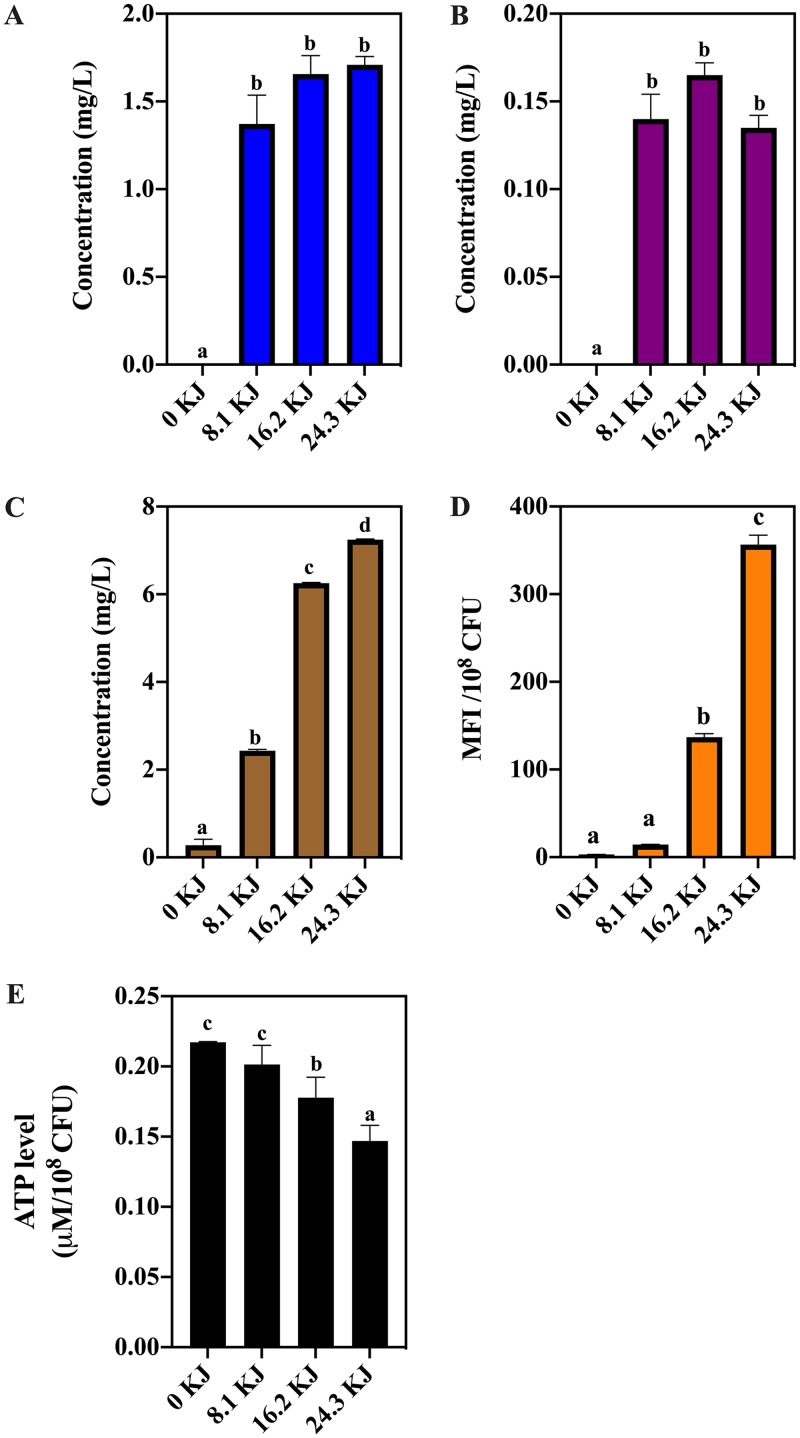
To evaluate the oxidative stress of NTP, we estimated the concentrations of three kinds of stable oxidant compounds produced by the NTP treatment. Significant (P < 0.05) increases of H2O2, ozone, and nitrate were found in the medium after 8.1, 16.2, and 24.3 kJ NTP exposure (Fig. 8A to C). The intracellular ROS was measured by the fluorescent probe dichlorofluorescein (DCF). The mean fluorescent intensity (MFI) of DCF increased as NTP energy increased (P < 0.05; Fig. 8D). To link oxidative stress to energy changes in S. aureus, we measured the intracellular ATP levels under various degrees of oxidative stress. The higher the intracellular and external RONS were, the lower was the ATP level found in S. aureus (Fig. 8A to E).
FIG 8.
The concentrations of hydrogen peroxide (A), ozone (B), and nitrate (C) produced by NTP treatment with 8.1, 16.2, and 24.3 kJ applied energies, respectively. (D) The intracellular ROS level and (E) ATP level in Staphylococcus aureus cells under NTP treatment with 8.1, 16.2, and 24.3 kJ applied energies. Different lowercase letters mean a significant difference at P < 0.05.
The role of ATP depletion in the formation of VBNC S. aureus cells.
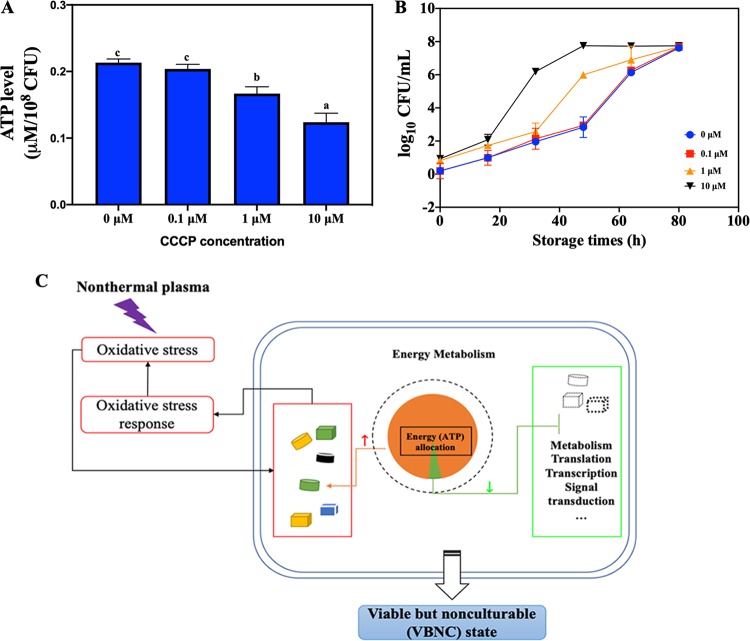
To address the effect of ATP depletion on the formation of VBNC S. aureus cells, we employed various concentrations (0 to 10 μM) of carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to decrease the initial ATP level in S. aureus, which was subsequently treated with NTP. As shown in Fig. 9A and B, when the initial ATP level in S. aureus was decreased by nearly half, the time it took for all the S. aureus cells to move into a VBNC state was decreased from 80 to 48 h. The lower the intracellular ATP level was, the faster the S. aureus cells entered the VBNC state.
FIG 9.
(A) The intracellular ATP level in Staphylococcus aureus cells treated with various carbonyl cyanide m-chlorophenyl hydrazone (CCCP) concentrations (0, 0.1, 1, and 10 μM). Different lowercase letters mean a significant difference at P < 0.05. (B) The VBNC counts for S. aureus pretreated with various CCCP concentrations after NTP exposure at 8.1 kJ. (C) The proposed model for the mechanism underlying the NTP-induced VBNC S. aureus cells.
DISCUSSION
In this study, NTP treatment was found to induce the formation of VBNC S. aureus, which was consistent with some previous studies (20, 26). However, as yet, no data are available to clarify the role of NTP in inducing S. aureus cells into a VBNC state.
Several studies have confirmed that the NTP produced RONS, which caused detrimental oxidative damage to DNA, lipid, and proteins of bacterial cells and was thought to be the major cause of bacterial inactivation (27). Han et al. (28) employed a 2,7-dichlorodihydrofluorescein (DCFH) fluorescence probe and found that the intracellular ROS in S. aureus cells increased significantly (P < 0.05) after the dielectric barrier discharge NTP exposure. From RNA-seq results, our study observed the significant upregulation of oxidative stress response genes in VBNC S. aureus (Fig. 7A). katA is responsible for catalase production, which detoxifies H2O2 into H2O and O2 and is considered the major contributor to staphylococcal resistance toward H2O2 (29) (Fig. 7B). Other antioxidation-related genes (dps, msrB, msrA2, and trxA) are also upregulated in VBNC S. aureus. dps encodes a nonspecific DNA-binding protein, which protects DNA against Fenton-mediated oxidative stress. The genes msrB and msrA2 are responsible for producing methionine sulfoxide reductase, which shields bacteria from oxidative and nitrosative stress (30–33). Thioredoxins are encoded by trxA, which acts as a key player in protein repair under oxidative stress through reducing disulfides (29). Similar results were found in the study of Mai-Prochnow et al. (34), who demonstrated that a 3-min plasma treatment provided oxidative stress, contributing to the production of oxidative stress regulons (OxyR, SoxR, and OspR) in Pseudomonas aeruginosa cells to develop resistance to the following plasma treatment. Therefore, the oxidative effect of NTP could invoke the antioxidative response in bacteria.
In this study, we also noticed that the production of RONS by NTP is inversely proportional to the intracellular ATP level of S. aureus (Fig. 8). ATP is the energy pool for protein production and other energy-consuming metabolic activities. The production of abundant antioxidant proteins in S. aureus consumed cellular energy and caused a drop in ATP, which required cells to generate energy. However, the starvation conditions (0.85% NaCl) limited nutrient acquisition for energy generation. In this study, RNA-seq data revealed a downregulation of energy generation pathways (carbohydrate and energy metabolism) in VBNC S. aureus. Glycolysis and the pentose phosphate pathway (PPP) in carbohydrate metabolism generate energy through carbohydrate degradation, and oxidative phosphorylation in energy metabolism produces ATP through a proton motive force (35). A reduction in carbohydrate and energy metabolism of VBNC microorganisms under starvation conditions was also reported in previous studies (36). Notably, upregulation was observed in the fatty acid degradation pathway, which mainly involved β-oxidation of fatty acids, which has been recognized as the major energy supplier for survival under long-term nutrient starvation (37). It indicated that VBNC S. aureus switched from carbohydrate to fatty acid metabolism to generate energy (ATP) for survival. On the other hand, energy-consuming activities, such as fatty acid and glycan biosynthesis, protein production, and membrane transport, were significantly repressed in VBNC S. aureus for energy saving (Fig. 4 and 6). Fatty acids and glycan (peptidoglycan) are used for constructing membrane and walls during cell growth and division (38, 39). The arrest in fatty acid and glycan biosynthesis led to the impediment of cell growth and division, which could cause failure of colony forming on the solid medium. In addition, RNA-seq data revealed the downregulation in ribosome proteins. Ribosomes are responsible for mRNA translation and protein production for normal physiological activities (40). The results agree with some previous studies, which also indicated low ribosome content in VBNC bacteria (41, 42). The ribosome inactivation decreased transcription and translation activities, subsequently lowering protein production to save cellular energy (43). Therefore, the cellular energy changed when S. aureus moved from the culturable to the VBNC state.
The reduction of ATP level in VBNC microorganisms has also been reported in previous studies (44–46). Ganesan et al. (44) observed that starvation-induced VBNC Lactococcus lactis exhibited downregulation of genes related to ATP synthase as well as a dramatic drop in intracellular ATP concentration. The authors hypothesized that the energy maintenance was required for bacteria to maintain culturability on solid agar. In the study of Meng et al. (45), transcriptome was employed to reveal that the ATP synthase subunits were significantly downregulated in VBNC Vibrio parahaemolyticus induced by alkaline peptone water (2% NaCl). Pu et al. (46) further confirmed that ATP depletion led to the formation of protein aggresomes, which retarded the regular metabolic activity of E. coli and induced the formation of VBNC states. Some studies, however, reported the contrary result that ATP production was enhanced in VBNC bacteria (16, 36). For example, Lai et al. (36) applied two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) protein analysis and found that ATP synthase F1 was upregulated in VBNC V. parahaemolyticus caused by the modified Morita mineral salt solution. The different results from various publications might be attributed to different VBNC induction conditions. In our case, ATP depletion seems to be important in the formation of VBNC S. aureus by NTP. In order to verify this hypothesis, we further employed an ionophore-CCCP to artificially decrease the initial ATP level in S. aureus. The results showed that ATP depletion could accelerate the formation of VBNC S. aureus by NTP.
Taking these results in aggregate, we proposed a model for the NTP-induced formation of VBNC S. aureus cells (Fig. 9C). The external stress, in this case, the oxidative stress from NTP, induced S. aureus to use a lot of energy for the oxidative stress response, which disrupts the balance of energy allocation in S. aureus. The limited energy contributes to growth and metabolism quiescence in S. aureus, which subsequently leads to the formation of a VBNC state. The energy allocation model could be employed to explain the different speeds at which S. aureus moves into a VBNC state under various applied NTP energies. Higher NTP energy produced higher levels of oxidant species and posed more serious oxidative stress on S. aureus cells, which can be indicated by the higher intracellular ROS level as the applied NTP energy increased (Fig. 8D). The higher oxidative stress requires S. aureus cells to allocate more energy to produce proteins related to the oxidative stress response, which decreases larger amounts of ATP (Fig. 8E). The energy provided for the rest of the energy-dependent physiological activities will be lesser when the applied NTP energy is higher, and it will therefore take a shorter time for S. aureus to change into a VBNC state.
In conclusion, our study clarified the role of NTP in the formation of VBNC S. aureus. The treatment of NTP provides the oxidative stress to trigger the S. aureus energy-dependent oxidative stress response and to change the intracellular energy allocation balance, which reduces the energy for bacterial metabolism and results in the final formation of a VBNC state.
MATERIALS AND METHODS
Bacterial strain preparation.
Staphylococcus aureus strain S15, isolated from fresh beef, was used in this study. The whole genome of strain S15 was sequenced using single-molecule real-time (SMRT) technology. SMRT Link v5.0.1 was used for filtering the low-quality reads, and the final filtered reads were assembled to generate one contig without gaps. The strain was cultured in tryptic soy broth (TSB; Base Biotech Co., Hangzhou, China), which was shaken at 180 rpm at 37°C for 12 h to reach the mid-exponential phase. The initial concentration of S. aureus was approximately 8 log10 CFU/ml.
Nonthermal plasma (NTP) treatment.
A dielectric barrier discharge (DBD) atmospheric nonthermal plasma system (Nanjing Suman Electronics Co., Ltd., Nanjing, China) consisting of a power supply (CTP-2000K) and a DBD reactor (DBD-50) was used in this study. A cell suspension (80 ml) was placed in a petri dish with a diameter of 150 mm. The maximum frequency of the power supply was 10 kHz, and the maximum applied power was 500 W. In this case, the applied power was set at 44.95 W, and the gas distance between the upper electrode and the bacterial surface was 8 mm. The treatment times were 0, 3, 6, and 9 min, corresponding to the applied energy of 0, 8.1, 16.2, and 24.3 kJ, respectively. After NTP treatment, S. aureus was stored at 4°C for the following measurements. The H2O2 and ozone production after NTP treatment were measured with a hydrogen peroxide assay kit (Beyotime, Shanghai, China) and a Spectroquant ozone test kit (Merck, Germany) (5), respectively. The nitrate level in the medium was measured by reduction to nitrite with cadmium and subsequently reacted with Griess reagent according to the method of Follett and Ratcliff (47). DCFH-DA (2,7-dichlorodihydrofluorescein diacetate) was applied to determine the intracellular ROS level (48). S. aureus suspension was mixed with DCFH-DA (10 μM) and incubated at 37°C for 20 min before NTP treatment. After NTP exposure, S. aureus was obtained by centrifugation at 2,320 × g for 10 min and washed three times with sterile saline solution to remove the excess DCFH-DA. The mean fluorescence intensity (MFI) was determined using a fluorescent microplate reader (49).
Enumeration of culturable S. aureus cells by culture-based method.
The bacterial samples were serially diluted with sterile saline solution (0.85% NaCl). The appropriate dilution was plated on tryptic soy agar (TSA; Qingdao Hope Bio-Technology Co., Ltd., Qingdao, Shandong, China), followed by incubation at 37°C for 24 h.
Enumeration of total counts and viable and culturable S. aureus cells.
Propidium monoazide (PMAxx) (20 mM in distilled water [dH2O]; Biotium, Hayward, CA) was used for selecting viable S. aureus according to the manufacturer’s instructions. An aliquot (400 μl) of S. aureus cells was mixed with PMAxx (5 mM), with incubation in a rocker in the dark for 10 min at 25 ± 2°C. The mixture was exposed to light in a PMA-Lite LED photolysis device (Biotium, Inc., California, USA) to cross-link PMAxx to DNA for 15 min. With a centrifugation of 2,320 × g at 4°C for 10 min, the collected cells were used for DNA extraction with a TIANamp bacterial DNA kit (Tiangen Biotech Co., Ltd., Beijing, China). The qPCR measurement was carried out on a Thermo QuantStudio 3 real-time PCR system using a Fast EvaGreen qPCR master mix (Biotium, Inc., California, USA). Each qPCR mixture (20 μl) contained 10 μl 2× master mix, 1 μl (each) nuc forward and reverse primers (10 μM; Table 1), 1 μl template DNA, 0.25 μl carboxy-X-rhodamine (ROX) solution (50 nM), and 6.75 μl sterile DNase-free water. A qPCR procedure included one cycle at 95°C for 2 min, 40 cycles at 95°C for 5 s, at the annealing temperature of 50.2°C for 15 s, and at 72°C for 60 s, followed by a melting curve step of 95°C for 15 s, 60°C for 1 min, and 95°C for 1 s to verify specificity. To determine the amplification efficiency (E), 10-fold dilutions of S. aureus were prepared to obtain concentrations of 2- to 8-log. PMA-treated and -untreated S. aureus was used to establish a PMA-qPCR and qPCR standard curve through linear fitting analysis with Origin 8.0 software. The fitted equations for PMA-qPCR and qPCR are CT (threshold cycle) = –3.367 × log10 CFU/ml + 43.455, with an E value of 98.2% and R2 of 0.999, and CT = –3.342 × log10 CFU/ml + 44.375, with an E value of 99.1% and R2 of 0.991, respectively.
TABLE 1.
Primers of nuc gene for qPCR and PMA-qPCR
| Gene | Sequence (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|
| nuc | GCGATTGATGGTGATACGGTT | 279 | 54 |
| AGCCAAGCCTTGACGAACTAAAGC |
The total viable counts and total counts of S. aureus were determined using the CT value according to the PMA-qPCR and qPCR standard curves, respectively. Culturable counts were measured using the plate count method, and VBNC counts were determined using the differences between total viable counts and culturable counts.
Measurement of intracellular ATP.
Intracellular ATP levels were quantified using a BacTiter-Glo microbial cell viability assay (Promega Biotech Co., Ltd., Beijing, China) (50). Briefly, the samples and standard ATP solution were mixed with BacTiter-Glo reagent (1:1). The luminescence was measured with a microplate reader (BioTek Instruments, Inc., Vermont, USA). ATP concentrations were finally normalized to log10 CFU/ml (46).
Analysis of interior structure by TEM.
TEM was used to observe the interior structure of S. aureus. S. aureus was fixed in 2.5% (vol/vol) glutaraldehyde solution at 4°C overnight. After fixation, the cells were washed in phosphate-buffered saline (PBS) (0.1 M, pH 7.0) and were further fixed in 1% osmic acid for 1 h. The samples were dehydrated in a series of ethanol solutions of 30 to 90% and then rinsed twice in 100% ethanol at 20-min intervals. The cells were infiltrated with a solution of acetone and Epon-Araldite for 24 h and polymerized at 60°C for 48 h. Next, the specimens were sectioned, and then the sections ~50 nm thick were placed on copper sieves and contrasted with uranyl acetate and lead citrate for 30 min. The sections were observed using a JEM-1230 TEM (JEOL Japan Electronics Co., Ltd., Japan).
Illumina high-throughput transcriptome sequencing (RNA-seq) and data analysis.
Triplicate biological samples of the CK and NTP-induced VBNC S. aureus were used for the extraction of the total RNA with TRIzol. The quality of the extracted RNA was confirmed using 1% agarose gels, NanoPhotometer (Implen, California, USA), and the Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). The cDNA library was constructed with the TruSeq RNA sample prep kit v2 (Illumina, San Diego, CA, USA). Purified fragmented cDNA was further processed with end reparation (30 min at 30°C) and purification with AMPure XP beads (Beckman Coulter, California, USA). Addition of the poly(A) tail was done with an A-tailing mix (30 min at 37°C) before ligating sequencing adapters (10 min at 30°C). The second-strand cDNA was degraded using the uracil-N-glycosylase enzyme at 37°C for 10 min, and the product was purified with AMPure XP beads. Several rounds of PCR amplification with PCR primer cocktail were performed to enrich the cDNA fragments, and the PCR products were purified with AMPure XP Beads. The clustering of the index-coded samples was performed on a cBot cluster generation system according to the manufacturer’s instructions. After cluster generation, the cDNA libraries were constructed and sequenced using the Illumina HiSeq TM 2500 platform with 150-bp paired-end reads. Raw data were filtered to remove nonsense sequences. The reads with trimming were mapped to the reference genome (CP040801) using Bowtie 2 v2.2.8 (51), and gene expression was calculated using RNA-Seq with expectation-maximization (RSEM) to identify known genes (52). To evaluate reproducibility between samples, the correlation coefficient among replicas was calculated. Values closer to 1 indicated better reproducibility. Principal-component analysis (PCA) was performed with the R package g models (http://www.r-project.org) to reveal the relationship between samples. The gene expression level of the control S. aureus and the NTP-induced VBNC S. aureus was further normalized by using the fragments per kilobase of the transcripts per million mapped reads method (53) to eliminate the influence of different gene lengths and the amount of sequencing data on the calculation of gene expression. DEGs across these two samples were determined using the threshold of log2 fold change >1 and a false discovery rate-adjusted P (q value) of <0.05. DEGs were then subjected to an enrichment analysis of KEGG pathways, and q values were corrected using <0.05 as the threshold.
Validation by quantitative reverse transcription-PCR (RT-qPCR).
To verify RNA-seq data, 11 DEGs were randomly selected for RT-qPCR. RNA was extracted using an RNA prep pure cell/bacterium kit (Tiangen Biotech Co., Ltd., China). The concentration, purity, and integrity of extracted RNA were confirmed using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Inc., Delaware, USA) and agarose gel electrophoresis. Total RNA was then reverse transcribed to cDNA with a Quantscript RT kit (Tiangen Biotech Co., Ltd., Beijing, China). Each qPCR mixture (20 μl) included 10 μl of 2× SuperReal PreMix Plus, 0.6 μl of forward and reverse primers (10 μM), 1 μl of 50× ROX reference dye, 1 μl of cDNA sample, and 6.8 μl RNase-free double-distilled water (ddH2O). The qPCR run consisted of 15 min at 95°C, 40 cycles at 95°C for 10 s and at 60°C for 32 s, followed by a melting curve. The housekeeping gene 16S rRNA was used as the reference gene. All the primers for the selected DEGs were designed with the Primer-BLAST tool (NCBI) and are exhibited in Table 2.
TABLE 2.
Primers for the random selected genes for the verification of RNA-seq by qPCR
| Gene | Sequence (5′–3′) | Product size (bp) |
|---|---|---|
| fadA | ACCATCAGAAAGTGGGGCAG | 234 |
| CATCAACGTGCCTATGACGC | ||
| ACX4 | ACAAAGCAATGGGGTTTGCC | 147 |
| TGCGGGAGGTCTTGAAACAG | ||
| serA | AATCAATGTGGCACGTGGTG | 121 |
| AGGCGAATCAGTTGCAGGTT | ||
| acsA | TTCTCAGTGCAACAAACGCC | 134 |
| AGCTGGTGAACGAGTTGGAC | ||
| ywdH | TGACACCAAATGTTGCACGAG | 228 |
| TGACTGGAGATTTACCGCCC | ||
| kdpD | TGAACAAGTCACCCTATCGTCC | 193 |
| ATGTAGGACCGCGTCCAAATG | ||
| atpG | TTCTGCTTGTCTCGCTCTGT | 118 |
| AGACGCAAAAGCAAGTGAGC | ||
| atpA | TGCAGATCCACCAACACGAG | 198 |
| AACGATGACTTAGGTGGCGG | ||
| atpE | TCTTGCTGTTGACGTGCTTG | 192 |
| CGCATGGGGTCCATTGAAAG | ||
| fadN | AGTACAAGCTCACAGCCACC | 152 |
| AAAAAGGCGCATGAAGACGG | ||
| crtO | CGGTCGATTATAACGTTGCACAAT | 152 |
| TAGAGACAAAGAGGGCAGAGTT |
ATP depletion.
When the S. aureus culture grew into the mid-exponential phase, cells were centrifuged and resuspended in TSB supplemented with 0.1 to 10 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP) in dimethyl sulfoxide (DMSO), followed by incubation at 37°C for 4 h (43).
Statistical analysis.
All experiments were carried out in triplicate. Statistical analysis was performed using a one-way analysis of variance with a Tukey’s test and Student’s t test using SPSS statistical software v20.0 (SPSS, Inc., Illinois, USA). Statistical significance was considered at a P value of <0.05.
Data availability.
The whole-genome information of strain S15 has been submitted to GenBank (accession number CP040801).
ACKNOWLEDGMENTS
We deeply appreciate Christopher A. Elkins’ professional comments on this work.
This study was supported by the National Natural Science Foundation of China (grant 31772079).
REFERENCES
- 1.Augustin MA, Riley M, Stockmann R, Bennett L, Kahl A, Lockett T, Osmond M, Sanguansri P, Stonehouse W, Zajac I, Cobiac L. 2016. Role of food processing in food and nutrition security. Trends Food Sci Tech 56:115–125. doi: 10.1016/j.tifs.2016.08.005. [DOI] [Google Scholar]
- 2.Liao XY, Liu DH, Xiang QS, Ahn J, Chen SG, Ye XQ, Ding T. 2017. Inactivation mechanisms of non-thermal plasma on microbes: a review. Food Control 75:83–91. doi: 10.1016/j.foodcont.2016.12.021. [DOI] [Google Scholar]
- 3.Bourke P, Ziuzina D, Boehm D, Cullen PJ, Keener K. 2018. The potential of cold plasma for safe and sustainable food production. Trends Biotechnol 36:615–626. doi: 10.1016/j.tibtech.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Muhammad AI, Liao X, Cullen PJ, Liu D, Xiang Q, Wang J, Chen S, Ye X, Ding T. 2018. Effects of nonthermal plasma technology on functional food components. Compr Rev Food Sci Food Saf 17:1379–1394. doi: 10.1111/1541-4337.12379. [DOI] [PubMed] [Google Scholar]
- 5.Liao X, Li J, Muhammad AI, Suo Y, Chen S, Ye X, Liu D, Ding T. 2018. Application of a dielectric barrier discharge atmospheric cold plasma (Dbd-Acp) for Escherichia coli inactivation in apple juice. J Food Sci 83:401–408. doi: 10.1111/1750-3841.14045. [DOI] [PubMed] [Google Scholar]
- 6.Korachi M, Ozen F, Aslan N, Vannini L, Guerzoni ME, Gottardi D, Ekinci FY. 2015. Biochemical changes to milk following treatment by a novel, cold atmospheric plasma system. Int Dairy J 42:64–69. doi: 10.1016/j.idairyj.2014.10.006. [DOI] [Google Scholar]
- 7.Niemira BA. 2012. Cold plasma reduction of Salmonella and Escherichia coli O157:H7 on almonds using ambient pressure gases. J Food Sci 77:M171–M175. doi: 10.1111/j.1750-3841.2011.02594.x. [DOI] [PubMed] [Google Scholar]
- 8.Misra N, Patil S, Moiseev T, Bourke P, Mosnier J, Keener K, Cullen P. 2014. In-package atmospheric pressure cold plasma treatment of strawberries. J Food Eng 125:131–138. doi: 10.1016/j.jfoodeng.2013.10.023. [DOI] [Google Scholar]
- 9.Scholtz V, Pazlarova J, Souskova H, Khun J, Julak J. 2015. Nonthermal plasma: a tool for decontamination and disinfection. Biotechnol Adv 33:1108–1119. doi: 10.1016/j.biotechadv.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Liao X, Muhammad AI, Chen S, Hu Y, Ye X, Liu D, Ding T. 2019. Bacterial spore inactivation induced by cold plasma. Crit Rev Food Sci Nutr 59:2562–2572. doi: 10.1080/10408398.2018.1460797. [DOI] [PubMed] [Google Scholar]
- 11.Gilmore BF, Flynn PB, O’Brien S, Hickok N, Freeman T, Bourke P. 2018. Cold plasmas for biofilm control: opportunities and challenges. Trends Biotechnol 36:627–638. doi: 10.1016/j.tibtech.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Misra N, Schlüter O, Cullen PJ. 2016. Cold plasma in food and agriculture: fundamentals and applications. Academic Press, San Diego, CA. [Google Scholar]
- 13.Manas P, Pagan R. 2005. Microbial inactivation by new technologies of food preservation. J Appl Microbiol 98:1387–1399. doi: 10.1111/j.1365-2672.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu D, Forghani F, Daliri EBM, Li J, Liao X, Liu D, Ye X, Chen S, Ding T. 2019. Microbial response to some nonthermal physical technologies. Trends Food Sci Tech 95:107–117. [Google Scholar]
- 15.Zhao X, Zhong J, Wei C, Lin CW, Ding T. 2017. Current perspectives on viable but non-culturable state in foodborne pathogens. Front Microbiol 8:580. doi: 10.3389/fmicb.2017.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao F, Bi X, Hao Y, Liao X. 2013. Induction of viable but nonculturable Escherichia coli O157:H7 by high pressure CO2 and its characteristics. PLoS One 8:e62388. doi: 10.1371/journal.pone.0062388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Said MB, Otaki M, Hassen A. 2012. Use of lytic phage to control Salmonella Typhi’s viability after irradiation by pulsed UV light. Ann Microbiol 62:107–111. doi: 10.1007/s13213-011-0234-5. [DOI] [Google Scholar]
- 18.Kramer B, Muranyi P. 2014. Effect of pulsed light on structural and physiological properties of Listeria innocua and Escherichia coli. J Appl Microbiol 116:596–611. doi: 10.1111/jam.12394. [DOI] [PubMed] [Google Scholar]
- 19.Liao H, Jiang L, Zhang R. 2018. Induction of a viable but non-culturable state in Salmonella Typhimurium by thermosonication and factors affecting resuscitation. FEMS Microbiol Lett 365:fnx249. doi: 10.1093/femsle/fnx249. [DOI] [PubMed] [Google Scholar]
- 20.Joaquin JC, Kwan C, Abramzon N, Vandervoort K, Brelles-Mariño G. 2009. Is gas-discharge plasma a new solution to the old problem of biofilm inactivation? Microbiology 155:724–732. doi: 10.1099/mic.0.021501-0. [DOI] [PubMed] [Google Scholar]
- 21.Cooper M, Fridman G, Fridman A, Joshi SG. 2010. Biological responses of Bacillus stratosphericus to floating electrode-dielectric barrier discharge plasma treatment. J Appl Microbiol 109:2039–2048. doi: 10.1111/j.1365-2672.2010.04834.x. [DOI] [PubMed] [Google Scholar]
- 22.Ziuzina D, Boehm D, Patil S, Cullen PJ, Bourke P. 2015. Cold plasma inactivation of bacterial biofilms and reduction of quorum sensing regulated virulence factors. PLoS One 10:e0138209. doi: 10.1371/journal.pone.0138209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Um H, Kong H, Lee H, Choi H, Park E, Kim S, Murugiyan S, Chung E, Kang K, Lee S. 2013. Altered gene expression and intracellular changes of the viable but nonculturable state in Ralstonia solanacearum by copper treatment. Plant Pathol J 29:374–385. doi: 10.5423/PPJ.OA.07.2013.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gründling A. 2013. Potassium uptake systems in Staphylococcus aureus: new stories about ancient systems. mBio 4:e00784-13. doi: 10.1128/mBio.00784-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alegado RA, Chin CY, Monack DM, Tan MW. 2011. The two-component sensor kinase KdpD is required for Salmonella Typhimurium colonization of Caenorhabditis elegans and survival in macrophages. Cell Microbiol 13:1618–1637. doi: 10.1111/j.1462-5822.2011.01645.x. [DOI] [PubMed] [Google Scholar]
- 26.Kvam E, Davis B, Mondello F, Garner AL. 2012. Nonthermal atmospheric plasma rapidly disinfects multidrug-resistant microbes by inducing cell surface damage. Antimicrob Agents Chemother 56:2028–2036. doi: 10.1128/AAC.05642-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi SG, Cooper M, Yost A, Paff M, Ercan UK, Fridman G, Friedman G, Fridman A, Brooks AD. 2011. Nonthermal dielectric-barrier discharge plasma-induced inactivation involves oxidative DNA damage and membrane lipid peroxidation in Escherichia coli. Antimicrob Agents Chemother 55:1053–1062. doi: 10.1128/AAC.01002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han L, Patil S, Boehm D, Milosavljevic V, Cullen PJ, Bourke P. 2016. Mechanisms of inactivation by high-voltage atmospheric cold plasma differ for Escherichia coli and Staphylococcus aureus. Appl Environ Microbiol 82:450–458. doi: 10.1128/AEM.02660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaupp R, Ledala N, Somerville GA. 2012. Staphylococcal response to oxidative stress. Front Cell Infect Microbiol 2:33. doi: 10.3389/fcimb.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh VK, Moskovitz J. 2003. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 149:2739–2747. doi: 10.1099/mic.0.26442-0. [DOI] [PubMed] [Google Scholar]
- 31.Drazic A, Winter J. 2014. The physiological role of reversible methionine oxidation. Biochim Biophys Acta 1844:1367–1382. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Lee WL, Gold B, Darby C, Brot N, Jiang X, De Carvalho LPS, Wellner D, St John G, Jacobs WR Jr, Nathan C. 2009. Mycobacterium tuberculosis expresses methionine sulphoxide reductases A and B that protect from killing by nitrite and hypochlorite. Mol Microbiol 71:583–593. doi: 10.1111/j.1365-2958.2008.06548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasindran SJ, Saikolappan S, Dhandayuthapani S. 2007. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol 2:619–630. doi: 10.2217/17460913.2.6.619. [DOI] [PubMed] [Google Scholar]
- 34.Mai-Prochnow A, Bradbury M, Ostrikov K, Murphy AB. 2015. Pseudomonas aeruginosa biofilm response and resistance to cold atmospheric pressure plasma is linked to the redox-active molecule phenazine. PLoS One 10:e0130373. doi: 10.1371/journal.pone.0130373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tretter L, Adam-Vizi V. 2005. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos Trans R Soc Lond B Biol Sci 360:2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai C, Chen S, Lin I, Chang C, Wong H. 2009. Change of protein profiles in the induction of the viable but nonculturable state of Vibrio parahaemolyticus. Int J Food Microbiol 135:118–124. doi: 10.1016/j.ijfoodmicro.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 37.Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A. 2003. The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 38.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund VA, Wacnik K, Turner RD, Cotterell BE, Walther CG, Fenn SJ, Grein F, Wollman AJ, Leake MC, Olivier N, Cadby A, Mesnage S, Jones S, Foster SJ, Lund VA, Wacnik K, Turner RD, Cotterell BE, Walther CG, Fenn SJ, Grein F, Wollman AJ, Leake MC, Olivier N, Cadby A, Mesnage S, Jones S, Foster SJ. 2018. Molecular coordination of Staphylococcus aureus cell division. Elife 7:e32057. doi: 10.7554/eLife.32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittmann H. 1982. Components of bacterial ribosomes. Annu Rev Biochem 51:155–183. doi: 10.1146/annurev.bi.51.070182.001103. [DOI] [PubMed] [Google Scholar]
- 41.Kim JS, Yamasaki R, Song S, Zhang WW, Wood TK. 2018. Single cell observations show persister cells wake based on ribosome content. Environ Microbiol 20:2085–2098. doi: 10.1111/1462-2920.14093. [DOI] [PubMed] [Google Scholar]
- 42.Linder K, Oliver JD. 1989. Membrane fatty acid and virulence changes in the viable but nonculturable state of Vibrio vulnificus. Appl Environ Microbiol 55:2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganesan B, Stuart MR, Weimer BC. 2007. Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl Environ Microbiol 73:2498–2512. doi: 10.1128/AEM.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng L, Alter T, Aho T, Huehn S. 2015. Gene expression profiles of Vibrio parahaemolyticus in viable but non-culturable state. FEMS Microbiol Ecol 91:fiv035. doi: 10.1093/femsec/fiv035. [DOI] [PubMed] [Google Scholar]
- 46.Pu Y, Li Y, Jin X, Tian T, Ma Q, Zhao Z, Lin S-Y, Chen Z, Li B, Yao G, Leake MC, Lo C-J, Bai F. 2019. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell 73:143–156.e4. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Follett MJ, Ratcliff PW. 1963. Determination of nitrite and nitrate in meat products. J Sci Food Agric 14:138–144. doi: 10.1002/jsfa.2740140302. [DOI] [Google Scholar]
- 48.Liao X, Xuan X, Li J, Suo Y, Liu D, Ye X, Chen S, Ding T. 2017. Bactericidal action of slightly acidic electrolyzed water against Escherichia coli and Staphylococcus aureus via multiple cell targets. Food Control 79:380–385. doi: 10.1016/j.foodcont.2017.03.050. [DOI] [Google Scholar]
- 49.Li J, Ma L, Liao X, Liu D, Lu X, Chen S, Ye X, Ding T. 2018. Ultrasound-induced Escherichia coli O157:H7 cell death exhibits physical disruption and biochemical apoptosis. Front Microbiol 9:2486. doi: 10.3389/fmicb.2018.02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 54.Fang H, Hedin G. 2003. Rapid screening and identification of methicillin-resistant Staphylococcus aureus from clinical samples by selective-broth and real-time PCR assay. J Clin Microbiol 41:2894–2899. doi: 10.1128/jcm.41.7.2894-2899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome information of strain S15 has been submitted to GenBank (accession number CP040801).