S. aureus is a leading agent of infections worldwide, and its main virulence characteristic is the ability to produce biofilms on surfaces such as medical devices. Biofilms are known to confer increased resistance to antimicrobials and to the host immune responses, requiring aggressive antibiotic treatment and removal of the infected surface. Here, we investigated a new source of antibiofilm compounds, the skin microbiome. Specifically, we found that a commensal strain of S. epidermidis produces molecules with antibiofilm activity, leading to a significant decrease of S. aureus biofilm formation and to a reduction of previously established biofilms. The molecules potentiated the activity of antibiotics and affected the expression of hundreds of S. aureus genes, including those associated with biofilm formation. Our research highlights the search for compounds that can aid us in the fight against S. aureus infections.
KEYWORDS: skin microbiota, antivirulence, Staphylococcus epidermidis, Staphylococcus aureus, biofilm
ABSTRACT
The microbiota influences host health through several mechanisms, including protecting it from pathogen colonization. Staphylococcus epidermidis is one of the most frequently found species in the skin microbiota, and its presence can limit the development of pathogens such as Staphylococcus aureus. S. aureus causes diverse types of infections ranging from skin abscesses to bloodstream infections. Given the increasing prevalence of S. aureus drug-resistant strains, it is imperative to search for new strategies for treatment and prevention. Thus, we investigated the activity of molecules produced by a commensal S. epidermidis isolate against S. aureus biofilms. We showed that molecules present in S. epidermidis cell-free conditioned media (CFCM) caused a significant reduction in biofilm formation in most S. aureus clinical isolates, including all 4 agr types and agr-defective strains, without any impact on growth. S. epidermidis molecules also disrupted established S. aureus biofilms and reduced the antibiotic concentration required to eliminate them. Preliminary characterization of the active compound showed that its activity is resistant to heat, protease inhibitors, trypsin, proteinase K, and sodium periodate treatments, suggesting that it is not proteinaceous. RNA sequencing revealed that S. epidermidis-secreted molecules modulate the expression of hundreds of S. aureus genes, some of which are associated with biofilm production. Biofilm formation is one of the main virulence factors of S. aureus and has been associated with chronic infections and antimicrobial resistance. Therefore, molecules that can counteract this virulence factor may be promising alternatives as novel therapeutic agents to control S. aureus infections.
IMPORTANCE S. aureus is a leading agent of infections worldwide, and its main virulence characteristic is the ability to produce biofilms on surfaces such as medical devices. Biofilms are known to confer increased resistance to antimicrobials and to the host immune responses, requiring aggressive antibiotic treatment and removal of the infected surface. Here, we investigated a new source of antibiofilm compounds, the skin microbiome. Specifically, we found that a commensal strain of S. epidermidis produces molecules with antibiofilm activity, leading to a significant decrease of S. aureus biofilm formation and to a reduction of previously established biofilms. The molecules potentiated the activity of antibiotics and affected the expression of hundreds of S. aureus genes, including those associated with biofilm formation. Our research highlights the search for compounds that can aid us in the fight against S. aureus infections.
INTRODUCTION
The human microbiota is considered a crucial factor that influences the state of health and disease of the host. In recent years, studies have correlated the microbiota composition with several human diseases, and its role in protection against infections has been addressed (1). Most of these studies have focused on the gut microbiota, and relatively little is known about the complexity of the skin microbiota and the interactions among its components. The skin is the most extensive and the most exposed organ of the human body and is considered a first line of defense against environmental conditions, harmful chemicals, and exogenous pathogens (2). It is currently known that the composition of the skin microbiota can vary according to the skin site and host characteristics, such as age, gender, and lifestyle (3). In general, Staphylococcus, Propionibacterium, Micrococcus, and Corynebacterium are the most abundant bacterial genera on the superficial layers of the human skin (4). Staphylococcus epidermidis is one of the most frequently isolated species from human epithelial samples, being considered ubiquitous in the human skin (5). Studies have shown that this species has an arsenal of self-defense mechanisms aiming to limit the establishment and growth of possible skin pathogens by the production of bacteriocins and proteases (5, 6). Moreover, it has been described that molecules produced by S. epidermidis, known as phenol-soluble modulins, have antimicrobial activity against pathogens such as Streptococcus pyogenes and Staphylococcus aureus (7, 8). Furthermore, during the treatment of atopic dermatitis patients, which includes a regimen of dilute bleach baths, topical steroids, bland emollients, and, in some cases, systemic antibiotics, a significant decrease in the numbers of S. aureus, the major pathogen related to the aggravation of skin lesions, with a concomitant increase of S. epidermidis colonization, has been observed (9). In addition, the introduction of S. epidermidis strains producing the protease Esp into the nasal cavities of S. aureus asymptomatic carriers resulted in clearance of S. aureus colonization (6). Therefore, a possible antagonistic relationship has been suggested to exist between these species, supporting the hypothesis that pathogens are kept under control by skin microbiota members.
S. aureus is associated with high morbidity and mortality rates, causing ailments that range from skin and soft tissue infections to invasive diseases such as pneumonia, bacteremia, and meningitis (10). Resistance to several antibiotics is a common trait of S. aureus clinical strains, and methicillin-resistant S. aureus (MRSA) isolates are a huge concern for public health due to the limited treatment options available (11). One of the virulence factors that contributes to the success of S. aureus infections is the ability to form biofilms on biotic (bones and heart valves) and abiotic (catheters, prostheses, and other medical devices) surfaces (12). The process of biofilm formation induces many phenotypic alterations, including reduced growth rate and higher tolerance to antibiotics and host immune responses (13, 14). Some antibiotics act efficiently on planktonic cells but not on cells grown in biofilms, leading to recurrent infections (15). Thus, biofilm formation is associated with chronic and recurrent infections caused by S. aureus and is currently considered a worldwide challenge for the treatment of infections caused by this pathogen (15).
Due to the ability of S. aureus to produce biofilms and the high antimicrobial resistance rates found in this pathogen, therapeutic alternatives are limited. Therefore, the search for new compounds to combat S. aureus infections is urgent. Recent studies showed that the human microbiota is a source of antivirulence molecules, indicating a potential novel source of substances against multidrug-resistant pathogens (16–18). The major advantage of employing an antivirulence approach is to impose lower selective pressure when compared to conventional antibiotics, delaying the development of resistant variants. In addition, antivirulence molecules will likely only cause a minor impact on host commensals, acting efficiently and specifically on the targeted virulence factors (19). Considering that the microbiota competes with exogenous pathogens for host colonization, we hypothesized that molecules produced by members of the skin microbiota would impact the virulence of skin pathogens. Here, we show that molecules produced by the commensal S. epidermidis have a drastic impact on S. aureus global gene expression and on its ability to form biofilms.
RESULTS
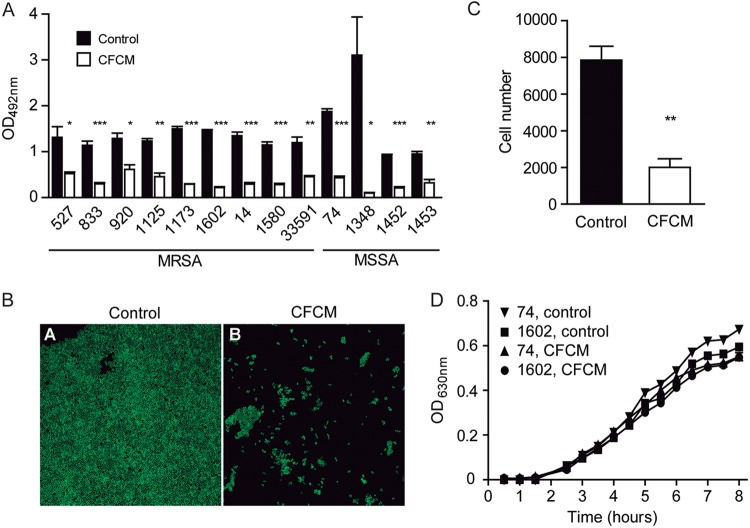
Cell-free conditioned media (CFCM) from different S. epidermidis isolates grown for 24 h in Trypticase soy broth (TSB) media were collected and concentrated, and their activity on S. aureus biofilm formation was tested. CFCM from all S. epidermidis strains tested showed significantly inhibited S. aureus biofilm formation, with CFCM from one commensal isolate (RF1) showing the strongest reduction (Fig. S1A). This isolate was chosen for further analysis. The activity of RF1 CFCM was tested against biofilm production using 29 previously characterized S. aureus clinical isolates, both methicillin-susceptible S. aureus (MSSA) and MRSA, belonging to the 4 agr types, and isolated from diverse types of infections (Table 1). When S. aureus was grown in the presence of CFCM, biofilm production was reduced over 30% in 20 (69%) strains, compared to biofilm formation with the addition of control CFCM. Biofilm formation without the addition of control CFCM or CFCM was also used for comparison, and the vast majority of strains tested were not affected by control CFCM. Nevertheless, CFCM significantly reduced biofilm formation for all strains tested, regardless of whether control CFCM or no-addition controls were used for comparison (Fig. S1B). A more pronounced reduction (over 50%) in biofilm formation was observed for 13 (65%) of these strains, which were selected for further analyses of CFCM activity (Fig. 1A). Curiously, among the strains showing an increased susceptibility to the effect of CFCM on biofilm formation (over 50%), 69.2% were MRSA. In contrast, 66.6% of the strains whose biofilm formation was not impacted (less than 30% reduction) by CFCM were MSSA (Table 1). We also analyzed the composition of biofilms formed by all 13 isolates that showed greater reduction (over 50%) with CFCM, as well as 3 isolates with reduction between 30 and 50% and 6 isolates with less than 30% reduction or no reduction. Among the 13 strains with greater biofilm reduction, 9 had biofilms composed mainly of protein, and 1 strain had a polysaccharide-based biofilm. Three other strains had mixed biofilms, with similar biofilm reduction caused by proteinase K or sodium metaperiodate. These data show that RF1 CFCM has activity against biofilms with different matrix compositions (Table 1). Among the isolates that showed intermediate reduction (30 to 50%) and little or no reduction (<30%), the composition of the biofilm matrix was also heterogeneous (5 mixed and 4 proteinaceous). We then selected the strain that showed the greatest reduction in biofilm formation by CFCM (strain 1602) and performed confocal microscopy analyses, the results of which confirmed the initial observation of the impact of CFCM on biofilm production and cell number (Fig. 1B and C). The impact of CFCM on S. aureus planktonic growth was verified, and no significant effects were observed (Fig. 1D).
TABLE 1.
Characteristics of S. aureus clinical strains used in this studya
| Strain | Isolation site | Methicillin susceptibility | Agr type | Biofilm production | Matrix type | Reduction of biofilm formation by CFCM (%)b |
|---|---|---|---|---|---|---|
| 1348 | Blood | MSSA | 3 | Strong | Mix | 96.8 |
| 1452 | Blood | MSSA | 1 | Strong | Ptn | 86.3 |
| 74 | Skin lesion | MSSA | 3 | Moderate | Ptn | 77.0 |
| 1453 | Blood | MSSA | 4 | Strong | Ptn | 66.0 |
| 1602 | Blood | MRSA | 3 | Moderate | Mix | 85.0 |
| 1173 | Blood | MRSA | 3 | Moderate | Ptn | 80.6 |
| 14 | Skin lesion | MRSA | 2 | Strong | Ptn | 78.1 |
| 1580 | Blood | MRSA | 3 | Moderate | Mix | 75.4 |
| 833 | Urine | MRSA | 3 | Strong | Ptn | 73.9 |
| 1125 | Prosthesis | MRSA | 3 | Strong | Pol | 63.2 |
| ATCC 33591 | — | MRSA | 1 | Strong | Ptn | 62.1 |
| 527 | BAL | MRSA | 3 | Strong | Ptn | 60.3 |
| 920 | BAL | MRSA | 3 | Strong | Ptn | 52.2 |
| 1035 | Skin lesion | MSSA | 3 | Strong | ND | 48.2 |
| 26 | Skin lesion | MSSA | 2 | Moderate | ND | 45.9 |
| 297 | Skin lesion | MSSA | 2 | Moderate | ND | 45.3 |
| 244 | Skin lesion | MSSA | 2 | Moderate | Mix | 40.4 |
| 1445 | Blood | MSSA | 1 | Moderate | Ptn | 32.7 |
| 326 | Skin lesion | MSSA | 1 | Moderate | Mix | 37.4 |
| 1166 | Blood | MSSA | 1 | Moderate | ND | 35.5 |
| 41 | Skin lesion | MSSA | 1 | Moderate | ND | 28.0 |
| 100 | Skin lesion | MSSA | 1 | Strong | Mix | 20.0 |
| 10 | Skin lesion | MSSA | 1 | Moderate | Ptn | 18.0 |
| 57 | Skin lesion | MSSA | 1 | Moderate | Mix | −7.5 |
| 112 | Skin lesion | MSSA | 2 | Moderate | Ptn | −4.0 |
| 1349 | Blood | MSSA | 3 | Strong | Mix | −7.9 |
| 1380 | Blood | MRSA | 3 | Moderate | Ptn | 23.0 |
| 1581 | Blood | MRSA | 3 | Moderate | ND | 14.0 |
| 255 | Skin lesion | MRSA | 2 | Moderate | ND | 9.6 |
MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; BAL, bronchoalveolar lavage; Mix, mixed matrix composition (similar reduction with proteinase K and sodium metaperiodate); Ptn, mostly protein-based matrix composition (major reduction with proteinase K); Pol, mostly polysaccharide matrix composition (major reduction with sodium periodate); bold, strains with over 50% reduction in biofilm formation; ND, not determined; —, information not provided in source references.
Reduction compared to control CFCM.
FIG 1.
Cell-free conditioned media (CFCM) from commensal S. epidermidis represses biofilm production of S. aureus clinical isolates. (A) Biofilm formation of selected S. aureus clinical isolates grown in the presence or absence of S. epidermidis CFCM. (B) Confocal microscopy of biofilms produced by S. aureus clinical isolate (MRSA isolate 1602) grown in the presence or absence of S. epidermidis CFCM. (C) Cell count on S. aureus (MRSA isolate 1602) biofilm cells grown in the presence or absence of S. epidermidis CFCM. (D) Growth curves of selected S. aureus clinical isolates with or without S. epidermidis CFCM. OD, optical density; upside-down triangles, MSSA isolate 74 control; squares, MRSA isolate 1602 control; triangles, MSSA isolate 74 with CFCM; circles, MRSA isolate 1602 with CFCM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
We then investigated if the effect of CFCM during the formation of biofilms could also be observed on established S. aureus biofilm. The molecules secreted by S. epidermidis showed activity on established S. aureus biofilms, a desirable feature for the treatment of biofilm-associated infections (Fig. 2A). The effect was variable, depending on the S. aureus strain tested, with biofilm reduction ranging from 7.2% to 58.8%. Most tested strains (12/13; 92.3%) had a reduction greater than 18% on the biofilm after addition of S. epidermidis CFCM (Fig. 2A).
FIG 2.
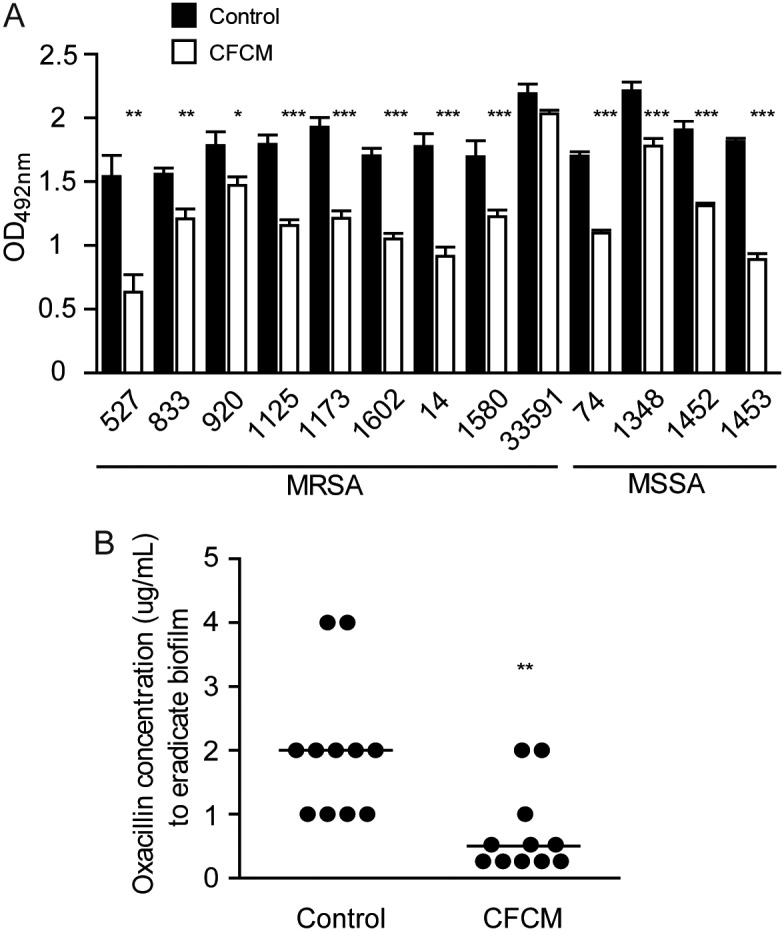
Activity of S. epidermidis cell-free conditioned media (CFCM) on preestablished S. aureus biofilm. (A) Optical density of biofilm matrix staining after treatment with either the control medium or S. epidermidis CFCM of 24-h preestablished biofilms. (B) Oxacillin concentration used to obtain nonviable preestablished biofilms (MSSA isolate 74) cells with or without S. epidermidis CFCM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Next, we tested the ability of CFCM to reduce the minimal inhibitory concentration (MIC) of oxacillin required to eradicate previously established S. aureus biofilms. The addition of CFCM alone did not affect the viability of biofilm cells. However, when CFCM was used in combination with oxacillin, there was a reduction of the concentration of the antibiotic necessary to eliminate viable biofilm cells. When oxacillin alone was used to treat biofilms, an average concentration of 2 μg/ml of the antibiotic was required to disrupt biofilms. On the other hand, when CFCM and oxacillin were used in combination, an average concentration of 0.25 μg/ml of the antibiotic was enough to achieve the same level of biofilm disruption (Fig. 2B), suggesting an additive effect between CFCM and oxacillin.
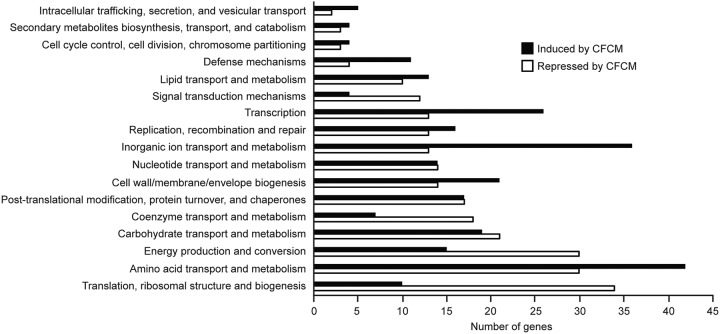
Transcriptomic analysis was then performed by transcriptome sequencing (RNA-seq) to determine the effect of CFCM on S. aureus gene expression. In particular, we sought to determine if the expression of genes involved in biofilm formation by S. aureus was modulated by S. epidermidis-secreted molecules. By doing so, we found 90.6% (2,601 out of 2,872) of the genes were expressed in at least one of the conditions (control or CFCM). Of these, 30.5% of all detected genes (876) had their expression levels modulated by CFCM. Of these, 407 genes were upregulated and 469 were downregulated when S. aureus was grown in the presence of S. epidermidis CFCM (Table S1; Fig. 3). Additionally, we found eight genes that had no identifiable orthologs in the reference strain as regulated by CFCM (2 activated and 6 repressed; Table S2). Within the group of genes that were differentially expressed in the presence of CFCM, nine have been previously associated with S. aureus biofilm formation (Table 2). Among these, expression of icaR, an important negative regulator of the icaADBC operon, which encodes the major S. aureus biofilm polysaccharide matrix PIA/PNAG [polysaccharide intercellular adhesin/poly-β(1-6)-N-acetylglucosamine], was upregulated (3.7-fold). Transcription of rsp, an AraC-type transcriptional regulator that inhibits attachment and biofilm formation in S. aureus, was also induced by CFCM (2.7-fold) (20, 21). Some of the genes associated with biofilm formation showed reduced expression, including sasG (4.1-fold), which encodes S. aureus surface protein G, hla (5.1-fold), which encodes an alpha-hemolysin, spa (4.4-fold), which encodes protein A, and the genes of the agr system (agrA [2.1-fold], agrB [2.4-fold], and hld [9.5-fold]). Transcription of clpC, encoding an ATPase required for stress tolerance and biofilm formation in S. aureus, was also repressed (10.5-fold) (22). In order to validate the RNA-seq results showing differential expression of genes involved in biofilm formation, we evaluated the effect of CFCM on the expression of icaR and icaA by real-time quantitative PCR (RT-qPCR). Our results confirmed the upregulation of icaR and also showed a significant reduction of icaA expression in response to CFCM, in agreement with the biofilm phenotype observed (Fig. 4).
FIG 3.
Metabolic pathways affected by S. epidermidis cell-free conditioned media (CFCM) on S. aureus biofilms, as determined by RNA-seq.
TABLE 2.
Transcriptional analysis of genes associated with S. aureus biofilm formation in response to S. epidermidis cell-free conditioned media
| Gene | Predicted function | Fold change | P value |
|---|---|---|---|
| icaR | ica operon transcriptional regulator | 3.74 | 0.00005 |
| rsp | AraC-type transcriptional regulator | 2.69 | 0.00005 |
| clpC | ATP-dependent Clp protease ATP-binding subunit ClpC | −10.52 | 0.00005 |
| hld | Delta-hemolysin | −9.50 | 0.00005 |
| hla | Alpha-hemolysin | −5.10 | 0.00005 |
| spa | Protein A | −4.45 | 0.00005 |
| agrB | AgrB transmembrane protein | −2.40 | 0.00015 |
| agrA | AgrA response regulator | −2.15 | 0.00005 |
| sasG | S. aureus surface protein G | −4.15 | 0.00005 |
FIG 4.
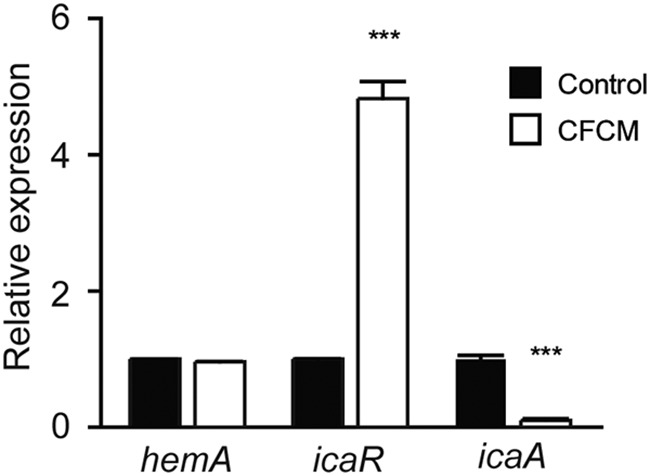
Expression of icaR and icaA genes in response to S. epidermidis cell-free conditioned media (CFCM) on S. aureus (MRSA isolate 1602) biofilms, as determined by RT-qPCR. ***, P < 0.0001.
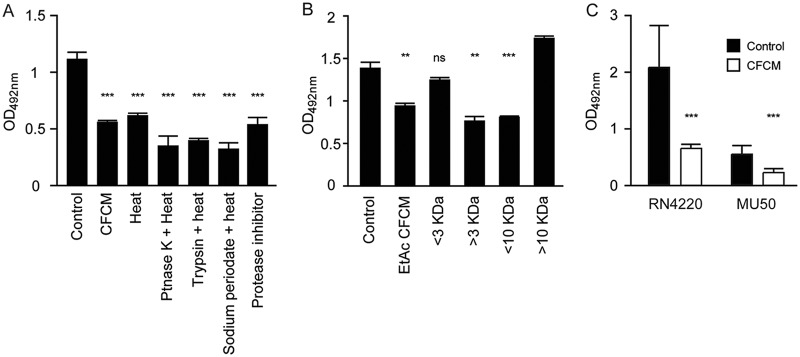
Next, we initiated the characterization of the active molecules present in the CFCM of S. epidermidis. Boiling the CFCM for 40 min did not reduce its effect on S. aureus biofilm formation, and neither did proteinase K, trypsin, and sodium periodate treatments followed by heat inactivation (Fig. 5A). We also treated the CFCM with a protease inhibitor cocktail since some S. epidermidis isolates are known to produce the serine protease Esp, which has antibiofilm activity (6). Treatment with this cocktail, which includes inhibitors of serine proteases, did not abolish CFCM antibiofilm activity. Additionally, extraction of the CFCM with ethyl acetate resulted in the recovery of a biologically active fraction, indicating that the active molecules are soluble in this solvent and therefore may be either nonpolar or only mildly polar (Fig. 5B). Fractionation of the ethyl acetate-soluble fraction of CFCM by use of centrifugal filters indicated that the active molecules are between 3 and 10 kDa (Fig. 5B). Altogether, these results indicate that the antibiofilm molecules present in the CFCM of commensal S. epidermidis are heat, proteinase K, trypsin, sodium periodate, and protease inhibitor resistant, between 3 and 10 kDa, and hydrophobic in nature.
FIG 5.
Characterization of the antibiofilm molecules present in S. epidermidis cell-free conditioned media (CFCM). (A) Biofilm formation of the S. aureus clinical isolate 1602 grown in the presence of S. epidermidis CFCM treated with heat, proteinase K, trypsin, sodium periodate, or protease inhibitors. (B) Biofilm formation of the S. aureus clinical isolate 1602 grown in the presence of S. epidermidis CFCM fractionated by molecular weight (kDa) after ethyl acetate extraction. (C) Biofilm formation of S. aureus RN4220 and MU50 agr-defective strains grown in the presence of S. epidermidis CFCM. **, P < 0.01; ***, P < 0.0001.
In order to determine if the agr quorum-sensing system is involved in the inhibition of biofilm formation by CFCM, we tested CFCM activity on two agr-deficient S. aureus strains, RN4220 and Mu50 (23, 24). CFCM significantly reduced biofilm formation on both strains; biofilm formation by Mu50 was reduced by 52.4% in the presence of CFCM, and a slightly more pronounced effect was observed in RN4220, where biofilm formation was reduced by 58.8%. Altogether, these data show that a functional agr system is not required for CFCM antibiofilm activity (Fig. 5C).
DISCUSSION
The skin is the largest organ in the human body and is directly exposed to the environment, where it is constantly challenged by potential exogenous pathogens. In a similar way to the intestinal microbiota, the skin microbiota is associated with protection against pathogens (5). In recent studies, our group showed that the intestinal microbiota produces compounds with antivirulence activity, which may lead to new alternative therapeutic strategies (17, 25). Moreover, in addition to the bacteriocins produced by S. epidermidis (5), Cutibacterium acnes, another commensal bacterium from skin, was shown to suppress the growth of MRSA isolates (26). These and other studies have produced a solid body of evidence that the skin microbiota can also be a potential source of new compounds that may aid the treatment of drug-resistant infections (27–29).
The ability of S. aureus to acquire antimicrobial resistance genes and to produce biofilms makes infections increasingly hard to treat (30). Some molecules that act on S. aureus virulence have been described. For instance, savirin, a small molecule that acts on the important quorum-sensing regulator AgrA (31), is currently in the preclinical phase, and other molecules that act on S. aureus toxins are currently part of ongoing phase II studies (19). Because of the importance of biofilms in the persistence and recurrence of S. aureus infections, some studies have looked for alternative therapies that act on this virulence trait with molecules from various sources (32–35). Antibiofilm compounds derived from marine microorganisms were described, and recently, butenolide, a nontoxic compound derived from a marine Streptomyces sp., was shown to be effective in inhibiting MRSA biofilm formation and eradicating established biofilms (35). Also, a cell-free supernatant from commensal Staphylococcus chromogenes was shown to have antibiofilm properties against coagulase-negative Staphylococcus clinical isolates and a limited impact on S. aureus clinical isolates (36). Unlike the bioactivity described herein, the authors found that the activity was lost when the supernatant was treated with heat or proteinase K (36).
Here, we describe that CFCM of a commensal S. epidermidis strain isolated from the skin of a healthy donor contains molecules with a significant impact on biofilm formation of S. aureus clinical isolates without affecting bacterial growth. The effect is not universal, since some S. aureus clinical isolates were not affected. However, our data suggest a certain degree of correlation between CFCM activity and resistance to methicillin, which could be due to the proteinaceous composition of the biofilm matrix of MRSA isolates. In the literature, MRSA strains are known to produce protein-based biofilms, whereas MSSA strains usually produce polysaccharide-based structures (37). We analyzed the matrix composition of S. aureus biofilms, focusing on the 13 strains that showed the greatest reduction by CFCM, alongside a few representatives of strains that showed little or no reduction. These analyses showed that biofilms of most MRSA strains showed greater reduction when treated with proteinase K. However, one MRSA isolate showed greater reduction with sodium periodate, and two isolates showed similar reduction with both treatments, suggesting that they produce mixed biofilms. Among MSSA strains, 6 showed protein-based biofilm matrices, whereas the other 6 produced mixed biofilm matrices. Therefore, our results did not show a correlation between methicillin susceptibility, biofilm matrix composition, and susceptibility to the antibiofilm effect of CFCM. Even though proteinase K and sodium periodate treatments have been used in many studies to define the major components of S. aureus biofilms, biofilm matrices contain a variety of macromolecules. For example, in a study with Staphylococcus lugdunensis, known to produce protein-based biofilms, researchers showed, using specific fluorescent dyes and confocal laser scanning microscopy, that its biofilm contains both PIA and proteins (38). Furthermore, another study showcased the presence of PIA/PNAG in MRSA biofilm matrices using staining with a fluorochrome associated with an anti-PNAG antibody (39).
In addition to the effect shown by CFCM during biofilm formation, the active molecules produced by S. epidermidis were also able to reduce preestablished S. aureus biofilms and, when tested in combination with oxacillin, decreased the antibiotic concentration necessary to eliminate biofilm cells, suggesting that the active molecules could be used in combination with antibiotics to improve their activity against S. aureus biofilms.
A transcriptomic analysis of biofilms grown in the presence of S. epidermidis CFCM showed an increase of icaR expression. IcaR is an important negative regulator of the icaADBC operon, binding to its promoter region and repressing PIA/PNAG, a major biofilm component of S. aureus (40). Even though icaABDC was not shown as significantly repressed in our RNA-seq data, RT-qPCR experiments showed a reduction of icaA expression by biofilm cells grown in the presence of S. epidermidis CFCM, supporting the notion that activation of icaR expression by CFCM results in icaADBC operon repression, leading to decreased biofilm formation. Together, these data suggest that molecules secreted by S. epidermidis can promote inhibition of S. aureus biofilm through the repression of PIA/PNAG in an icaR-dependent manner. Another transcriptional regulator induced by S. epidermidis CFCM was Rsp, which was shown to repress genes that affect biofilm formation, including icaABC (21). Recently, this gene was shown to be repressed in S. aureus biofilms (73). Furthermore, Rsp was shown to have proteolytic properties, which correlates with the biofilm reduction when expression of this gene is induced.
Another important molecule for biofilm formation in S. aureus is the SasG adhesin, relevant for the attachment and early accumulation steps of ica-independent biofilm formation (15). Our transcriptomic analysis showed significant repression of sasG by S. epidermidis CFCM during biofilm formation, suggesting that repression of icaADBC is not the only pathway involved in the biofilm disruption phenotype. Transcriptome analysis also showed repression of multifunctional proteins that have been related not only to the accumulation step in S. aureus biofilm formation but also to its virulence, such as protein A (spa) (15) and alpha-hemolysin (hla) (41). The role of protein A in S. aureus evasion is well-known due to its ability to bind the Fc chain of immunoglobulins (42). In addition, the proinflammatory properties of protein A contribute to inflammation in patients with atopic dermatitis, who are commonly colonized by S. aureus (43). Alpha-hemolysin can bind to erythrocytes, monocytes, platelets, endothelial cells, and lymphocytes, as well as epithelial cells (44), and increased expression of hla is an important marker of the shift from commensalism to pathogenic behavior in S. aureus (27, 45). Another relevant gene whose expression was modulated by CFCM is clpC; CFCM strongly repressed its expression. This gene encodes an ATPase that was previously shown to be required for stress tolerance and biofilm formation in S. aureus. In the absence of this gene, biofilm formation was shown to be significantly reduced (22).
In addition to the effects on icaR and sasG, our transcriptomic analysis showed that molecules secreted by S. epidermidis affected the S. aureus agr system; addition of CFCM to S. aureus cultures inhibited the expression of agrA, agrB, and hld. The agr system is a major S. aureus virulence regulator; it can induce the expression of several toxins, such as α-, β-, and γ-hemolysin; leukotoxins; and toxic shock syndrome toxins (TSSTs), as well as proteases (46). Some studies have proposed compounds with the ability to inhibit agr as a new therapeutic approach against S. aureus infections (47). However, besides causing a decrease in toxin expression, inactivation of agr has been associated with increased biofilm formation in some S. aureus strains (46, 48, 49). Compounds that not only decrease agr activity but also impair biofilm formation, like the ones present in S. epidermidis CFCM described here, may have important therapeutic potential.
Altogether, our data show that the antibiofilm activity displayed by S. epidermidis CFCM is not due to an autoinducing peptide (AIP) effect on the S. aureus agr system. Canovas et al. showed that AIPs from some Staphylococcus spp., including one out of the four S. epidermidis isolates tested, downregulate the S. aureus agr system (50). However, as mentioned earlier, data from several papers show that inhibition of the S. aureus agr system does not lead to biofilm reduction; rather, agr inhibition is associated with a significant increase in biofilm formation (46, 48, 49). Vuong et al. showed that AIP from S. epidermidis significantly improved biofilm formation by S. aureus (51). When the study was published, the occurrence of different agr types in S. epidermidis had not yet been described, and therefore, we do not know what type of AIP was tested. However, other studies have since shown that only AIP I of S. epidermidis can inhibit the agr system of S. aureus in all but one (type IV) agr types (52, 53). Our data show that an S. epidermidis agr type II strain (RF1) can inhibit biofilm formation by several S. aureus strains from all agr types, including previously characterized agr-defective strains (MU50 and RN4220) (23, 24). Therefore, CFCM antibiofilm activity does not appear to be due to AIP regulation. Nevertheless, the activity of CFCM on S. aureus gene expression appears to act through multiple pathways, including agr signaling, as, according to transcriptomic data, CFCM inhibits agr expression, along with downstream virulence genes regulated by quorum sensing.
Recently, phenol-soluble modulins (PSMs) present in the supernatants of S. epidermidis and S. aureus cultures were identified as having a negative impact on S. aureus persister cell frequency after treatment with antibiotics (54). It has been suggested that biofilms may be formed by a significant proportion of persister cells, and therefore, the effect of PSMs on persister cell frequency could be extrapolated to an effect on biofilm formation as the one described herein. However, it is unlikely that the active molecule present in RF1 CFCM is a PSM due to its molecular size and the protease-resistant nature of its activity on biofilm formation. Other studies investigating the effect of bioactive compounds on S. aureus biofilm formation have been published. While studying molecules produced by C. acnes (formerly Propionibacterium acnes), Wollenberg et al. have been able to identify coproporphyrin III, a molecule present in a C. acnes-conditioned medium that increases S. aureus biofilm formation (55). Altogether, studies in this area will aid in our understanding of the intricate chemical interplay between different species inhabiting the human body.
The goal of the present study was to shed light on the interactions between a major human skin commensal and one of the most important human skin pathogens, which is currently a global health issue. To do this, we focused on a specific trait of S. aureus that is directly linked to its virulence and ability to resist antimicrobial therapy, namely, the development of biofilms. Additionally, the analysis of the impact of S. epidermidis-secreted molecules on the S. aureus transcriptome enabled us to look at how this commensal can affect this important pathogen at the whole-genome level. However, it is safe to assume that the gene expression changes observed are not caused solely by the molecule responsible for the antibiofilm activity of CFCM. Thousands of molecules are likely present in CFCM, and many of these molecules may have specific effects on S. aureus physiology. Therefore, despite our interest in seeking antibiofilm molecules as new therapeutic strategies, the activity of CFCM on the S. aureus transcriptome may be a result of multifactorial events that occur between these species. Although the use of CFCM allowed us to probe the global effects that may occur during these interactions, it also represents a limitation of our study with regard to the identification of potential therapeutic agents. Nevertheless, we have generated some significant data that will aid in the identification of the active molecule, and our work opens an avenue for further bioprospection of the human skin microbiome for bioactive molecules. Future work will focus on determining the exact chemical nature of the bioactive compound and on describing its mode of action. In the future, this and other bioactive molecules from the human microbiome could be used as alternatives to the currently available antibiotics, with the goal of circumventing the phenomenon of antimicrobial drug resistance. Additionally, and perhaps in a more short-term approach, such molecules may be used in combination with antibiotics in order to disturb bacterial biofilms and decrease the antimicrobial concentrations required to treat biofilm-related infections.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in accordance with the recommendations and approval of the Research Ethics Committee of the National School of Public Health, Oswaldo Cruz Foundation, Brazil (protocol number, 2.919.895). All subjects provided written informed consent.
Bacterial strains.
For two volunteer subjects who were included in the study, swabs previously moistened with sterile saline (0.85%) were rubbed against the heel area and cultured on mannitol salt agar and blood agar. Three commensal S. epidermidis isolates were obtained and named RF1 (one subject), RF3, and RF5 (another subject). Initial experiments were performed with these three strains as well as other clinical S. epidermidis isolates, and strain RF1 was selected for further studies. All S. epidermidis strains were identified by conventional biochemical tests (56) and matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF microflex LT; Bruker Daltonics, Leipzig, Germany). The RF1 strain was characterized as agr type II by PCR (57), and it is a nonbiofilm producer. S. aureus clinical isolates categorized as strong or moderate biofilm producers (29 isolates) (58), including 13 MRSA and 16 MSSA, were selected from a previously characterized collection of strains of the Laboratório de Infecção Hospitalar at Universidade Federal do Rio de Janeiro (Table 1). The agr type of each of the strains was determined by PCR (59), and all 4 agr types were represented (type 1, 9 strains; type 2, 6 strains; type 3, 13 strains; and type 4, 1 strain). Two agr-defective strains, MU50 (23) and RN4220 (24), were also used in this study.
Biofilm matrix composition assay.
To determine if biofilms were composed mainly of proteins or polysaccharides, established biofilms were treated with either proteinase K (100 μg/ml) or sodium periodate (40 mM) for 2 h at 37°C. After incubation, biofilms were washed with phosphate-buffered saline (PBS) and stained with 0.1% safranin as described for biofilm assays (21). The relative biofilm reduction elicited by each treatment compared to PBS (control) indicated the role of each component (proteins or polysaccharides) on biofilm matrix composition.
Cell-free conditioned medium preparations.
After growth on Trypticase soy agar (TSA), the RF1 strain was inoculated in 50 ml of TSB (BD Diagnostics, Oxford, UK) for 24 h at 37°C with shaking (250 rpm). To obtain the CFCM, cultures were centrifuged at 3,100 × g for 15 min and filtered using 0.22-μm filters. CFCM was then evaporated to dryness using a Speed Vac Concentrator plus (Eppendorf, Hamburg, Germany), and the dried residue was resuspended in sterile 0.85% saline at a relative concentration of 20×. This concentrated CFCM was used at 1:10 in the experiments described herein. Therefore, the final concentration of the CFCM in the experiments, relative to the original bacterial cultures, was 2×. The same procedures were performed with culture media without bacterial inoculum, and the extract produced was used as a control for all experiments. For preliminary characterization of the active molecules, the stability of active molecules in CFCM was assayed after boiling for 40 min. CFCM was also treated with proteinase K (20 mg/ml) (Invitrogen, CA, USA), trypsin (20 μg/ml) (Promega, WI, USA), and sodium periodate (40 mM) (Sigma-Aldrich, MO, USA) for 1 h at 37°C, and all the treatments were inactivated by boiling the mixture for 40 min. In order to exclude effects of proteases that can be secreted by S. epidermidis, we also added 0.2% of a protease inhibitor cocktail P8340 (Sigma-Aldrich, MO, USA) to the CFCM. Ethyl acetate extraction was performed by adding ethyl acetate (1:1) to RF1 filtered supernatants and manually agitating and incubating the solution at room temperature for up to 20 min for phase separation. The soluble phase was collected, and ethyl acetate was evaporated using a Speed Vac. Dried residues were resuspended in saline, and the solution was fractionated by centrifugation using Amicon filters (Merck Millipore, MA, USA) with 3- and 10-kDa cutoff membranes for 30 min at 5,000 × g. Each of the fractions was also concentrated 20× using a Speed Vac.
Bacterial planktonic growth.
Growth curves of S. aureus clinical isolates in the presence of CFCM or the control medium were performed on 96-well plates in triplicate. Overnight growth on TSB was diluted to an optical density at 630 nm (OD630) of 0.05 in TSB with 10% CFCM or a control medium. The microplate was incubated with agitation at 37°C, and the OD630 was recorded every 30 min with an enzyme-linked immunosorbent assay (ELISA) auto reader (Thermoplate TP Reader, Thermoplate, MN, USA).
Biofilm formation.
S. aureus biofilm formation was assessed by the microtiter plate test, as described elsewhere with modifications (58). Bacterial colonies were used to prepare cell suspensions in sterile distilled water with densities adjusted to the 0.5 McFarland standard. For analysis of the impact of the CFCM on biofilm formation, 120 μl of TSB 1% glucose was supplemented with 15 μl of CFCM, or control CFCM and 15 μl of the bacterial suspension were added. Additionally, S. aureus biofilm formation was also assayed without any additions. After incubation for 24 h at 37°C, the content of each well was removed, and the wells were carefully washed 3 times with 200 μl of PBS (pH 7.2) (Laborclin, Paraná, Brazil). The plates were then incubated at 60°C for 1 h and stained with 150 μl per well of 0.1% safranin for 15 min at room temperature. Excess stain was removed by rinsing the wells twice with PBS. The dye was then solubilized using 150 μl of a 95% ethanol solution, and the OD of each well was measured at 492 nm with an ELISA plate reader after 30 min. Results were obtained by subtracting the average ODs of the negative controls (uncultured media) from the average ODs of the experimental wells. Experiments were performed using three biological replicates and repeated at least three times.
Activity of the CFCM on established biofilms.
The wells of a sterile 96-well flat-bottomed tissue culture plate (manufacturer no. TPP 92096; Trasadingen, Switzerland) were filled with 135 μl of TSB 1% glucose, and 15 μl of bacterial suspension, prepared as described earlier, was added into each well, and the plate was incubated for 24 h at 37°C. After two washing steps with 200 μl of sterile PBS (pH 7.2), 10% CFCM or control (in PBS) was added, and the plate was incubated for an additional 24 h at 37°C (60). Safranin staining and measurement were then performed as described above. Experiments were performed in triplicate at least three times.
Combined activity of the CFCM and oxacillin on the viability of established biofilm cells.
Established biofilms of a selected MSSA strain (isolate 74) were inoculated as described above. After 24 h of incubation at 37°C and two washing steps with 200 μl of sterile PBS (pH 7.2), 10% CFCM or control with or without increasing concentrations of oxacillin (0.25 to 256 μg/ml) were added, and plates were incubated for 24 h at 37°C. After a single washing step with PBS, the viability of biofilm cells was revealed using resazurin 0.02% for 3 h. A blue color indicated dead cells, whereas pink indicated live cells (60).
Confocal microscopy and image processing.
Biofilms of a selected MRSA strain (isolate 1602) were formed on Nunc Lab-Tek Chambered Coverglass (Thermo Scientific, MA, USA) with either the CFCM or control under the same conditions used for biofilm formation on microplate assays. After incubation, chambers were rinsed three times with 200 μl of 0.1 M HEPES buffer (pH 7), fixed with 100 μl of 2.5% glutaraldehyde, and incubated overnight at 4°C. Following three washes with HEPES buffer, 150 μl of DAPI (4’,6-diamidino-2-phenylindole dihydrochloride) in HEPES (1:200) was added, and biofilms were incubated at room temperature for 10 min. Two washes with HEPES buffer were performed, and 300 μl of a solution of 20% acrylamide/bis-acrylamide, 0.5 μl of TEMED (N,N,N′,N′-tetramethylethylenediamine), and 0.5 μl of 10% ammonium persulfate was added. Biofilms were then observed using a Leica TCS-SPE microscope (Leica Microsystems, Germany) on fluorescence mode using a 63× objective lens and a 1.5× digital zoom. For excitation, a 405-nm laser line was used, and emission was collected using a 580-nm bandpass filter. All images were collected and processed using the Leica Application Suite X (LAS X) software. For cell quantification, the first 19 sections of each confocal series were selected, representing a total volume of 8.8 × 104 μm3. The bacterial cells present in each biofilm volume were automatically segmented and quantified using the daime software (61).
RNA extraction.
Biofilms of a selected MRSA strain (isolate 1602) were formed in 24-well flat-bottomed tissue culture plates (TPP; Trasadingen, Switzerland) with either the CFCM or control medium (the CFCM was prepared without bacterial inoculum) under the same conditions used for biofilm formation on microplate assays. After incubation, wells were rinsed three times with 500 μl of PBS, 1 ml of RNAprotect bacteria reagent (Qiagen, Hilden, Germany) was added in each well, and biofilms were scraped. After 5 min at room temperature, the sessile cell suspension was centrifuged, and pellets were treated with a lysis solution (20 mg/ml lysozyme, 40 μg/ml lysostaphin, and 4 mg/ml proteinase K). Purification was performed with High Pure RNA isolation kit (Roche, Basel, Switzerland), and RNA was treated with the RNase-Free DNase set (Qiagen, Hilden, Germany).
RNA sequencing.
rRNA depletion was performed with the Ribo-Zero rRNA removal kit (Illumina) according to the manufacturer's recommendations (74). Removal of rRNA is achieved with magnetic spheres that bind to removal probes that hybridize to rRNA, producing an RNA sample ready for library construction. Library construction was performed with the TruSeq Stranded mRNA sample preparation kit according to the manufacturer´s recommendations (catalog no. RS-122-9004; Illumina). This protocol enables the acquisition of a suitable library for subsequent cluster generation for nucleotide sequencing. First, poly(A) RNA molecules were purified using magnetic spheres bound to oligonucleotide poly(T). After purification, mRNA was fragmented in small pieces using divalent cations under high temperature. Cleaved RNA fragments were reverse transcribed to cDNA (first chain) using reverse transcriptase and arbitrary primers. Chain specificity was achieved by substitution of dTTP for dUTP on the second strand marking mix (SMM), following the synthesis of the second cDNA chain using DNA polymerase I and RNase H. This results in removal of the RNA template with concomitant synthesis of the second strand. The presence of actinomycin D on the first strand synthesis act D mix (FSA) prevents spurious DNA-dependent synthesis, allowing only RNA-dependent synthesis, improving chain specificity. The 3´ ends of cDNA fragments were then adenylated with a single A base, and indexing adapters were ligated. Products were then purified using complementary adapters and beads and were enriched by PCR to create the final cDNA library.
The RNA obtained was used for transcriptome analyses using an Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) at the Plataforma de Sequenciamento de Ácidos Nucleicos de Nova Geração–RPT01J of the Fundação Oswaldo Cruz (Rio de Janeiro, Brazil). Data processing and analyses were performed at the Plataforma de Bioinformática–RPT04A of the Fundação Oswaldo Cruz. The bcl files containing the raw sequencing reads were converted to fastq files using the bcl2fastq software version 2.17 (Illumina). Technical and low-quality sequences were removed using Trimmomatic with the following parameters: ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:40 (62). Read quality was evaluated with FastQC (Babraham Bioinformatics). Filtered reads were mapped with Bowtie2 (–no-mixed –no-discordant) to the S. aureus NCTC8325 genome (GenBank accession no. NC_007795.1) as a reference (63). The SAMtools package was used to extract and sort only the mapped reads (-G 77, -G 141) (64). Finally, the Cufflinks suite was used to compare global gene expression between both conditions (65–67). Genes that showed a differential expression of 2-fold with P < 0.05 between conditions (CFCM and control) were considered significantly regulated by CFCM. Unmapped reads were extracted from the original files into two files, one containing forward reads and another containing reverse reads, and sorted using SAMtools. Forward and reverse reads from all samples were concatenated into two files and then used for de novo transcriptome assembly using Trinity 2.8.5 (68, 69). Using the downstream scripts available within Trinity, the reads used to assemble the transcriptome (originally unmapped to the reference genome) were then mapped to the newly assembled transcriptome using Salmon (70), and differential expression was calculated using DESeq2 (71). Genes that showed a differential expression of 4-fold with P < 0.001 between conditions (CFCM and control) were selected. These genes were extracted and annotated against the NCBI RefSeq genome database for bacteria (taxid, 2) using online BLASTn with the default parameters.
For a final annotation, the top 10 genomes for each entry were downloaded and used for a local BLASTn using the individual gene sequences as queries and the most differentially expressed transcripts as subjects. Metabolic pathways for each of the affected genes were determined using the EggNOG pathway database v. 4.5.1 (http://eggnogdb.embl.de) (72). To do this, gene IDs were searched manually in the database, and genes were categorized based on the resulting classification.
Real-time quantitative PCR.
RT-qPCR was performed to confirm the effect of the CFCM on the expression of genes related to biofilm formation obtained by RNA-seq. To obtain cDNA, RNA extracted as described above, with either the CFCM or control medium, was subjected to a reverse transcriptase reaction using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). Primers were designed for this study and are listed in Table 3. The relative expression levels of tested genes were normalized according to the expression levels obtained with the housekeeping gene hemA, which encodes the glutamyl tRNA reductase.
TABLE 3.
Primers used in gene expression analyses by RT-qPCR
| Target gene | Primer | Sequence (5′–3′) |
|---|---|---|
| icaR | icaRfw | CAGAGAAGGGGTATGACGGTAC |
| icaRrev | GGTAATCAAAACAACATTTAACACTTTGTTCG | |
| icaA | icaAF | GTGCAGTTGTCGACGTTGGC |
| icaAR | CACATGGCAAGCGGTTCATAC | |
| hemA | hemAfw | GAGATGATGCCTTACGAATTGCC |
| hemArev | GTAACGACCTGTGTGAATTTGATCAAC |
Statistical analysis.
All comparisons were performed using unpaired Student's t tests (Prism 5, GraphPad Software, CA, USA). Differences were considered statistically significant when values of P < 0.05 were obtained.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to the Plataforma de Sequenciamento de Ácidos Nucléicos de Nova Geração–RJ–RPT01J and to the Plataforma de Bioinformática–RPT04A–Rede de Plataformas Tecnológicas FIOCRUZ.
We thank Pedro Leão at the Federal University of Rio de Janeiro and UniMicro for his valuable contributions with confocal microscopy data processing and acquisition.
This study was supported by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento Pessoal de Nível Superior-Brasil (CAPES) (Finance Code 001), as well as the Fundação Oswaldo Cruz Inova Fiocruz/VPPCB Program.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Swain Ewald HA, Ewald PW. 2018. Natural selection, the microbiome, and public health. Yale J Biol Med 91:445–455. [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenthal M, Goldberg D, Aiello A, Larson E, Foxman B. 2011. Skin microbiota: microbial community structure and its potential association with health and disease. Infect Genet Evol 11:839–848. doi: 10.1016/j.meegid.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabó K, Erdei L, Bolla BS, Tax G, Bíró T, Kemény L. 2017. Factors shaping the composition of the cutaneous microbiota. Br J Dermatol 176:344–351. doi: 10.1111/bjd.14967. [DOI] [PubMed] [Google Scholar]
- 4.Byrd AL, Belkaid Y, Segre JA. 2018. The human skin microbiome. Nat Rev Microbiol 16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 5.Christensen GJ, Brüggemann H. 2014. Bacterial skin commensals and their role as host guardians. Benef Microbes 5:201–215. doi: 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 6.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 7.Cogen AL, Yamasaki K, Muto J, Sanchez KM, Crotty Alexander L, Tanios J, Lai Y, Kim JE, Nizet V, Gallo RL. 2010. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin-gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS One 5:e8557. doi: 10.1371/journal.pone.0008557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, Macleod DT, Torpey JW, Otto M, Nizet V, Kim JE, Gallo RL. 2010. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol 130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong HH, NISC Comparative Sequence Program, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, Turner ML, Segre JA. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohadian Moghadam S, Modoodi Yaghooti M, Pourramezan N, Pourmand MR. 2017. Molecular characterization and antimicrobial susceptibility of the CA-MRSA isolated from healthcare workers, Tehran, Iran. Microb Pathog 107:409–412. doi: 10.1016/j.micpath.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 12.Otto M. 2018. Staphylococcal biofilms. Microbiol Spectr 6. doi: 10.1128/microbiolspec.GPP3-0023-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherr TD, Heim CE, Morrison JM, Kielian T. 2014. Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front Immunol 5:37. doi: 10.3389/fimmu.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Günther F, Blessing B, Tacconelli E, Mutters NT. 2017. MRSA decolonization failure—are biofilms the missing link? Antimicrob Resist Infect Control 6:32. doi: 10.1186/s13756-017-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moormeier DE, Bayles KW. 2017. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol 104:365–376. doi: 10.1111/mmi.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Wang W, Xu SX, Magarvey NA, McCormick JK. 2011. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc Natl Acad Sci U S A 108:3360–3365. doi: 10.1073/pnas.1017431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antunes LC, McDonald JA, Schroeter K, Carlucci C, Ferreira RB, Wang M, Yurist-Doutsch S, Hira G, Jacobson K, Davies J, Allen-Vercoe E, Finlay BB. 2014. Antivirulence activity of the human gut metabolome. mBio 5:e01183-14. doi: 10.1128/mBio.01183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemfack MC, Ravella SR, Lorenz N, Kai M, Jung K, Schulz S, Piechulla B. 2016. Novel volatiles of skin-borne bacteria inhibit the growth of Gram-positive bacteria and affect quorum-sensing controlled phenotypes of Gram-negative bacteria. Syst Appl Microbiol 39:503–515. doi: 10.1016/j.syapm.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PubMed] [Google Scholar]
- 20.Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. 2011. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol 193:5231–5241. doi: 10.1128/JB.05454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, He L, Song Y, Villaruz AE, Joo HS, Liu Q, Zhu Y, Wang Y, Qin J, Otto M, Li M. 2015. AraC-type regulator Rsp adapts Staphylococcus aureus gene expression to acute infection. Infect Immun 84:723–734. doi: 10.1128/IAI.01088-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frees D, Chastanet A, Qazi S, Sørensen K, Hill P, Msadek T, Ingmer H. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol 54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 23.Sakoulas G, Eliopoulos GM, Moellering RC Jr, Wennersten C, Venkataraman L, Novick RP, Gold HS. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 46:1492–1502. doi: 10.1128/aac.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traber K, Novick R. 2006. A slipped‐mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate δ‐ and α‐haemolysins. Mol Microbiol 59:1519–1530. doi: 10.1111/j.1365-2958.2006.04986.x. [DOI] [PubMed] [Google Scholar]
- 25.Peixoto RJM, Alves ES, Wang M, Ferreira RBR, Granato A, Han J, Gill H, Jacobson K, Lobo LA, Domingues RMCP, Borchers CH, Davies JE, Finlay BB, Antunes LCM. 2017. Repression of Salmonella host cell invasion by aromatic small molecules from the human fecal metabolome. Appl Environ Microbiol 83:e01148-17. doi: 10.1128/AEM.01148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, Gallo RL, Huang CM. 2013. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One 8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. 2016. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Sullivan JN, Rea MC, O’Connor PM, Hill C, Ross RP. 2019. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol 95:fiy241. doi: 10.1093/femsec/fiy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Goh BN, Teh WK, Jiang Z, Goh JPZ, Goh A, Wu G, Hoon SS, Raida M, Camattari A, Yang L, O'Donoghue AJ, Dawson TL Jr. 2018. Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J Invest Dermatol 138:1137–1145. doi: 10.1016/j.jid.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Kamaruzzaman NF, Tan LP, Mat Yazid KA, Saeed SI, Hamdan RH, Choong SS, Wong WK, Chivu A, Gibson AJ. 2018. Targeting the bacterial protective armour; challenges and novel strategies in the treatment of microbial biofilm. Materials (Basel) 11:1705. doi: 10.3390/ma11091705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sully EK, Malachowa N, Elmore BO, Alexander SM, Femling JK, Gray BM, DeLeo FR, Otto M, Cheung AL, Edwards BS, Sklar LA, Horswill AR, Hall PR, Gresham HD. 2014. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog 10:e1004174. doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pompilio A, Pomponio S, Di Vincenzo V, Crocetta V, Nicoletti M, Piovano M, Garbarino JA, Di Bonaventura G. 2013. Antimicrobial and antibiofilm activity of secondary metabolites of lichens against methicillin-resistant Staphylococcus aureus strains from cystic fibrosis patients. Future Microbiol 8:281–292. doi: 10.2217/fmb.12.142. [DOI] [PubMed] [Google Scholar]
- 33.De Cremer K, Delattin N, De Brucker K, Peeters A, Kucharíková S, Gerits E, Verstraeten N, Michiels J, Van Dijck P, Cammue BP, Thevissen K. 2014. Oral administration of the broad-spectrum antibiofilm compound toremifene inhibits Candida albicans and Staphylococcus aureus biofilm formation in vivo. Antimicrob Agents Chemother 58:7606–7610. doi: 10.1128/AAC.03869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrestha L, Kayama S, Sasaki M, Kato F, Hisatsune J, Tsuruda K, Koizumi K, Tatsukawa N, Yu L, Takeda K, Sugai M. 2016. Inhibitory effects of antibiofilm compound 1 against Staphylococcus aureus biofilms. Microbiol Immunol 60:148–159. doi: 10.1111/1348-0421.12359. [DOI] [PubMed] [Google Scholar]
- 35.Yin Q, Liang J, Zhang W, Zhang L, Hu ZL, Zhang Y, Xu Y. 2019. Butenolide, a marine-derived broad-spectrum antibiofilm agent against both Gram-positive and Gram-negative pathogenic bacteria. Mar Biotechnol 21:88–98. doi: 10.1007/s10126-018-9861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaac P, Bohl LP, Breser ML, Orellano MS, Conesa A, Ferrero MA, Porporatto C. 2017. Commensal coagulase-negative Staphylococcus from the udder of healthy cows inhibits biofilm formation of mastitis-related pathogens. Vet Microbiol 207:259–266. doi: 10.1016/j.vetmic.2017.05.025. [DOI] [PubMed] [Google Scholar]
- 37.O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O'Gara JP. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol 45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. 2015. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol 5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson MJ, Parks PJ, Peterson ML. 2013. A mucosal model to study microbial biofilm development and anti-biofilm therapeutics. J Microbiol Methods 92:201–208. doi: 10.1016/j.mimet.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cue D, Lei MG, Lee CY. 2012. Genetic regulation of the intercellular adhesion locus in staphylococci. Front Cell Infect Microbiol 2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson MJ, Lin YC, Gillman AN, Parks PJ, Schlievert PM, Peterson ML. 2012. Alpha-toxin promotes Staphylococcus aureus mucosal biofilm formation. Front Cell Infect Microbiol 2:64. doi: 10.3389/fcimb.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Missiakas D, Schneewind O. 2016. Staphylococcus aureus vaccines: deviating from the carol. J Exp Med 213:1645–1653. doi: 10.1084/jem.20160569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geoghegan JA, Irvine AD, Foster TJ. 2018. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol 26:484–497. doi: 10.1016/j.tim.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Berube BJ, Bubeck Wardenburg J. 2013. Staphylococcus aureus α-toxin: nearly a century of intrigue. Toxins (Basel) 5:1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenkins A, Diep BA, Mai TT, Vo NH, Warrener P, Suzich J, Stover CK, Sellman BR. 2015. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio 6:e02272-14. doi: 10.1128/mBio.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. 2014. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol 22:676–685. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Tan L, Li SR, Jiang B, Hu XM, Li S. 2018. Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front Microbiol 9:55. doi: 10.3389/fmicb.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/jb.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suligoy CM, Lattar SM, Noto Llana M, González CD, Alvarez LP, Robinson DA, Gómez MI, Buzzola FR, Sordelli DO. 2018. Mutation of Agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front Cell Infect Microbiol 8:18. doi: 10.3389/fcimb.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canovas J, Baldry M, Bojer MS, Andersen PS, Grzeskowiak PK, Stegger M, Damborg P, Olsen CA, Ingmer H. 2016. Cross-talk between Staphylococcus aureus and other staphylococcal species via the agr quorum sensing system. Frontiers Microbiol 7:1733. doi: 10.3389/fmicb.2016.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vuong C, Saenz HL, Gotz F, Otto M. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 52.Otto M, Echner H, Voelter W, Götz F. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 69:1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, Sanford JA, O’Neill AM, Liggins MC, Nakatsuji T, Cech NB, Cheung AL, Zengler K, Horswill AR, Gallo RL. 2019. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med 11:eaat8329. doi: 10.1126/scitranslmed.aat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bojer MS, Lindemose S, Vestergaard M, Ingmer H. 2018. Quorum sensing-regulated phenol-soluble modulins limit persister cell populations in Staphylococcus aureus. Front Microbiol 20:255. doi: 10.3389/fmicb.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wollenberg MS, Claesen J, Escapa IF, Aldridge KL, Fischbach MA, Lemon KP. 2014. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio 5:e01286-14. doi: 10.1128/mBio.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bannerman TL, Peacock SJ. 2007. Staphylococcus, Micrococcus, and other catalase-positive cocci, p 390–411. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC. [Google Scholar]
- 57.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. 2003. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol 69:18–23. doi: 10.1128/aem.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Cirković I, Ruzicka F. 2007. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 59.Gilot P, Lina G, Cochard T, Poutrel B. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol 40:4060–4067. doi: 10.1128/jcm.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodrigues A, Gomes A, Marçal PH, Dias-Souza MV. 2017. Dexamethasone abrogates the antimicrobial and antibiofilm activities of different drugs against clinical isolates of Staphylococcus aureus and Pseudomonas aeruginosa. J Adv Res 8:55–61. doi: 10.1016/j.jare.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daims H, Lücker S, Wagner M. 2006. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 62.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goff L, Trapnell C, Kelley D. 2018. cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R package version 2.24.0. doi: 10.18129/B9.bioc.cummeRbund. [DOI] [Google Scholar]
- 68.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeswanth S, Chaudhury A, Sarma PVGK. 2017. Quantitative expression analysis of SpA, FnbA and Rsp genes in Staphylococcus aureus: actively associated in the formation of biofilms. Curr Microbiol 74:1394–1403. doi: 10.1007/s00284-017-1331-x. [DOI] [PubMed] [Google Scholar]
- 74.Illumina. 2016. Ribo-Zero rRNA removal kit reference guide. Document #15066012 v02. Illumina, San Diego, CA: https://jp.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/ribosomal-depletion/ribo-zero/ribo-zero-reference-guide-15066012-02.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.