MRSA is an important human pathogen; thus, its increasing prevalence in livestock over the last decade has a potentially large impact on public health. Farmers and veterinarians are especially at risk due to their close contact with animals. Our work demonstrates a dramatic increase in MRSA prevalence in Swiss pigs, from 2% in 2009 to 44% in 2017. Whole-genome sequencing allowed us to show a close association between farmer and pig strains as well as veterinarian and horse strains, indicating that the respective animals are a likely source of human colonization. Furthermore, we could demonstrate that pig spa t011 strains cluster separately and are probably less likely to colonize humans than are pig spa t034 strains. This research may provide a basis for a more substantiated risk assessment and preventive measures.
KEYWORDS: Staphylococcus aureus, human, methicillin, antimicrobial resistance, sequencing, DNA sequencing
ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged over the last few decades as a One Health problem with an increasing prevalence in various animal species. The most notable animals are pigs, as asymptomatic carriers, and horses, where there is often an association with infections. The current study looked at the course of MRSA prevalence in Swiss livestock since 2009, with a special focus on pigs, followed by screening of veterinarians and farmers. Livestock isolates were obtained from the Swiss monitoring program and then characterized by spa typing. Concentrating on the year 2017, we analyzed the prevalence of MRSA in Swiss veterinarians and farmers, followed by whole-genome sequencing of selected human and animal strains. The phylogeny was assessed by applying core-genome multilocus sequence typing (MLST) and single-nucleotide polymorphism (SNP) analyses, followed by screening for resistance genes and virulence factors. The prevalence of MRSA in Swiss pigs showed a dramatic increase from 2% in 2009 to 44% in 2017. Isolates typically belonged to clonal complex 398 (CC398), split between spa t011 and t034. The higher prevalence was mainly due to an increase in t011. spa t034 strains from farmers were found to be closely associated with porcine t034 strains. The same could be shown for spa t011 strains from horses and veterinarians. spa t034 strains had a high number of additional resistance genes, and two strains had acquired the immune evasion cluster. However, all but one of the pig spa t011 strains clustered in a separate group. Thus, the increase in pig spa t011 strains does not directly translate to humans.
IMPORTANCE MRSA is an important human pathogen; thus, its increasing prevalence in livestock over the last decade has a potentially large impact on public health. Farmers and veterinarians are especially at risk due to their close contact with animals. Our work demonstrates a dramatic increase in MRSA prevalence in Swiss pigs, from 2% in 2009 to 44% in 2017. Whole-genome sequencing allowed us to show a close association between farmer and pig strains as well as veterinarian and horse strains, indicating that the respective animals are a likely source of human colonization. Furthermore, we could demonstrate that pig spa t011 strains cluster separately and are probably less likely to colonize humans than are pig spa t034 strains. This research may provide a basis for a more substantiated risk assessment and preventive measures.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) was first discovered in the 1960s and is thought to have developed independently from several clones of methicillin-susceptible Staphylococcus aureus (MSSA) (1). In human medicine, it was initially recognized as a nosocomial infection; however, in the 1990s, an increasing incidence of hospital-independent human MRSA infections was observed (2). These so-called community-acquired MRSA (CA-MRSA) cases have since been reported in many countries.
With the detection of MRSA in pigs and farmers, MRSA has become a One Health issue (3). These livestock-associated MRSA (LA-MRSA) strains are not only associated with disease in animals, particularly horses, but also human infections (4, 5). Pigs, in particular, can be heavily colonized with MRSA and are thought to constitute the main reservoir (4, 6–8). Therefore, humans with regular close contact with these animals, e.g., farmers, slaughterhouse workers, and veterinarians, have a higher risk of being colonized and thus of developing infections (6, 9–12). Furthermore, LA-MRSA can be found in horses, veal calves, and poultry (1). In horses, LA-MRSA can also be an agent of nosocomial infection in an animal clinic environment and can be transmitted to clinical staff in this context (13, 14).
LA-MRSA strains mainly belong to clonal complex 398 (CC398). They are thought to have emerged from ancestral human MSSA, gaining resistance genes but losing human specific virulence factors in the process (15). The human innate immunomodulatory genes (chemotaxis inhibitory protein [chp], staphylococcal complement inhibitor [scn], and the plasminogen activator staphylokinase [sak]) often carried on a phiSa3 prophage are usually not present in animal isolates (15). However, it is possible for CC398 MRSA to regain these virulence factors, which might enhance its potential as a human pathogen. The acquisition can occur by lateral gene transfer during cocolonization in the host (16). Other known virulence factors, like leucocidins, e.g., Panton-Valentine leucocidin (PVL) or enterotoxins, are also rarely found in LA-MRSA (1, 17).
MRSA strains are defined by their methicillin resistance, mainly mediated by mecA, but they frequently also carry additional resistance genes. For LA-MRSA, this is most frequently a tetracycline resistance [e.g., tet(M)] gene which is present in virtually all strains, probably reflecting the widespread use of this antibiotic on farms (17, 18). The vga(E) gene conferring resistance to streptogramin A, pleuromutilin, and lincosamide was also originally detected in an LA-MRSA isolate from a Swiss pig (19). It was found to be located on Tn6133 together with erm(A) (macrolide, lincosamide, and streptogramin B resistance) and ant(9)-Ia (spectinomycin resistance) (19, 20).
Various typing methods have been used to analyze the phylogeny and epidemiology of MRSA. A classical multilocus sequence typing (MLST) scheme based on seven housekeeping genes was developed by Enright et al. in 2000 (21). spa typing based on repeats in the staphylococcal protein A sequence allows higher discrimination than does MLST and remains a popular typing method (22). However, whole-genome sequencing-based methods are now rapidly becoming standard since they allow much higher discrimination as well as detection of virulence and resistance genes (23). The MLST scheme has been expanded to core-genome MLST (cgMLST), which allows a more detailed analysis, while single-nucleotide polymorphism (SNP)-based methods allow the highest discrimination and are most useful for closely related isolates (23).
The current study aims to evaluate the spread of LA-MRSA (with a special focus on pig isolates) to farmers and veterinarians. In this framework, traditional spa typing was compared to cgMLST and SNP analyses of selected isolates. A previous MRSA prevalence study in Swiss veterinarians in 2012 revealed a prevalence of 3.8%, with LA-MRSA only detected in veterinarians treating farm animals (24). Since then, routine monitoring data indicated a greater than 100% increase in MRSA prevalence in pigs (25). Therefore, we wanted to analyze the impact of this development on MRSA carriage in farmers and veterinarians. To address this question, farmers and veterinarians were sampled at two major Swiss conventions. The collected MRSA isolates (n = 23), as well as selected isolates from pigs (n = 12), pork (n = 2), poultry meat (n = 3), and horses (n = 3), were subjected to whole-genome sequencing.
RESULTS
MRSA prevalence in livestock at the slaughterhouse.
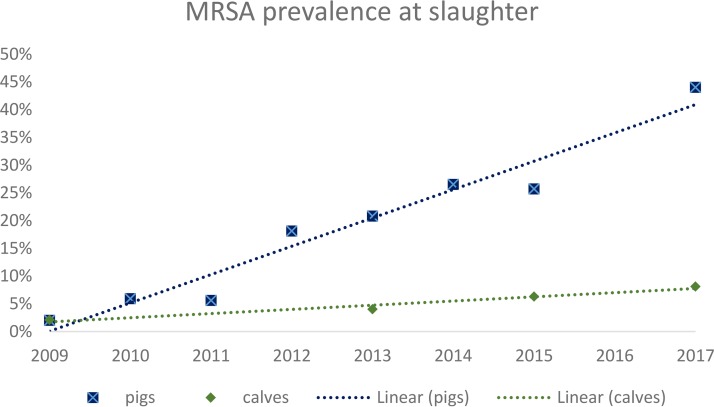
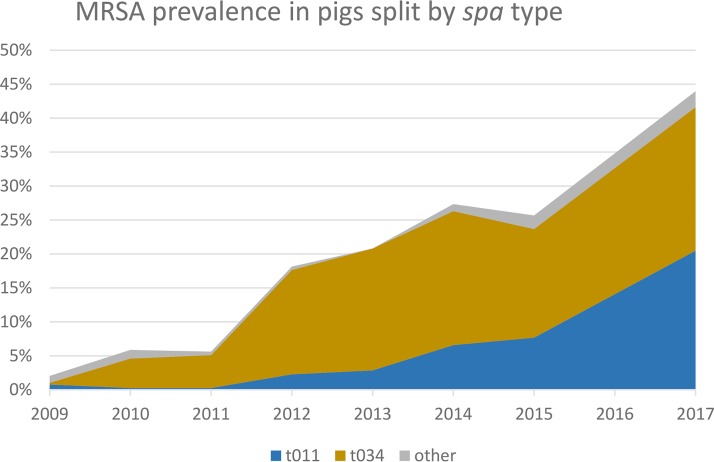
In Switzerland, the occurrence of MRSA in fattening pigs at slaughter increased continuously and significantly from 2009 to 2017 (Fig. 1 and Table 1). From 131 MRSA isolates obtained in 2017, 48.1% (n = 63) belonged to spa t034 and 46.6% (n = 61) to spa t011, while the remainder belonged to rare spa types (t1451 [n = 3], t899 [n = 2], t2330 [n = 1], and t2876 [n = 1]). spa t011 was mainly responsible for the recent increase in prevalence (Fig. 2).
FIG 1.
MRSA prevalence in Swiss livestock at slaughter between 2009 and 2017 with linear trendlines indicated (Microsoft Excel 2016). The steep increase for pigs is clearly visible compared to a slow increase for calves.
TABLE 1.
Prevalence of MRSA in Swiss pigs and calves 2009 to 2017a
| Yr | No. of porcine nasal swabs (n) | No. of MRSA-positive porcine nasal swabs | Prevalence of MRSA (%) (95% CI) | No. of calf nasal swabs | No. of MRSA-positive calf nasal swabs | Prevalence of MRSA (%) (95% CI) |
|---|---|---|---|---|---|---|
| 2009 | 393 | 8 | 2.0 (0.9–4.0) | NA | NA | NA |
| 2010 | 392 | 23 | 5.9 (3.8–8.7) | 240 | 5 | 2.1 (0.7–4.8) |
| 2011 | 392 | 22 | 5.6 (3.6–8.4) | NA | NA | NA |
| 2012 | 397 | 72 | 18.1 (14.5–22.3) | NA | NA | NA |
| 2013 | 351 | 73 | 20.8 (16.7–25.4) | 253 | 10 | 4.0 (1.9–7.2) |
| 2014 | 289 | 79 | 27.3 (22.3–32.9) | NA | NA | NA |
| 2015 | 300 | 77 | 25.7 (20.8–31.0) | 300 | 19 | 6.3 (3.9–9.7) |
| 2017 | 298 | 131 | 44.0 (38.2–49.8) | 297 | 24 | 8.1 (5.3–11.8) |
MRSA, methicillin-resistant Staphylococcus aureus; 95% CI, 95% confidence interval; NA, not analyzed.
FIG 2.
MRSA prevalence in Swiss pigs at slaughter according to spa type. It is clearly visible that spa t011 is mainly responsible for the recent increase in prevalence.
MRSA prevalence in Swiss veal calves remains comparatively low, at 8.1% in 2017 (n = 24). Nevertheless, the prevalence also shows a slight but steady increase since 2009 (Fig. 1 and Table 1). Of the 24 MRSA isolates in 2017, the majority also belonged to spa t011 and t034 (t011 [n = 14], t034 [n = 7], t17339 [n = 2], and t127 [n = 1]).
MRSA prevalence in meat at retail outlets.
The MRSA prevalence in pig meat remained extremely low in 2015 and 2017 (Table 2). In 2017, one MRSA isolate belonged to the LA-MRSA type (spa t034), whereas the other MRSA isolate was categorized as hospital-acquired MRSA (HA-MRSA) (spa t002). No MRSA strains were detected in Swiss beef in 2015 or 2017 (Table 2). From 2014 to 2016, a decrease in MRSA prevalence in chicken meat produced abroad was detected (Table 2). Eight isolates from 2016 were typed as LA-MRSA (spa t034 [n = 3], spa t1430 [n = 3], and spa t2123 [n = 2]). One isolate belonged to spa t153, which is not categorized.
TABLE 2.
Prevalence of MRSA in fresh meat from 2014 to 2017a
| Yr | No. of meat samples (source) | No. of MRSA-positive meat samples (source) | Prevalence of MRSA (%) (95% CI) |
|---|---|---|---|
| 2014 | 319 (chicken) | 22 (chicken) | 6.9 (chicken) (4.4–10.3) |
| 2015 | 302 (pork), 298 (beef) | 2 (pork), 0 (beef) | 0.7 (pork) (0.1–2.4), 0 (beef) (0–1.2) |
| 2016 | 302 (chicken) | 9 (chicken) | 3.0 (chicken) (1.4–5.6) |
| 2017 | 301 (pork), 299 (beef) | 2 (pork), 0 (beef) | 0.7 (pork) (0.1–2.4), 0 (beef) (0–1.2) |
MRSA, methicillin-resistant Staphylococcus aureus; 95% CI, confidence interval.
Prevalence in veterinarians and farmers.
From a total of 212 participating veterinarians, 102 veterinarians reported treating only small animals, 62 reported treating only farm animals, 13 were equine specialists, 28 treated a combination of species (eight included horses), four did not work clinically, and three did not report their area of practice. Fourteen veterinarians tested positive for MRSA; for three, both the nose and hand swabs were positive, resulting in a total of 17 isolates (15 thereof were available for sequencing). Concerning the farmers, eight out of 156 tested positive with nose swabs. The participating farmers worked mostly (n = 97 [62%]) on mixed farms with pigs, dairy or beef cattle, poultry, and/or horses. Fifty-three farmers kept only one livestock species, five only poultry, 19 only dairy cattle, 21 only beef cattle, six only pigs, and two only horses. Another 97 farmers kept two to five of the above-mentioned livestock animal species on their farms. Six farmers did not answer the questionnaire. MRSA-positive farmers were found three times within the group of farmers keeping solely beef cattle, and another five MRSA-positive farmers kept poultry, pigs, cattle, and horses in diverse combinations. In total, the prevalence for veterinarians was 6.6% (95% confidence interval [CI], 3.7 to 10.8%) and for farmers was 5.1% (95% CI, 2.2 to 9.9%). More than half (54% [n = 7]) of the veterinarian isolates belonged to CC398, and interestingly, all of those were spa t011. The proportion of CC398 isolates for farmers was 75% (n = 6). Thereof, five isolates were spa t034, while one isolate was spa t899.
All but one veterinarian carrying a CC398 strain reported contact with horses, whereas there was no clear pattern for farmers (Table 3). The odds ratio for LA-MRSA carriage for veterinarians treating horses (excluding own pets) was 6.2 (95% CI, 1.5 to 23.9), indicating horses as a clear risk factor.
TABLE 3.
All isolates included for whole-genome sequencinga
| Isolate | Source | Date of sampling (day/mo/yr) | Human specialtyb | spa type | ST | Additional resistance genes | Virulence genes (IEC, tsst, lukED) |
|---|---|---|---|---|---|---|---|
| 17KM0012 | Horse (lung) | 01/01/2017 | NA | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | chp, sak, scn |
| 17KM0743 | Horse (wound) | 08/03/2017 | NA | t011 | 398 | aac(6')-aph(2″), dfrK, str, tet(M) | None |
| 17KM2889 | Horse (wound) | 13/11/2017 | NA | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Msa0181 | Pork | 21/02/2017 | NA | t011 | 752 | aac(6')-aph(2″), tet(K), tet(M) | None |
| 17Msa1165 | Pork | 05/12/2017 | NA | t002 | 2626 | aac(6')-aph(2″) | chp, sak, scn, lukED |
| 17Msa0110 | Pig nose | 06/02/2017 | NA | t011 | 398 | tet(K), tet(M) | None |
| 17Msa0134 | Pig nose | 12/02/2017 | NA | t034 | 398 | dfrG, tet(K), tet(M) | None |
| 17Msa0215 | Pig nose | 07/03/2017 | NA | t034 | 398 | dfrG, erm(A), erm(C), spc, str, tet(K), tet(M), vga(E) | None |
| 17Msa0259 | Pig nose | 20/03/2017 | NA | t011 | 398 | tet(K), tet(M) | None |
| 17Msa0460 | Pig nose | 09/05/2017 | NA | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Msa0764 | Pig nose | 07/08/2017 | NA | t034 | 398 | dfrG, erm(A), spc, str, tet(K), tet(M), vga(E) | None |
| 17Msa0826 | Pig nose | 04/09/2017 | NA | t011 | 398 | tet(K), tet(M) | None |
| 17Msa0990 | Pig nose | 09/10/2017 | NA | t011 | 398 | tet(K), tet(M) | None |
| 17Msa1021 | Pig nose | 30/10/2017 | NA | t034 | 398 | dfrG, erm(A), erm(C), spc, str, tet(K), tet(M), vga(E) | None |
| 17Msa1042 | Pig nose | 11/09/2017 | NA | t034 | 398 | dfrG, erm(A), spc, str, tet(K), tet(M), vga(E) | None |
| 17Msa1053 | Pig nose | 06/11/2017 | NA | t011 | 398 | tet(K), tet(M) | None |
| 17Msa1148 | Pig nose | 04/12/2017 | NA | t011 | 398 | str, tet(K), tet(M) | None |
| 16Msa0021 | Poultry meat | 25/01/2016 | NA | t034 | 398 | dfrG, erm(A), spc, tet(K), tet(M), vga(E) | None |
| 16Msa0083 | Poultry meat | 21/03/2016 | NA | t034 | 398 | dfrG, erm(A), spc, tet(K), tet(M), vga(E) | None |
| 16Msa0143 | Poultry meat | 31/05/2016 | NA | t034 | 398 | dfrG, lnu(B), lsa(E) spc, tet(K), tet(M) | None |
| 17Gst002 | Vet | 11/5/2017–12/5/2017 | Small animals | t003 | 225 | aadD, erm(A), spc | sak, scn, lukED |
| 17Gst003 | Vet | 11/5/2017–12/5/2017 | Small animals | t003 | 225 | aadD, erm(A), spc | sak, scn, lukED |
| 17Gst018 | Vet | 11/5/2017–12/5/2017 | Horses | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Gst094_hand | Vet | 11/5/2017–12/5/2017 | Horses | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Gst094_nose | Vet | 11/5/2017–12/5/2017 | Horses | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Gst112 | Vet | 11/5/2017–12/5/2017 | Horses | t011 | 398 | aac(6')-aph(2″), dfrK, str, tet(M) | None |
| 17Gst165 | Vet | 11/5/2017–12/5/2017 | Small and farm animals (pet horse) | t011 | 398 | aac(6')-aph(2″), dfrK, str, tet(M) | None |
| 17Gst166 | Vet | 11/5/2017–12/5/2017 | Small and farm animals and horses | t011 | 398 | aac(6')-aph(2″), dfrK, str, tet(M) | None |
| 17Gst174_hand | Vet | 11/5/2017–12/5/2017 | Farm animals | t118 | 8 | None | chp, sak, scn, lukED, tsst-1 |
| 17Gst174_nose | Vet | 11/5/2017–12/5/2017 | Farm animals | t118 | 8 | None | chp, sak, scn, lukED, tsst-1 |
| 17Gst176 | Vet | 11/5/2017–12/5/2017 | Small animals | t17424 | 22 | None | chp, sak, scn |
| 17Gst187 | Vet | 11/5/2017–12/5/2017 | Small animals | t038 | 45 | None | chp, sak, scn |
| 17Gst193 | Vet | 11/5/2017–12/5/2017 | Small animals and horses | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Gst196 | Vet | 11/5/2017–12/5/2017 | Unknown | t133 | 45 | None | chp, sak, scn |
| 17Gst231 | Vet | 11/5/2017–12/5/2017 | Farm animals | t011 | 398 | aac(6')-aph(2″), dfrK, tet(M) | None |
| 17Gst312 | Farmer | 24/11/2017–26/11/2017 | Cattle, horse, sheep | t034 | 398 | dfrG, erm(A), spc, str, tet(M), vga(E) | None |
| 17Gst329 | Farmer | 24/11/2017–26/11/2017 | Poultry, cattle, horses | t034 | 398 | dfrG, erm(A), spc, tet(K), tet(M), vga(E) | None |
| 17Gst354 | Farmer | 24/11/2017–26/11/2017 | Cattle | t899 | 398 | fexA, str | chp, sak, scn |
| 17Gst358 | Farmer | 24/11/2017–26/11/2017 | Pigs, cattle | t034 | 398 | dfrG, erm(A), erm(C), spc, tet(K), tet(M), vga(E) | None |
| 17Gst374 | Farmer | 24/11/2017–26/11/2017 | Poultry, pigs | t5634 | 22 | None | chp, sak, tsst-1 |
| 17Gst388 | Farmer | 24/11/2017–26/11/2017 | Cattle | t034 | 398 | dfrG, erm(A), spc, str, tet(K), tet(M), vga(E) | chp, sak, scn |
| 17Gst426 | Farmer | 24/11/2017–26/11/2017 | Poultry, cattle | t1510 | 45 | None | chp, sak, scn |
| 17Gst450 | Farmer | 24/11/2017–26/11/2017 | Cattle | t034 | 398 | dfrG, erm(A), spc, str, tet(K), tet(M), vga(E) | None |
All detected resistance genes other than blaZ and mecA (which were present in all isolates) are given. In addition, assemblies were checked for the presence of the immune evasion cluster (IEC) containing the genes chp, sak, and scn, toxic shock syndrome toxin-1 gene (tsst-1), Panton-Valentine leucocidin (not present in any strain), and leucotoxin gene lukED.
NA, not applicable.
Phylogenetic analyses.
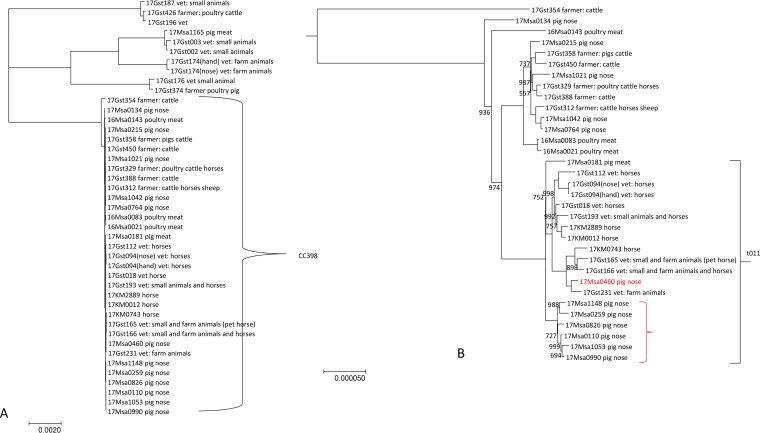
A very high congruence could be observed between core-genome SNP (cgSNP) and cgMLST phylogenies, indicating a clonal population structure (Fig. 3 and 4). When looking more closely at the CC398 cluster, a separation in two clusters which correspond with the spa t011 and t034 can be observed. The spa t034 cluster contains isolates from pigs, farmers, and poultry meat. However, the spa t011 is clearly split into two subgroups visible both in the SNP and cgMLST tree. One subgroup contains only pig isolates, while the other is mainly composed of horse and veterinarian isolates (with the exception of one pig isolate). Interestingly, all but one of the vets in that cluster reported contact with horses, and the isolate of this one vet was most closely linked to the pig isolate (Fig. 3 and 4).
FIG 3.
cgMLST tree, neighbor-joining algorithm, and square root scaling. Branch labels indicate distance (number of loci) and resampling support (where it is less than 100%). The detected resistance genes other than blaZ and mecA are shown in black. Selected virulence factors are shown in gray. CC398 isolates form two clusters according to spa type. Note the higher prevalence of resistance genes in CC398 isolates in contrast to the lower prevalence of virulence genes.
FIG 4.
Core-genome SNP tree of all isolates. (A) The close relationship of CC398 isolates is visualized. (B) The CC398 cluster is enlarged, with the spa t011 cluster marked. Note the subcluster composed of most pig isolates (marked in red). The scale indicates the number of substitutions per site. The numbers indicate bootstraps where they are less than 1,000.
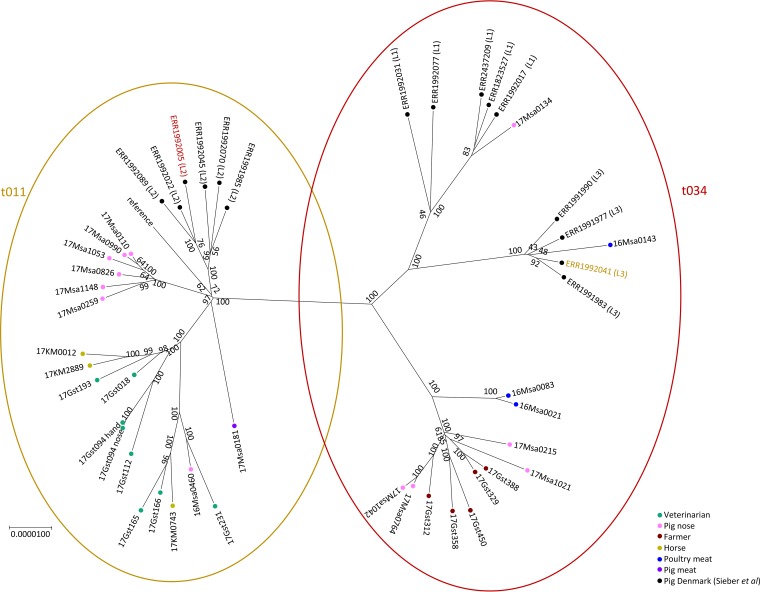
In a comparison of the Swiss spa t011 and spa t034 isolates to selected isolates from Denmark (see Table S2 in the supplemental material), we again see a clear split between the two spa types (Fig. 5). Additionally, the Danish isolates clearly cluster according to the lineage described by Sieber et al. (26). However, the majority of Swiss isolates cluster separately, with the Swiss pig spa t011 strains in particular again forming their own cluster.
FIG 5.
Core-genome SNP tree of isolates with spa t011 and spa t034 supplemented with strains from Denmark (Sieber et al. [26]). The strains can be seen to cluster according to spa type, with two exceptions. The Danish strains also cluster in our analysis according to the described lineages but for the most part separately from the Swiss strains. The scale bar indicates numbers of substitutions per site, and the numbers indicate bootstrap support.
Resistance genes.
Except for blaZ and mecA, which were present in all strains, additional resistance genes (Table 3) could be detected in all strains of CC398 but not all HA/CA-MRSA strains (Fig. 3), indicating a higher exposure to antibiotics among LA-MRSA. This is especially marked for tetracycline resistance, with tet(M) being present in all but one (human) isolate, while none of the HA/CA-MRSA strains showed tetracycline resistance. Resistance patterns also show a clear association with phylogenetic clusters (Fig. 3). The t034 cluster contains more resistance genes than the spa t011 cluster, with the pig spa t011 subcluster being the least resistant. The phenotypic resistance patterns corresponded well with the presence of resistance genes (Table S1).
Virulence factors.
The genes chp, encoding chemotaxis inhibitory protein of Staphylococcus (CHIPS), sak, encoding staphylokinase, and scn, encoding staphylococcal complement inhibitor (SCIN), belonging to the immune evasion cluster, were only present in three strains of LA-MRSA (two from farmers and one from a horse) (Fig. 3), while at least two of them were present in all HA/CA-MRSA strains. None of the LA-MRSA strains were found to carry known enterotoxin genes or PVL. In contrast, some of HA/CA-MRSA strains carried the toxic shock syndrome toxin-1 gene (tsst-1), as well as the leucocidin gene lukED (Fig. 3 and Table 3). All HA/CA-MRSA strains had various enterotoxin genes.
DISCUSSION
In the current study, the prevalence of MRSA in Swiss slaughter pigs was investigated, and the isolates were compared to those from Swiss veterinarians, farmers, and horses.
MRSA sequence type 398 (ST398)-spa t011 is responsible for the steep increase in MRSA in Swiss slaughter pigs in recent years. This spa type has also been reported from pigs in other countries. A recent study from Spain looking at indoor-housed pigs found over 80% of strains belonging to this type (27), while a study looking at isolates in Cameroon and South Africa found only this type (28). In Spain, an MRSA ST398-t011 isolate was even found in a wild boar (29). spa typing results for MRSA strains from pigs at slaughter for 2017 were available from Finland (MRSA prevalence, 77%, with 43% from spa t034 and 12% from spa t011), where spa t034 is dominant, and Spain (MRSA prevalence, 90%, with 11% from spa t034 and 70% from spa t011), where there is a clear dominance of spa t011 (30). Sieber et al. (26) found that CC398 isolates from Danish pigs clustered into three different lineages. Our analysis, which included selected strains from this study, showed that the Swiss strains, for the most part, form separate clusters, which may reflect the low number of imports in the Swiss pork industry. Interestingly, Swiss MRSA ST398-t011 from pigs forms a distinct cluster separate from that of farmers and veterinarians in contrast to porcine MRSA ST398-t034 (Fig. 3). Hence, porcine MRSA ST398-t011 seems to be a successful colonizer of pigs but not humans. Antimicrobial pressure as a driver can be excluded, as these strains harbor, in contrast to porcine MRSA ST398-t034, a very low number of resistance genes in addition to the beta-lactam resistance genes. The underlying molecular features need further research.
An obvious divergence between the CC398 MRSA and the others in the examined sample is the difference in resistance and virulence gene distribution. While LA-MRSA strains, especially those of spa t034, contain a higher number of resistance genes, HA/CA-MRSA strains harbor more virulence genes. At least two out of three genes of the immune evasion cluster (IEC) (chp, sac, and scn) were detected in all HA/CA-MRSA strains but only in three LA-MRSA strains. Two of these were spa t034 strains from farmers, while one was a spa t011 strain from a horse. The IEC genes are thought to be a marker of human adaptation and are rarely present in LA-MRSA (31, 32). CC398 MRSA strains are thought to have lost the IEC during their evolution from human MSSA and adaptation to pigs (15). It is thus conceivable that by regaining these virulence genes, they might readapt to the human host and thus increase their pathogenic potential. Since the current study looked at human carriers, not infections, we cannot draw conclusions about the pathogenicity of our isolates. Horizontal gene transfer during cocolonization has been demonstrated between different CC398 strains in pigs under experimental conditions (16) and might therefore also occur in the noses of humans. Isolates from horses have been described to be more frequent carriers of IEC genes (33). Since we included only three horse isolates, we cannot give an estimate of the IEC prevalence in Swiss horse strains. However, since one of them was positive but none of the 12 pig isolates was, it might be more frequent in horse than in pig isolates.
A prevalence of 6.6% in veterinarians was higher than in a comparable study from 2012, where it was 3.8% (24). Treating horses clearly emerged as a risk factor for LA-MRSA positivity in veterinarians, with an odds ratio of 6.2. This observation is corroborated by the phylogenetic analyses clustering all but one veterinary isolate with horse strains. Contact with horses is a known risk factor for MRSA transmission to veterinary personnel (14, 34). Recent studies in other European countries also found ST398-t011 to be the dominant type in horses (14, 33), while older studies found other types, mainly CC8 (35). Abdelbary et al. (36) looked into the phylogeny of horse-associated LA-MRSA strains from different countries and found a CC398 subclone associated with equine hospitals. They used denaturing high-pressure liquid chromatography for mutation discovery as opposed to our whole-genome sequencing; therefore, our results are not directly comparable. However, the equine subclone found in their study was also associated with spa t011. The emergence of ST398-t011 in horses coincides with the rise of this type in pigs. Considering the cgMLST and cgSNP analyses, there appears to be no epidemiological link between the two, as the majority of pig isolates clearly formed a separate cluster.
In the present study, we also included some strains isolated from pork and poultry meat. The prevalence in pork was found to be extremely low (<1%), meaning that it is a very unlikely source for human infection. One of the two pork strains was not even LA-MRSA, which could indicate that the meat was contaminated by human handling. All three sequenced poultry strains were isolated from imported meat since there were no isolates from Swiss meat; this could explain their separation from the main cluster of t034 strains. Overall, there is no indication that either poultry meat or pork plays a major role in human colonization with MRSA in Switzerland.
LA-MRSA strains are also found in hospital patients, meaning they not only colonize patients but also cause infections. While the incidence in Europe is still low (Goerge et al. estimated <3 infections/100,000 inhabitants per year in the German and Danish populations), it can be higher in areas with high livestock density (37). Among the total MRSA isolates from two Swiss hospitals in 2017, only about 1% were LA-MRSA (two isolates, both spa t011) (25), indicating that the incidence seems to be low. LA-MRSA strains might reacquire more human-associated virulence factors and thus evolve into more virulent strains. Since LA-MRSA strains already harbor more additional resistance genes than do other types, these infections are also more difficult to treat.
Moreover, MRSA prevalence and the role of cattle have to be considered. Due to the comparatively low prevalence and the focus on pigs, they were not included in the sequencing study. The prevalence in dairy cattle is not systematically monitored in Switzerland; however, it is probably very low, as MRSA strains are rarely isolated from milk or other clinical samples. In 2012, 200 bulk milk samples were analyzed for the occurrence of MRSA, and three MRSA isolates were found (1.5%) (38). The prevalence in veal calves, though still comparatively low (8% versus 44% in pigs), is on the rise and might also become a factor in transmission to humans. The majority of veal calf isolates in 2017 were spa t011 (58%) which makes them an unlikely source for the farmers in the present study. However, they should be included in future studies, especially if the prevalence continues to rise.
In conclusion, LA-MRSA is a serious emerging problem in the pig industry, indicating possible antimicrobial overuse. However, the recent rise in spa t011 strains from pigs does not directly translate to a higher prevalence in humans. This type appears to be less likely to colonize humans or acquire resistance genes. Further research is necessary to confirm this finding and elucidate the underlying cause of the successful spread of this clone.
MATERIALS AND METHODS
Samples from livestock at the slaughterhouse.
Stratified random nasal swabs samples from fattening pigs and calves were taken and analyzed for the presence of MRSA in the framework of the national antimicrobial resistance monitoring program. The sampling plan is based on a given number of samples/year, which depends on the prevalence of assumed/detected MRSA. Samples were collected evenly throughout the year at the largest pig and cattle slaughterhouses, which encompassed at least 75% of all slaughtered animals each year. Every slaughterhouse taking part in the program collected samples proportional to the number of animals of the species slaughtered per year. For calves and fattening pigs, samples from one animal selected at random per farm were taken. It was ensured that farms were not repeatedly sampled within 1 year. The representativeness of the sampling was shown by Overesch et al. (18). Nasal swabs from fattening pigs were taken in 2009 (n = 393), 2010 (n = 392), 2011 (n = 392), 2012 (n = 397), 2013 (n = 351), 2014 (n = 289), 2015 (n = 300), and 2017 (n = 298). Nasal swabs from calves were taken in 2010 (n = 240), 2013 (n = 253), 2015 (n = 300), and 2017 (n = 297).
Samples from retail meat.
Meat samples (minimum 50 g) were taken from fresh, skinless, chilled, packed, and untreated meat sold through retail outlets. Samples were collected in all Swiss cantons spread throughout the year. Each canton’s population density and market shares of retailers were considered for the sampling plan. Samples were collected from domestic production, which consists of 302 pork and 298 beef samples in 2015 and 301 pork and 299 beef samples in 2017. Approximately half of the chicken meat consumed in Switzerland is imported. Hence, imported and domestic chicken meat accounted for around one-third and two-thirds, respectively, of the chicken meat samples. In 2014 and 2016, 319 and 302 chicken samples, respectively, were collected and analyzed.
Samples from humans.
Samples from veterinarians were obtained at the 2017 annual conference of the Swiss Veterinary Society (GST). Attendants interested in participating in the study were asked to take nasal and hand swabs from themselves and to indicate which animals they routinely treated (horses, farm animals, and companion animals). The same was done for farmers attending the 2017 Suisse Tier (national farming exhibition), except that they were only asked for nose swabs. The studies were approved by the ethics commission of the canton of Bern (Req-2017-00793). A total of 212 vets and 156 farmers participated in the study.
Samples from horses.
Horse isolates were obtained during routine diagnostics and stored at −80°C until use.
MRSA detection.
MRSA detection in swabs from livestock and humans and meat samples was performed according to a method published previously (18), with small adjustments. Swabs or 5 g of meat were transferred into tubes containing 10 ml or 45 ml, respectively, of Mueller-Hinton broth supplemented with 6.5% NaCl. Samples were incubated aerobically at 37°C ± 1°C for 24 h under agitation. One milliliter from each preenrichment was inoculated into 9 ml tryptone soy broth containing 3.5 mg/liter cefoxitin and 75 mg/liter aztreonam, followed by further aerobic incubation at 37°C ± 1°C for 24 h. Subsequently, a loopful was spread onto MRSA selective agar plates (BBL CHROMagar MRSA; Becton, Dickinson, Franklin Lakes, NJ), which were incubated at 37°C ± 1°C for 24 h. Pink- to mauve-colored colonies were regarded as suspicious, and if present, one such colony was cultivated onto tryptone soy agar plates containing 5% sheep blood (TSA-SB; Oxoid Ltd., Basingstoke, England) at 37°C ± 1°C for 24 h. Staphylococcus aureus was identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Biotyper 3.0; Bruker). The obtained MRSA isolates were subsequently stored at −80°C in glycerol buffer for further analysis.
Phenotypic resistance.
The MICs of the antimicrobials were determined by broth microdilution in cation-adjusted Mueller-Hinton broth, using Sensititre susceptibility plates (EUST; Trek Diagnostics Systems, Thermo Fisher, UK), according to CLSI guidelines (2).
Molecular characterization.
The presence of the mecA gene, as well as attribution to CC398, was tested for each isolate by a duplex PCR (39). Additionally, spa typing was performed for all strains, according to Harmsen et al. (22).
Statistical analyses.
Confidence intervals were calculated using the R (version 3.4.1) function binom.test(), and odds ratios were calculated using the R function fisher.test () (8). Prevalence trendlines were obtained using Excel 2016 (Microsoft).
Selection of MRSA strains for sequencing.
An overview of isolates selected for whole-genome sequencing can be seen in Table 3. Twelve isolates from pig nasal swabs were chosen to represent the main spa t011 (n = 7) and spa t034 (n = 5) and to be spread over time and area during 2017. The isolates were chosen according to the most prevalent resistance phenotypes (Table S1). From a total of 61 MRSA spa t011 isolates, 47 (77%) isolates exhibit in addition to resistance to beta-lactam antibiotics only one to three additional resistances. In contrast, 49 out of 63 MRSA spa t034 isolates exhibit uniform resistance patterns with up to eight additionally phenotypic resistances. Furthermore, two pig meat isolates also obtained in the national monitoring program in 2017 and 3 poultry meat isolates with spa t034 from 2016 were sequenced (no poultry meat isolates were available from 2017).
Fifteen MRSA isolates recovered from veterinarians and eight isolates from farmers were included, resulting in 23 human isolates from 2017. Three isolates from horses that were isolated in routine diagnostics in the same year were also included; two isolates were from wounds, and one isolate was from bronchoalveolar lavage fluid.
In addition, 15 isolates from a recent study on pig CC398 in Denmark (26) were included. Isolates were selected to represent the three described lineages (L1 to L3) and to be spa t011 or spa t034 (Table S2).
Sequencing.
The whole genomes of all selected isolates were sequenced using the NextSeq platform (Illumina Nextera XT library) to obtain paired 150-bp reads. Sequencing was performed by the external companies Microsynth (Balgach, Switzerland) and Eurofins Genomics Germany GmbH (Ebersberg, Germany) (horse isolates).
Phylogenetic analyses.
Reads were mapped to a reference genome (GenBank accession no. NC_017333.1) using the bwa 0.7.13 (40) aln command with –q option set to 15, followed by merging of forward and reverse alignments with sampe. SAMtools 1.3 (41) was then used to convert the SAM file to a BAM file. SNP calling was then performed using samtools 0.1.19 mpileup and bcftools. The obtained mapping alignments were converted to FASTA files for use in downstream analyses.
To obtain an SNP tree, the mapping assemblies were loaded into MEGA 7 (42) to remove all gaps and thus obtain a core-genome alignment. The tree was calculated with PhyML 3.3.20180214 (43, 44) after first determining the best model according to the Bayesian information criterion with modeltest-ng 0.1.3 (https://github.com/ddarriba/modeltest).
Core-genome MLST analyses were performed using chewBBACA version 1.0 (https://github.com/B-UMMI/chewBBACA) for allele calling and employing the schema published on PubMLST (https://pubmlst.org/, accessed 3 August 2018).
To detect virulence and resistance genes, a de novo assembly was performed for each strain using SPAdes 3.10.1 (45). Then, virulence and resistance genes were called with abricate (https://github.com/tseemann/abricate) using the databases VFDB and ResFinder (https://cge.cbs.dtu.dk/services).
Data availability.
All reads were submitted to the Sequence Read Archive under BioProject number PRJNA556204.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the University of Bern, the Swiss Federal Food Safety and Veterinary Office, and the Gesellschaft Schweizer Tierärztinnen und Tierärzte.
We thank Manon Wider, Sandra Zumwald, and Susanne Rickli for their work isolating the strains and Simon Feyer for technical support. We also thank all participating veterinarians and farmers.
Calculations were performed on UBELIX (http://www.id.unibe.ch/hpc), the high-performance computing (HPC) cluster at the University of Bern.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL. 2016. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J Glob Antimicrob Resist 6:95–101. doi: 10.1016/j.jgar.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Köck R, Becker K, Cookson B, van Gemert-Pijnen JE, Harbarth S, Kluytmans J, Mielke M, Peters G, Skov RL, Struelens MJ, Tacconelli E, Navarro Torné A, Witte W, Friedrich AW. 2010. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill 15:pii=19688 https://www.eurosurveillance.org/content/10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- 3.Wulf M, Voss A. 2008. MRSA in livestock animals-an epidemic waiting to happen? Clin Microbiol Infect 14:519–521. doi: 10.1111/j.1469-0691.2008.01970.x. [DOI] [PubMed] [Google Scholar]
- 4.Weese JS. 2010. Methicillin-resistant Staphylococcus aureus in animals. ILAR J 51:233–244. doi: 10.1093/ilar.51.3.233. [DOI] [PubMed] [Google Scholar]
- 5.van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis 13:1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catry B, Van Duijkeren E, Pomba MC, Greko C, Moreno MA, Pyorala S, Ruzauskas M, Sanders P, Threlfall EJ, Ungemach F, Torneke K, Munoz-Madero C, Torren-Edo J, Scientific Advisory Group On A. 2010. Reflection paper on MRSA in food-producing and companion animals: epidemiology and control options for human and animal health. Epidemiol Infect 138:626–644. doi: 10.1017/S0950268810000014. [DOI] [PubMed] [Google Scholar]
- 7.Smith TC, Pearson N. 2011. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis 11:327–339. doi: 10.1089/vbz.2010.0072. [DOI] [PubMed] [Google Scholar]
- 8.Wagenaar JA, Yue H, Pritchard J, Broekhuizen-Stins M, Huijsdens X, Mevius DJ, Bosch T, Van Duijkeren E. 2009. Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet Microbiol 139:405–409. doi: 10.1016/j.vetmic.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol 128:298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Mulders MN, Haenen AP, Geenen PL, Vesseur PC, Poldervaart ES, Bosch T, Huijsdens XW, Hengeveld PD, Dam-Deisz WD, Graat EA, Mevius D, Voss A, Van De Giessen AW. 2010. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol Infect 138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- 11.Verkade E, van Benthem B, den Bergh MK, van Cleef B, van Rijen M, Bosch T, Kluytmans J. 2013. Dynamics and determinants of Staphylococcus aureus carriage in livestock veterinarians: a prospective cohort study. Clin Infect Dis 57:e11–e17. doi: 10.1093/cid/cit228. [DOI] [PubMed] [Google Scholar]
- 12.Witte W, Strommenger B, Stanek C, Cuny C. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis 13:255–258. doi: 10.3201/eid1302.060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieber S, Gerber V, Jandova V, Rossano A, Evison JM, Perreten V. 2011. Evolution of multidrug-resistant Staphylococcus aureus infections in horses and colonized personnel in an equine clinic between 2005 and 2010. Microb Drug Resist 17:471–478. doi: 10.1089/mdr.2010.0188. [DOI] [PubMed] [Google Scholar]
- 14.Albert E, Biksi I, Nemet Z, Csuka E, Kelemen B, Morvay F, Bakos Z, Bodo G, Toth B, Collaud A, Rossano A, Perreten V. 2019. Outbreaks of a methicillin-resistant Staphylococcus aureus clone ST398-t011 in a Hungarian equine clinic: emergence of rifampicin and chloramphenicol resistance after treatment with these antibiotics. Microb Drug Resist 25:1219–1226. doi: 10.1089/mdr.2018.0384. [DOI] [PubMed] [Google Scholar]
- 15.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM, Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, et al. . 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol 6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirolo M, Gioffre A, Visaggio D, Gherardi M, Pavia G, Samele P, Ciambrone L, Di Natale R, Spatari G, Casalinuovo F, Visca P. 2019. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol 19:51. doi: 10.1186/s12866-019-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overesch G, Buttner S, Rossano A, Perreten V. 2011. The increase of methicillin-resistant Staphylococcus aureus (MRSA) and the presence of an unusual sequence type ST49 in slaughter pigs in Switzerland. BMC Vet Res 7:30. doi: 10.1186/1746-6148-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55:4900–4904. doi: 10.1128/AAC.00528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. 2013. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J Dairy Sci 96:2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- 21.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/jcm.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphreys H, Coleman DC. 2019. Contribution of whole-genome sequencing to understanding of the epidemiology and control of meticillin-resistant Staphylococcus aureus. J Hosp Infect 102:189–199. doi: 10.1016/j.jhin.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Wettstein Rosenkranz K, Rothenanger E, Brodard I, Collaud A, Overesch G, Bigler B, Marschall J, Perreten V. 2014. Nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA) among Swiss veterinary health care providers: detection of livestock- and healthcare-associated clones. Schweiz Arch Tierheilkd 156:317–325. doi: 10.1024/0036-7281/a000601. [DOI] [PubMed] [Google Scholar]
- 25.Heim D, Kronenberg A, Overesch G, Pluss-Suard C, Schupbach G, Bless P, Burgmann H, Dubuis O, Egli A, Frei R, Gaia V, Gasser M, Gotz C, Hardegger M, Hilty M, Kittl S, McArdell CS, Nordmann P, Carmo LP, Reinhardt M, Riedo J, Saam M, Schrenzel J, Sinreich M, Stephan R, Widmer A, Zanetti G, Zimmerman-Steffens S. 2018. Swiss antibiotic resistance report 2018. Usage of antibiotics and occurrence of antibiotic resistance in bacteria from humans and animals in Switzerland. Federal Office of Public Health, Bern, Switzerland: https://www.dora.lib4ri.ch/eawag/islandora/object/eawag:17936. [Google Scholar]
- 26.Sieber RN, Skov RL, Nielsen J, Schulz J, Price LB, Aarestrup FM, Larsen AR, Stegger M, Larsen J, Sieber RN, Skov RL, Nielsen J, Schulz J, Price LB, Aarestrup FM, Larsen AR, Stegger M, Larsen J. 2018. Drivers and dynamics of methicillin-resistant livestock-associated Staphylococcus aureus CC398 in pigs and humans in Denmark. mBio 9:e02142-18. doi: 10.1128/mBio.02142-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Flores A, Potel-Alvarellos C, Francisco-Tome M, Constenla-Carames L, Perez-Roth E, Lopez-Coton C, Comesana-Da Vila E, Eiroa-de la Puente L, Alvarez-Fernandez M. 2019. Methicillin-resistant Staphylococcus aureus in swine housed indoors in Galicia, Spain. Enferm Infecc Microbiol Clin, in press. doi: 10.1016/j.eimc.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Founou LL, Founou RC, Allam M, Ismail A, Finyom Djoko C, Essack SY. 2019. Genome analysis of methicillin-resistant Staphylococcus aureus isolated from pigs: detection of the clonal lineage ST398 in Cameroon and South Africa. Zoonoses Public Health 66:512. doi: 10.1111/zph.12586. [DOI] [PubMed] [Google Scholar]
- 29.Mama OM, Ruiz-Ripa L, Fernandez-Fernandez R, Gonzalez-Barrio D, Ruiz-Fons JF, Torres C. 2019. High frequency of coagulase-positive staphylococci carriage in healthy wild boar with detection of MRSA of lineage ST398-t011. FEMS Microbiol Lett 366:fny292. doi: 10.1093/femsle/fny292. [DOI] [PubMed] [Google Scholar]
- 30.European Food Safety Authority. 2019. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J 17:5598. doi: 10.2903/j.efsa.2019.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goerke C, Wirtz C, Fluckiger U, Wolz C. 2006. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol Microbiol 61:1673–1685. doi: 10.1111/j.1365-2958.2006.05354.x. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy AJ, Lindsay JA, Loeffler A. 2012. Are all meticillin-resistant Staphylococcus aureus (MRSA) equal in all hosts? Epidemiological and genetic comparison between animal and human MRSA. Vet Dermatol 23:267–275, e53–e54. doi: 10.1111/j.1365-3164.2012.01072.x. [DOI] [PubMed] [Google Scholar]
- 33.Walther B, Klein KS, Barton AK, Semmler T, Huber C, Merle R, Tedin K, Mitrach F, Lubke-Becker A, Gehlen H. 2018. Equine methicillin-resistant sequence type 398 Staphylococcus aureus (MRSA) harbor mobile genetic elements promoting host adaptation. Front Microbiol 9:2516. doi: 10.3389/fmicb.2018.02516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuny C, Witte W. 2017. MRSA in equine hospitals and its significance for infections in humans. Vet Microbiol 200:59–64. doi: 10.1016/j.vetmic.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Walther B, Monecke S, Ruscher C, Friedrich AW, Ehricht R, Slickers P, Soba A, Wleklinski CG, Wieler LH, Lubke-Becker A. 2009. Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J Clin Microbiol 47:704–710. doi: 10.1128/JCM.01626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelbary MM, Wittenberg A, Cuny C, Layer F, Kurt K, Wieler LH, Walther B, Skov R, Larsen J, Hasman H, Fitzgerald JR, Smith TC, Wagenaar JA, Pantosti A, Hallin M, Struelens MJ, Edwards G, Bose R, Nubel U, Witte W. 2014. Phylogenetic analysis of Staphylococcus aureus CC398 reveals a sub-lineage epidemiologically associated with infections in horses. PLoS One 9:e88083. doi: 10.1371/journal.pone.0088083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goerge T, Lorenz MB, van Alen S, Hübner N-O, Becker K, Köck R. 2017. MRSA colonization and infection among persons with occupational livestock exposure in Europe: prevalence, preventive options and evidence. Vet Microbiol 200:6–12. doi: 10.1016/j.vetmic.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 38.Büttner S, Overesch G. 2012. Bulk tank milk sampling to monitor trends in antimicrobial resistance on dairy farms—a pilot-study. Poster. 3rd ASM Conf Antimicrob Resist Zoonotic Bact Foodborne Pathog Anim Hum Environ, Aix-en-Provence, France, 26 June 2012. [Google Scholar]
- 39.Stegger M, Lindsay JA, Moodley A, Skov R, Broens EM, Guardabassi L. 2011. Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J Clin Microbiol 49:732–734. doi: 10.1128/JCM.01970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reads were submitted to the Sequence Read Archive under BioProject number PRJNA556204.